Abstract
Microbial archaeology is flourishing in the era of high-throughput sequencing, revealing the agents behind devastating historical plagues, identifying the cryptic movements of pathogens in prehistory, and reconstructing the ancestral microbiota of humans. Here, we introduce the fundamental concepts and theoretical framework of the discipline, then discuss applied methodologies for pathogen identification and microbiome characterization from archaeological samples. We give special attention to the process of identifying, validating, and authenticating ancient microbes using high-throughput DNA sequencing data. Finally, we outline standards and precautions to guide future research in the field.
Keywords: ancient DNA, metagenomics, microbiology, bacteria, pathogens, microbiome, high-throughput sequencing
1. INTRODUCTION
In 2011, the first fully reconstructed ancient bacterial genome sequence was published—that of Yersinia pestis—which confirmed at least one of the etiological agents of the Black Death pandemic (16) and put to rest years of controversy that had dogged polymerase chain reaction (PCR)–based attempts to identify the pathogen in archaeological samples (41, 57, 58, 154, 155). Other ancient microbial genome sequences quickly followed, including those from other plague epidemics (14, 47, 158, 178, 192) and additional pathogens, such as Mycobacterium leprae (170), Mycobacterium tuberculosis (13), Tannerella forsythia (196), Brucella melitensis (85), and Helicobacter pylori (112). The key turning point was the availability of high-throughput sequencing (HTS), a transformative innovation in DNA sequencing (114, 115), and sequence capture enrichment methods (16, 20, 62, 75)—two techniques that increased both sample throughput and data output by orders of magnitude. These advancements revolutionized ancient DNA (aDNA) research and, more broadly, ushered in the era of genomics nearly overnight (92, 140).
However, with these technological advances come new challenges. Tools and techniques are needed to sort, evaluate, authenticate, and interpret the hundreds of millions of DNA sequences that have now become the standard output of genomics and paleogenomics laboratories alike. Numerous protocols, scripts, pipelines, and computational environments are available, as are a myriad of genetic and genomic databases, but the rapid proliferation of these tools has left many uncertain about which ones to use and when to use them. For example, the decision to use either alignment-based or alignment-free taxonomic classifiers can have a strong impact on microbial community reconstruction (106). Likewise, the choice of reference databases can greatly affect taxonomic assignment (149) and, consequently, the false positive and false negative rates of pathogen detection. Similar but nonequivalent choices in parameter settings can introduce systematic biases, leading to spurious sequence alignments and false claims, and failure to statistically account for both biological and taphonomic factors in the selection of appropriate analysis pipelines and statistical tests can result in inaccurate conclusions.
As the complexity of paleogenomic data analysis increases, standards and guidelines for best practices are required to ensure not only high-quality data generation, but also accurate and meaningful data interpretation. Numerous challenges face the growing field of microbial archaeology— some stemming from the way microbes reproduce and recombine during life, others shared with genomics more generally, and still others specific to ancient and degraded samples. Concerted effort will be required by the research community to identify and address these challenges in order to achieve a robust and established scientific discipline.
In March 2016, the Max Planck Institute for the Science of Human History hosted the first Standards, Precautions, and Advances in Ancient Metagenomics conference in order to identify and discuss the challenges involved in analyzing ancient microbial metagenomic data. Here, we present the outcomes of this meeting and outline a series of precautions and best practices for the emerging field of microbial archaeology.
2. RESEARCH DIRECTIONS IN MICROBIAL ARCHAEOLOGY
Research directions within the growing field of microbial archaeology can generally be divided into two paths: pathogenomics and microbiome studies. The former focuses on understanding pathogen evolution and host-microbe interactions involved in disease states (138), whereas the latter focuses on understanding the diversity, structure, and function of endogenous microbial communities and their interactions with the host during both health and disease states (78, 79). In general, pathogenomics is concerned primarily with individual disease-causing microorganisms, such as those causing plague (Y. pestis), tuberculosis (M. tuberculosis), or leprosy (M. leprae), whereas microbiome studies focus more on the distribution and diversity of the microbes native to a given host and their role in host functions, such as digestion, immune system stimulation, and chronic inflammation.
There is a great deal of overlap between these two disciplines in practice, as pathogenomics may include polymicrobial infections (e.g., dental caries) or mixed coinfections (e.g., pneumonia and tuberculosis), and microbiome studies may focus on keystone taxa that disproportionately drive community behavior (e.g., Streptococcus mutans or Porphyromonas gingivalis). Additionally, both disciplines rely heavily on metagenomic sequence data, and thus many of their analytical tools are shared or similar.
2.1. The Growth of the Field
Microbial archaeology can trace its origins back several decades, and early research in the field focused on targeted PCR amplification of short specific loci, followed by electrophoretic characterization or Sanger DNA sequencing. Mycobacterial spoligotyping of skeletal lesions (208) and sequencing of amplified 16S ribosomal RNA (rRNA) gene clones from paleofeces (23) are characteristic of paleomicrobiology approaches in the pre-HTS era. However, these low-throughput techniques, which were adapted from protocols originally developed for clinical and ecological applications, have several drawbacks when applied to ancient and degraded samples from environmental contexts. First, targeted PCR typically requires long (>100 base pairs), well-preserved DNA templates, which are not characteristic of the vast majority of authentic aDNA fragments (64, 181); second, ancient samples typically require a large number (>35) of PCR cycles for successful target amplification, which makes this approach particularly sensitive to background and environmental contamination; third, cloning and Sanger sequencing do not allow efficient investigation of template damage patterns in order to authenticate aDNA sequences; fourth, targeted PCR is particularly susceptible to amplification biases, including both off-target and skewed PCR amplification, as well as taxonomic dropout; and finally, the experimental replicability of studies using these techniques is generally low, and the results have proven to be difficult to independently authenticate or validate (57, 64, 199, 207). Such problems reached a critical point in 2005, when a prominent review of aDNA research summed up the field of microbial archaeology as “the microbial problem” and largely dismissed it as a discipline (200).
The advent of HTS technologies in the mid-2000s presented a powerful solution to the inherent shortcomings of conventional PCR-based approaches, and this new technology has dramatically influenced the field of microbial archaeology. Today, nearly all ancient microbial research utilizes HTS-based techniques, and multiple sequencing platforms and analytical strategies are available. The situation mirrors that of genetic research on ancient humans, which at first was hampered by contamination concerns resulting from PCR amplification and Sanger sequencing–based approaches but is now flourishing in the post-HTS era (65, 157, 159).
2.2. Definition of Terms
This article focuses on the analysis of metagenomic (all available DNA) data obtained from HTS shotgun-sequenced (untargeted) or sequence-captured (target-enriched) genes and genomes obtained from a microbiota (an assemblage of microorganisms) present within a microbiome (a defined microbial ecosystem) (113). Archaeological samples typically contain mixtures of endogenous (antemortem) and exogenous (postmortem) microbial DNA that may include host-associated commensal taxa (e.g., oral microbes in dental calculus), epidemic pathogens (e.g., Y. pestis in the pulp cavity of teeth), and environmental bacteria (e.g., soil microbes involved in decomposition). Additionally, contaminating DNA sequences from handling (e.g., skin microbes), storage conditions (e.g., bacteria and fungi overgrowth), and laboratory sources (e.g., reagents contaminated with enzyme expression vectors) may also be present.
This addition of both ancient and modern exogenous microbial DNA in archaeological remains makes ancient pathogen and microbiome studies more complicated than investigations of fresh samples. For example, in contrast to a freshly cultured clinical specimen, which would typically contain a single clonal pathogen and no other major DNA sources, analysts of ancient pathogens must grapple with complex host and environmental backgrounds, potentially including nonpathogenic, soil-derived relatives of the pathogen of interest. Postmortem colonization and contamination also present challenges for microbiome analysis by skewing diversity metrics and inflating community membership. It is thus important to note the distinction between ancient endogenous microbiota, which are the host-associated microbes that were present during life, and exogenous microbiota, which include both decomposition-related and recent contaminant taxa.
3. WHAT IS A MICROBIAL SPECIES?
Before endogenous microbiota can be analyzed, it is first important to define what microbes are. For the purposes of this review, we define microbes as members of the prokaryotic domains Bacteria and Archaea. Microeukaryotes and viruses are thus beyond the scope of this review, even though noteworthy achievements have been made in the successful genetic characterization of potato late blight evolution (116, 205), barley stripe mosaic virus (176), Spanish influenza strains (189), early simian immunodeficiency virus (150), and seventeenth- and eighteenth-century smallpox strains (12, 42).
Although species annotations are routinely applied to microbial taxa, there is relatively little consensus on what a microbial species actually is (2, 40). Unlike Ernst Mayr’s birds, microbes adhere to few, if any, of the tenets of the biological species concept (35, 72, 117), and although many microbial species concepts have been proposed, none have been widely accepted (2). This is largely because although microbes reproduce asexually by binary fission at a cellular level, they also exchange genetic information horizontally—including across taxonomically divergent groups. The discovery of such microbial mating systems earned the 1958 Nobel Prize in Physiology or Medicine for molecular biologist Joshua Lederberg (82), who, incidentally, later went on to popularize the term microbiome in 2001 (101).
At the heart of the microbial species problem is a tension between methods-based and methods-free species definitions, in part reflecting philosophical differences in the fields of microbial systematics and evolutionary biology. At a crude level, methods-based approaches are objectively measurable, but they suffer from the fact that methods continuously change with new technologies and that the species criteria that have been established are largely arbitrary. Methods-free definitions are more grounded in evolutionary theory but are often unmeasurable (2). In the genomics era, methods-based definitions currently prevail as pragmatic solutions to allow researchers to name and discuss taxonomic groups using Linnaean taxonomy, a stopgap measure that is both unsatisfying and at times misleading but is also necessary to allow investigations of what are essentially phenotypic and genetic clusters (29) of metapopulation lineages (2) that defy easy classification.
3.1. Pragmatic Definitions
For historical reasons, the gold standard of pragmatic solutions to the microbial species problem is the characterization of genome similarity based on reciprocal, pairwise DNA reassociation values under controlled conditions. Microbes whose reassociation values are ≥70% in DNA hybridization experiments are generally considered to belong to the same species, in part because this threshold generally recapitulates classical species distinctions based on phenotypic traits (2). Because the method is empirical and requires purified genomic DNA from both microbes being tested, it can be applied only to cultivable microbes. Given that only a small fraction of microbial taxa can currently be cultivated using known techniques (147, 193), this definition is poorly suited to the identification of most microbial species. Moreover, because of the highly fragmented nature of aDNA, this method cannot be applied to ancient samples.
Alternatively, the 16S rRNA gene can be PCR amplified from a pool of noncultured microbes, and the resulting sequence identities can be calculated as a proxy for DNA reassociation values. A cutoff of roughly 97–99% sequence identity for the full gene generally correlates with species boundaries determined by DNA reassociation (118, 190). Taxa defined by their 16S rRNA gene sequence alone are not described as species, but rather as operational taxonomic units (OTUs)— convenient measurable proxies for microbes that are related by descent. Although the term OTU is generally used to refer to a species-like unit, it can theoretically represent microbial biodiversity at any level as long as its definition is clear and consistent (77).
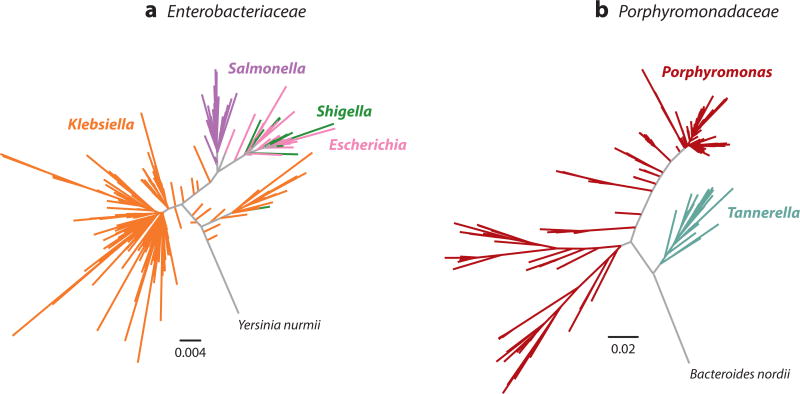
Because 16S rRNA gene amplification and sequencing can be performed on mixed microbial communities without cultivation, it is a powerful method for the discovery of novel taxa; however, this method also has important limitations. Current short-read HTS technologies, such as Illumina sequencing by synthesis, do not allow for deep sequencing of the full ~1,540-basepair-long 16S rRNA gene; instead, maximum achievable read lengths typically limit analysis to one or more of the gene’s nine shorter hypervariable regions. However, even these short regions are generally longer than most aDNA fragments (207). Taxonomic resolution varies across these regions (207), effectively reducing confident taxonomic assignment to the level of genus or family for many groups. This reduction in resolution is not consistent across microbial phyla and tends to disproportionately affect certain groups (207). Emerging technologies, such as Pacific Biosciences’ single-molecule real-time sequencing, are capable of sequencing full genes and may soon replace hypervariable-region short-read sequencing in metataxonomic studies of modern samples (167); however, the highly fragmented nature of aDNA strongly limits the benefit of this technology for ancient samples. Nevertheless, even with full-length 16S rRNA gene sequences, taxonomic assignment can be problematic for some microbial groups (Figure 1). For example, gut bacteria belonging to the family Enterobacteriaceae are generally poorly resolved by 16S rRNA sequences, with the clinically distinctive genera Escherichia, Salmonella, Shigella, and Klebsiella essentially forming one 16S rRNA gene sequence cluster (Figure 1a), whereas other groups, such as the oral genera Porphyromonas and Tannerella, are monophyletic and can be easily distinguished on the basis of 16S rRNA sequences alone (Figure 1b).
Figure 1.
Evolutionary relationships of taxa within the bacterial families (a) Enterobacteriaceae and (b) Porphyromonadaceae based on full-length 16S rRNA gene sequences. Taxonomy and phylogeny are incongruent for the gut-associated genera Klebsiella, Salmonella, Escherichia, and Shigella, which are not monophyletic, but rather exhibit polyphyletic and paraphyletic clade structure. By contrast, taxonomy and phylogeny are congruent for the oral-associated genera Tannerella and Porphyromonas, which form distinct monophyletic clades with high bootstrap support (38). Trees are shown relative to outgroup taxa within the same bacterial family. Note the difference in scales between the two trees. Supplemental Appendix 1 provides the specific parameters used in tree construction.
Finally, unlike most microbial genes, the number of 16SrRNA gene copies per genome is highly variable, ranging from 1 to 5 in archaea and from 1 to 15 or more in bacteria (3, 102). Microbial rRNA (rrn) genes are typically colocated into an operon, and operon copy number is associated with microbial habitat and lifestyle (102, 180). Operon copy number is only weakly correlated with taxonomic ranks of genus and higher, and in some cases copy number even varies within species (190). Among archaea, >60% of taxa have a single rrn operon, but among bacteria, >60% of taxa have three or more copies, and up to seven copies are commonly found (3). Although 16S rRNA gene copies may undergo homogenization through gene conversion (68), different sequences are observed within a single species and even within a single genome. Fewer than 40% of taxa with multiple 16S rRNA genes have identical 16S rRNA sequences in each operon, although sequence divergence between copies is typically < 1% (3, 190). 16S rRNA gene reference databases generally do not take this into account, and instead contain composite or consensus sequences obtained from simultaneous PCR amplification and pooled sequencing of all 16S gene copies on a genome (90). The combined effects of multiple rrn operons per genome and different 16S rRNA gene sequences per operon result in systematic skewing of relative taxonomic abundance and overestimation of microbial diversity in mixed microbial communities (190). The Ribosomal RNA Operon Copy Number Database (rrnDB) maintains updated, annotated lists of rRNA operon copy numbers (180).
Other widely used methods for defining species boundaries include multilocus sequence analysis and multilocus sequence typing, which are similar to the method described above but rely on a panel of usually seven to ten core genes rather than focusing on a single gene (60, 146), as well as genome-wide average nucleotide identity, which compares the sequences of all orthologous genes in the complete genomes of species pairs (87, 160). Average nucleotide identity is in some ways simply a methodological update of the DNA reassociation approach, in which a 95–96% average nucleotide identity is roughly equivalent to a 70% DNA reassociation value (87, 160); however, it therefore also suffers from the same problem that complete genomes are required either from cultivated isolates or from genomes painstakingly assembled in silico from deep-sequenced shotgun or sequence-enriched metagenomic data sets, making it difficult to apply in general, but especially in the context of microbial archaeology.
When assigning taxonomy to metagenomic data, many popular tools use a combination of core gene–focused and whole-genome data, and such an approach is favored in emerging taxonomic tools such as the Metagenomic Intra-Species Diversity Analysis System (MIDAS) pipeline (129), which seeks to characterize strain-level differences in mixed microbial communities. Departing from these approaches are those based on k-mer binning, such as algorithms implemented by Kraken (201), which differ from a gene-centered focus and instead mine taxonomic information from reference databases containing frequency distributions of short sequence fragments (k-mers) across a range of known taxa. Because both of these tools rely on relatively short DNA sequences for taxonomic classification, they are particularly amenable to studies of ancient microbes.
3.2. Complicating Factors
Although microbes reproduce asexually, they do not transmit genetic information in a strictly vertical manner. Microbes are capable of—and frequently do—horizontally transfer genes, plasmids, transposons, and other genetic elements by a wide range of means, including transformation (uptake of DNA from the environment), conjugation (direct transfer of DNA between cells via a pilus), and transduction (transfer of DNA by viruses). Collectively, these processes are referred to as horizontal gene transfer or lateral gene transfer (132, 185), and the transferred DNA can subsequently gain enhanced permanence in the cell through homologous recombination or insertion into the host chromosome.
Although most horizontal gene transfer occurs between related taxa, DNA can also be transferred across higher taxonomic ranks, and even across domains (63). Horizontal gene transfer can also transcend time through the uptake of short, degraded aDNA fragments into living cells (136). Within the context of the microbiome, some bacterial members of a biofilm are prolific producers of extracellular DNA, which they use as a scaffold to anchor themselves in space (61, 67, 198). Given the close proximity and metabolic cooperation of diverse taxa within biofilms, such extracellular DNA serves as an important source of genetic material for horizontal gene transfer via transformation and is thought to be a major factor in the spread of virulence and antibiotic resistance genes within host-associated microbiota (144).
The fluidity by which microbes can acquire—and also lose—large portions of their genomes has no parallel among macroorganisms. Within a given microbial species, the number of genes frequently varies by as much as 20% across strains. For example, genome size among 17 strains of the periopathogen P. gingivalis ranges from 2.2 to 2.4 Mb, a difference of 8%, but the number of genes differs by 22%, ranging from 1,870 genes in strain F0569 to 2,405 genes in strain JCVI SC001. This is true even though these strains exhibit >99.4% sequence identity in the 16S rRNA gene and >98.8% sequence identity across a panel of 11 housekeeping genes (coa, dnaK, ef-tu, ftsQ, gdpxJ, hagB, mcmA, nah, pga, recA, and pepO) (93) [analysis performed on all complete or nearly complete (scaffolded) genomes available in GenBank as of November 2016; for details, see Supplemental Appendix 1]. By contrast, all members of a eukaryotic species carry a nearly identical gene set, and 75% of human genes have homologs in the genome of the puffer fish Takifugu rubripes, which diverged from mammals more than 450 Mya (8, 148).
To account for these vast differences in genome size and gene content among strains, the collective genomes of all members of a microbial species-level clade (a monophyletic group of related taxa) are conceptualized as having two parts: a core genome and a pan-genome. The pan-genome, a term first introduced in 2005, consists of all genes within all strains of a species-level clade (184), whereas the core genome represents a subset of genes that are generally shared among strains. The core genome is variably defined, but the National Center for Biotechnology Information defines it as comprising the genes that are present in >80% of all genomes within a species-level clade (183). By contrast, the minimum core genome is defined as the number of genes shared by all genomes within a special-level clade or equivalent. By either definition, the core genome comprises primarily housekeeping genes involved in replication, transcription, translation, and other basic cell functions required for life (118). In general, core genes of well-studied clades make up ~70–80% of the pan-genome (183).
Despite being relatively central to cell function, core genes can undergo homologous recombination, a process known as core genome transfer. Core genes involved in transcription and translation, such as rRNAs, recombine only rarely (28, 89, 99), but recombination rates of other core genes can be high (202). Streptococcus is an important host-associated genus known for high levels of genome plasticity and recombination. In one study of streptococcal human and agricultural pathogens, core genome recombination was detected in all investigated streptococcal lineages, and 18–37% of the core genome was estimated to be recombinant (103). Core genome transfer rates vary considerably among taxa. H. pylori, Salmonella enterica, Streptococcus pneumoniae, Neisseria meningitidis, and Neisseria lactamica are human-associated bacteria with unusually high core genome recombination rates, in which nucleotide changes resulting from recombination exceed those arising from mutation by more than fivefold; by contrast, recombination rates in Staphylococcus aureus, Lactobacillus casei, Bartonella henselae, and Bordetella pertussis are fivefold lower than mutation rates (191). Core genome recombination occurs most frequently in taxa that are naturally competent (genetically capable of transformation), but it has also been documented in noncompetent cells at genetic loci in proximity to mobile elements (44, 145, 191).
Noncore genes of the pan-genome include many gene types that may be involved in adaptation to various nutrient sources or environmental conditions, and they may or may not be carried on mobile elements. Noncore genes that are found within >20% of strains are called accessory genes. Those found in 1–20% of strains and in <1% of strains are called dispensable and unique genes, respectively, and they are more common than accessory genes (122, 183).
The nonvertical transfer of DNA among microbes serves as a mechanism to increase genetic diversity beyond that introduced through mutation alone, and it plays a major role in microbial evolution (34, 132). This fundamental process, however, complicates attempts to define species boundaries and to trace the evolutionary history of microbial lineages, and it has led some to argue that no natural classification system can be described for microbes because their evolutionary relationships are web-like rather than tree-like (10, 39). However, not all taxa freely exchange genetic information (191), and not all genes transfer easily or frequently (202, 203). For example, monomorphic pathogens that reproduce primarily by clonal expansion show little evidence of recombination over broad timescales (1, 203), and core housekeeping genes that are informational in nature rarely transfer or recombine (99). Consequently, the ancient genomes of monomorphic pathogens, such as M. leprae and M. tuberculosis, are easier to reconstruct than those of commensal taxa, such as H. pylori or T. forsythia (13, 112, 170, 196). Despite the messiness of microbial phylogenies (66), however, microbes generally behave as ecologically coherent entities at the levels of species, genus, family, and order, as currently defined by 16S rRNA gene sequence cutoffs (148).
4. THE POWER AND PITFALLS OF NAMES
Names are powerful entities that allow microbial taxa to be discussed and analyzed in a meaningful way. However, given the heterogeneous phenotypic, genetic, genomic, and metagenomic means by which microbial taxa are detected and observed, it is difficult to devise a single nomenclature system. Instead, overlapping systems of both formal and provisional schemes are currently in use, which both facilitate and limit the study of individual microbes and communities, as well as the reconstruction of ancient microbial genomes and microbiota.
4.1. Valid Species Names and Microbial Systematics
Despite the difficulty of defining what a microbial species is, methods for granting valid microbial species names are outlined by the International Code of Nomenclature of Bacteria set forth by the International Committee on Systematics of Prokaryotes (ICSP; http://www.the-icsp.org) (98, 186). This code governs all microbial taxonomic assignments at and below the Linnaean rank of class (141); however, only the rank of species has a formal definition: “[A] species is a category that circumscribes a (preferably) genomically coherent group of individual isolates/strains sharing a high degree of similarity in (many) independent features, comparatively tested under highly standardized conditions” (179, p. 1044; see also 162). The ICSP requires all new taxa to be published in the International Journal of Systematic and Evolutionary Microbiology, and minimal standards for the description of new species have been established by ICSP subcommittees (51). These standards include (a) isolation of the new species in pure culture, (b) 16S rRNA gene sequencing to establish phylogenetic position, (c) morphological description, (d) chemotaxonomic characterization to establish genus affiliation, (e) explanation of the genotypic and phenotypic basis for species differentiation, and ( f ) deposition of the type strain in at least two permanently established culture collections in two different countries (84). Genome sequencing is not currently required for the establishment of new microbial species, nor is genome sequencing alone sufficient to establish a new species. The List of Prokaryotic Names with Standing in Nomenclature (LPSN; http://www.bacterio.net) maintains an updated list of valid taxa (141).
4.2. Naming the Nameless
Given the emphasis placed on structural and functional properties of microbial isolates, taxa that cannot be grown in pure culture—either because their growth conditions are unknown or because they are parasitic and require the presence of other microbes to grow—are typically limited to candidatus (candidate) status. For example, the candidate phylum Saccharibacteria (formerly TM7), which includes at least 12 members in the human oral cavity, has proven very difficult to isolate in pure culture (21). The only successfully cultivated phylotype to date, provisionally named TM7x, was determined to be an epibiont (an organism that lives on the surface of another organism) of the host bacterium Actinomyces odontolyticus, suggesting that oral Saccharibacteria may play an important role in bacterial predation in the oral cavity (70). However, the apparent parasitic lifestyle of such taxa precludes attempts to classify them using conventional systematics criteria. Similar challenges face other microbial groups that are resistant to isolation in pure culture, making them difficult to discuss and study (21, 50). Additionally, such a standard could never be applied to ancient microbes, effectively shutting the door to the possibility of discovering and naming extinct species.
As a consequence of the high bar set by the ICSP for obtaining a valid species name, comparatively few microbial species have been officially named and validated—15,974 as of 2014 (141), compared with the >645,000 for which there is currently 16S rRNA gene sequence evidence (OTUs clustered at 99% in the SILVA SSU Ref NR 99 database, release 128; https://www.arbsilva.de) (152, 204). As a result, most microbial taxa are currently nameless but not necessarily unknown.
The challenge of how to devise a functioning nomenclature scheme for such a situation is clearly illustrated by the taxon table maintained by the Human Oral Microbiome Database (HOMD), a public scientific resource that curates an up-to-date list of human oral microbes (27). As of November 2016, the HOMD included 687 species-level oral taxa, of which 335 had both a valid species name and at least one sequenced genome, 36 had a valid species name and no sequenced genome, 88 had no valid species name but at least one sequenced genome, and 228 had no valid species name and no sequenced genome. The HOMD addressed this problem by developing a provisional naming scheme based on binning 16S rRNA gene sequences into unique phylotypes that are then assigned a Human Oral Taxon number. This number then allows phenotypic, phylogenetic, genomic, clinical, and other data types to be linked within the HOMD portal. This scheme, however, is limited to microbes of the human oral cavity and is not generalizable to other microbiomes, such as those of the human gut, soil, or ocean.
Alternatively, as of March 2017, GenBank (11, 131) contains 14,022 sequenced microbial genomes and maintains a taxonomy common tree of 23,653 named and candidatus microbial species (993 archaea and 22,660 bacteria) that “does not follow a single taxonomic treatise but rather attempts to incorporate phylogenetic and taxonomic knowledge from a variety of sources” (127). By taking this pragmatic approach, they are able to utilize a diverse range of existing phenotypic, genetic, and genomic microbial data in a common phylogenetic framework (165).
4.3. Taxonomy Versus Phylogeny
Although species names are practical entities that allow microbial taxa to be discussed and analyzed in a meaningful way, they can also be misleading. Ideally, taxonomy (microbial classification) should reflect phylogeny (evolutionary history), and species are periodically renamed to reflect improved understanding of phylogenetic relationships. However, there are also many well-known examples of named microbes for which the taxonomy is in congruent with phylogeny. In some cases, such discrepancies apply to clinically important taxa that differ from nonpathogenic taxa mainly because of horizontally transferred virulence factors that result in major phenotypic changes, as in the case of Yersinia pseudotuberculosis and Y. pestis (25). In other cases, genera that are clearly polyphyletic or paraphyletic, such as Klebsiella (Figure 1a), Clostridium, and Ruminococcus, persist despite repeated attempts at taxonomic reorganization (100, 133, 153). The ICSP has procedures for correcting such problems (98), and many taxa have been reclassified under this scheme. For example, Bacteroides forsythus, a gram-negative anaerobic rod first isolated from periodontal patients (182), was reclassified into its own genus as Tannerella forsythensis after major differences with other Bacteroides taxa were discovered (163) and was later renamed Tannerella forsythia to adhere to ICSP guidelines regarding gender consistency in Latin genus-species name combinations (111). Given that microbial classifications frequently change as new information becomes available, it is important to be aware of nomenclature history, especially when conducting literature searches and comparing results generated using different databases.
5. METATAXONOMICS: WHO IS THERE?
The first step in any analysis of metagenomic data, either modern or ancient, is often addressing the question “Who is there?” The question is simple and yet surprisingly difficult to answer, in part because of our incomplete knowledge about biological diversity in general. The number of bacterial species on earth is unknown but is likely massive. The number of bacterial cells is estimated at 1030 (108), and microbes are thought to make up half of the earth’s total biomass (175). Estimates ranging from millions to a trillion microbial species have been proposed (108, 166), in stark contrast with current estimates of just over 8 million eukaryotic species (123). Suffice it to say that relatively few of these microbes have been characterized, genetically or otherwise.
Rather than providing definitive answers, a variety of techniques have been developed to computationally approximate the taxonomic composition of a given microbial community from HTS data, an approach known as metataxonomics (113). Establishing reliable methods for accurate taxonomic assignment and placement of DNA sequences within a phylogenetic tree is a necessary first step in most modern and ancient microbial analyses. For ancient pathogenomic studies, it is the basis for identifying the proverbial pathogen needle in a haystack of background DNA, and for ancient microbiome studies, it is essential for identifying community membership and calculating ecological diversity metrics (77). It is also the input data for the Bayesian-based tool SourceTracker (91), which can, for example, model the relative contributions of ancient and archaeological microbiota sources in a given sample.
5.1. Amplicon Metataxonomics
Many efforts to characterize “who is there” in soils, oceans, and host-associated microbiota focus on targeted amplification and deep sequencing of the 16S rRNA gene (56, 79). This gene, universally present in all microbes, encodes the 16S small subunit of rRNA and is essential for life. Amplicon-based metataxonomics typically targets this gene because its sequence is similar enough across microbial taxa to be amplified using universal PCR primers and yet distinct enough among species to be useful for taxonomic classification. This approach, which can also be applied to other genetic loci, is also known as phylotyping (187) and metabarcoding (7, 46), and it has been widely used in aDNA studies (e.g., 126, 187).
The ability to rapidly and inexpensively sequence tens or hundreds of thousands of partial 16S rRNA gene sequences simultaneously using HTS was essential to the success of the Human Microbiome Project (79), and bioinformatics tools such as Quantitative Insights into Microbial Ecology (QIIME) (24, 128) and mothur (168) were developed specifically to analyze the data resulting from large sequencing projects. However, it is now known that this amplicon-based approach cannot be straightforwardly applied to ancient microbial studies because the targeted hypervariable regions far exceed the average length of aDNA molecules and additionally contain length polymorphisms that contribute to biased amplification of degraded DNA (207). For this reason, targeted amplification of the 16S rRNA gene is not recommended for aDNA samples, although short-read mapping of shotgun-filtered 16S rRNA gene sequences can still be performed. Despite its limitations for aDNA research, the technique remains important owing to the extensive comparative data from modern microbiomes that have been generated using this approach.
5.2. Metagenomics
As an alternative to single-gene amplicon-based approaches, genome-wide information can be generated using metagenomics, an approach that is highly amenable to aDNA research. When applied in an untargeted manner to all DNA recovered from a given environment, this approach is called shotgun metagenomics, and sequences are generated from a random subset of DNA from the collective genomes of all of the organisms present in a sample (113). This technique is used to characterize microbiota in microbiome studies and as a screening tool in pathogenomics.
Alternatively, a targeted subset of the DNA in a sample may be selected before sequencing using bait hybridization techniques in order to enrich the HTS library for the genomes of one or more organisms of interest, a process known as sequence capture. This technique can be performed either on chip (20) or in solution (9, 52), and the goal is to remove unwanted DNA from a sample prior to sequencing. Bait sizes are typically less than 120 base pairs and are suitable for capturing short aDNA molecules. If this technique is highly efficient, analysis of the resulting captured DNA might qualify as genomics (the study of specific genomes), but in most cases post-duplicate-removal enrichment success is modest, and many unintended off-target sequences are also captured (31). The efficacy of sequence capture is nonetheless sufficient to merit its use in ancient pathogenomic studies for genome reconstruction (14).
Metagenomic approaches have many advantages. They are better suited to aDNA studies than PCR-based approaches because they are not affected by length or sequence variants in the underlying genome, and even very short and highly degraded DNA fragments can be successfully sequenced using HTS platforms. The resulting sequences allow not only metataxonomic characterization, but functional inferences as well. Finally, if untargeted, all four domains of life—bacteria, archaea, eukarya, and viruses—can be investigated simultaneously.
When deciding on strategies for the analysis of metagenomic data, the research question is the most relevant factor. For the characterization of ancient microbiota, the goal is not only to identify the microbes present, but also to reveal the microbial community structure and functional potential. For this purpose, a nontargeted, shotgun metagenomics approach is often most suitable. Ancient pathogen screening strategies, by contrast, focus on specific taxa and can be divided into targeted and nontargeted approaches. Targeted approaches are applied with a particular pathogen species in mind, often because skeletal lesions indicate the presence of a specific disease, such as leprosy (170) or tuberculosis (13), or because a particular burial context suggests the presence of a pathogen, as in the case of Y. pestis and medieval mass graves (16, 178). Unfortunately, however, relatively few bacterial pathogens cause skeletal lesions, and burials often provide little disease context. In this case, nontargeted screening approaches are applied in order to detect any bacterial pathogen that might be present in a sample. Shotgun metagenomics is typically used to screen for unknown pathogens. However, enrichment techniques are available that can target multiple pathogenic species at once (15).
For any of these approaches, the metagenomic context of the sample must be considered during the validation of the findings. For this reason, specialized computational tools for analyzing metagenomic data are often applied not only to characterize microbiota but also in the context of pathogen screening.
6. METATAXONOMIC TOOLS AND DATABASES
Numerous tools and databases are available that can facilitate metataxonomic analyses of both modern and ancient samples. Here, we provide an overview of different approaches and strategies commonly used in microbial archaeology studies, as well as a selection of frequently used software (for a detailed description and systematic comparison, see 106, 142). Supplemental Appendix 2 provides a list of tools and databases mentioned in this article, along with website links and references.
6.1. Tools of the Trade: Factors to Consider
When selecting a tool for a particular research question based on genetic comparison, the basic strategies employed by that tool must be considered, including the reference strategy, which defines the content and structure of the database to which the input data are compared; the query strategy, which is the concept by which this comparison is facilitated; and the data type upon which the reference database is built, which is either DNA or protein sequences.
6.1.1. The reference strategy
The way a reference database is constructed and queried by a software tool has a strong influence on the sensitivity and specificity of the analysis, as well as on the potential biases that can occur. Commonly applied reference strategies can be divided into three classes: single-locus approaches, which focus on rRNA genes or other genes that show orthologs in all species represented in the database; multilocus approaches, which represent an extension of the previous approach applied to multiple genes; and whole-genome approaches, which use nearly all available genomic information.
6.1.1.1. Single-locus approaches
Tools that focus on single loci, such as the 16S rRNA gene, have the advantage that single-locus databases contain more taxonomic entries than those covering multiple loci or complete genomes. For investigations of microbial diversity, no other gene has been studied as extensively as the 16S rRNA gene. Several large-scale, publicly available 16S rRNA gene databases are available, each of which contains more than 1 million aligned reference sequences: the Ribosomal Database Project (RDP) (3,356,809 sequences) (30), Greengenes (2013 release, 1,262,986 sequences) (37), and SILVA (SSU Ref, 1,922,213 sequences) (152), the last of which contains entries for more than 645,000 distinct bacterial taxa (as defined by a 99% identity clustering cutoff for OTU assignment; the number of database entries is reported as of December 1, 2016).
As a highly conserved gene, however, the 16S rRNA gene generally contains insufficient sequence diversity to differentiate between closely related taxa, such as different bacterial strains of the same species, and rrn operon copy number variation contributes to skewed estimates of taxonomic abundance and diversity (180, 190). Additionally, a relatively large number of reads covering the rRNA locus is needed for a reliable taxonomic classification, and although this is easily achievable using an amplicon approach, only a small proportion (~0.2–0.6%) of shotgun microbial aDNA reads typically map to the 16S rRNA gene, which is simply a function of the relatively short length (~1,540 base pairs) of the multicopy (four-copy average) gene compared with the average size of a microbial genome (1–3 Mb). This presents a limitation for the detection of low-abundance taxa. Finally, when working with short-read metagenomic data, we have found that minor changes in closed reference OTU picking parameters (such as UCLUST - -max_accepts and - -max_rejects values) can dramatically alter diversity estimates; consequently, default settings should be adjusted in microbial archaeology studies to optimize performance for very short sequences, which are typical of aDNA.
Two widely used software pipelines that use 16S rRNA reference databases are QIIME (24) and mothur (168). Metagenome Analyzer (MEGAN) (80) can also be used in connection with rRNA gene reference data.
6.1.1.2. Multilocus approaches
An alternative technique for metataxonomic assessment focuses on multiple loci, usually a small set of (single-copy) housekeeping genes. Here, the advantage is that more multilocus sequence analysis and multilocus sequence typing data sets are available in comparison with full genomes, and the sequence divergence level can be higher in comparison with rRNA genes, potentially allowing for the differentiation of bacterial strains. Another advantage is that genes can be selected that occur only as single copies in each reference genome sequence.
One widely used software pipeline that uses a variant of a multilocus sequence analysis–based approach is the Metagenomic Phylogenetic Analysis (MetaPhlAn) pipeline (172). The current MetaPhlAn database (version 2) includes ~1 million marker genes identified from ~17,000 reference genome sequences obtained from >7,000 species-level taxa; the marker genes were selected to define specific microbial clades (mainly bacterial and archaeal, but also including viruses and eukaryotes) and are spread across all functional annotation classes (188). The latter is essential because the microorganisms present in biological or environmental samples may differ in, for example, their metabolic pathways and can be detected only if they are represented in the reference database. In contrast to whole-genome alignment, read alignment against clade-specific markers is fast, comparing favorably to the Basic Local Alignment Search Tool (BLAST) (172), although not quite as fast as Kraken (201). Once normalized by the sequence length of each individual marker, the total number of high-quality hits offers a simple measure for calculating taxon abundances. Because gene elements that are shared among clades do not fulfill the definition of markers, MetaPhlAn assignment is robust to horizontal gene transfer. Quasi markers, which are informative only in the absence of particular taxa sharing closely related sequences, can also improve sensitivity and enable strain detection. Typically, MetaPhlAn analyses achieve high specificity, owing to the marker selection procedure, but relatively limited sensitivity, owing to the incompleteness of the marker database. Ongoing sequencing efforts aimed at characterizing microbial diversity will continue to improve the approach’s sensitivity. In addition to being available as a stand-alone package, MetaPhlAn is integrated into the metaBIT metagenomic pipeline (110).
6.1.1.3. Whole-genome approaches
The third reference strategy is to use complete genomes as a target database. Using whole genomes has the advantage that known taxa present in low abundances can potentially be identified because any sequenced DNA fragment that originates from a species contained in the target database can be (theoretically) assigned. Therefore, this strategy can be beneficial in ancient pathogen detection studies, where only traces of DNA from a pathogenic organism are expected to be present. Additionally, because variation across the entire genome is considered, this method maximizes the power to make fine-scale distinctions among species and strains.
Challenges, however, can arise from horizontal gene transfer, bacterial recombination, or mobile elements, which can influence the precision of assignments. Additionally, genome sizes of free-living taxa vary by nearly an order of magnitude, ranging from the tiny 0.16-Mb genome of Candidatus Carsonella ruddii to the massive 10-Mb genome of Solibacter usitatus (118); as a result, taxa with larger genomes contribute proportionally more fragment reads per cell to a metagenome, potentially making them appear more abundant if genome size differences are not taken into account. Whole-genome databases are also far less complete in comparison with rRNA databases, and thus, for a given sample, far fewer taxa may have a close representative in the reference database. This may lead to a larger number of false negative (unidentified) or false positive (identified as the closest representative in the database) assignments, a phenomenon often referred to as database bias.
Whole-genome databases are rapidly growing, however. Since the publication of the first genome sequence of a free-living bacterium, Haemophilus influenzae, in 1995 (49), the number of sequenced microbial genomes has increased exponentially (137), with 129,346 sequencing projects at various stages of completion registered in the Genomes Online Database (GOLD) as of March 2017 (125). Currently, microbial genomes are available through multiple—often overlapping and cross-referenced—databases that have been customized for different purposes, ranging from those that aim to encompass all microbes, such as GOLD and the National Center for Biotechnology Information’s RefSeq database (131) to those that focus on specific groups of bacteria, such as EnteroBase (http://enterobase.warwick.ac.uk) and the HOMD (27). Pipelines that utilize whole-genome data for taxonomic assignments include the MEGAN Alignment Tool (MALT) (71) and MIDAS (129).
6.1.2. The query strategy
Metagenomic tools differ not only in the way that their reference databases are constructed but also in the way those databases are queried, which depends on whether read alignment is performed. As such, query strategies can be divided into alignment-based and alignment-free approaches.
6.1.2.1. Alignment-based approaches
One important strategy for the assignment of query sequences to large reference databases is based on classical sequence alignments. The algorithms applied are usually modifications of the Needleman-Wunsch (130) or Smith-Waterman (177) algorithms designed to perform semiglobal or local alignments, respectively. However, the calculation of precise alignments is time consuming, even for more efficient variants of these algorithms. For this reason, most software tools use so-called seed-and-extend approaches. Here, candidate matches are first determined in a time-effective but imprecise manner, and precise alignments are then calculated only for these candidates. Well-known examples of this algorithm type include BLAST (5) and its faster variant MEGABLAST (124), as well as UBLAST and USEARCH (43). Novel tools such as MALT (71) and lambda (69) also belong in this category, as do read mappers such as the Burrows-Wheeler Alignment (BWA) tool (104) and Bowtie 2 (97). Additionally, read mappers that are able to account for high nucleotide misincorporation rates, such as the Burrows-Wheeler Alignment–Position-Specific Scoring Matrix (BWA-PSSM) tool (86), may improve method sensitivity for aDNA.
The calculation of precise alignments has multiple advantages. It allows, for example, for a sophisticated evaluation of every match and a statistical assessment of all matches for a given taxon. This enables the calculation of aDNA damage patterns in order to authenticate the aligned matches as representing aDNA fragments (17, 83). Furthermore, edit distance or percent identity distributions can be assessed in order to verify the assignment of query sequences. The distribution of query sequences across the reference sequence can also be inspected in detail, and spurious taxa can be detected by their uneven genome coverage. The primary disadvantage of alignment-based methods is that they are time consuming, even when using time-saving seed-and-extend approaches.
6.1.2.2. Alignment-free approaches
Not all methods for metagenomic analysis rely on calculating precise alignments. Rather than taxonomically assigning sequencing reads or other sequences, alignment-free methods determine the relative abundances of taxa based on other features. Kraken (201), for example, uses an approach that is based on exactly matching k-mers. Both the sequences present in the reference database and the query sequences are split into overlapping subsequences of length k. This allows fast matching of queries to the database by hashing algorithms, a concept that is often applied in the seeding step of seed-and-extend alignment-based methods. Other methods are based on similarities and dissimilarities of nucleotide composition (74), comparison of Burrows-Wheeler transformed sequence information (6), or machine learning approaches (156).
An advantage of many alignment-free methods is their speed. However, a precise evaluation of taxonomic assignments is more difficult compared with alignment-based methods. Particularly in the context of ancient metagenomic analyses, this poses challenges because it does not allow for detailed authentication unless further alignment steps are taken in order to evaluate DNA damage patterns or other alignment-based features.
6.1.3. Data type
The input data type for metagenomic analysis is DNA sequencing reads or assembled DNA sequences, such as contigs. The data type for the reference database can be either DNA or amino acid sequences. In the case of a protein database, the DNA input sequences must be translated from all six possible reading frames, which is analogous to BLASTX (5).
An advantage of using a protein database is its higher level of sequence conservation, which allows the exploration of deeper evolutionary relationships. Furthermore, tools such as DIAMOND (19), lambda (69), and RapSearch2 (206) provide extremely fast algorithms for performing DNA-to-protein alignments. However, protein alignments can be challenging to use for ancient metagenomic data because they are incompatible with DNA alignment–based authentication procedures. Furthermore, short aDNA reads translate into even shorter peptide sequences, and DNA damage can lead to in silico errors during translation and therefore to misassignments.
6.2. Algorithms, Pipelines, and Environments
A versatile selection of software is available for metagenomic analysis. In the context of aDNA, these software packages are often not directly comparable; some of them represent specific algorithms that perform specific steps of an analysis, whereas others combine algorithms into analysis pipelines that perform multiple steps of the metagenomic analysis.
Algorithms can be divided into different categories according to the analytical step they are performing. If they apply an alignment-based concept, then the first step is alignment, or mapping. In principle, any DNA or protein alignment tool can be used, including sequence similarity search tools such as BLAST (5), as well as read mappers. The next step is to classify the alignments using a taxonomy in order to assign the query sequences to specific taxa, a process known as binning. Examples of binning algorithms include UCLUST (43), RDP Classifier (194), naive lowest common ancestor (LCA), and weighted LCA (80).
MALT (71) is a tool that combines alignment and binning. It is based on a seed-and-extend approach and uses spaced seeds for higher sensitivity. Seeds are matched to a DNA sequence database that can contain single or multiple loci or whole genomes. MALT calculates precise local or semiglobal alignments and uses (weighted) LCA for binning. It integrates well with the MEGAN environment, which does not contain alignment functionality on its own.
MetaPhlAn (172) is an example of a metagenomic analysis pipeline. It uses a selection of multiple loci as a target database. The selection of loci varies for different taxa, thus ensuring specificity on different taxonomic levels and for different parts of the taxonomy. For its alignment algorithms, MetaPhlAn integrates MEGABLAST or Bowtie 2; however, other alignment algorithms could be integrated as well. The taxonomic assignment algorithm is specific to MetaPhlAn owing to its unique database structure. Another example of a metagenomic pipeline is MIDAS (129), which combines multiple databases, aligners, and binning algorithms into an integrated pipeline.
In some cases, single algorithms and more complex analytical procedures are made accessible in interactive environments that also provide an integrated visualization of the results. MEGAN (80) is an example of such an environment. In other cases, multiple pipelines are made accessible through an environment, as is the case with QIIME (96) and metaBIT (110). MetaBIT, for example, implements MetaPhlAn-based microbial profiling and provides built-in modules for (a) visualizing profiles, including Krona plots (134); (b) calculating diversity indices (e.g., alpha and beta diversity); and (c) performing a range of statistical analyses. These analyses include principal coordinate analysis, hierarchical clustering, and linear discriminant analysis, the latter of which is used to identify microbes that drive differentiation among user-defined groups, such as archaeological sites, samples, and/or DNA libraries (171). MetaBIT also provides preprocessed microbial profiles from the Human Microbiome Project (78, 79) and cross-biome soil communities (48), allowing users to place their (ancient) microbial communities within the known diversity of human and soil microbiomes.
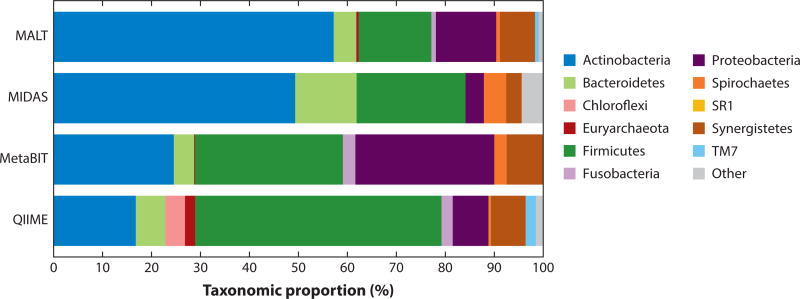
Because each software strategy operates differently and makes different assumptions based on the algorithms and databases incorporated into the pipeline, the results generated by different software are not identical. Figure 2 illustrates the oral microbial community reconstructed from an ancient dental calculus sample as performed by five common metataxonomics pipelines: QIIME, metaBIT, MIDAS, and MALT. Although the five pipelines generally return profiles containing similar taxa, systematic differences in taxon relative abundance and phylum dropout are apparent owing to differences in pipeline parameters, algorithms, databases, and other factors.
Figure 2.
Reconstructed taxonomic profile for archaeological dental calculus using the QIIME, metaBIT, MIDAS, and MALT pipelines. Analysis was performed on 1,967,941 shotgun metagenomic DNA sequences obtained from a previously described dental calculus sample from the Spanish Chalcolithic site of Camino del Molino (approximately 2340–2920 BCE) (207). Although all four pipelines generally identify the same phyla, predictable biases are also readily apparent. For example, QIIME estimates the largest proportion to be Firmicutes, a phylum known to have a high rrn operon copy number (190). Euryarchaeota is absent in the MIDAS analysis because there are no reference genome sequences for this phylum in the database. MIDAS and metaBIT, which rely on genome-scale databases, also fail to detect the largely uncultivated phyla Saccharibacteria (TM7), Chloroflexi, and SR1. Explanations for other differences in phylum frequency abundance, such as the high proportion of Actinobacteria estimated by MALT and the absence of Fusobacterium detected by MIDAS, are not as clear. Supplemental Appendix 1 provides the specific parameters used for each analysis. Abbreviations: MALT, MEGAN Alignment Tool; MIDAS, Metagenomic Intra-Species Diversity Analysis System; QIIME, Quantitative Insights into Microbial Ecology.
7. THE CHALLENGE OF RELATED TAXA
Although numerous tools and growing databases are available for taxonomic identification, care must be taken to avoid taxonomic misassignment, especially for the short, damaged sequences typical of aDNA. Accurate taxonomic assignment can be particularly challenging if closely related environmental contaminants are also present. Additionally, clades of closely related species within endogenous microbiota can be difficult to resolve if database coverage is poor or sequencing effort is insufficient. Validation efforts focusing on the evenness of genome mapping coverage, sequence identity distributions, and sequence monomorphism (haploidy) are powerful approaches for identifying and eliminating false positive assignments in microbial archaeology studies.
7.1. All in the Family: The Rise of Pathogens
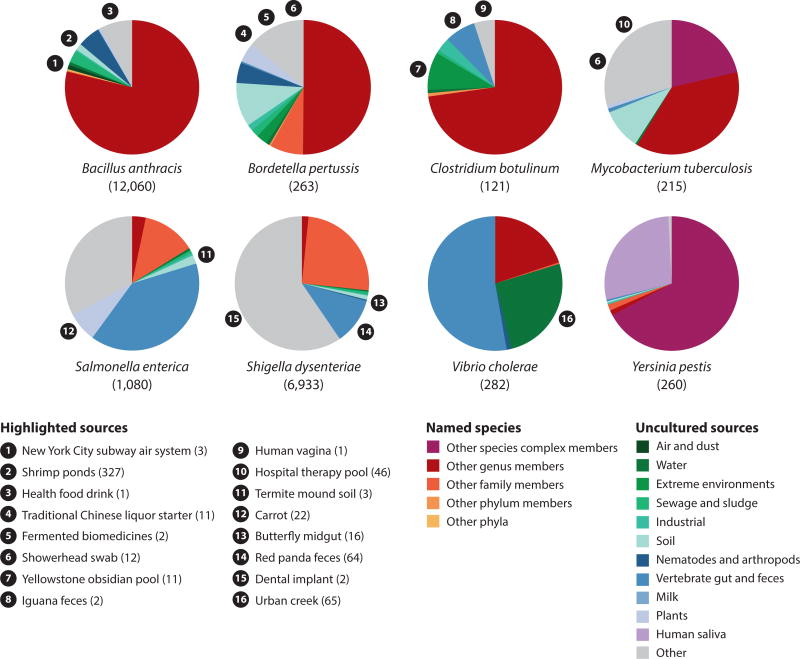
Many pathogens have close relatives in soil. Y. pestis, for example, is a close relative (if not subspecies) of Y. pseudotuberculosis (25), a soil bacterium that has also been found in the gastrointestinal tracts of nematodes and insects (55, 209) and those of birds and mammals (53, 109) as well as on improperly washed vegetables (81). Likewise, a majority of the nearly 200 named Mycobacterium species (and thousands of unnamed ribosomal phylotypes) are soil inhabitants (22, 45, 76), with the pathogenic M. tuberculosis complex, M. leprae, and to some degree M. avium making up a minority of species that regularly infect humans. During analysis, sequences from these environmental relatives can misalign to the pathogen genome, producing false positives (Supplemental Table 1). Other pathogens with close, nonpathogenic environmental relatives include B. pertussis, S. enterica, Bacillus anthracis, Clostridium botulinum, Shigella dysenteriae, and Vibrio cholerae (see Supplemental Appendix 3). Figure 3 shows the extent of closely related cultured and uncultured relatives for each of these eight pathogens based on a 16S rRNA gene sequence identity of >97% in the RDP database of more than 3 million sequences.
Figure 3.
Pathogens and their close environmental relatives. Many obligate pathogens share close 16S rRNA gene sequences with environmental microbes. Bacillus anthracis, Bordetella pertussis, Clostridium botulinum, and Mycobacterium tuberculosis have close relatives in soil, sewage, and extreme environments, whereas Salmonella enterica, Shigella dysenteriae, and Vibrio cholerae have close relatives in vertebrate gut and feces. V. cholerae relatives are also abundant in water sources, and B. anthracis and B. pertussis share close relatives found in association with nematodes and arthropods. By contrast, few environmental relatives outside of the Yersinia pseudotuberculosis complex were observed for Yersinia pestis; however, Y. pestis shows strong similarity to 16S rRNA sequences obtained from a study of global diversity in human saliva, indicating that human saliva in some parts of the world may harbor a previously undetected Yersinia relative. The total number of RDP database matches for targets other than the respective pathogen is shown in parentheses. Of all the obligate pathogens investigated, B. anthracis and S. dysenteriae had the highest number of hits to environmental sources. A subset of sources are highlighted (along with the number of RDP database matches for these targets, shown in parentheses) to illustrate the diversity of environments from which close matches were observed. Supplemental Appendix 3 provides a detailed list of taxa and sources. Abbreviation: RDP, Ribosomal Database Project.
When working with very large data sets, as is typical for HTS, it is important to be aware of—and have a plan for handling—false positives. In pathogen studies, shotgun metagenomic sequence data generated from bone lesions or dental pulp (typically a few million sequence reads) are often initially mapped against a panel of pathogen genomes using standard mapping tools in order to screen for putative taxa of interest. Although an important first step, this approach yields many false positives, and in fact, nearly any large metagenomic data set is expected to yield false positives under such an approach. For example, mapping metagenomic data from three sources—soil, ocean, and healthy human saliva—against a panel of 14 pathogen reference genome sequences resulted in numerous false positive hits to all 14 pathogens, ranging from a low of 2 Treponema pallidum hits per million soil reads to a high of 173 B. anthracis hits per million saliva reads (Supplemental Figure 1). Overall, healthy saliva yielded the highest number of false positives, with a mean of 67 false positive hits per million metagenomic sequences. None of these reads were true positives; rather, they resulted from mismapping at highly conserved regions and in regions of low complexity, mapping of mobile elements and other horizontal gene transfer sites, and mapping at orthologous sites in related taxa because of database bias. Any alignment-based screening study requires further validation in order to identify true positives against a background of spurious hits.
7.2. Close but Not Close Enough
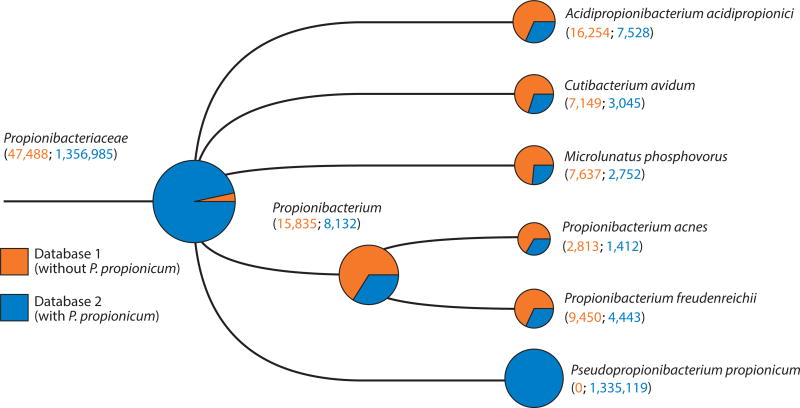
In addition to mistaking environmental contaminants for ancient pathogens, it is possible to mistake one endogenous taxon for another, especially in ancient microbiome samples. Within the oral cavity, for example, many genera include multiple closely related members, and the number of named species and known phylotypes for the five largest genera in the HOMD are as follows: Prevotella, 50; Treponema, 49, Streptococcus, 36; Actinomyces, 30; and Lactobacillus, 26. The oral cavity contains numerous pathobionts, or endogenous potential pathogens, such as S. pneumoniae, N. meningitidis, and H. influenzae, and it may also become colonized with obligate pathogens, such as M. tuberculosis, during respiratory infections (195). Additionally, nearby skin pathobionts, such as S. aureus and Propionibacterium acnes, and food microbes, such as Propionibacterium freudenreichii and Lactobacillus rhamnosus, may also be present. Correctly assigning sequences within these genera is therefore important for understanding the dietary and potential health implications of past microbial communities. Database bias, however, can be a major challenge because of the limited number of species with sequenced genomes, and it can result in both the underestimation of important taxa in ancient microbiota and sequence misassignment to the closest sequenced genome (Figure 4).
Figure 4.
Lack of signal for highly abundant endogenous bacteria resulting from database bias. A medieval dental calculus sample (G12) (196) was screened using MALT (71) and visualized in MEGAN6 (80) using two databases, one without the genome sequence of the oral bacterium Pseudopropionibacterium propionicum (database 1, red) and another with that genome sequence included (database 2, blue). The tree visualizes the results of both analyses, and nodes are scaled based on the summed number of hits to a log scale. The inclusion of P. propionicum results in hits being shifted away from related dietary (Propionibacterium freudenreichii ), skin (Propionibacterium acnes), and other species, as well previously nonaligned hits, toward the oral bacterial genome, revealing the presence of a previously unseen, highly abundant species. This highly abundant species, with more than 1 million assigned reads, would not have been detected in metagenomic screening methods before 2012, when the genome sequence was published. Supplemental Appendix 1 provides additional details. Abbreviations: MALT, MEGAN Alignment Tool; MEGAN, Metagenome Analyzer.
7.3. Validation Is Essential
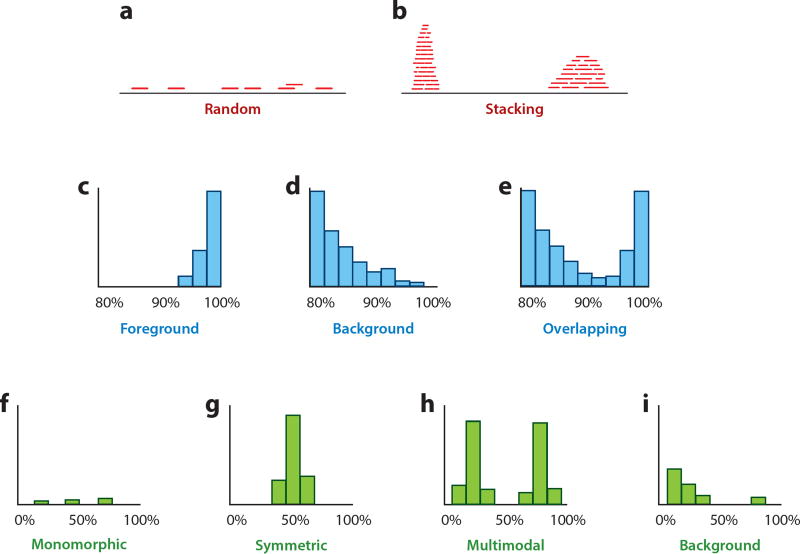
If the number of reads supporting a species identification is low and robust phylogenetic confirmation is not possible, further validation steps should be taken. There are multiple measures that can reduce the number of false positive hits, including evaluating coverage evenness, sequence identity, and haploidy (Figure 5, Supplemental Figure 2).
Figure 5.
Schematic overview of different measures for the validation of species assignments in metagenomic data analysis. (a, b) Evenness of coverage. Correctly assigned reads are expected to distribute randomly across the reference (panel a); accumulation of reads in regions of high sequence conservation indicates misassigned reads originating from different closely related species (panel b). (c–e) Percent identity distributions. In panel c, most reads show a high similarity to the reference, which indicates a correct assignment. In panel d, most reads are highly dissimilar to the reference, which suggests that they originate from different related species. In some cases, as in panel e, a mixture of correctly assigned and misassigned reads can be observed. (f–i ) Haploidy. Because bacteria are haploid organisms, only one allele is expected for each genomic position. Only a small number of multiallelic sites are expected, which can result from a few misassigned or incorrectly aligned reads (panel f). A large number of multiallelic sites indicates that the assigned reads originate from more than one species or strain, which can result in symmetric allele frequency distributions (e.g., if two species or strains are present in equal abundance) (panel g) or asymmetric distributions (e.g., if two species or strains are present in unequal abundance) (panel h). A large number of misassigned reads from closely related species can result in a large number of multiallelic sites with low frequencies of the derived allele (panel i). Supplemental Figure 2 provides examples with empirical data from microbial archaeology studies.
7.3.1. Evenness of coverage
One measure that can validate species assignments is the evenness of coverage across the reference sequence. If all reads that are assigned to a species represent DNA fragments originating from the same source, they should be distributed randomly across the reference sequence (Figure 5a). An accumulation of reads in distinct regions, by contrast, suggests that they do not stem from the species they have been assigned to, but originate from one or more related species and map to regions that are conserved among this group of species (Figure 5b). Regions that commonly attract read assignments from other taxa include rRNA and tRNA loci, mobile elements, repetitive regions, and any highly conserved protein-coding sequences.
7.3.2. Percent identity distributions
Another measure for the evaluation of read assignments is their similarity to the reference. Of course, the percent identity values of assigned reads will vary, but a large amount of information can be gained by investigating the distribution of values across all read alignments for a given taxon. It can be assumed that, with the exception of damaged sites, most reads will map to the reference genome sequence with zero mismatches if the species assignment is correct, and the number of reads with an increasing number of mismatches will decrease progressively. However, the expected diversity of strains varies depending on the species, which should be taken into account. For example, H. pylori, the causative agent of gastritis and some forms of gastric cancer, exhibits an extremely high level of strain diversity, with nearly every unrelated isolate possessing a distinct genome sequence (107). Ancient strains of H. pylori are thus expected to differ from modern strains but should still fall within expected models of variation (112). By contrast, other microbes fall at the other end of the scale, exhibiting little or no sequence variation across hundreds or thousands of isolates. These genetically monomorphic microbes include many well-known epidemic pathogens, such as B. anthracis, Burkholderia mallei, Escherichia coli O157:H7, M. leprae, the M. tuberculosis complex, S. enterica serovar Typhi, Shigella sonnei, Chlamydia pneumoniae, T. pallidum, and Y. pestis (1). In these taxa, sequence differences between ancient and modern strains are expected to be minimal and limited to a small number of sites (16).
Overall, a high similarity between most assigned reads and the reference supports a true positive assignment (Figure 5c). Conversely, if most reads are dissimilar to the reference, the species assignment is likely incorrect (Figure 5d), although it may be accurate at the genus level. A clear distinction between these two cases is not always possible because correct and incorrect assignments can be present at the same time, resulting in multimodal distributions in which the background and foreground cannot be clearly distinguished (Figure 5e). In some cases, edit distances (number of mismatches) are used instead of percent identity values because they are more suitable for distinguishing very closely related taxa, such as Y. pestis and Y. pseudotuberculosis (158).
7.3.3. Haploidy
Haploidy is another measure that provides an indication of whether reads from multiple taxa have been misassigned to a single species. This approach can be applied to haploid bacterial and archaeal genomes as well as eukaryotic organelle genomes. In haploid genomes, nearly all variable sites should be monoallelic (Figure 5f), so a large number of multiallelic sites suggests that the aligned reads originate from multiple taxa. In the case of two taxa present in equal abundance, all multiallelic sites will show a frequency of the two alleles at ~50% (Figure 5g). This does not necessarily mean that the species assignment is wrong. It could, for example, indicate the presence of two different strains of the same bacterium, as has been observed in cases of multiple M. tuberculosis infections (26). Reads that originate from two or more sources in unequal abundance result in a multimodal distribution (13) (Figure 5h). Even if the vast majority of the reads originate from a single species, a small number of damaged or misassigned reads may still produce a few low-frequency single-nucleotide polymorphism (SNP) calls of the derived allele (Figure 5i). Note that this analysis is possible only with sufficient coverage for reliable SNP calls.
8. DNA PRESERVATION AND CONTAMINATION
After the validation of taxonomic assignments, it is important to test whether the DNA sequences of interest could be explained by exogenous contamination. Authentic aDNA undergoes predictable forms of damage and decay, which can result in nucleotide misincorporation during library repair and amplification. Such damage can be estimated by analyzing nucleotide substitution patterns as well as strand breakage as a function of lambda (λ, a proxy for DNA fragmentation; see Section 8.2). Additionally, with respect to ancient microbiota, Bayesian source estimation tools can test for potential mixing of endogenous microbiota with exogenous sources. Finally, rigorous standards set forth to prevent laboratory contamination and instrument sequence bleed-through should be followed in order to ensure that the findings are not the result of poor laboratory or data hygiene.
8.1. Damage Patterns
Early studies of HTS data from Late Pleistocene animals revealed that molecular damage accumulating after death is a common feature of aDNA molecules (17, 18). In particular, depurination, nick formation, and cytosine deamination are the most important DNA decay reactions in subfossil material (32). Although such reactions limit the amount of aDNA material amenable to sequencing, they also provide important molecular signatures for data authentication (94, 151, 181). For instance, nucleotide misincorporation profiles, in which the probability of sequence mismatches between reads and the reference genome used in sequence alignment is traced from read starts to read ends, provide a visual way to assess levels of inferred cytosine deamination within a given data set. Statistical DNA damage models, such as the one implemented in mapDamage (59, 83), exploit such features to quantify DNA damage patterns. Here, the preferential accumulation of deaminated cytosine (uracil), a chemical analog of thymine, within the overhanging ends of aDNA templates typically results in increasing C→T misincorporation rates toward read starts (complementary G→A misincorporation rates are expected to increase in an almost symmetric fashion toward read ends in standard double-stranded library preparations) (17). In addition, the amount of nucleotide misincorporations displayed by DNA from a sample correlates with the age and depositional environment (4, 164). Demonstrating such signatures in ancient HTS data sets has thus been considered key to data authentication (151), especially because such signatures have been found in all sample types tested to date, including skeletal remains, dental calculus, plant remains, and even DNA retrieved from permafrost (135, 143, 207).
However, when sequence data are scanned for such deamination footprints, the expected nucleotide misincorporation profiles can vary depending on the molecular tools used during library construction, especially according to the endonuclease (161) and exonuclease (33, 54, 95) activities of the end repair enzymes, the type of DNA ligase used during adapter ligation (121, 174), and the polymerase employed for library amplification (33, 73, 173). In particular, amplification using AccuPrime Pfx DNA polymerase results in typical misincorporation profiles following 454-like DNA library types (120) but removes the elevated C→T pattern toward read starts on TruSeq DNA library constructs (173). Differences in HTS-library preparation protocols also contribute to the characteristically different damage patterns observed in double-stranded (120) and single-stranded (54) aDNA libraries (Supplemental Figure 3). Additionally, minimal sequencing efforts are necessary in order to assess the presence of cytosine deamination, and only a few thousand sequences from the genome of interest are typically needed to recover reliable estimates of cytosine deamination (Supplemental Figure 4). For smaller data sets from very-low-abundance taxa containing on the order of hundreds of reads, alternative strategies can be used to assess terminal damage patterns (197).
Finally, note that the presence of genuine nucleotide misincorporation profiles is not sufficient to rule out environmental contamination and authenticate the aDNA data as endogenous. First, the data produced will likely comprise a mixture of both ancient and modern DNA contaminants in varying proportions. Second, body decomposition occurs rapidly after death, and microbes that invade skeletal tissues during early decomposition (119) will also accumulate significant DNA damage levels. This can be critical for studies of pathogens with closely related soil members. Conversely, authentic pathogens with reduced kinetics of DNA decay—e.g., owing to the presence of particular chemical components in their bacterial wall, as proposed for M. leprae (170)—may show less damage than other ancient taxa. Lastly, even small conserved regions within the genomes of different bacterial species can result in sufficient read alignment to show what looks like genuine damage profiles; in the case illustrated in Supplemental Figure 5, for example, 0.07–0.11% of simulated aDNA reads from the environmental Mycobacterium smegmatis reference genome sequence can be successfully aligned on the M. leprae and M. tuberculosis genome sequences while still showing the expected damage profiles. Therefore, unidentified or misidentified bacterial sources for the aDNA data can drive expected DNA damage profiles when using another bacterial genome for read alignment, which therefore cannot be used as the sole criterion for data authentication. Correct phylogenetic placement and/or edit distance distributions compatible with the levels of variation observed within the group of candidate bacteria are thus mandatory. The simulated data in Supplemental Figure 4 show basal G→A (C→T) levels at read starts (ends) below 1% only when aligned against the genome of the correct species.
8.2. DNA Fragmentation
aDNA is highly fragmented through a process that is driven by depurination followed by hydrolysis of the DNA backbone (105). This process, initially described in vitro, is easily identifiable using HTS by looking at DNA breakpoints, which in aDNA are enriched in purines (both A and G) (17). Fragmentation patterns can be visually inspected and are produced by default in software such as mapDamage (83).
The length distribution of aDNA can be approximated by a lognormal distribution (4) and shows an exponential decline in the tail, which results from random fragmentation of DNA (36). Upon logarithmic transformation of fragment length frequencies, the decline can be characterized by a linear function with slope λ. The fragmentation constant λ therefore represents the fraction of bonds broken in the DNA backbone (4, 36) and is a summary statistic of the magnitude of DNA fragmentation. If multiple samples are available, it is possible to assess whether DNA from different organisms retrieved from the same samples is fragmented in a correlated way, e.g., sequencing reads originating from host and pathogen, or different taxa in a given microbiome.
This analysis is possible only if all samples were prepared (DNA extraction and library preparation) and sequenced (sequencing platform) in the same way. The correlation in the magnitude of DNA fragmentation among different organisms from the same samples has been proposed as an additional way to authenticate aDNA findings (158) because DNA stored in the same tissue should generally fragment in a correlated way at similar (Figure 6a) or different (Figure 6b, c) magnitudes. The interpretation of the results should take into account the peculiarities of the DNA sources and tissues analyzed, because there are cases where the correlation is not expected—e.g., when DNA from a given organism is differentially exposed to antemortem processes that fragment DNA (Figure 6d).
Figure 6.
DNA fragmentation from pairs of organisms retrieved from the same historic and prehistoric samples. The x and y axes show the fragmentation constant lambda (λ), which describes the fraction of broken bonds in the DNA backbone. The value of this constant can be assessed directly from the length distribution of high-throughput sequencing reads. The dashed red line indicates the linear regression. (a) DNA fragmentation of host and pathogen in DNA fragments retrieved from Solanum tuberosum herbarium samples infected with Phytophthora infestans (205). DNA from both organisms is fragmented in a correlated way and at a similar magnitude. (b) DNA fragmentation of host and pathogen in DNA fragments retrieved from Homo sapiens teeth samples infected with Yersinia pestis (158). DNA from both organisms is fragmented in a correlated way, but the Y. pestis DNA shows a higher magnitude of fragmentation than the H. sapiens DNA. (c) Fragmentation of oral archaeal (Methanobrevibacter sp.) and bacterial (Streptococcus sp.) DNA retrieved from Chalcolithic-era (approximately 2340–2920 BCE) human dental calculus (207). DNA from both organisms fragments in a correlated way but at different magnitudes: The Methanobrevibacter sp. DNA is less fragmented than the Streptococcus sp. DNA, which may be related to the more robust ether linkages in archaeal cell membranes and the protective action of histones in archaeal genomes. (d) DNA fragmentation of Methanobrevibacter sp. and H. sapiens retrieved from dental calculus (the same samples as in panel c). The fragmentation of H. sapiens DNA is not correlated with either that of Methanobrevibacter sp. (data shown here) or that of Streptococcus sp. (data not shown here). Human DNA often exhibits a higher magnitude of fragmentation in dental calculus compared with microbial DNA, a pattern consistent with an inflammation-driven entry of acellular human DNA into dental plaque biofilms that are rich in extracellular nucleases (139).
8.3. Estimating Source Contribution
In addition to profiling DNA damage and decay patterns, source modeling is a powerful tool for authenticating well-preserved microbiome samples. Originally developed to detect contamination in metagenomic studies, the software tool Source Tracker (91) models the composition of a metagenome sink (i.e., sample) as the product of different contributing metagenome sources using Bayesian inference. For example, the microbial composition of a given sample, such as archaeological dental calculus, can be modeled as a mixture of DNA originating from dental plaque, skin bacteria, soil, and other sources. Using reference metagenomic data sets selected for each of these sources, the algorithm then estimates the proportion of the dental calculus sample originating from each source (Figure 7). SourceTracker is a useful screening tool and has been used to estimate the microbiome preservation of both paleofeces (187) and dental calculus (196, 207) before further analysis of microbiome structure and function. Archaeological samples that produce poor SourceTracker profiles are likely altered by decomposition processes or contamination and are thus not suitable for inferring ancient microbial ecology; however, they may still contain traces of endogenous microbial, dietary, and host DNA that can be studied individually. SourceTracker is implemented in the QIIME pipeline and is most often applied to 16S rRNA gene sequence data, but it can be used with any data set organized as a frequency table.
Figure 7.
Bayesian source estimation of the microbial composition of dental calculus and laboratory samples. Using SourceTracker (91) with a panel of modern dental plaque, skin, and soil reference sources indicates that the modern dental calculus (GU30C) contains a large proportion of microbial DNA (>60%) originating from human dental plaque. Likewise, >70% of microbial DNA in an ancient calculus sample from a high-altitude tomb in Nepal (37.UM2010.9, dating from approximately 400–650 CE) originates from dental plaque. By contrast, microbial DNA from nineteenth-century dental calculus from the West African island of St. Helena (STH16) derives nearly entirely from soil (80%) and human skin (3%). Microbial DNA collected from the surfaces of an osteological laboratory originates from human skin and unknown sources. Consistent with these results, the modern and Nepalese dental calculus samples are dominated by phylotypes belonging to known oral-associated genera (77–80%); by contrast, only 2% of phylotypes in the St. Helena calculus are consistent with oral genera, and 64% are classified as environmental Mycobacteria spp. Data are from Reference 207.
8.4. Sample and Data Hygiene
Standards and precautions for preventing and managing modern DNA contamination are now well established in most aDNA laboratories. These include isolated and dedicated aDNA facilities with high-efficiency particulate arrestance (HEPA) air filtration, regular UV irradiation, and NaOCl sterilization of workspaces; consistent use of personal protection (such as full-body suits, double gloving, and eye shields); continuous contamination monitoring using reagent blanks and nontemplate negative controls in all experiments; and unidirectional workflows. Although these best practices were initially developed for applications related to ancient human genome research, they are equally—if not more—important in ancient microbial research.
Microbial DNA is everywhere, and it is the dominant source of nonendogenous genetic material in all aDNA data sets. Ancient microbial research should never be carried out in a laboratory that cultures, amplifies, or processes samples of living microorganisms. Full laboratory separation with independent ventilation is critical to prevent cross-contamination, and simply providing an isolated workspace within a microbiology laboratory is insufficient to prevent contamination of ancient samples during extraction and library preparation steps. In addition, many common molecular biology reagents, such as DNA polymerases and other enzymes, deoxynucleotide triphosphates (dNTPs), oligonucleotides, and some buffers, are contaminated with microbial DNA, often deriving from expression vectors (169). Levels of contamination are often batch and brand specific and result from different manufacturing procedures. Rigorous testing of all reagents should be performed prior to their use on ancient samples.
Finally, cross-contamination is a serious but preventable challenge for highly multiplexed HTS. So-called barcode bleeding is a phenomenon where by a portion of sample-specific index sequences (barcodes) appear to switch between samples, forming chimeras, likely as a result of jumping PCR. Samples are at most risk for barcode bleeding during pooled amplification steps (e.g., during pooled sequence capture or during reamplification of a final library pool) and during clustering steps prior to Illumina sequencing. Empirical studies have found that up to 1% of HTS reads may be affected by barcode bleeding on early-generation Illumina sequencing platforms, such as the GAIIx (88). By that estimate, if two single-indexed samples are pooled and sequenced together to a depth of 200 million reads, thousands to millions of chimeric sequences can be generated and assigned to the wrong sample. Fortunately, this problem can be overcome by using a multiple-indexing strategy, which allows the rapid identification and removal of sequences with chimeric index pairs (88, 161).
9. CONCLUSIONS
Microbial archaeology is a rapidly growing, multidisciplinary field that has the potential to reveal the complex evolutionary history of humans and their microbes. Over the past five years, great advances have been made in revealing the agents behind devastating historical plagues, tracing the cryptic movements of pathogens in prehistory, and reconstructing the ancestral microbiota of humans. However, many challenges remain. Here we have discussed the foundational concepts of the discipline and suggest standards and precautions to support future research. In the sidebar titled Recommended Authenticity Guidelines, we present seven guiding principles aimed at establishing a common research standard and framework for the genetic investigation of ancient microbes.
Through this framework, we seek to provide a robust empirical and theoretical foundation to support the growing field of microbial archaeology.
Supplementary Material
RECOMMENDED AUTHENTICITY GUIDELINES.
To guide future research in microbial archaeology, we propose the following standards and precautions:
Dedicated aDNA laboratories are necessary to minimize and manage contamination.
Metataxonomic approaches differ in their assumptions and biases, and the results of these analyses should be interpreted with these factors in mind. Parameters, protocols, and databases should be kept as consistent as possible to minimize technical variation. Approaches that have been demonstrated to produce taxonomically biased results for degraded DNA, such as 16S rRNA gene amplicon sequencing, are not recommended for quantitative analyses.
Mapping to a reference genome sequence alone is not sufficient for species-level identification. Mapping should be competitive (to more than one candidate reference) and must take the metagenomic nature of the sample into account. Some genomic loci are more phylogenetically informative than others. In general, mobile elements are not recommended for taxonomic identification and should be used only with caution on a case-by-case basis.
Microbial species identification requires multiple lines of validation, which may include, but are not limited to, demonstrations of coverage evenness, genetic similarity, and haploidy.
DNA damage must be assessed. Microbial aDNA should exhibit patterns of DNA damage and fragmentation; however, the magnitude of damage may vary depending on the source context and species, and the damage pattern itself depends on the workflow and enzymes used during library preparation.
For ancient microbiome samples, such as dental calculus and paleofeces, microbiome community composition must be assessed and tested for biological plausibility before analysis. Diversity analyses and community comparisons should be undertaken with caution and performed only on well-preserved samples; otherwise, the results will be more informative about the process of decomposition than about the original microbiome of the individual.
Finally, research questions and hypotheses involving ancient microbes should be biologically informed. Microbial archaeology is an emerging field drawing on the expertise of researchers who have been trained in multiple disciplines, including archaeology, anthropology, microbiology, evolutionary biology, computational biology, population genetics, and medicine, to name a few. Cross-disciplinary communication and collaboration are critical to develop a robust framework for microbial archaeology research.
Acknowledgments
This work was supported by the Max Planck Society (C.W., A.H., and J.K.), its Donation Award (C.W.), and its Presidential Innovation Fund (H.A.B.); the US National Science Foundation (grants BCS-1516633 and BCS-1643318 to C.W.); the US National Institutes of Health (grant 2R01GM089886 to C.W.); the European Research Council (starting grant APGREID to J.K. and consolidator grant PEGASUS to L.O.); the Danish Research Foundation (grant DNRF94 to L.O.); and the Villum Fonden (grant miGENEPI to L.O.). The authors thank the participants of the 2016 Standards, Precautions, and Advances in Ancient Metagenomics (SPAAM) conference for their thoughtful comments, ideas, and suggestions, as well as Floyd Dewhirst at the Forsyth Institute for providing helpful information about the Human Oral Microbiome Database.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Achtman M. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu. Rev. Microbiol. 2008;66 62:53–70. doi: 10.1146/annurev.micro.62.081307.162832. [DOI] [PubMed] [Google Scholar]
- 2.Achtman M, Wagner M. Microbial diversity and the genetic nature of microbial species. Nat. Rev. Microbiol. 2008;6:431–40. doi: 10.1038/nrmicro1872. [DOI] [PubMed] [Google Scholar]
- 3.Acinas SG, Marcelino LA, Klepac-Ceraj V, Polz MF. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 2004;186:2629–35. doi: 10.1128/JB.186.9.2629-2635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allentoft ME, Collins M, Harker D, Haile J, Oskam CL, et al. The half-life of DNA in bone: measuring decay kinetics in 158 dated fossils. Proc. Biol. Sci. 2012;279:4724–33. doi: 10.1098/rspb.2012.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 6.Ander C, Schulz-Trieglaff OB, Stoye J, Cox AJ. metaBEETL: high-throughput analysis of heterogeneous microbial populations from shotgun DNA sequences. BMC Bioinform. 2013;14(Suppl. 5):S2. doi: 10.1186/1471-2105-14-S5-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersen K, Bird KL, Rasmussen M, Haile J, Breuning-Madsen H, et al. Meta-barcoding of ‘dirt’ DNA from soil reflects vertebrate biodiversity. Mol. Ecol. 2012;21:1966–79. doi: 10.1111/j.1365-294X.2011.05261.x. [DOI] [PubMed] [Google Scholar]
- 8.Aparicio S, Chapman J, Stupka E, Putnam N, Chia JM, et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–10. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- 9.Avila-Arcos MC, Cappellini E, Romero-Navarro JA, Wales N, Moreno-Mayar JV, et al. Application and comparison of large-scale solution-based DNA capture-enrichment methods on ancient DNA. Sci. Rep. 2011;1:74. doi: 10.1038/srep00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bapteste E, O’Malley MA, Beiko RG, Ereshefsky M, Gogarten JP, et al. Prokaryotic evolution and the tree of life are two different things. Biol. Direct. 2009;4:34. doi: 10.1186/1745-6150-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, et al. GenBank. Nucleic Acids Res. 2017;45:D37–42. doi: 10.1093/nar/gkw1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biagini P, Theves C, Balaresque P, Geraut A, Cannet C, et al. Variola virus in a 300-year-old Siberian mummy. N. Engl. J. Med. 2012;367:2057–59. doi: 10.1056/NEJMc1208124. [DOI] [PubMed] [Google Scholar]
- 13.Bos KI, Harkins KM, Herbig A, Coscolla M, Weber N, et al. Pre-Columbian mycobacterial genomes reveal seals as a source of New World human tuberculosis. Nature. 2014;514:494–97. doi: 10.1038/nature13591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bos KI, Herbig A, Sahl J, Waglechner N, Fourment M, et al. Eighteenth century Yersinia pestis genomes reveal the long-term persistence of an historical plague focus. eLife. 2016;5:e12994. doi: 10.7554/eLife.12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bos KI, Jager G, Schuenemann VJ, Vagene AJ, Spyrou MA, et al. Parallel detection of ancient pathogens via array-based DNA capture. Philos. Trans. R. Soc. Lond. B. 2015;370:20130375. doi: 10.1098/rstb.2013.0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bos KI, Schuenemann VJ, Golding GB, Burbano HA, Waglechner N, et al. A draft genome of Yersinia pestis from victims of the Black Death. Nature. 2011;478:506–10. doi: 10.1038/nature10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Briggs AW, Stenzel U, Johnson PL, Green RE, Kelso J, et al. Patterns of damage in genomic DNA sequences from a Neandertal. PNAS. 2007;104:14616–21. doi: 10.1073/pnas.0704665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brotherton P, Endicott P, Sanchez JJ, Beaumont M, Barnett R, et al. Novel high-resolution characterization of ancient DNA reveals C >U-type base modification events as the sole cause of post mortem miscoding lesions. Nucleic Acids Res. 2007;35:5717–28. doi: 10.1093/nar/gkm588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 20.Burbano HA, Hodges E, Green RE, Briggs AW, Krause J, et al. Targeted investigation of the Neandertal genome by array-based sequence capture. Science. 2010;328:723–25. doi: 10.1126/science.1188046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camanocha A, Dewhirst FE. Host-associated bacterial taxa from Chlorobi, Chloroflexi, GN02, Synergistetes, SR1, TM7, and WPS-2 Phyla/candidate divisions. J. Oral Microbiol. 2014;6:25468. doi: 10.3402/jom.v6.25468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cambier CJ, Falkow S, Ramakrishnan L. Host evasion and exploitation schemes of Mycobacterium tuberculosis. Cell. 2014;159:1497–509. doi: 10.1016/j.cell.2014.11.024. [DOI] [PubMed] [Google Scholar]
- 23.Cano RJ, Tiefenbrunner F, Ubaldi M, Del Cueto C, Luciani S, et al. Sequence analysis of bacterial DNA in the colon and stomach of the Tyrolean Iceman. Am. J. Phys. Anthropol. 2000;112:297–309. doi: 10.1002/1096-8644(200007)112:3<297::AID-AJPA2>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 24.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–36. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chain PS, Carniel E, Larimer FW, Lamerdin J, Stoutland PO, et al. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. PNAS. 2004;101:13826–31. doi: 10.1073/pnas.0404012101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan JZ, Sergeant MJ, Lee OY, Minnikin DE, Besra GS, et al. Metagenomic analysis of tuberculosis in a mummy. N. Engl. J. Med. 2013;369:289–90. doi: 10.1056/NEJMc1302295. [DOI] [PubMed] [Google Scholar]
- 27.Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. The Human Oral Mi-crobiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database. 2010;2010 doi: 10.1093/database/baq013. baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–87. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 29.Cohan FM. What are bacterial species? Annu. Rev. Microbiol. 2002;56:457–87. doi: 10.1146/annurev.micro.56.012302.160634. [DOI] [PubMed] [Google Scholar]
- 30.Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, et al. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–96. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cruz-Dávalos DI, Llamas B, Gaunitz C, Fages A, Gamba C, et al. Experimental conditions improving in-solution target enrichment for ancient DNA. Mol. Ecol. Resour. 2017 doi: 10.1111/1755-0998.12595. In press. https://doi.org/10.1111/1755-0998.12595. [DOI] [PubMed]
- 32.Dabney J, Knapp M, Glocke I, Gansauge MT, Weihmann A, et al. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. PNAS. 2013;110:15758–63. doi: 10.1073/pnas.1314445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dabney J, Meyer M, Pääbo S. Ancient DNA damage. Cold Spring Harb. Perspect. Biol. 2013;5:a012567. doi: 10.1101/cshperspect.a012567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de la Cruz F, Davies J. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 2000;8:128–33. doi: 10.1016/s0966-842x(00)01703-0. [DOI] [PubMed] [Google Scholar]
- 35.de Meeus T, Durand P, Renaud F. Species concepts: What for? Trends Parasitol. 2003;19:425–27. doi: 10.1016/s1471-4922(03)00195-8. [DOI] [PubMed] [Google Scholar]
- 36.Deagle BE, Eveson JP, Jarman SN. Quantification of damage in DNA recovered from highly degraded samples—a case study on DNA in faeces. Front. Zool. 2006;3:11. doi: 10.1186/1742-9994-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, et al. The human oral microbiome. J. Bacteriol. 2010;192:5002–17. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–29. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 40.Doolittle WF, Papke RT. Genomics and the bacterial species problem. Genome Biol. 2006;7:116. doi: 10.1186/gb-2006-7-9-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drancourt M, Raoult D. Molecular detection of Yersinia pestis in dental pulp. Microbiology. 2004;150:263–64. doi: 10.1099/mic.0.26885-0. [DOI] [PubMed] [Google Scholar]
- 42.Duggan AT, Perdomo MF, Piombino-Mascali D, Marciniak S, Poinar D, et al. 17th century variola virus reveals the recent history of smallpox. Curr. Biol. 2016;26:3407–12. doi: 10.1016/j.cub.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–61. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 44.Everitt RG, Didelot X, Batty EM, Miller RR, Knox K, et al. Mobile elements drive recombination hotspots in the core genome of Staphylococcus aureus. Nat. Commun. 2014;5:3956. doi: 10.1038/ncomms4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Falkinham JO., III Environmental sources of nontuberculous mycobacteria. Clin. Chest Med. 2015;36:35–41. doi: 10.1016/j.ccm.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 46.Fantini E, Gianese G, Giuliano G, Fiore A. Bacterial metabarcoding by 16S rRNA gene ion torrent amplicon sequencing. Methods Mol. Biol. 2015;1231:77–90. doi: 10.1007/978-1-4939-1720-4_5. [DOI] [PubMed] [Google Scholar]
- 47.Feldman M, Harbeck M, Keller M, Spyrou MA, Rott A, et al. A high-coverage Yersinia pestis genome from a sixth-century Justinianic Plague victim. Mol. Biol. Evol. 2016;33:2911–23. doi: 10.1093/molbev/msw170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, et al. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. PNAS. 2012;109:21390–95. doi: 10.1073/pnas.1215210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 50.Fodor AA, DeSantis TZ, Wylie KM, Badger JH, Ye Y, et al. The “most wanted” taxa from the human microbiome for whole genome sequencing. PLOS ONE. 2012;7:e41294. doi: 10.1371/journal.pone.0041294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Freney J, Kloos WE, Hajek V, Webster JA, Bes M, et al. Recommended minimal standards for description of new staphylococcal species. Int. J. Syst. Bacteriol. 1999;49:489–502. doi: 10.1099/00207713-49-2-489. [DOI] [PubMed] [Google Scholar]
- 52.Fu Q, Meyer M, Gao X, Stenzel U, Burbano HA, et al. DNA analysis of an early modern human from Tianyuan Cave, China. PNAS. 2013;110:2223–27. doi: 10.1073/pnas.1221359110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fukushima H, Gomyoda M. Intestinal carriage of Yersinia pseudotuberculosis by wild birds and mammals in Japan. Appl. Environ. Microbiol. 1991;57:1152–55. doi: 10.1128/aem.57.4.1152-1155.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gansauge MT, Meyer M. Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat. Protoc. 2013;8:737–48. doi: 10.1038/nprot.2013.038. [DOI] [PubMed] [Google Scholar]
- 55.Gengler S, Laudisoit A, Batoko H, Wattiau P. Long-term persistence of Yersinia pseudotuberculosis in entomopathogenic nematodes. PLOS ONE. 2015;10:e0116818. doi: 10.1371/journal.pone.0116818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gilbert JA, Jansson JK, Knight R. The Earth Microbiome project: successes and aspirations. BMC Biol. 2014;12:69. doi: 10.1186/s12915-014-0069-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gilbert MT, Cuccui J, White W, Lynnerup N, Titball RW, et al. Absence of Yersinia pestis-specific DNA in human teeth from five European excavations of putative plague victims. Microbiology. 2004;150:341–54. doi: 10.1099/mic.0.26594-0. [DOI] [PubMed] [Google Scholar]
- 58.Gilbert MT, Cuccui J, White W, Lynnerup N, Titball RW, et al. Response to Drancourt and Raoult. Microbiology. 2004;150:264–65. doi: 10.1099/mic.0.26594-0. [DOI] [PubMed] [Google Scholar]
- 59.Ginolhac A, Rasmussen M, Gilbert MT, Willerslev E, Orlando L. mapDamage: testing for damage patterns in ancient DNA sequences. Bioinformatics. 2011;27:2153–55. doi: 10.1093/bioinformatics/btr347. [DOI] [PubMed] [Google Scholar]
- 60.Glaeser SP, Kampfer P. Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst. Appl. Microbiol. 2015;38:237–45. doi: 10.1016/j.syapm.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 61.Gloag ES, Turnbull L, Huang A, Vallotton P, Wang H, et al. Self-organization of bacterial biofilms is facilitated by extracellular DNA. PNAS. 2013;110:11541–46. doi: 10.1073/pnas.1218898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 2009;27:182–89. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat. Rev. Microbiol. 2005;3:679–87. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- 64.Green RE, Briggs AW, Krause J, Prüfer K, Burbano HA, et al. The Neandertal genome and ancient DNA authenticity. EMBO J. 2009;28:2494–502. doi: 10.1038/emboj.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–22. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gribaldo S, Brochier C. Phylogeny of prokaryotes: Does it exist and why should we care? Res. Microbiol. 2009;160:513–21. doi: 10.1016/j.resmic.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 67.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 68.Hashimoto JG, Stevenson BS, Schmidt TM. Rates and consequences of recombination between rRNA operons. J. Bacteriol. 2003;185:966–72. doi: 10.1128/JB.185.3.966-972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hauswedell H, Singer J, Reinert K. Lambda: the local aligner for massive biological data. Bioinformatics. 2014;30 doi: 10.1093/bioinformatics/btu439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.He X, McLean JS, Edlund A, Yooseph S, Hall AP, et al. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. PNAS. 2015;112:244–49. doi: 10.1073/pnas.1419038112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Herbig A, Maixner F, Bos KI, Zink A, Krause J, Huson DH. MALT: fast alignment and analysis of metagenomic DNA sequence data applied to the Tyrolean Iceman. 2016 bioRxiv:050559 https://doi.org/10.1101/050559.
- 72.Hey J. On the failure of modern species concepts. Trends Ecol. Evol. 2006;21:447–50. doi: 10.1016/j.tree.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 73.Heyn P, Stenzel U, Briggs AW, Kircher M, Hofreiter M, Meyer M. Road blocks on paleogenomes—polymerase extension profiling reveals the frequency of blocking lesions in ancient DNA. Nucleic Acids Res. 2010;38:e161. doi: 10.1093/nar/gkq572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Higashi S, Barreto AMS, Cantão ME, de Vasconcelos ATR. Analysis of composition-based metagenomic classification. BMC Genom. 2012;13(Suppl. 5):S1. doi: 10.1186/1471-2164-13-S5-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, et al. Genome-wide in situ exon capture for selective resequencing. Nat. Genet. 2007;39:1522–27. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- 76.Hruska K, Kaevska M. Mycobacteria in water, soil, plants and air: a review. Vet. Med. 2012;57:623–79. [Google Scholar]
- 77.Hughes J, Bohannan BJ. Application of ecological diversity statistics in microbial ecology. In: Kowalchuk GA, de Bruijn FJ, Head IM, Akkermans ADL, van Elsas JD, editors. Molecular Microbial Ecology Manual. 2 Dordrecht, Neth: Kluwer Acad; 2004. pp. 1321–44. [Google Scholar]
- 78.Hum. Microbiome Proj. Consort. A framework for human microbiome research. Nature. 2012;486:215–21. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hum. Microbiome Proj. Consort. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huson DH, Beier S, Flade I, Górska A, El-Hadidi M, et al. MEGAN Community Edition— interactive exploration and analysis of large-scale microbiome sequencing data. PLOS Comput. Biol. 2016;12:e1004957. doi: 10.1371/journal.pcbi.1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jalava K, Hakkinen M, Valkonen M, Nakari UM, Palo T, et al. An outbreak of gastrointestinal illness and erythema nodosum from grated carrots contaminated with Yersinia pseudotuberculosis. J. Infect. Dis. 2006;194:1209–16. doi: 10.1086/508191. [DOI] [PubMed] [Google Scholar]
- 82.Johnston M. Joshua Lederberg on bacterial recombination. Genetics. 2016;203:613–14. doi: 10.1534/genetics.116.190637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jonsson H, Ginolhac A, Schubert M, Johnson PL, Orlando L. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013;29:1682–84. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kampfer P, Buczolits S, Albrecht A, Busse HJ, Stackebrandt E. Towards a standardized format for the description of a novel species (of an established genus): Ochrobactrum gallinifaecis sp. nov. Int. J. Syst. Evol. Microbiol. 2003;53:893–96. doi: 10.1099/ijs.0.02710-0. [DOI] [PubMed] [Google Scholar]
- 85.Kay GL, Sergeant MJ, Giuffra V, Bandiera P, Milanese M, et al. Recovery of a medieval Brucella melitensis genome using shotgun metagenomics. mBio. 2014;5:e01337–14. doi: 10.1128/mBio.01337-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kerpedjiev P, Frellsen J, Lindgreen S, Krogh A. Adaptable probabilistic mapping of short reads using position specific scoring matrices. BMC Bioinform. 2014;15:1. doi: 10.1186/1471-2105-15-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim M, Oh H-S, Park S-C, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014;64:346–51. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 88.Kircher M, Sawyer S, Meyer M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 2012;40:e3. doi: 10.1093/nar/gkr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kitahara K, Miyazaki K. Revisiting bacterial phylogeny: natural and experimental evidence for horizontal gene transfer of 16S rRNA. Mob. Genet. Elem. 2013;3:e24210. doi: 10.4161/mge.24210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Klappenbach JA, Saxman PR, Cole JR, Schmidt TM. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 2001;29:181–84. doi: 10.1093/nar/29.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, et al. Bayesian community-wide culture-independent microbial source tracking. Nat. Methods. 2011;8:761–63. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Koboldt DC, Steinberg KM, Larson DE, Wilson RK, Mardis ER. The next-generation sequencing revolution and its impact on genomics. Cell. 2013;155:27–38. doi: 10.1016/j.cell.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Koehler A, Karch H, Beikler T, Flemmig TF, Suerbaum S, Schmidt H. Multilocus sequence analysis of Porphyromonas gingivalis indicates frequent recombination. Microbiology. 2003;149:2407–15. doi: 10.1099/mic.0.26267-0. [DOI] [PubMed] [Google Scholar]
- 94.Krause J, Briggs AW, Kircher M, Maricic T, Zwyns N, et al. A complete mtDNA genome of an early modern human from Kostenki, Russia. Curr. Biol. 2010;20:231–36. doi: 10.1016/j.cub.2009.11.068. [DOI] [PubMed] [Google Scholar]
- 95.Kucera RB, Nichols NM. DNA-dependent DNA polymerases. Curr. Protoc. Mol. Biol. 2008;84:3.5.1–19. doi: 10.1002/0471142727.mb0305s84. [DOI] [PubMed] [Google Scholar]
- 96.Kuczynski J, Stombaugh J, Walters WA, González A, Caporaso JG, Knight R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Bioinform. 2011;36:10.7.1–20. doi: 10.1002/0471250953.bi1007s36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–59. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lapage SP, Sneath PHA, Lessel EF, Skerman VBD, Seeliger HPR, Clark WA. International Code of Nomenclature of Bacteria: Bacteriological Code, 1990 Revision. Washington, DC: ASM Press; 1992. [PubMed] [Google Scholar]
- 99.Lawrence JG, Hendrickson H. Lateral gene transfer: When will adolescence end? Mol. Microbiol. 2003;50:739–49. doi: 10.1046/j.1365-2958.2003.03778.x. [DOI] [PubMed] [Google Scholar]
- 100.Lawson PA, Citron DM, Tyrrell KL, Finegold SM. Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O’Toole 1935) Prévot 1938. Anaerobe. 2016;40:95–99. doi: 10.1016/j.anaerobe.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 101.Lederberg J, McCray AT. ‘Ome sweet ‘omics—a genealogical treasury of words. Scientist. 2001;15:8. [Google Scholar]
- 102.Lee ZM, Bussema C, III, Schmidt TM. rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res. 2009;37:D489–93. doi: 10.1093/nar/gkn689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lefebure T, Stanhope MJ. Evolution of the core and pan-genome of Streptococcus:positive selection, recombination, and genome composition. Genome Biol. 2007;8:R71. doi: 10.1186/gb-2007-8-5-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–15. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 106.Lindgreen S, Adair KL, Gardner PP. An evaluation of the accuracy and speed of metagenome analysis tools. Sci. Rep. 2016;6:19233. doi: 10.1038/srep19233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Linz B, Balloux F, Moodley Y, Manica A, Liu H, et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445:915–18. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Locey KJ, Lennon JT. Scaling laws predict global microbial diversity. PNAS. 2016;113:5970–75. doi: 10.1073/pnas.1521291113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Long C, Jones TF, Vugia DJ, Scheftel J, Strockbine N, et al. Yersinia pseudotuberculosis and Y. enterocolitica infections, FoodNet, 1996–2007. Emerg. Infect. Dis. 2010;16:566–67. doi: 10.3201/eid1603.091106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Louvel G, Der Sarkissian C, Hanghoj K, Orlando L. metaBIT, an integrative and automated metagenomic pipeline for analysing microbial profiles from high-throughput sequencing shotgun data. Mol. Ecol. Resour. 2016;16:1415–27. doi: 10.1111/1755-0998.12546. [DOI] [PubMed] [Google Scholar]
- 111.Maiden MFJ, Cohee P, Tanner ACR. Proposal to conserve the adjectival form of the specific epithet in the reclassification of Bacteroides forsythus Tanner et al. 1986 to the genus Tannerella Sakamoto et al. 2002 as Tannerella forsythia corrig., gen. nov., comb. nov. Request for an Opinion. Int. J. Syst. Evol. Microbiol. 2003;53:2111–12. doi: 10.1099/ijs.0.02641-0. [DOI] [PubMed] [Google Scholar]
- 112.Maixner F, Krause-Kyora B, Turaev D, Herbig A, Hoopmann MR, et al. The 5300-year-old Helicobacter pylori genome of the Iceman. Science. 2016;351:162–65. doi: 10.1126/science.aad2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Marchesi JR, Ravel J. The vocabulary of microbiome research: a proposal. Microbiome. 2015;3:31. doi: 10.1186/s40168-015-0094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mardis ER. Next-generation DNA sequencing methods. Annu. Rev. Genom. Hum. Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 115.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–80. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Martin MD, Cappellini E, Samaniego JA, Zepeda ML, Campos PF, et al. Reconstructing genome evolution in historic samples of the Irish potato famine pathogen. Nat. Commun. 2013;4:2172. doi: 10.1038/ncomms3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mayr E. Systematics and the Origin of Species from the Viewpoint of a Zoologist. New York: Columbia Univ. Press; 1942. [Google Scholar]
- 118.Medini D, Serruto D, Parkhill J, Relman DA, Donati C, et al. Microbiology in the post-genomic era. Nat. Rev. Microbiol. 2008;6:419–30. doi: 10.1038/nrmicro1901. [DOI] [PubMed] [Google Scholar]
- 119.Metcalf JL, Xu ZZ, Weiss S, Lax S, Van Treuren W, et al. Microbial community assembly and metabolic function during mammalian corpse decomposition. Science. 2016;351:158–62. doi: 10.1126/science.aad2646. [DOI] [PubMed] [Google Scholar]
- 120.Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010;2010 doi: 10.1101/pdb.prot5448. pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 121.Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–26. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mira A, Martin-Cuadrado AB, D’Auria G, Rodriguez-Valera F. The bacterial pan-genome: a new paradigm in microbiology. Int. Microbiol. 2010;13:45–57. doi: 10.2436/20.1501.01.110. [DOI] [PubMed] [Google Scholar]
- 123.Mora C, Tittensor DP, Adl S, Simpson AG, Worm B. How many species are there on Earth and in the ocean? PLOS Biol. 2011;9:e1001127. doi: 10.1371/journal.pbio.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, Schäffer AA. Database indexing for production MegaBLAST searches. Bioinformatics. 2008;24:1757–64. doi: 10.1093/bioinformatics/btn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mukherjee S, Stamatis D, Bertsch J, Ovchinnikova G, Verezemska O, et al. Genomes OnLine Database (GOLD) v.6: data updates and feature enhancements. Nucleic Acids Res. 2017;45:D446–56. doi: 10.1093/nar/gkw992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Murray DC, Haile J, Dortsch J, White NE, Haouchar D, et al. Scrapheap Challenge: a novel bulk-bone metabarcoding method to investigate ancient DNA in faunal assemblages. Sci. Rep. 3:3371. doi: 10.1038/srep03371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Natl. Cent. Biotechnol. Inf. How to reference the NCBI taxonomy database. 2017 https://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/index.cgi?chapter=howcite.
- 128.Navas-Molina JA, Peralta-Sanchez JM, Gonzalez A, McMurdie PJ, Vazquez-Baeza Y, et al. Advancing our understanding of the human microbiome using QIIME. Methods Enzymol. 2013;531:371–444. doi: 10.1016/B978-0-12-407863-5.00019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nayfach S, Rodriguez-Mueller B, Garud N, Pollard KS. An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res. 2016;26:1612–25. doi: 10.1101/gr.201863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–53. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 131.O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D45. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 133.Octavia S, Lan R. The family Enterobacteriaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes: Gammaproteobacteria. 4 Berlin: Springer-Verlag; 2014. pp. 225–86. [Google Scholar]
- 134.Ondov BD, Bergman NH, Phillippy AM. Interactive metagenomic visualization in a Web browser. BMC Bioinform. 2011;12:385. doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Orlando L, Ginolhac A, Zhang G, Froese D, Albrechtsen A, et al. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature. 2013;499:74–78. doi: 10.1038/nature12323. [DOI] [PubMed] [Google Scholar]
- 136.Overballe-Petersen S, Harms K, Orlando LA, Mayar JV, Rasmussen S, et al. Bacterial natural transformation by highly fragmented and damaged DNA. PNAS. 2013;110:19860–65. doi: 10.1073/pnas.1315278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pagani I, Liolios K, Jansson J, Chen IM, Smirnova T, et al. The Genomes OnLine Database (GOLD) v.4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2012;40:D571–79. doi: 10.1093/nar/gkr1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pallen MJ, Wren BW. Bacterial pathogenomics. Nature. 2007;449:835–42. doi: 10.1038/nature06248. [DOI] [PubMed] [Google Scholar]
- 139.Palmer LJ, Chapple IL, Wright HJ, Roberts A, Cooper PR. Extracellular deoxyribonuclease production by periodontal bacteria. J. Periodontal Res. 2012;47:439–45. doi: 10.1111/j.1600-0765.2011.01451.x. [DOI] [PubMed] [Google Scholar]
- 140.Parkhill J. What has high-throughput sequencing ever done for us? Nat. Rev. Microbiol. 2013;11:664–65. doi: 10.1038/nrmicro3112. [DOI] [PubMed] [Google Scholar]
- 141.Parte AC. LPSN—List of Prokaryotic Names with Standing in Nomenclature. Nucleic Acids Res. 2014;42:D613–16. doi: 10.1093/nar/gkt1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Peabody MA, Van Rossum T, Lo R, Brinkman FSL. Evaluation of shotgun metagenomics sequence classification methods using in silico and in vitro simulated communities. BMC Bioinform. 2015;16:363. doi: 10.1186/s12859-015-0788-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pedersen MW, Ruter A, Schweger C, Friebe H, Staff RA, et al. Postglacial viability and colonization in North America’s ice-free corridor. Nature. 2016;537:45–49. doi: 10.1038/nature19085. [DOI] [PubMed] [Google Scholar]
- 144.Penders J, Stobberingh EE, Savelkoul PH, Wolffs P. The human microbiome as a reservoir of antimicrobial resistance. Front. Microbiol. 2013;4:87. doi: 10.3389/fmicb.2013.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Perez-Losada M, Browne EB, Madsen A, Wirth T, Viscidi RP, Crandall KA. Population genetics of microbial pathogens estimated from multilocus sequence typing (MLST) data. Infect. Genet. Evol. 2006;6:97–112. doi: 10.1016/j.meegid.2005.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Perez-Losada M, Cabezas P, Castro-Nallar E, Crandall KA. Pathogen typing in the genomics era: MLST and the future of molecular epidemiology. Infect. Genet. Evol. 2013;16:38–53. doi: 10.1016/j.meegid.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 147.Pham VH, Kim J. Cultivation of unculturable soil bacteria. Trends Biotechnol. 2012;30:475–84. doi: 10.1016/j.tibtech.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 148.Philippot L, Andersson SG, Battin TJ, Prosser JI, Schimel JP, et al. The ecological coherence of high bacterial taxonomic ranks. Nat. Rev. Microbiol. 2010;8:523–29. doi: 10.1038/nrmicro2367. [DOI] [PubMed] [Google Scholar]
- 149.Pignatelli M, Aparicio G, Blanquer I, Hernández V, Moya A, Tamames J. Metagenomics reveals our incomplete knowledge of global diversity. Bioinformatics. 2008;24:2124–25. doi: 10.1093/bioinformatics/btn355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Poinar H, Kuch M, Pääbo S. Molecular analyses of oral polio vaccine samples. Science. 2001;292:743–44. doi: 10.1126/science.1058463. [DOI] [PubMed] [Google Scholar]
- 151.Prufer K, Meyer M. Comment on “Late Pleistocene human skeleton and mtDNA link Paleoamericans and modern Native Americans”. Science. 2015;347:835. doi: 10.1126/science.1260617. [DOI] [PubMed] [Google Scholar]
- 152.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–96. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rajilic-Stojanovic M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol. Rev. 2014;38:996–1047. doi: 10.1111/1574-6976.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Raoult D, Aboudharam G, Crubezy E, Larrouy G, Ludes B, Drancourt M. Molecular identification by “suicide PCR” of Yersinia pestis as the agent of medieval black death. PNAS. 2000;97:12800–3. doi: 10.1073/pnas.220225197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Raoult D, Drancourt M, Fournier PE, Ogata H. Yersinia pestis genotyping—response. Emerg. Infect. Dis. 2005;11:1318–19. [Google Scholar]
- 156.Rasheed Z, Rangwala H. Metagenomic taxonomic classification using extreme learning machines. J. Bioinform. Comput. Biol. 2012;10:1250015. doi: 10.1142/S0219720012500151. [DOI] [PubMed] [Google Scholar]
- 157.Rasmussen M, Li Y, Lindgreen S, Pedersen JS, Albrechtsen A, et al. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature. 2010;463:757–62. doi: 10.1038/nature08835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rasmussen S, Allentoft ME, Nielsen K, Orlando L, Sikora M, et al. Early divergent strains of Yersinia pestis in Eurasia 5,000 years ago. Cell. 2015;163:571–82. doi: 10.1016/j.cell.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Reich D, Green RE, Kircher M, Krause J, Patterson N, et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–60. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Richter M, Rosselló-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. PNAS. 2009;106:19126–31. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rohland N, Harney E, Mallick S, Nordenfelt S, Reich D. Partial uracil-DNA-glycosylase treatment for screening of ancient DNA. Philos. Trans. R. Soc. Lond. B. 2015;370:20130624. doi: 10.1098/rstb.2013.0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rossello´-Mora R, Amann R. The species concept for prokaryotes. FEMS Microbiol. Rev. 2001;25:39–67. doi: 10.1111/j.1574-6976.2001.tb00571.x. [DOI] [PubMed] [Google Scholar]
- 163.Sakamoto M, Suzuki M, Umeda M, Ishikawa I, Benno Y. Reclassification of Bacteroides forsythus (Tanner et al. 1986) as Tannerella forsythensis corrig., gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 2002;52:841–49. doi: 10.1099/00207713-52-3-841. [DOI] [PubMed] [Google Scholar]
- 164.Sawyer S, Krause J, Guschanski K, Savolainen V, Pääbo S. Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PLOS ONE. 2012;7:e34131. doi: 10.1371/journal.pone.0034131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Schloss PD, Girard RA, Martin T, Edwards J, Thrash JC. Status of the archaeal and bacterial census: an update. mBio. 2016;7:e00201–16. doi: 10.1128/mBio.00201-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Schloss PD, Jenior ML, Koumpouras CC, Westcott SL, Highlander SK. Sequencing 16S rRNA gene fragments using the PacBio SMRT DNA sequencing system. Peer J. 2016;4:e1869. doi: 10.7717/peerj.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Schuenemann VJ, Bos K, DeWitte S, Schmedes S, Jamieson J, et al. Targeted enrichment of ancient pathogens yielding the pPCP1 plasmid of Yersinia pestis from victims of the Black Death. PNAS. 2011;108:E746–52. doi: 10.1073/pnas.1105107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Schuenemann VJ, Singh P, Mendum TA, Krause-Kyora B, Jager G, et al. Genome-wide comparison of medieval and modern Mycobacterium leprae. Science. 2013;341:179–83. doi: 10.1126/science.1238286. [DOI] [PubMed] [Google Scholar]
- 171.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods. 2012;9:811–14. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Seguin-Orlando A, Hoover CA, Vasiliev SK, Ovodov ND, Shapiro B, et al. Amplification of TruSeq ancient DNA libraries with AccuPrime Pfx: consequences on nucleotide misincorporation and methylation patterns. STAR. 2015;1:1–9. [Google Scholar]
- 174.Seguin-Orlando A, Schubert M, Clary J, Stagegaard J, Alberdi MT, et al. Ligation bias in Illumina next-generation DNA libraries: implications for sequencing ancient genomes. PLOS ONE. 2013;8:e78575. doi: 10.1371/journal.pone.0078575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sleator RD. The human superorganism—of microbes and men. Med. Hypotheses. 2010;74:214–15. doi: 10.1016/j.mehy.2009.08.047. [DOI] [PubMed] [Google Scholar]
- 176.Smith O, Clapham A, Rose P, Liu Y, Wang J, Allaby RG. A complete ancient RNA genome: identification, reconstruction and evolutionary history of archaeological barley stripe mosaic virus. Sci. Rep. 2014;4:4003. doi: 10.1038/srep04003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–97. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 178.Spyrou MA, Tukhbatova RI, Feldman M, Drath J, Kacki S, et al. Historical Y. pestis genomes reveal the European Black Death as the source of ancient and modern plague pandemics. Cell Host Microbe. 2016;19:874–81. doi: 10.1016/j.chom.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 179.Stackebrandt E, Frederiksen W, Garrity GM, Grimont PA, Kampfer P, et al. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 2002;52:1043–47. doi: 10.1099/00207713-52-3-1043. [DOI] [PubMed] [Google Scholar]
- 180.Stoddard SF, Smith BJ, Hein R, Roller BR, Schmidt TM. rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 2015;43:D593–98. doi: 10.1093/nar/gku1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Stoneking M, Krause J. Learning about human population history from ancient and modern genomes. Nat. Rev. Genet. 2011;12:603–14. doi: 10.1038/nrg3029. [DOI] [PubMed] [Google Scholar]
- 182.Tanner ACR, Listgarten MA, Ebersole JL, Strzempko MN. Bacteroides forsythus sp. nov., a slowgrowing, fusiform Bacteroides sp. from the human oral cavity. Int. J. Syst. Bacteriol. 1986;36:213–21. [Google Scholar]
- 183.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–24. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. PNAS. 2005;102:13950–55. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005;3:711–21. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 186.Tindall BJ, Kampfer P, Euzeby JP, Oren A. Valid publication of names of prokaryotes according to the rules of nomenclature: past history and current practice. Int. J. Syst. Evol. Microbiol. 2006;56:2715–20. doi: 10.1099/ijs.0.64780-0. [DOI] [PubMed] [Google Scholar]
- 187.Tito RY, Knights D, Metcalf J, Obregon-Tito AJ, Cleeland L, et al. Insights from characterizing extinct human gut microbiomes. PLOS ONE. 2012;7:e51146. doi: 10.1371/journal.pone.0051146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods. 2015;12:902–3. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 189.Tumpey TM, Basler CF, Aguilar PV, Zeng H, Solorzano A, et al. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science. 2005;310:77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 190.Vetrovsky T, Baldrian P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLOS ONE. 2013;8:e57923. doi: 10.1371/journal.pone.0057923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Vos M, Didelot X. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 2009;3:199–208. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- 192.Wagner DM, Klunk J, Harbeck M, Devault A, Waglechner N, et al. Yersinia pestis and the plague of Justinian 541–543 AD: a genomic analysis. Lancet Infect. Dis. 2014;14:319–26. doi: 10.1016/S1473-3099(13)70323-2. [DOI] [PubMed] [Google Scholar]
- 193.Walker AW, Duncan SH, Louis P, Flint HJ. Phylogeny, culturing, and metagenomics of the human gut microbiota. Trends Microbiol. 2014;22:267–74. doi: 10.1016/j.tim.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 194.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–67. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Warinner C. Dental calculus and the evolution of the human oral microbiome. J. Calif. Dent. Assoc. 2016;44:411–20. [PubMed] [Google Scholar]
- 196.Warinner C, Rodrigues JF, Vyas R, Trachsel C, Shved N, et al. Pathogens and host immunity in the ancient human oral cavity. Nat. Genet. 2014;46:336–44. doi: 10.1038/ng.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Weiss CL, Dannemann M, Prüfer K, Burbano HA. Contesting the presence of wheat in the British Isles 8,000 years ago by assessing ancient DNA authenticity from low-coverage data. eLife. 2015;4:e10005. doi: 10.7554/eLife.10005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 199.Wilbur AK, Bouwman AS, Stone AC, Roberts CA, Pfister L-A, et al. Deficiencies and challenges in the study of ancient tuberculosis DNA. J. Archaeol. Sci. 2009;36:1990–97. [Google Scholar]
- 200.Willerslev E, Cooper A. Ancient DNA. Proc. Biol. Sci. 2005;272:3–16. doi: 10.1098/rspb.2004.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Yahara K, Didelot X, Ansari MA, Sheppard SK, Falush D. Efficient inference of recombination hot regions in bacterial genomes. Mol. Biol. Evol. 2014;31:1593–605. doi: 10.1093/molbev/msu082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yahara K, Didelot X, Jolley KA, Kobayashi I, Maiden MC, et al. The landscape of realized homologous recombination in pathogenic bacteria. Mol. Biol. Evol. 2016;33:456–71. doi: 10.1093/molbev/msv237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, et al. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014;42:D643–48. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yoshida K, Schuenemann VJ, Cano LM, Pais M, Mishra B, et al. The rise and fall of the Phytophthora infestans lineage that triggered the Irish potato famine. eLife. 2013;2:e00731. doi: 10.7554/eLife.00731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Zhao Y, Tang H, Ye Y. RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics. 2012;28:125–26. doi: 10.1093/bioinformatics/btr595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ziesemer KA, Mann AE, Sankaranarayanan K, Schroeder H, Ozga AT, et al. Intrinsic challenges in ancient microbiome reconstruction using 16S rRNA gene amplification. Sci. Rep. 2015;5:16498. doi: 10.1038/srep16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Zink AR, Sola C, Reischl U, Grabner W, Rastogi N, et al. Characterization of Mycobacterium tuberculosis complex DNAs from Egyptian mummies by spoligotyping. J. Clin. Microbiol. 2003;41:359–67. doi: 10.1128/JCM.41.1.359-367.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Zurek L, Denning SS, Schal C, Watson DW. Vector competence of Musca domestica (Diptera: Muscidae) for Yersinia pseudotuberculosis. J. Med. Entomol. 2001;38:333–35. doi: 10.1603/0022-2585-38.2.333. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.