Abstract
The characterization of glycosylation is critical for obtaining a comprehensive view of the regulation and functions of glycoproteins of interest. Due to the complex nature of oligosaccharides, due to variable compositions and linkages, and ion suppression effects, the chromatographic separation of glycans, including isomeric structures, is necessary for exhaustive characterization by mass spectrometry (MS). This review introduces the fundamental principles underlying the techniques in liquid chromatography (LC) utilized by modern day glycomics researchers. Recent advances in porous graphitized carbon, reverse phase, ion exchange and HILIC LC utilized in conjunction with MS, for the characterization of protein glycosylation, are described with an emphasis on methods capable of resolving isomeric glycan structures.
Keywords: glycan analysis, isomeric separation, liquid chromatography, mass spectrometry, LC-MS/MS
1. Introduction
Glycosylation of proteins, as a post-translational modification (PTM), represents a process critical for the proper function of many cellular processes. Cell surface glycans commonly interact with integral glycan-binding proteins that possess carbohydrate recognition domains specific for particular glycan structures [1]. Glycans and their interactions mediate a broad range of cellular processes including cell-cell recognition [2], cell adhesion [3, 4], immune cell trafficking [5] and protein solubility and stability [6], among others. In humans, the primary attachment sites for glycans are asparagine residues (N-linked glycosylation) localized in consensus sequences of asparagine-xxx-threonine/serine, where xxx can be any amino acid except proline, or serine or threonine residues (O-linked glycosylation), where no consensus sequence is required.
An enormous amount of stereochemical information that is essential for the diverse functions exhibited by glycans is encompassed within N- and O-linked structures. The high complexity of glycan structures is attributed to many features, including (i) different monosaccharide residues compositions, (ii) various linkage sites with potential α- or β-stereochemistries, (iii) branching possibilities, and (iv) the potential modification of glycans with additional functional groups (e.g., methyl, phosphate and sulfate functional groups). Changes in the glycosylation of proteins modulate the functions of glycoproteins in biological systems. Aberrant glycosylation has been implicated in numerous mammalian diseases, including congenital disorders of glycosylation [7], autoimmune disorders [8], neurological disease [9], cancer [10], and cardiovascular disease [11].
The biological relevance of different isomeric forms of glycans, and the importance of their characterization, is illustrated by Alley Jr. et al. [12] and Chung et al. [13]who investigated sialic acid linkage isomers in breast cancer patients and hypersensitive reactions to the monoclonal antibody (mAb) cancer drug Cetuximab®, respectively. Alley Jr. et al. reported a general trend of increased α-2,6 linked sialic acid residues in acidic glycans derived from the blood serum of stage IV breast cancer patients [12], demonstrating the diagnostic relevance of N-linked glycan isomers. Chung et al. screened serum samples of cancer patients treated with Cetuixmab® for IgE antibodies against the drug and found that most subjects who exhibited hypersensitive reactions had preexisting IgE antibodies specific for galactose-α-1,3-galactose [13]. This finding highlights the importance of methods for the exhaustive isomeric structural characterization of glycan moieties of therapeutic antibodies, as galactose linkage isomers can be the difference between successful treatment and anaphylaxis.
Mass spectrometry (MS) is widely regarded as the method of choice for providing data rich with structural information for glycoconjugate analysis [14–16]. However, MS and tandem MS alone do not permit comprehensive characterization of isomeric glycan structures. High order tandem MS, in some cases, is capable of complete elucidation of one or a few structures of interest, yet high concentrations are required which is a need not often attainable in biological systems. Therefore, it is necessary to utilize MS in conjunction with separation strategies, such as chromatographic, to achieve unequivocal characterization of pools of glycans containing isomeric structures. A variety of liquid chromatography (LC) separation modes has been employed to analyze glycans, including hydrophilic liquid interaction chromatography (HILIC) [17–19], porous graphitized carbon (PGC) chromatography [20–23], high-pH anion-exchange chromatography (HPAEC) [24, 25], and reversed-phase (RP) chromatography [26].
In LC-ESI-MS glycan analysis, ions are commonly detected as adducts. The formation of these adducts are influenced by the composition of the mobile phase and the sample solution, as well as glycan structures. In general, positive mode ESI is commonly used for the analysis of glycans. Glycan ions observed in ESI include protonated ions, alkali metal (sodium, potassium, etc) and ammonium adduct ions [27–29]. The latter are particularly common when ammonium acetate or ammonium formate are included in the HILIC mobile phases. Reductive amination of glycans is attained through the inclusion of sodium cynoborohydride reagent which prompts the formation of sodium adducts in HILIC [30]. As for PGC, protonated ions are often the most intense followed by alkali metal adducts for both native and derivatized glycans [31, 32]. When ion exchange chromatography is used, and desalters are employed, protonated ions are most abundant and are usually accompanied by weak sodium adducts [33]. Though RPLC is not used for native glycan analysis, derivatization technique such as permethylation which involves excess amount of sodium hydroxide can induce the formation of sodium adduct [31, 34, 35]. However, vast majority of adducted ion is dominated by protonated adducts as shown by Higel et al [29]. While metal adducts do not undergo rearrangement reactions, protonated glycans (native and reducing end labeled) are prone to the rearrangement of monosaccharides, such as the migration of fucose between core and branched structures [27, 28].
The primary aim of this review is to discuss and evaluate the abovementioned LC separation modes, coupled with MS, for the separation and analysis of glycan structures, with an emphasis on the isomeric resolution of glycans.
2. HILIC LC Separation of Glycans
Although reversed-phase liquid chromatography (RPLC) is the most commonly used chromatographic technique, the inability to retain highly hydrophilic and uncharged species, such as glycans, remains a major shortcoming. To overcome these problems, HILIC was proposed, developed and introduced by Alpert in 1990 [17], a technique which is widely used in glycomic studies. Unlike RPLC, HILIC utilizes a stationary phase that is more polar than the mobile phase [36]. Thus, it is considered a variant of normal phase liquid chromatography (NPLC). However, the primarily aqueous mobile phases used for HILIC largely differ from the traditional non-polar NPLC eluents, which contributes to solving the problems encountered when using totally organic eluents in ESI [37]. The retention of analytes on HILIC stationary phases is prompted by hydrogen bonding, ionic interactions and dipole-dipole interactions [19]. The separation of carbohydrates by HILIC is achieved through partitioning between an acetonitrile-rich mobile phase and a thin water layer immobilized on a polar stationary phase. In addition to this liquid/liquid partitioning mechanism, dipole-dipole and electrostatic interactions may also contribute to the separation mechanism [17, 36–39]. The salt concentration and pH can result in a charged stationary phase surface [17, 36–39]. A gradient shifting from organic phase (usually an aqueous acetonitrile solution) to aqueous phase is most often used to achieve glycan separations [40].
When this type of separation system is employed, a dextran ladder is often utilized for the calibration of retention time in glucose units (GU), which relies on certain conditions such as pH, salt concentration, temperature, etc. However, glycans are most commonly eluted in order of glycan size, from smallest to largest [19, 38]. To detect glycans and improve ionization efficiency, reducing end labeling approaches are frequently used in the HILIC analysis [41]. Reducing end labeling is the most widely-used technique in HILIC glycomics due to its compatibility and enhancement of separation and the convenience of quantitation using optical detection methods, made possible by the introduction of reducing end tags. The development of UPLC also largely facilitates the applications of HILIC separation with the particle size of the stationary phase reduced [42]. The mechanism of HILIC and its applications in the field of glycomics have been comprehensively reviewed by Hemström et al. [37] and Zauner et al. [43]. This review will only focus on the applications of HILIC towards glycan isomeric separation.
2.1 Chromatographic Packing Materials
2.1.1 Zwitterionic (ZIC®)-HILIC
A ZIC®-HILIC column consists of a zwitterionic functional group which is covalently attached to porous silica. The first application of ZIC®-HILIC (HILIC column with sulfobetaine group) in the glycomics field was introduced by Takegawa et al. in 2006; however, this promising technique had been used for years for the separation of small molecules in an aqueous phase [44]. 2-amino-pyridine (2-AP) labeled IgG glycans, derived from human serum, were analyzed by ZIC®-HILIC, allowing the isomeric separation of biantennary monogalactosylated and biantennary monogalactosylated bisecting glycans. The mechanism underlying the isomeric separation of such N-glycans is thought to be based on hydrophilic and electrostatic interactions between the glycans and the sulfobetaine stationary phase. However, since tandem MS experiments were not performed, branched isomers were not unassigned.
Recently, Mancera-Arteu et al. analyzed aniline labeled sialylated glycans from human AGP by a capillary ZIC®-HILIC column [45]. Although the N-glycans were only partially resolved, especially in the case of the highly sialylated species, the linkages of both fucose and sialic acid residues were possible to assign by exoglycosidase digestion experiments. Similarly, Mauko et al. isomerically separated and confirmed N-glycan structures using 2-aminobenzamide (2-AB) labeling in conjunction with LC-FL, LC-MS, and exoglycosidase digestion [46].
2.1.2 Amide/Amine Columns
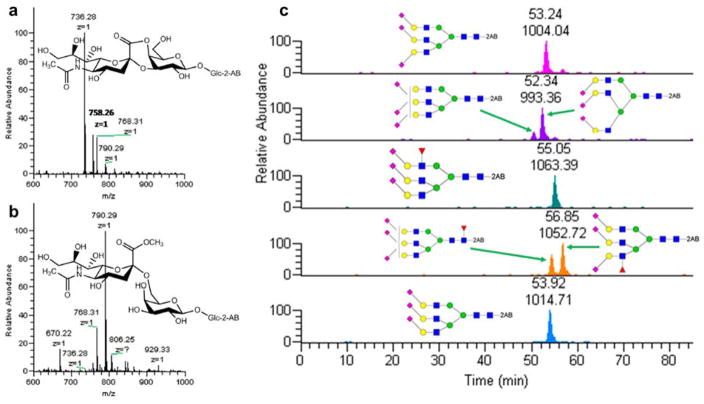
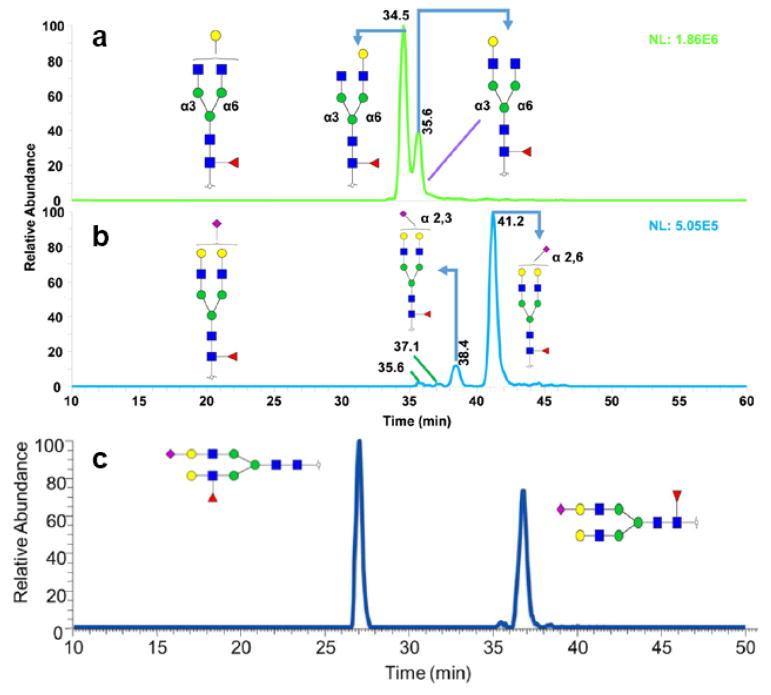
Most isomeric separation studies performed on amide/amine columns have utilized labeling with aniline [47], 2-AP [48] or 2-AB [38, 49–51]. As they are fluorescently labeled, the quantitative results of the glycans can be generated by using a fluorescence detector. Although it is possible to assign isomeric structures based on retention order and the results of exoglycosidase treatment, the time required for digestion and the inefficient ionization of sialylated species represent significant drawbacks. 4-(4,6-Dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride (DMT-MM)/methanol has been reported as a specific derivatization method for the distinction between α-2,3 and α-2,6 sialic acid linkages [52]. The products of the reaction with α-2,3 and α-2,6 linked sialic acid in the glycans are shown in Figure 1a and Figure 1b, respectively. In such products, the carboxyl group of α-2,3 linked sialic acid is methylated while that of α-2,6 sialic acid becomes cyclic lactones and result in a 32 Da shift in molecular weight [52].
Figure 1.
Scheme of (a) α-2,3 and (b) α-2,6 linked sialic acid molecular structures after DMT/MM derivatization. (c) EIC of the identified compositions in haptoglobin trisialylated N-glycans after DMT-MM/methanol derivatization. Reprinted and modified from [53], with permission.
Tousi et al. integrated the merits of both DMT-MM/methanol derivatization and 2 -AB labeling to identify and quantify the sialic acid linkages of haptoglobin derived from patients with different types of cancer [53]. The triantennary trisialylated glycans were resolved on an ethylene bridged hybrid amide column with sialic acid linkages distinguished by DMT-MM/methanol derivatization, as depicted in Figure 1c. The derivatized sialic acid linkages can be assigned by the m/z value shifts resulting from the derivatization. Additionally, linkages can be assigned by MS/MS facilitated by the detection of unique diagnostic ions for each linkage. Although there was some overlapping of peaks, which is problematic for fluorescence quantitation, the combination of derivatization and HILIC isomeric separation shows promise for future studies.
2.1.3 Hydroxyl Group HILIC Columns
Advanced Materials Technology recently introduced hydroxyl group Halo columns as a new format for HILIC. The columns were based on a novel fused-silica based Penta-HILIC column using proprietary chemistry. Tao et al. employed such columns in the separation of procainamide labeled fetuin and human serum N-glycans [54]. Structural information of sialylated glycan isomers derived from bovine fetuin was generated from sialidase digestion experiments, where the sialidase S only had the ability to cleave α-2,3 linked sialic acids and leave the α-2,6 linked sialic acids intact. The relative abundances of glycans in fetuin and human serum were both illustrated by selected reaction monitoring, which overcame the problem of coeluting glycans that could not be resolved by fluorescence detection. Moreover, to address ionization discrepancy of the sialylated species, a normalization to the summed response with the same composition were used.
3. RPLC Separation of Glycans Using MS
RPLC, which achieves separations through noncovalent interactions between the analyte and packing particle in the column, is one of the most frequently used separation techniques [26]. Unlike NPLC, RPLC uses nonpolar stationary phase such as hydrocarbons and polar solvents as mobile phases. The use of a polar solvent as a mobile phase is one of the advantages of RPLC because of the low cost and safety of the solvents. Retention times and elution orders in RPLC are dependent on both stationary phase materials and mobile phase composition. This section of the review discusses analytical RPLC-MS techniques including, not only, isomeric separation of glycans but the use of different mobile phases, column specifications, and derivatizing reagents.
3.1 LC Conditions and Derivatization Enabling Reversed-phase LC-MS
For glycan analysis, the majority of RPLC columns used to achieve separations utilize C18 as the stationary phase, with few C8 methods developed [26, 55, 56]. C18 columns can have a variety of specifications such as internal diameter, length, and particle size, all of which have a major impact on separation efficiency [26]. Compared to traditional LC columns, nano-LC systems have demonstrated improved sensitivity, separation efficiency, and resolution for glycan analysis [57–60]. Because of these advantages, C18 nano-LC coupled with MS has become one of the most commonly used methods. 2-D LC techniques combining RPLC and HILIC have been shown to separate isomeric glycans, and demonstrate improved resolution over 1-D strategies [26, 61–63].
Along with the column packing materials, mobile phase plays a major role in achieving separation. The organic solvent of the mobile phase frequently consists of methanol and acetonitrile, and occasionally the aqueous and organic phases are composed of a mixture of solvents. All of these solutions have acidic pH values due to the addition of mass spectrometer-compatible acids such as formic and acetic acid. However, buffered solutions can also be used as mobile phases. These buffers are prepared with various salts including sodium acetate, triethylammonium acetate, ammonium acetate, ammonium formate and sodium phosphate. In few cases, to increase the formation of sodium adducts, it has been demonstrated that introducing a small amount of NaOH to the aqueous phase can be effective [64, 65]. The formation of sodium adducts can also be achieved by suspending samples in sodium chloride solution at high concentration as reported by Zhou et al. [66]. The use of such sample solution was necessary to improve the intensity of MS/MS reporter ions of aminoxy TMT labeled glycans and provide more reliable glycomics quantitation results [66]. For glycans that contain acidic monosaccharides, such as sialic acids, it has been shown that ion-pairing reagents in the aqueous phase can improve peak shape [67, 68].
Because RPLC operates on hydrophobic stationary phases, most derivatizations increase hydrophobicity to enhance retention [26]. Increased hydrophobicity was demonstrated to increase sensitivity for large biomolecules when working with electrospray ionization [69]. To increase hydrophobicity in glycans, permethylation is the most common method. Instead of attaching a reducing end tag, permethylation replaces the hydrogen atom on every hydroxyl and amino group, of glycans, with methyl groups; hence increasing hydrophobicity [26, 70]. Thus, permethylated glycan analysis on C18 columns has been widely applied to complex biological sample analysis for candidate glycan biomarker discovery and disease state diagnosis. Mechref and co-workers have utilized the above LC-MS/MS method to profile the glycome derived from serum samples representing liver [63, 71, 72] and esophagus diseases [73]. The same method was also used to profile glycans from mouse brain tissue [74]. Glycomics chnages associated with trumatic brain injury was also assessed using the above LC-MS/MS analysis of permethylated glycans [75]. Glycans derived from glioma stem cell xenografts [76], neuroblastoma [77] and breast cancer [71, 78] cell lines, IgG [79], prostate specific antigen [80], and human milk, bovine milk and goat milk [23] were also analyzed using RPLC-MS/MS of reduced and permethylated glycans.
Additionally, MS analysis of glycans has been used in conjunction with many derivatization methods on the reducing end of glycans. Vreeker et al. provide a comprehensive list of all glycan derivatization reagents that have been used for the analysis of glycans [26]. Different derivatization reagents have been used with RPLC for the analysis of glycans, including anthranilic acid (AA) [81], 2-AB [81], 2-amino-5-bromopyridine (ABP) [55], 4-aminobenzoic acid butyl ester (ABBE) [57], 4-aminobenzoic acid methyl ester (ABME) [57], aminobenzoic acid ethyl ester (ABEE) [57], 4-n-heptyloxyaniline (HOA) [57], 8-aminonaphthalene-1,3,6-trisulfonic acid (ANTS) [82], 1-phenyl-3-methyl-5-pyrazolone (PMP) [83], and phenylhydrazine [84]. Reducing end tags directly influence the retention times of glycans, not only in the case of RPLC but also for PGC. While there are some differences in retention mechanism between RPLC and PGC, the hydrophobicity of reducing end tags prompt differences in retention times. For example, retention time differences between AB, PA and ABEE tags suggest differences in hydrophobicity as shown by Pabst et al. [85]. Besides reduction, the introduction of reducing end tags account for the rest of the methods. Reducing ends of glycans with open rings can be reduced to eliminate α and β anomers. Reduced glycans appear on chromatograms as one peak, rather than two α and β anomer peaks.
Sensitive quantitative glycomics for low abundance structures in complex biological samples is still challenging [86]. The reliability and sensitivity of glycan analysis can be improved by selected reaction monitoring (SRM) or multiple-reaction monitoring (MRM) using a triple quadrupole mass spectrometer [70, 77]. SRM or MRM quantitation is based on using transition ions that are observed in the tandem mass spectrum of the analyte of interest, not full mass scans. The increased signal-to-noise ratios observed in these methods enable more reliable and sensitive quantitation of low abundance structures. MRM provides higher selectivity and more precise quantitation than SRM because MRM selects multiple transition ions, instead of targeting a single transition ion. MRM was first used for the analysis of glucose tetra-oligosaccharides (Glcα1–6Glcα1–4Glcα1–4Glc) in urine, urine spots, and plasma [87]. Thus far, MRM has been applied to the analysis of reduced native glycans [88], PMP labeled glycans [89] and permethylated glycans [77]. We recently applied the RPLC method described above in conjunction with MRM to sensitively quantify glycans derived from biological samples such as human blood serum [77]. In this study, transition ions for 88 glycan structures were defined and employed to quantify such glycans present in blood serum. The increased sensitivity attained through MRM experiment enabled effective quantitation of permethylated glycans derived from 0.1μl injection of blood serum.
3.2 Isomeric Separation
One of the many challenges in resolving isomeric and isobaric glycan structures using RPLC is the fact that there are such small differences between isomers in complex glycan structures to prompt hydrophobic changes that could facilitate isomeric separation. Sample complexity and minimal structural differences between the isomers result in isomeric glycans interacting with the stationary phase in a similar fashion which has the consequence of low resolution between isomers or elution of all of the isomers at once. One of the simplest approaches is to have a long linear gradient to separate. This method, however, has a long analysis time. To reduce the analysis time, it is viable to use ultra-high-performance liquid chromatography which increases resolution [26]. As mentioned above, a 2-D LC approach with HILIC has been demonstrated to separate isomeric glycans. Although the separation of isomeric glycans with sialic acids has not been well demonstrated with RPLC, the separation of simpler glycans such as high mannose structures was achieved. However, such separation was only achieved through the use of an ion-pairing reagent and an ANTS tag [82].
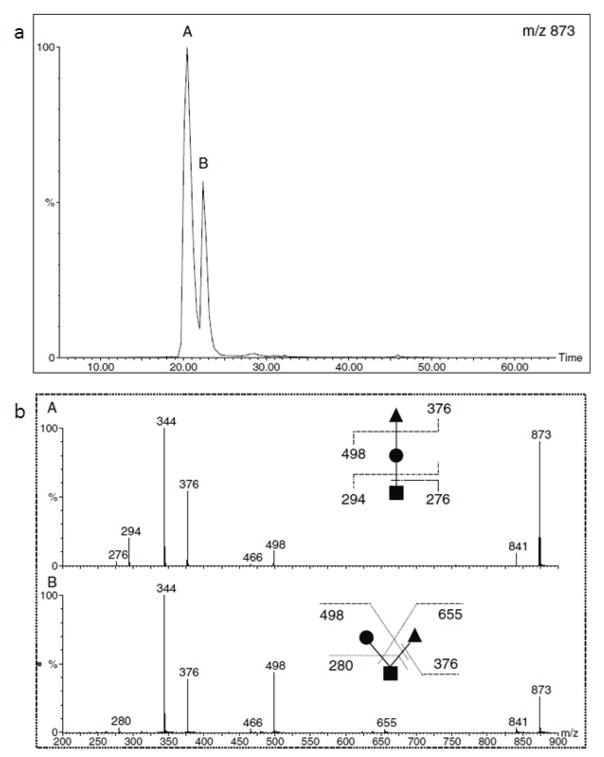
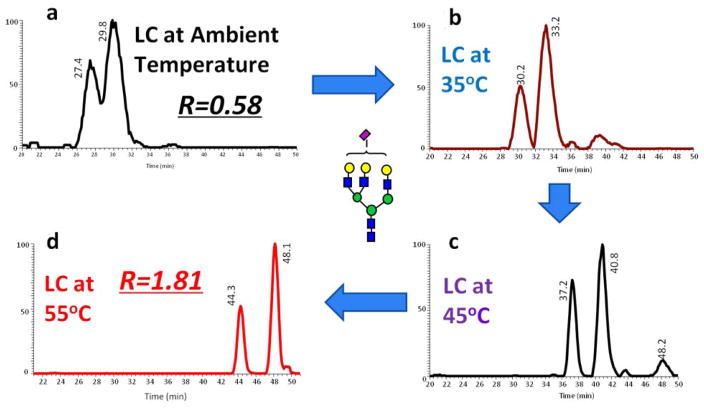
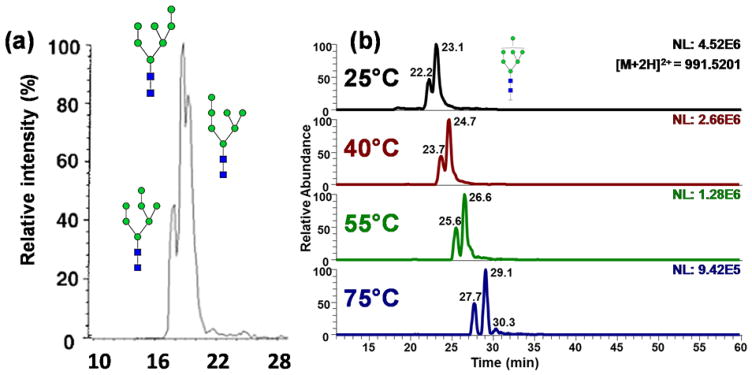
Isomeric separation of simpler oligosaccharides such as O-glycan trisaccharides with RPLC was demonstrated by Hanisch and Müller (Figure 2) [90]. Additionally, using a C18 column, separation of isomers of more complex N-glycans was shown at high temperature (55°C) by Zhou et at. [71]. Figure 3 illustrates the separation of a complex N-glycan on a C18 column at different column temperatures. Extracted ion chromatograms revealed an increase in the separation resolution at higher temperatures with a complete baseline separation attained at 55°C. Although RPLC is much more robust and reproducible than PGC and HILIC columns, the isomeric separation of more complex glycan structures is more viable using PGC or HILIC techniques [26].
Figure 2.
(a) RPLC separation of permethylated isomeric O-glycan trisaccharides. (b) MS/MS spectra of isomers in peaks A and B (upper panel). Reprinted from [90], with permission.
Figure 3.
EICs of tri-antennary mono-sialylated glycan derived from pooled human blood serum eluted by a gradient of 2% ACN, 98% H2O and 100% ACN (each containing 0.1% formic acid) at (a) ambient temperature, (b) 35 °C, (c) 45 °C, (d) 55 °C. Symbols in accordance with the Consortium for Functional Glycomics (CFG) notation. [71].
4. Ion Exchange-LC Separation of Glycans
Glycans are weak acids which make it possible to use ion exchange techniques for their separation [91]. The widely used mode of ion exchange chromatography for the analysis of glycans is high-pH anion-exchange chromatography (HPAEC) coupled with pulsed amperometric detection (PAD) and MS. Using a polymer-based resin, which is stable within a wider pH range than silica, HPAEC has been used to analyze aberrant glycosylation in disease development studies [92, 93]. HPAEC-PAD or –MS enable the detection of glycans at high sensitivity without any derivatization [94].
HPAEC is efficient in resolving glycans under alkaline conditions. Monosaccharides can be ionized to form oxyanions at high pH (0.03 M NaOH) solutions [95]. A monosaccharide contains a series of hydroxyl groups in which ionization capacity decreases as follows: 1-OH>2-OH≥6-OH>3-OH>4-OH [96]. Based on which hydroxyl group is ionized, glycans with different monosaccharides have slightly different interactions with the strong anion exchange stationary phase [95]. Thus the basic conditions of HPAEC allow efficient separation of glycans based on their formal charges, sizes, monosaccharide compositions and intramolecular linkages [97, 98]. Oligosaccharides and glycoforms of glycoproteins have been demonstrated to be essential for many biological functions, medicines and diseases [23, 99, 100]. Therefore, there is a continued need for better approaches to analyze glycans. Although PGC, HILIC, and RPLC have been used in the research of glycosylation, HPAEC still can be utilized as an alternative to these methods due to its unique separation mechanism and selectivity [101]. Grey et al. achieved the development of an HPAEC method for the mapping of glycans [102]. They compared HPAEC-PAD and MALDI-Time of Flight (TOF) MS for the analysis of N-glycoforms of recombinant protein and mAbs and introduced an optimized HPAEC method which was comparable with MALDI-TOF, but had the advantages of higher precision and easier sample preparation.
4.1 HPAEC-MS/MS
Although HPAEC-PAD can quantify glycans at fmol levels [103], the lack of structural information makes it difficult for the identification of glycans from complex biological samples in the absence of reliable standards. Recently, HPAEC has been coupled with tandem MS to acquire a structural information with high sensitivity [104]. The issue that has limited the interfacing between HPAEC and MS has been the high concentration of salts used by HPAEC which significantly inhibits the ionization efficiency of analytes in MS. Nevertheless, this issue can be resolved by an online suppressor that exchanges Na+ with H+. Maier et al. applied HPAEC-MS/MS with a prototype 1 mm online carbohydrate membrane desalter (CMD 300, Thermo Fischer Scientific, Sunnyvale, CA) to characterize and quantify the Fc N-glycans from heterogeneously glycosylated IgGs [33]. By using HPAEC-MS/MS, they concluded this method was an attractive and stable way for the characterization and quantification of complex biological samples. However, a rational for observing highly heterogeneous glycan mixture (specifically hybrid structures) was not adequately addressed.
Recently, Thermo Fisher Scientific has introduced a new desalter, namely Dionex Electrolytically Regenerated Desalter (Dionex ERD 500), which is more pressure tolerant (information related to this desalter are available at https://www.thermofisher.com/). Coulier et al. performed HPAEC-MS to analyze oligosaccharides in lignocellulosic hydrolysates and demonstrated this method was efficient for the separation and identification of many oligosaccharides in biomass hydrolysates and classified them through their degree of polymerization in a single run [105]. Li et al. applied HPAEC-PAD-MS/MS to qualitatively and quantitatively analyze the branches of dextran [106]. They used HPAEC coupled with PAD and QTOF-MS in parallel, where PAD provided quantitation while MS/MS provided the structural information.
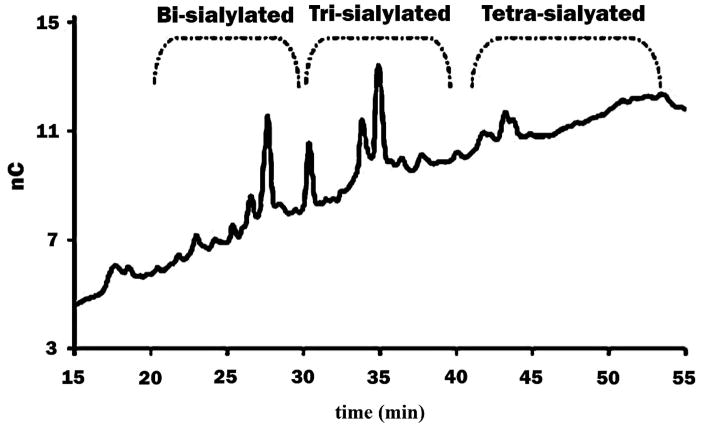
When analyzing complex samples, complete separation of all glycans is still a significant challenge. Therefore 2-D chromatography has been utilized as one way to achieve better separation [107]. In glycosylation studies, sialylated glycans have been shown to play a major role in diseases and biological functions [108, 109]. Sialic acid, generally present at the non-reducing termini of N- or O-glycans, increases glycan acidity. The properties of acidic glycans make it possible to separate them by their number of sialic acids and size, using anion exchange [95]. Figure 4 depicted the separation of sialylated glycans derived from AGP. Glycans elution order was dependent on the number of sialic acid moieties. The Zamze group applied weak anion exchange chromatography to separate glycans released from rat brain tissue and classified them into several major regions of elution based on mono-, di-, tri- and tetra- sialylation and their sizes [110]. With its advantages of specific selectivity based on formal charges, sizes, compositions, and linkages of analytes, ion-exchange chromatography can be combined with other techniques to achieve 2-D analysis. Bones et al. used an anion-exchange column as the first-dimensional separation based on the number of sialic acids and glycan sizes [111]. They then used HILIC as a second-dimensional separation, after enzymatic treatment, to obtain glycan structural information.
Figure 4.
Separation of glycans from AGP by HPAEC. Glycans can be separated by the number of sialic acid residues in their structure. Reprinted and modified from [95], with permission.
4.2 Isomeric Separation
Ion-exchange, specifically anion-exchange, techniques provide possible choices for the isomeric separation of monosaccharides and oligosaccharides. Townsend et al. found that glycans with α-2,3 and α-2,6 sialic acid linkage could be successfully separated through HPAEC, with the presence of α-2,6 linkage resulting in a shorter retention time [112]. They postulated that the reason for the earlier retention time was the blocking of 6-OH, in the α-2,6 linkage, resulting in an overall reduction of charge [113]. Maier et al. demonstrated an isomeric separation by HPAEC-MS/MS when analyzing N-glycans from IgGs [33]. As shown in Figure 5, six glycan isomers were successfully separated including two neutral structures and four sialylated structures. Also, peak patterns were similar between HPAEC-PAD and HPAEC-MS, which suggested that interfacing to MS not adversely influence the efficiency of separation. HPAEC can also provide isomeric separation of linear oligosaccharides. In research by Lin Yi et al. [106], the separation of linkage and branch isomers for dextran samples was achieved. Grey et al. concluded that they separated many structural isomers of glycans released from monoclonal antibodies by HPAEC-PAD [102]. Rohrer et al. also reported that they achieved linkage and branch isomer separation of IgG samples [114].
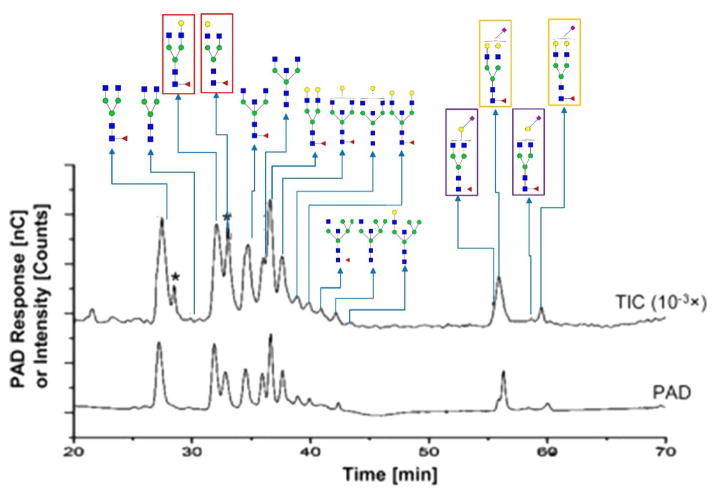
Figure 5.
Isomeric separation performed by HPAEC. HPAEC-PAD chromatograms and total ion current chromatograms from positive-mode HPAEC-MS (intensity attenuation: 10−3×) demonstrating the isomeric separation of PNGase F-released N-glycans from a purified human IgG (h-IgG, 99%). Six branch isomers are highlighted by frames with different colors. The same frame color denotes the isomers from the same glycan composition. Symbols in accordance with CFG notation. A spike peak is indicated by an asterisk. Reprinted and modified from [33].
As an alternative method for glycan studies, HPAEC-MS/MS has been proven to be as efficient as other separation techniques and can provide different selectivity for glycan samples, while achieving good isomeric separation based on the charge and linkage of glycans. However, the lack of nano-scale systems limits its utility for samples of which quantity is limited. Although isomeric separation when using HPAEC has been reported, few studies have focused on this field, which makes further optimization of the technique for this purpose a possibility.
5. PGC-LC Separation of Glycans
PGC columns have been used as a powerful tool to achieve separation of glycans. In contrast to RPLC, PGC provides stronger retention for native glycan species and can retain small glycans such as O-glycans [20]. In recent years, PGC has been commonly used in non-derivatized glycan analysis, yet labeling strategies have also been employed for the separations on PGC [24]. Separation on PGC is facilitated by the combination of RP behavior, based on the hydrophobicity of analytes, and a polar retention effect of graphite, based on the high polarizability of graphitic carbon material. Furthermore, since the column material is planar, the 3-D structure of the analyte also influences retention [115]. Thus, PGC is highly sensitive to structural as well as linkage isomers and able to resolve isomeric glycans [21]. In the past decades, isomeric glycan structures were found to be related to several diseases and considered as potential biomarkers for early diagnosis [116–118]. Therefore, the ability to separate and identify structural and linkage isomers makes PGC-LC-MS a promising analytical method in glycomic studies. Although PGC columns, relative to RPLC, lack robustness and reproducibility of retention time and resolution, retention time shifting can be corrected by regenerating PGC columns by washing with acidic and basic acetonitrile as reported by Pabst et al [26, 32, 119]
5.1 Native Glycans Separated by PGC-LC
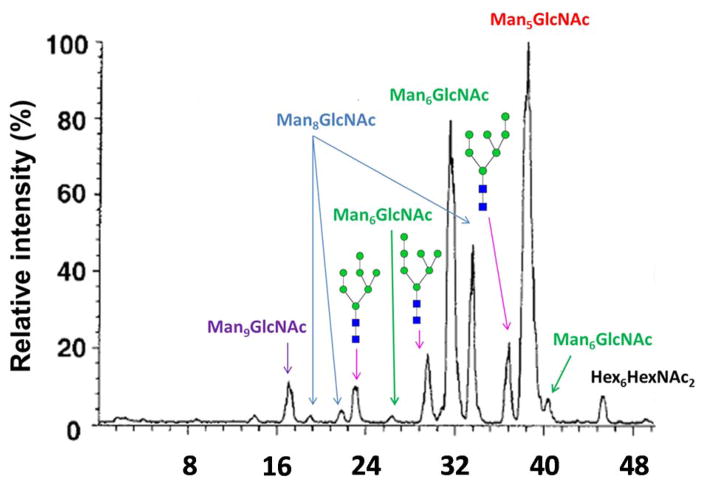
Separations using graphitized carbon chromatography (GCC) dates back to the 1990s. Kawasaki et al. first separated N-glycans released from bovine ribonuclease B (RNase B) and erythropoietin (EPO) in 1999 [120] and 2000 [121], respectively. In the former study, RNase B glycans were released by endoglycosidase H and separated using GCC. Figure 6 depicts the base peak chromatography of RNase B oligosaccharide alditols, where the peaks were assigned to their corresponding structures. Accordingly, Man6, Man7, and Man8 individually had 3 baseline-resolved isomers. MS/MS experiments were used to assign the isomeric structures of Man7 peaks. Man7 standards were utilized to confirm the assignment, by comparing retention time. In the EPO glycan profiling, the isomers were well resolved, yet remained unassigned.
Figure 6.
Base peak chromatogram of RNase B oligosaccharide alditols. Reprinted from [120], with permission.
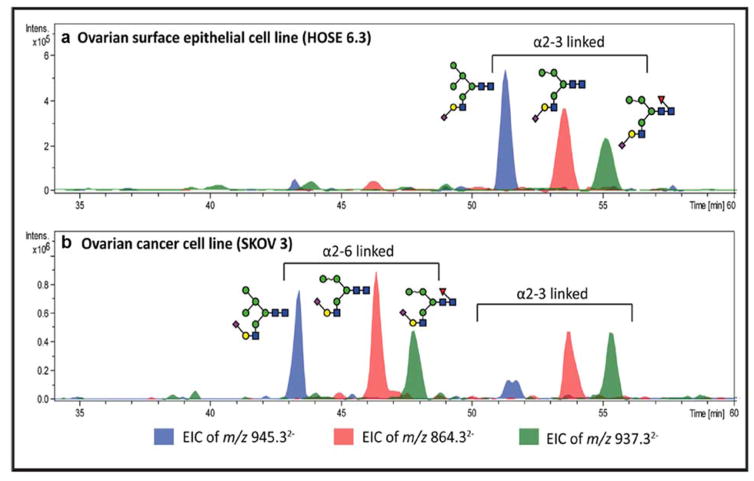
A nano-scaled column packed with PGC used to separate reduced native oligosaccharides as well as O-glycans derived from mucin glycoproteins was introduced by Milady Niñonuevo et al. in 2005. Although the Hex4HexNAc2Fuc1 glycan was separated into several peaks which were determined to be isomers, the structural information of each isomer was not provided or defined [122]. Pabst et al. further separated reduced N-glycans derived from mammalian fibrin on PGC [123]. The linkage isomers of biantennary disialylated structures were well-resolved. The linkage of both sialic acids and galactose were confirmed by applying exoglycosidase digestion towards the corresponding glycans. The voltage influence on the PGC column from the ESI source, solvent gradient, temperature and ionic strength as well as pH within the elution buffer were further studied [32]. This strategy was applied in a study of membrane proteins from ovarian cancer cell lines [124]. The sialic acid linkages of the glycans were identified, with α-2,3 linkage found to have a stronger affinity for PGC than the corresponding α-2,6 sialylated glycans. A representative result is shown in Figure 7.
Figure 7.
EIC of 3 hybrid monosialylated structures in (a) an ovarian surface epithelial cell line and (b) an ovarian cancer cell line. The sialic acid linkage isomers were separated with α-2,6 linked structures eluting earlier than α-2,3 linked species. Reprinted and modified from [124].
Hua et al. achieved the separation and quantitative analysis of non-reduced native glycans derived from two groups of prostate cancer patients with different prognoses [125]. The Agilent HPLC-Chip/TOF-MS was utilized with both enrichment and analytical columns packed with PGC. The extracted ion chromatograms (EIC) are shown in Figure 8. Accordingly, over 100 N-glycans with distinct molecular compositions were identified. The concentration of the identified glycans extended over 5 orders of magnitude. The peaks with the same m/z were considered as glycan isomers and quantified based on ion abundances. 15 isomers from 9 N-glycan compositions were identified as having statistically significant differences between patients with poor prognoses and good prognoses. Since the released N-glycans were not reduced prior to LC-MS analysis, the α and β anomers of native glycans should have been observed. However, several glycan structures were only assigned one peak without anomeric labeling. The same technique was more recently emplyed to profile glycans derived from dried blood spots. In total, 44 N-glycan compositions representing 150 isomers were reported [126]. In both studies, cartoons were utilized to designate peaks.
Figure 8.
ECC of identified glycans from a representative serum sample. Different charge states and adducts were all taken into account. Reprinted and modified from [125], with permission.
5.2 Derivatized Glycans Separated by PGC-LC
5.2.1. Reducing End Labeled Glycans
Although commonly used in HILIC and RPLC, reducing end-labeling techniques have rarely been used in PGC-LC-MS because native glycans can be well retained and separated. A reducing end-labeling approach utilizing N,N-dimethylation with iodomethane after reductive amination was introduced by Broberg et al. [127]. DMBA-derivatized human milk oligosaccharides were analyzed by PGC-LC-MS [128]. Compositional isomers were well resolved and assigned by MS/MS.
5.2.2 Permethylated Glycans
It has been previously demonstrated that permethylation of glycans can significantly increase the ionization efficiency of sialylated species, up to two orders of magnitude [129, 130]. The first isomeric separation of permethylated glycans on PGC was achieved by Costello et al. [131]. Man-7 and Man-8 glycans derived from RNase B were isomerically separated, and isomers were assigned based on fragment patterns in MS/MS experiments. However, the LC resolution of Man-7 was around 0.4 as shown in Figure 9a, which means the peaks were not baseline-resolved. Also, separation of the sialylated species was not shown and considered to generate broader peaks.
Figure 9.
(a) EIC of permethylated Man-7 glycans at [M+2Na]2+ m/z 1013.49. Although the 3 peaks were separated, the resolution was below 1. Reprinted from [131], with permission. (b) EIC of permethylated Man-7 glycans at [M+2H]2+ m/z 991.52. Man7 isomers were base line separated. Reprinted from [132], with permission.
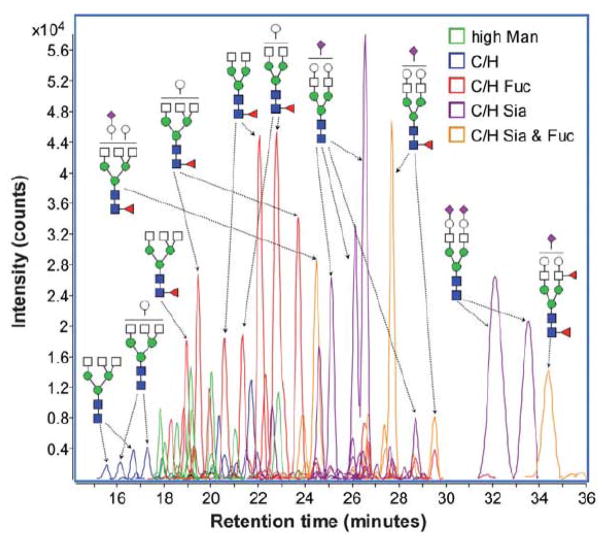
Recently, we achieved an efficient separation of permethylated glycans on PGC under high-temperature conditions [23, 78, 132]. Compared to the previous work, increasing the separation temperature not only enabled a baseline resolution of high mannose glycans (Figure 9b) but also resulted in the separation of glycan isomers originating from the linkage and position of sialic acids and fucose, respectively (Figure 10). It is suggested that the high temperature eliminated the intramolecular interaction of permethylated glycan, thus making the glycans structure interaction with PGC column more homogeneous [132]. However, due to the strong retention of PGC material, the highly sialylated glycans may suffer from elution issues, which can be overcome through the use of buffer additives such as ammonium formate or acetate.
Figure 10.
Extracted ion chromatogram (EIC) of permethylated Hex4HexNAc4Fuc1 glycans at m/z 1017.5413 (a), Hex5HexNAc4Fuc1NeuAc1 glycans at m/z 867.1213 (b) derived from human blood serum, and Hex5HexNAc4Fuc1NeuAc1 glycans at m/z 867.1213 (c) from human milk. Reprinted and modified from [23], with permission.
5.2.3 Methylamidated Glycans
Methylamidation has also been used as a neutralization method to stabilize sialic acid residues and enhance the ionization of sialylated glycans [133, 134]. Recently, Zhang et al. reported an isomeric separation of methyl amidated glycans on PGC [135]. Accordingly, an enhancement of resolution was gained by methyl amidation. Also, it exhibited the ability to separate seven isomers of biantennary disialylated glycans and resulted in the identification of structural isomers based on MS/MS. Three out of the seven peaks had similar tandem MS spectra and were considered as new fetuin glycans, reported for the first time [135].
6. Biomedical Applications
The potential biomedical applications for the isomeric separation of oligosaccharides are far reaching. While the various chromatographic methods and substrates discussed in this review have differing capabilities for the isomeric separation of glycans, ranging from partial to complete baseline separation, examples of biomedical applications will be provided for each chromatographic technique discussed.
The application of HILIC for the isomeric separation of glycans broadly falls into two categories depending on the type of column used, ZIC®-HILIC or Amide/Amine. While some exceptions do exist, such as the novel porous fused silica column with hydroxyl groups mentioned in section 2.1.3, this holds true in the majority of cases. The biological application of ZIC®-HILIC has been shown by Takegawa et al. [44], Mancera-Arteu et al.[45] and Mauko et al.[46]. Takegawa et al. demonstrated the isomeric separation of 2-AP labeled IgG glycans from human serum [44]. Mancera-Arteu et al.[45] utilized the technology for the separation of aniline labeled sialylated glycans from human AGP. While Mauko et al. [46] performed separations of 2-AB labeled N-glycans from recombinant mAbs. A comparable application of an amide/amine column can be found in the work of Tousi et al. [53] utilized a dual labeling strategy where both DMT-MM/methanol derivatization and 2-AB labeling was utilized to characterize and quantify the sialic acid linkages of haptoglobin and human plasma, focusing on trisialylated species. The developed methodology was employed to investigate potential structural alterations in the N-linked oligosaccharide pool enriched from the sera of limited number of samples representating different cancers, including lung (N=4), breast (N=4), ovarian (N=3), pancreatic (N=4) and gastric cancer (N=4) (Figure 1) [53].
Examples of RPLC, utilizing C18 columns, coupled with MS being applied for the isomeric separation of glycans from biologically relevant samples are provided by Dong et al. [23], Zhou et al. [23] and Hanish et al. [90]. In a publication by the Mechref group, Dong et al. analyzed milk oligosaccharides from several mammalian sources and reported partial isomeric separation of permethylated Gal-GlcNAc-lactose isomers derived from human milk [23]. Further, in a subsequent publication by the same group, Zhou et al. achieved baseline resolution of permethylated tri-antennary mono-sialylated glycans derived from pooled human blood serum, with the assistance of elevated temperatures [23]. In the study by Hanish et al. permethylated isomeric O-glycan trisaccharides from fusion protein MUC1-S, expressed in EBNA-293 cells, were separated. However, baseline resolution was not achieved [90]. Additionally, ion exchange chromatography, in the form of HPAEC, coupled with MS/MS has been successfully applied for the separation and characterization of isomeric Fc N-linked glycans of the heterogeneously glycosylated monoclonal antibody (mAb) IgG1 (Figure 5) [33]. Characterization of the type and distribution of N-glycans is of the utmost importance for the pharmacology of mAbs due to the potential impact glycosylation may have on variables such as immunogenicity, protein-protein interactions, dynamics and pharmaco-kinetics [136]. Further examples of ion exchange chromatography being successfully utilized for the isomeric separation of IgG type mAb oligosaccharides are provided by both Grey et al. [102] and Rohrer et al. [114].
PGC represents another chromatographic substrate for the isomeric separation of both native and derivatized glycans from biologically relevant sources. The isomeric separation of reduced native glycans derived from RNase B and EPO [120, 121], mucin glycoproteins [122], mammalian fibrin [32, 123] and the membrane proteins from epithelial ovarian cancer cell lines [124] has been reported. PGC has also been utilized for the isomeric separation of non-reduced native glycans. Examples include the characterization and quantification of glycans from prostate cancer patients with different prognoses, where 15 isomers were identified as biomarkers correlating, in a statistically significant fashion, with good and poor prognoses [125] and the analysis of dried blood spots where 150 isomers of 44 N-glycans were demonstrated [126]. In addition to native glycans, PGC has also been effffeeectively utilized in the isomeric separation of derivatized glycans such as DMBA-labeled free oligosaccharides from human milk [128], methylamidated glycans from fetuin and human blood serum [135] and permethylated glycans released from RNase B [23, 131], IgG and pooled human blood serum [23] as well as free oligosaccharides and N-glycans from human, bovine and goat milk samples [23]. The biomedical applications of the LC-MS methodologies described herein, along with additional examples of the application of these technologies are summarized in Table 1.
Table 1.
Biomedical Applications of LC separation strategies for MS based glycomics
| LC-Strategy | Biomedical Application | Ref. |
|---|---|---|
| HILIC | ||
| ZIC®-HILIC | 2-AP human serum IgG N-glycans | [44] |
| ZIC®-HILIC | Aniline labeled sialylated N-glycans from human AGP | [45] |
| ZIC®-HILIC | 2-AB labeled N-glycans from recombinant mAbs | [46] |
| Amide/Amine | Aniline labeled N-glycoforms of chicken and pheasant ovotransferrin | [47] |
| Amide/Amine | 2-AP labeled IgG N-glycans | [48] |
| Amide/Amine | 2-AB labeled serum N-glycans for identification of cancer-associated alterations in glycosylation | [49] |
| Amide/Amine | 2-AB labeled N-glycans from keyhole limpet hemocyanin, asialofetuin and RNase B | [58] |
| Amide/Amine | DMT/MM and 2-AB labeled N-glycans released from haptoglobin and human plasma (development), and sera from patients with lung, breast, ovarian, pancreatic, or gastric cancer. | [53] |
| fused-silica penta | Procainamide labeled N-glycans from fetuin and human serum | [54] |
| RPLC | ||
| C18 | ANTS derivatized N-glycans from fetuin, RNase B and chicken ovalbumin | [82] |
| C18 | Permethylated glycan alditols from fusion protein MUC1-S expressed in EBNA-293 cells | [90] |
| C18/PGC | Reduced and permethylated free oligosaccharides and N-glycans from human, bovine, and goat milk | [23] |
| C18 | Reduced and permethylated N-glycans from human blood serum for the identification of biomarkers associated with hepatocellular carcinoma | [63] |
| C18 | Reduced and permethylated N-glycans released from RNase B, porcine thyroglobulin, and pooled male human blood serum | [66] |
| C18 | Reduced and permethylated N-glycans released from neuroblastoma cell lines (MYCN-nonamplified SY5Y and MYCN-amplified NLF) | [35] |
| C18 | Reduced and permethylated N-glycans released from ipsilateral cortical brain tissues | [75] |
| C18 | Reduced and permethylated N-glycans released from glioma stem cell xenografts | [76] |
| C18 | Reduced and permethylated N-glycans released from brain-targeting breast carcinoma cells (MDA-MB-231BR) and metastatic breast cancer cells (MDA-MB-231) | [77] |
| HPAEC | ||
| CarboPac PA200 | N-glycans released from glycoproteins of the human SKOV3 ovarian carcinoma cell line | [104] |
| CarboPac PA200 | Isomeric Fc N-linked glycans of the heterogeneously glycosylated mAb IgG1 | [33] |
| CarboPac PA1 | Oligosaccharides in lignocellulosic biomass hydrolysates | [105] |
| CarboPac PA-10 | Qualitative and quantitative analysis of dextran branching patterns | [106] |
| Vydac301VHP57/NPLC/C18 | Sialylated N-glycans in adult rat brain tissue | [137] |
| GlycoSep C/Amine/Amine | Complex N-glycans present on the European Biological Reference Preparation 3 EPO standard | [111] |
| PGC | ||
| PGC | N-glycans released from RNase B | [120] |
| PGC/C18 | Reduced and permethylated N-glycans derived from serum haptoglobin of hepatocellular carcinoma and cirrhosis patients | [78] |
| PGC | N-glycans released from human EPO from CHO cells | [121] |
| PGC | Reduced native N- and O-linked oligosacchides derived from mucin glycoproteins | [122] |
| PGC | Reduced N-glycans derived from mammalian fibrin | [123] |
| PGC | N-glycans derived from bovine thyroid stimulating hormone, fibrin, fetuin, and recombinant EPO and 4E10 from CHO cells | [32] |
| PGC | N-glycan alditols released from membrane proteins from serous ovarian cancer cell lines (SKOV 3, IGROV 1, A2780, and OVCAR 3) and non-cancerous ovarian surface epithelial cell lines (HOSE 6.3 and HOSE 17.1) | [124] |
| PGC | Human milk DMBA oligosaccharides | [128] |
| PGC | Kidney N-glycans in a mouse model of systemic lupus erythematosus | [138] |
| PGC | Reduced and permethylated malto-oligosaccharides, N-glycans released from RNase B and articular cartilage and O-glycans released from C. elegans | [131] |
| PGC | Reduced and permethylated N-glycans released from fetuin, RNase B, IgG and pooled human blood serum. | [23] |
| PGC | Reduced and permethylated N-glycans released from fetuin, RNase B and the two breast cancer cell lines MDA-MB-231 and MDA-MB-231BR. | [132] |
| PGC | Methylamidated sialylated N-glycans released from bovine and human serum | [135] |
| PGC chip | Native glycans derived from two groups of prostate cancer patients with different prognoses | [125] |
| PGC chip | N-glycans released from dried human blood spots | [126] |
7. Concluding Remarks
Recent advancements in chromatographic separation technologies, coupled with MS, have enabled the isomeric separation of glycans, resulting in a greater appreciation of the biological importance of individual isomers. Moving forward, it is essential that future efforts seek the continued improvement of LC technologies, to further understand the biological and biomedical roles glycans play in the diverse functions in which they are involved. Presently, HILIC, C18-RPLC, HPAEC and PGC all represent LC strategies capable of isomeric separation, to varying degrees. The needs for high sensitivity and efficient isomeric separation dictate the development of methods that can address both of these requirements. Permethylation and other recently developed derivatization tagging procedures represent methods capable of providing the high detection sensitivity required. To date, separation on PGC has proven to be the best approach for achieving the efficient isomeric separation required for native, permethylated and derivatized glycans. As the number of laboratories implementing the application of LC methodologies capable of isomeric separation increases, so will our understanding of the rich field of glycobiology.
Acknowledgments
This review paper was supported by CPRIT grant (RP130624) and NIH grant (1R01GM112490-01).
Abbreviations
- NPLC
normal phase liquid chromatography
- ZIC®
Zwitterionic
- PGC
porous graphitic carbon
- HPAEC
high-performance anion-exchange chromatography
- AGP
alpha-1-acid glycoprotein
- GCC
graphitized carbon chromatography
- RNase B
ribonuclease B
- EPO
erythropoietin
- EIC
extracted ion chromatographs
- SRM
selected reaction monitoring
- MRM
multiple-reaction monitoring
- 2-AP
2-amino-pyridine
- AA
anthranilic acid
- 2-AB
2-aminobenzamide
- ABP
2-amino-5-bromopyridine
- ABBE
4-aminobenzoic acid butyl ester
- ABME
4-aminobenzoic acid methyl ester
- ABEE
aminobenzoic acid ethyl ester
- HOA
4-n-heptyloxyaniline
- ANTS
8-aminonaphthalene-1,3,6-trisulfonic acid
- PMP
1-phenyl-3-methyl-5-pyrazolone
- DMT-MM
4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride
Footnotes
Conflict of interest statement
The authors declare no conflict of interest.
References
- 1.Taylor ME, Drickamer K. Current opinion in structural biology. 2014;28:14–22. doi: 10.1016/j.sbi.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohtsubo K, Marth JD. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 3.Yoshida-Moriguchi T, Yu L, Stalnaker SH, Davis S, Kunz S, Madson M, Oldstone MB, Schachter H, Wells L, Campbell KP. Science (New York, NY) 2010;327:88–92. doi: 10.1126/science.1180512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phillips ML, Nudelman E, Gaeta FC, Perez M, Singhal AK, Hakomori S, Paulson JC. Science (New York, NY) 1990;250:1130–1132. doi: 10.1126/science.1701274. [DOI] [PubMed] [Google Scholar]
- 5.Sperandio M, Gleissner CA, Ley K. Immunological reviews. 2009;230:97–113. doi: 10.1111/j.1600-065X.2009.00795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sola RJ, Griebenow K. Journal of pharmaceutical sciences. 2009;98:1223–1245. doi: 10.1002/jps.21504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freeze HH. Current molecular medicine. 2007;7:389–396. doi: 10.2174/156652407780831548. [DOI] [PubMed] [Google Scholar]
- 8.Goulabchand R, Vincent T, Batteux F, Eliaou JF, Guilpain P. Autoimmunity reviews. 2014;13:742–750. doi: 10.1016/j.autrev.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Lefebvre T, Caillet-Boudin ML, Buee L, Delacourte A, Michalski JC. Advances in experimental medicine and biology. 2003;535:189–202. doi: 10.1007/978-1-4615-0065-0_12. [DOI] [PubMed] [Google Scholar]
- 10.Christiansen MN, Chik J, Lee L, Anugraham M, Abrahams JL, Packer NH. Proteomics. 2014;14:525–546. doi: 10.1002/pmic.201300387. [DOI] [PubMed] [Google Scholar]
- 11.Marsh SA, Collins HE, Chatham JC. The Journal of biological chemistry. 2014;289:34449–34456. doi: 10.1074/jbc.R114.585984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alley WR, Novotny MV. Journal of Proteome Research. 2010;9:3062–3072. doi: 10.1021/pr901210r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung CH, Mirakhur B, Chan E, Le QT, Berlin J, Morse M, Murphy BA, Satinover SM, Hosen J, Mauro D, Slebos RJ, Zhou Q, Gold D, Hatley T, Hicklin DJ, Platts-Mills TAE. The New England journal of medicine. 2008;358:1109–1117. doi: 10.1056/NEJMoa074943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dell A, Morris HR. Science (New York, NY) 2001;291:2351–2356. doi: 10.1126/science.1058890. [DOI] [PubMed] [Google Scholar]
- 15.Mechref Y, Novotny MV. Chemical reviews. 2002;102:321–369. doi: 10.1021/cr0103017. [DOI] [PubMed] [Google Scholar]
- 16.Novotny MV, Mechref Y. Journal of separation science. 2005;28:1956–1968. doi: 10.1002/jssc.200500258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alpert AJ. Journal of chromatography. 1990;499:177–196. doi: 10.1016/s0021-9673(00)96972-3. [DOI] [PubMed] [Google Scholar]
- 18.Anumula KR. Analytical biochemistry. 2006;350:1–23. doi: 10.1016/j.ab.2005.09.037. [DOI] [PubMed] [Google Scholar]
- 19.Wuhrer M, de Boer AR, Deelder AM. Mass spectrometry reviews. 2009;28:192–206. doi: 10.1002/mas.20195. [DOI] [PubMed] [Google Scholar]
- 20.Ruhaak LR, Deelder AM, Wuhrer M. Analytical and bioanalytical chemistry. 2009;394:163–174. doi: 10.1007/s00216-009-2664-5. [DOI] [PubMed] [Google Scholar]
- 21.Stavenhagen K, Kolarich D, Wuhrer M. Chromatographia. 2015;78:307–320. doi: 10.1007/s10337-014-2813-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolarich D, Windwarder M, Alagesan K, Altmann F. Glyco-Engineering: Methods and Protocols. In: Castilho A, editor. Methods Mol Biol. Springer; New York: 2015. pp. 427–435. [DOI] [PubMed] [Google Scholar]
- 23.Zhou S, Dong X, Veillon L, Huang Y, Mechref Y. Analytical and bioanalytical chemistry. 2016 doi: 10.1007/s00216-016-9996-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruhaak LR, Zauner G, Huhn C, Bruggink C, Deelder AM, Wuhrer M. Analytical and bioanalytical chemistry. 2010;397:3457–3481. doi: 10.1007/s00216-010-3532-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rohrer JS, Thayer J, Weitzhandler M, Avdalovic N. Glycobiology. 1998;8:35–43. doi: 10.1093/glycob/8.1.35. [DOI] [PubMed] [Google Scholar]
- 26.Vreeker GC, Wuhrer M. Analytical and bioanalytical chemistry. 2016 doi: 10.1007/s00216-016-0073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wuhrer M. Glycoconj J. 2013;30:11–22. doi: 10.1007/s10719-012-9376-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wuhrer M, Koeleman CAM, Hokke CH, Deelder AM. Rapid Communications in Mass Spectrometry. 2006;20:1747–1754. doi: 10.1002/rcm.2509. [DOI] [PubMed] [Google Scholar]
- 29.Higel F, Demelbauer U, Seidl A, Friess W, Sörgel F. Analytical and bioanalytical chemistry. 2013;405:2481–2493. doi: 10.1007/s00216-012-6690-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruhaak LR, Zauner G, Huhn C, Bruggink C, Deelder AM, Wuhrer M. Analytical and bioanalytical chemistry. 2010;397:3457–3481. doi: 10.1007/s00216-010-3532-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou S, Dong X, Veillon L, Huang Y, Mechref Y. Analytical and bioanalytical chemistry. 2017;409:453–466. doi: 10.1007/s00216-016-9996-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pabst M, Altmann F. Analytical chemistry. 2008;80:7534–7542. doi: 10.1021/ac801024r. [DOI] [PubMed] [Google Scholar]
- 33.Maier M, Reusch D, Bruggink C, Bulau P, Wuhrer M, Molhoj M. Journal of chromatography B, Analytical technologies in the biomedical and life sciences. 2016;1033–1034:342–352. doi: 10.1016/j.jchromb.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Hu Y, Zhou S, Yu CY, Tang H, Mechref Y. Rapid communications in mass spectrometry : RCM. 2015;29:135–142. doi: 10.1002/rcm.7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu Y, Mechref Y. Electrophoresis. 2012;33:1768–1777. doi: 10.1002/elps.201100703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buszewski B, Noga S. Analytical and bioanalytical chemistry. 2012;402:231–247. doi: 10.1007/s00216-011-5308-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hemstrom P, Irgum K. Journal of separation science. 2006;29:1784–1821. doi: 10.1002/jssc.200600199. [DOI] [PubMed] [Google Scholar]
- 38.Ahn J, Bones J, Yu YQ, Rudd PM, Gilar M. Journal of chromatography B, Analytical technologies in the biomedical and life sciences. 2010;878:403–408. doi: 10.1016/j.jchromb.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 39.Alpert AJ. Analytical chemistry. 2008;80:62–76. doi: 10.1021/ac070997p. [DOI] [PubMed] [Google Scholar]
- 40.Yoshida T. Journal of biochemical and biophysical methods. 2004;60:265–280. doi: 10.1016/j.jbbm.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 41.Marino K, Bones J, Kattla JJ, Rudd PM. Nature chemical biology. 2010;6:713–723. doi: 10.1038/nchembio.437. [DOI] [PubMed] [Google Scholar]
- 42.Grumbach ES, Diehl DM, Neue UD. Journal of separation science. 2008;31:1511–1518. doi: 10.1002/jssc.200700673. [DOI] [PubMed] [Google Scholar]
- 43.Zauner G, Deelder AM, Wuhrer M. Electrophoresis. 2011;32:3456–3466. doi: 10.1002/elps.201100247. [DOI] [PubMed] [Google Scholar]
- 44.Takegawa Y, Deguchi K, Keira T, Ito H, Nakagawa H, Nishimura S. Journal of chromatography A. 2006;1113:177–181. doi: 10.1016/j.chroma.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 45.Mancera-Arteu M, Gimenez E, Barbosa J, Sanz-Nebot V. Analytica chimica acta. 2016;940:92–103. doi: 10.1016/j.aca.2016.07.043. [DOI] [PubMed] [Google Scholar]
- 46.Mauko L, Lacher NA, Pelzing M, Nordborg A, Haddad PR, Hilder EF. Journal of chromatography B, Analytical technologies in the biomedical and life sciences. 2012;911:93–104. doi: 10.1016/j.jchromb.2012.10.043. [DOI] [PubMed] [Google Scholar]
- 47.Jiang K, Wang C, Sun Y, Liu Y, Zhang Y, Huang L, Wang Z. Journal of agricultural and food chemistry. 2014;62:7245–7254. doi: 10.1021/jf501352j. [DOI] [PubMed] [Google Scholar]
- 48.Yamaguchi Y, Nishima W, Re S, Sugita Y. Rapid communications in mass spectrometry : RCM. 2012;26:2877–2884. doi: 10.1002/rcm.6412. [DOI] [PubMed] [Google Scholar]
- 49.Bones J, Mittermayr S, O’Donoghue N, Guttman A, Rudd PM. Analytical chemistry. 2010;82:10208–10215. doi: 10.1021/ac102860w. [DOI] [PubMed] [Google Scholar]
- 50.Zhao J, Li S, Li C, Wu SL, Xu W, Chen Y, Shameem M, Richardson D, Li H. Analytical chemistry. 2016;88:7049–7059. doi: 10.1021/acs.analchem.6b00636. [DOI] [PubMed] [Google Scholar]
- 51.Melmer M, Stangler T, Schiefermeier M, Brunner W, Toll H, Rupprechter A, Lindner W, Premstaller A. Analytical and bioanalytical chemistry. 2010;398:905–914. doi: 10.1007/s00216-010-3988-x. [DOI] [PubMed] [Google Scholar]
- 52.Wheeler SF, Domann P, Harvey DJ. Rapid communications in mass spectrometry : RCM. 2009;23:303–312. doi: 10.1002/rcm.3867. [DOI] [PubMed] [Google Scholar]
- 53.Tousi F, Bones J, Hancock WS, Hincapie M. Analytical chemistry. 2013;85:8421–8428. doi: 10.1021/ac4018007. [DOI] [PubMed] [Google Scholar]
- 54.Tao S, Huang Y, Boyes BE, Orlando R. Analytical chemistry. 2014;86:10584–10590. doi: 10.1021/ac5020996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li M, Kinzer JA. Rapid communications in mass spectrometry : RCM. 2003;17:1462–1466. doi: 10.1002/rcm.1064. [DOI] [PubMed] [Google Scholar]
- 56.Lattova E, Perreault H. Journal of Chromatography B. 2003;793:167–179. doi: 10.1016/s1570-0232(03)00374-x. [DOI] [PubMed] [Google Scholar]
- 57.Schmid D, Behnke B, Metzger J, Kuhn R. Biomed Chromatogr. 2002;16:151–156. doi: 10.1002/bmc.152. [DOI] [PubMed] [Google Scholar]
- 58.Wuhrer M, Koeleman CAM, Hokke CH, Deelder AM. International Journal of Mass Spectrometry. 2004;232:51–57. [Google Scholar]
- 59.Wuhrer M, Deelder AM, Hokke CH. Journal of chromatography B, Analytical technologies in the biomedical and life sciences. 2005;825:124–133. doi: 10.1016/j.jchromb.2005.01.030. [DOI] [PubMed] [Google Scholar]
- 60.Stephan Kirsch LB. Bioanalysis. 2009;1:1307–1327. doi: 10.4155/bio.09.110. [DOI] [PubMed] [Google Scholar]
- 61.Wuhrer M, Koeleman CA, Deelder AM, Hokke CH. FEBS J. 2006;273:347–361. doi: 10.1111/j.1742-4658.2005.05068.x. [DOI] [PubMed] [Google Scholar]
- 62.Wuhrer M, Koeleman CA, Fitzpatrick JM, Hoffmann KF, Deelder AM, Hokke CH. Glycobiology. 2006;16:991–1006. doi: 10.1093/glycob/cwl020. [DOI] [PubMed] [Google Scholar]
- 63.Wuhrer M, Koeleman CA, Deelder AM. In: Glycomics: Methods and Protocols. Packer NH, Karlsson NG, editors. Humana Press; Totowa, NJ: 2009. pp. 79–91. [Google Scholar]
- 64.Ritamo I, Rabina J, Natunen S, Valmu L. Analytical and bioanalytical chemistry. 2013;405:2469–2480. doi: 10.1007/s00216-012-6680-5. [DOI] [PubMed] [Google Scholar]
- 65.Robijn ML, Koeleman CA, Hokke CH, Deelder AM. Mol Biochem Parasitol. 2007;151:162–172. doi: 10.1016/j.molbiopara.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 66.Zhou S, Hu Y, Veillon L, Snovida SI, Rogers JC, Saba J, Mechref Y. Analytical chemistry. 2016;88:7515–7522. doi: 10.1021/acs.analchem.6b00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Melmer M, Stangler T, Premstaller A, Lindner W. Journal of chromatography A. 2011;1218:118–123. doi: 10.1016/j.chroma.2010.10.122. [DOI] [PubMed] [Google Scholar]
- 68.Royle L, Mattu TS, Hart E, Langridge JI, Merry AH, Murphy N, Harvey DJ, Dwek RA, Rudd PM. Analytical biochemistry. 2002;304:70–90. doi: 10.1006/abio.2002.5619. [DOI] [PubMed] [Google Scholar]
- 69.Williams DK, Meadows CW, Bori ID, Hawkridge AM, Comins DL, Muddiman DC. Journal of the American Chemical Society. 2008;130:2122–2123. doi: 10.1021/ja076849y. [DOI] [PubMed] [Google Scholar]
- 70.Zhou S, Wooding KM, Mechref Y. Methods Mol Biol. 2017:83–96. doi: 10.1007/978-1-4939-6493-2_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yin H, Zhu J, Wu J, Tan Z, An M, Zhou S, Mechref Y, Lubman DM. Electrophoresis. 2016;37:2624–2632. doi: 10.1002/elps.201600176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang Y, Zhou S, Zhu J, Lubman DM, Mechref Y. Electrophoresis. doi: 10.1002/elps.201700025. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hu Y, Desantos-Garcia JL, Mechref Y. Rapid Communications in Mass Spectrometry. 2013;27:865–877. doi: 10.1002/rcm.6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu Y, Zhou S, Khalil SI, Renteria CL, Mechref Y. Analytical chemistry. 2013;85:4074–4079. doi: 10.1021/ac400106x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abou-Abbass H, Bahmad H, Abou-El-Hassan H, Zhu R, Zhou S, Dong X, Hamade E, Mallah K, Zebian A, Ramadan N, Mondello S, Fares J, Comair Y, Atweh S, Darwish H, Zibara K, Mechref Y, Kobeissy F. Electrophoresis. 2016;37:1562–1576. doi: 10.1002/elps.201500583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wildburger NC, Zhou S, Zacharias LG, Kroes RA, Moskal JR, Schmidt M, Mirzaei P, Gumin J, Lang FF, Mechref Y, Nilsson CL. J Proteome Res. 2015;14:3932–3939. doi: 10.1021/acs.jproteome.5b00549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hu Y, Mayampurath A, Khan S, Cohen JK, Mechref Y, Volchenboum SL. J Proteome Res. 2015;14:2074–2081. doi: 10.1021/pr5011718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou S, Hu Y, DeSantos-Garcia JL, Mechref Y. J Am Soc Mass Spectrom. 2015;26:596–603. doi: 10.1007/s13361-014-1054-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou S, Tello N, Harvey A, Boyes B, Orlando R, Mechref Y. Electrophoresis. 2016;37:1489–1497. doi: 10.1002/elps.201600013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Song E, Hu Y, Hussein A, Yu CY, Tang H, Mechref Y. Journal of Proteome Research. 2015;14:2872–2883. doi: 10.1021/acs.jproteome.5b00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Higel F, Demelbauer U, Seidl A, Friess W, Sorgel F. Analytical and bioanalytical chemistry. 2013;405:2481–2493. doi: 10.1007/s00216-012-6690-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gennaro LA, Harvey DJ, Vouros P. Rapid communications in mass spectrometry : RCM. 2003;17:1528–1534. doi: 10.1002/rcm.1079. [DOI] [PubMed] [Google Scholar]
- 83.Mason KE, Meikle PJ, Hopwood JJ, Fuller M. Analytical chemistry. 2006;78:4534–4542. doi: 10.1021/ac052083d. [DOI] [PubMed] [Google Scholar]
- 84.Lattova E, Perreault H. Journal of Chromatography A. 2003;1016:71–87. doi: 10.1016/s0021-9673(03)01297-4. [DOI] [PubMed] [Google Scholar]
- 85.Pabst M, Kolarich D, Pöltl G, Dalik T, Lubec G, Hofinger A, Altmann F. Analytical biochemistry. 2009;384:263–273. doi: 10.1016/j.ab.2008.09.041. [DOI] [PubMed] [Google Scholar]
- 86.Veillon L, Zhou S, Mechref Y. Methods Enzymol. 2017;585:431–477. doi: 10.1016/bs.mie.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Young SP, Stevens RD, An Y, Chen YT, Millington DS. Analytical biochemistry. 2003;316:175–180. doi: 10.1016/s0003-2697(03)00056-3. [DOI] [PubMed] [Google Scholar]
- 88.Hong Q, Ruhaak LR, Totten SM, Smilowitz JT, German JB, Lebrilla CB. Analytical chemistry. 2014;86:2640–2647. doi: 10.1021/ac404006z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sowell J, Wood T. Analytica chimica acta. 2011;686:102–106. doi: 10.1016/j.aca.2010.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hanisch F-G, Müller S. Glycomics: Methods and Protocols. In: Packer NH, Karlsson NG, editors. Methods Mol Biol. Humana Press; 2009. pp. 107–115. [Google Scholar]
- 91.Cataldi TRI, Campa C, De Benedetto GE. Fresenius’ Journal of Analytical Chemistry. 2000;368:739–758. doi: 10.1007/s002160000588. [DOI] [PubMed] [Google Scholar]
- 92.Anderson N, Pollacchi A, Hayes P, Therapondos G, Newsome P, Boyter A, Smith K. Biomed Chromatogr. 2002;16:365–372. doi: 10.1002/bmc.167. [DOI] [PubMed] [Google Scholar]
- 93.Mooney P, Hayes P, Smith K. Biomed Chromatogr. 2006;20:1351–1358. doi: 10.1002/bmc.704. [DOI] [PubMed] [Google Scholar]
- 94.Yamamoto S, Kinoshita M, Suzuki S. J Pharm Biomed Anal. 2016;130:273–300. doi: 10.1016/j.jpba.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 95.Behan JL, Smith KD. Biomed Chromatogr. 2011;25:39–46. doi: 10.1002/bmc.1514. [DOI] [PubMed] [Google Scholar]
- 96.Rendleman JA. Carbohydrates in Solution. In: Isbell HS, editor. Advances in Chemistry. American Chemical Society; 1973. pp. 51–69. [Google Scholar]
- 97.Hardy MR, Townsend RR, Lee YC. Analytical biochemistry. 1988;170:54–62. doi: 10.1016/0003-2697(88)90089-9. [DOI] [PubMed] [Google Scholar]
- 98.Stadheim TA, Li H, Kett W, Burnina IN, Gerngross TU. Nature Protocols. 2008;3:1026–1031. doi: 10.1038/nprot.2008.76. [DOI] [PubMed] [Google Scholar]
- 99.Wooding KM, Peng W, Mechref Y. Current Pharmaceutical Biotechnology. 2016;17:788–801. doi: 10.2174/1389201017666160401145012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Alley WR, Jr, Vasseur JA, Goetz JA, Svoboda M, Mann BF, Matei DE, Menning N, Hussein A, Mechref Y, Novotny MV. J Proteome Res. 2012;11:2282–2300. doi: 10.1021/pr201070k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zheng T, Rohrer J, Rao S. Genetic Engineering & Biotechnology News. 2010:30. [Google Scholar]
- 102.Grey C, Edebrink P, Krook M, Jacobsson SP. Journal of chromatography. B, Analytical technologies in the biomedical and life sciences. 2009;877:1827–1832. doi: 10.1016/j.jchromb.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 103.Davies MJ, Hounsell EF. Biomedical Chromatography. 1996;10:285–289. doi: 10.1002/(SICI)1099-0801(199611)10:6<285::AID-BMC616>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 104.Kandzia S, Costa J. Ovarian Cancer: Methods and Protocols. In: Malek A, Tchernitsa O, editors. Methods in Molecular Biology. Springer Science; 2013. pp. 301–312. [Google Scholar]
- 105.Coulier L, Zha Y, Bas R, Punt PJ. Bioresour Technol. 2013;133:221–231. doi: 10.1016/j.biortech.2013.01.085. [DOI] [PubMed] [Google Scholar]
- 106.Yi L, Ouyang Y, Sun X, Xu N, Linhardt RJ, Zhang Z. Journal of chromatography A. 2015;1423:79–85. doi: 10.1016/j.chroma.2015.10.064. [DOI] [PubMed] [Google Scholar]
- 107.Pucic M, Pinto S, Novokmet M, Knezevic A, Gornik O, Polasek O, Vlahovicek K, Wang W, Rudd PM, Wright AF, Campbell H, Rudan I, Lauc G. Glycobiology. 2010;20:970–975. doi: 10.1093/glycob/cwq052. [DOI] [PubMed] [Google Scholar]
- 108.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, Massague J. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kolbl AC, Andergassen U, Jeschke U. Front Oncol. 2015;5:219. doi: 10.3389/fonc.2015.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zamze S, Harvey DJ, Chen Y-J, Guile GR, Dwek RA, Wing DR. Eur J Biochem. 1998;258:243–270. doi: 10.1046/j.1432-1327.1998.2580243.x. [DOI] [PubMed] [Google Scholar]
- 111.Bones J, McLoughlin N, Hilliard M, Wynne K, Karger BL, Rudd PM. Analytical chemistry. 2011;83:4154–4162. doi: 10.1021/ac200406z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Townsend RR, Hardy MR, Hindsgaul O, Lee YC. Analytical biochemistry. 1988;174:459–470. doi: 10.1016/0003-2697(88)90044-9. [DOI] [PubMed] [Google Scholar]
- 113.Townsend RR, Hardy MR, Cumming DA, Carver JP, Bendia KB. Analytical biochemistry. 1989;182:1–8. doi: 10.1016/0003-2697(89)90708-2. [DOI] [PubMed] [Google Scholar]
- 114.Rohrer JS, Basumallick L, Hurum DC. Glycobiology. 2016;26:582–591. doi: 10.1093/glycob/cww006. [DOI] [PubMed] [Google Scholar]
- 115.West C, Elfakir C, Lafosse M. Journal of chromatography A. 2010;1217:3201–3216. doi: 10.1016/j.chroma.2009.09.052. [DOI] [PubMed] [Google Scholar]
- 116.Hedlund M, Ng E, Varki A, Varki NM. Cancer research. 2008;68:388–394. doi: 10.1158/0008-5472.CAN-07-1340. [DOI] [PubMed] [Google Scholar]
- 117.Rudd PM, Endo T, Colominas C, Groth D, Wheeler SF, Harvey DJ, Wormald MR, Serban H, Prusiner SB, Kobata A, Dwek RA. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:13044–13049. doi: 10.1073/pnas.96.23.13044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Saldova R, Wormald MR, Dwek RA, Rudd PM. Disease markers. 2008;25:219–232. doi: 10.1155/2008/601583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jansen RS, Rosing H, Schellens JHM, Beijnen JH. Journal of Chromatography A. 2009;1216:3168–3174. doi: 10.1016/j.chroma.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 120.Kawasaki N, Ohta M, Hyuga S, Hashimoto O, Hayakawa T. Analytical biochemistry. 1999;269:297–303. doi: 10.1006/abio.1999.4026. [DOI] [PubMed] [Google Scholar]
- 121.Kawasaki N, Ohta M, Hyuga S, Hyuga M, Hayakawa T. Analytical biochemistry. 2000;285:82–91. doi: 10.1006/abio.2000.4739. [DOI] [PubMed] [Google Scholar]
- 122.Ninonuevo M, An H, Yin H, Killeen K, Grimm R, Ward R, German B, Lebrilla C. Electrophoresis. 2005;26:3641–3649. doi: 10.1002/elps.200500246. [DOI] [PubMed] [Google Scholar]
- 123.Pabst M, Bondili JS, Stadlmann J, Mach L, Altmann F. Analytical chemistry. 2007;79:5051–5057. doi: 10.1021/ac070363i. [DOI] [PubMed] [Google Scholar]
- 124.Anugraham M, Jacob F, Nixdorf S, Everest-Dass AV, Heinzelmann-Schwarz V, Packer NH. Molecular & cellular proteomics : MCP. 2014;13:2213–2232. doi: 10.1074/mcp.M113.037085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hua S, An HJ, Ozcan S, Ro GS, Soares S, DeVere-White R, Lebrilla CB. The Analyst. 2011;136:3663–3671. doi: 10.1039/c1an15093f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ruhaak LR, Miyamoto S, Kelly K, Lebrilla CB. Analytical chemistry. 2012;84:396–402. doi: 10.1021/ac202775t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Broberg S, Broberg A, Duus JO. Rapid communications in mass spectrometry : RCM. 2000;14:1801–1805. doi: 10.1002/1097-0231(20001015)14:19<1801::AID-RCM96>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 128.Broberg A. Carbohydrate research. 2007;342:1462–1469. doi: 10.1016/j.carres.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 129.Mechref Y, Kang P, Novotny MV. Methods Mol Biol. 2009;534:53–64. doi: 10.1007/978-1-59745-022-5_4. [DOI] [PubMed] [Google Scholar]
- 130.Kang P, Mechref Y, Novotny MV. Rapid communications in mass spectrometry : RCM. 2008;22:721–734. doi: 10.1002/rcm.3395. [DOI] [PubMed] [Google Scholar]
- 131.Costello CE, Contado-Miller JM, Cipollo JF. J Am Soc Mass Spectrom. 2007;18:1799–1812. doi: 10.1016/j.jasms.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhou S, Dong X, Huang Y, Vellion L, Mechref Y. Analytical chemistry. Submitted. [Google Scholar]
- 133.Liu X, Qiu H, Lee RK, Chen W, Li J. Analytical chemistry. 2010;82:8300–8306. doi: 10.1021/ac101831t. [DOI] [PubMed] [Google Scholar]
- 134.Nishikaze T, Kawabata S, Tanaka K. Analytical chemistry. 2014;86:5360–5369. doi: 10.1021/ac500340t. [DOI] [PubMed] [Google Scholar]
- 135.Zhang Q, Feng X, Li H, Liu BF, Lin Y, Liu X. Analytical chemistry. 2014;86:7913–7919. doi: 10.1021/ac501844b. [DOI] [PubMed] [Google Scholar]
- 136.Reusch D, Tejada ML. Glycobiology. 2015;25:1325–1334. doi: 10.1093/glycob/cwv065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zamze S, Harvey DJ, Chen Y, Guile GR, Dwek RA, Wing DR. Eur J Biochem. 1998;258:243–270. doi: 10.1046/j.1432-1327.1998.2580243.x. [DOI] [PubMed] [Google Scholar]
- 138.Hashii N, Kawasaki N, Itoh S, Nakajima Y, Kawanishi T, Yamaguchi T. Immunology. 2009;126:336–345. doi: 10.1111/j.1365-2567.2008.02898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]