Abstract
Connexin37 (Cx37) is a gap junction protein involved in cell-to-cell communication in the vasculature and other tissues. Cx37 suppresses proliferation of vascular cells involved in tissue development and repair in vivo, as well as tumor cells. Global deletion of Cx37 in mice leads to enhanced vasculogenesis in development, as well as collateralgenesis and angiogenesis in response to injury, which together support improved tissue remodeling and recovery following ischemic injury. Here we report the 1H, 15N, and 13C resonance assignments for an important regulatory domain of Cx37, the carboxyl terminus (CT; C233-V333). The predicted secondary structure of the Cx37CT domain based on the chemical shifts is that of an intrinsically disordered protein. In the 1H-15N HSQC, N-terminal residues S254-Y259 displayed a second weaker peak and residues E261-Y266 had significant line broadening. These residues are flanked by prolines (P250, P258, P260, and P268), suggesting proline cis-trans isomerization. Overall, these assignments will be useful for identifying the binding sites for intra- and inter-molecular interactions that affect Cx37 channel activity.
Keywords: Cx37, gap junction, carboxyl terminus, intrinsically disordered protein
Biological context
Connexins are the integral membrane proteins that comprise gap junction channels. These channels provide a pathway for direct cytoplasmic exchange of ions and low molecular weight metabolites (<1 kDa) between adjacent cells. As such, they provide a pathway for the propagation and/or amplification of signal transduction cascades triggered by cytokines, growth factors, and other cell signaling molecules involved in growth, development, and response to injury and disease. Gap junction channels are formed by the apposition of two connexons from adjacent cells, where each connexon is formed of six connexin proteins. Connexins are tetraspan transmembrane domain proteins with intracellular amino- and carboxyl-termini. There are 21 different connexin genes in the human genome with differential spatial-temporal expression throughout the body. In the arterial vasculature, gap junctions couple the vascular smooth muscle cells (VSMCs), the endothelial cells (ECs), and also connect ECs to underlying smooth muscle cells via myoendothelial gap junctions (Haefliger et al., 2004). The major connexins in arterial vessels are connexin45 (Cx45), connexin43 (Cx43), connexin40 (Cx40), and connexin37 (Cx37). The expression of these connexins is not uniform and varies with vessel size and vascular territory (for review, see (Brisset et al., 2009)). The intercellular communication in the vasculature contributes to the regulation of resistance in different vascular beds according to metabolic needs, and therefore ultimately participates in the regulation of blood pressure. Pathological conditions, such as atherosclerosis, diabetes, and hypertension are associated with changes in connexin regulation and expression (for review, see (Morel, 2014)).
Though there is significant sequence homology among connexins in the transmembrane and extracellular loops (see Figure 2, (Harris, 2001)), the major divergence in primary structures occurs in the cytoplasmic loop and carboxyl terminal (CT) domains. The CT plays a role in the trafficking, localization, and turnover of gap junction channels, as well as the level of gap junction intercellular communication via numerous post-translational modifications and protein-protein interactions (Herve, 2007; Laird, 2010; Lampe and Lau, 2004; Thevenin et al., 2013). The CT is also important for regulating junctional conductance, pH sensitivity, and voltage sensitivity (Anumonwo et al., 2001; Moreno et al., 2002; Morley et al., 1996; Revilla et al., 1999). Structural studies from our labs revealed that the CT domain of connexins is predominately unstructured (Bouvier et al., 2008; Nelson et al., 2013; Sorgen et al., 2004b; Stauch et al., 2012). Intrinsically disordered domains are now well recognized to be loci for regulation of protein function because their conformations can readily be modulated by the local environment, phosphorylation, and by interaction with proteins and small-molecule binding partners. In the case of membrane proteins, like connexins, intrinsically disordered domains play an important role in cell signaling events by allowing many different binding partners with both high specificity and low affinity to rapidly switch between molecular partners, thus activating alternative signaling pathways (Dunker et al., 2002). We and others have shown that the connexin CT can bind multiple proteins (>20 known for Cx43), some of which, where tested, modulate channel function (for review see (Gilleron et al., 2012)).
Figure 2.
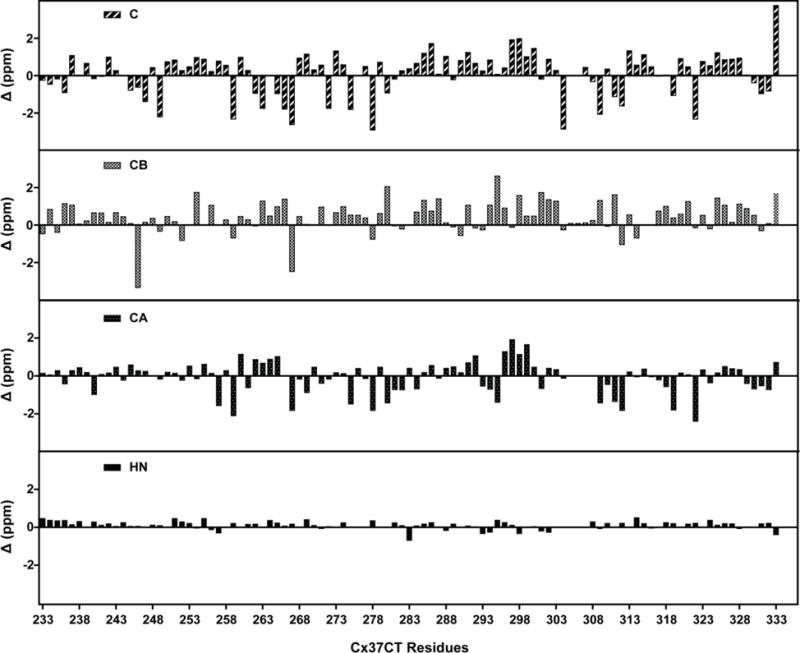
Chemical shift index values for Cx37 carboxyl terminal domain residues. Threshold deviations from random coil 1HN, 13Cα, 13Cβ, and 13CO chemical shifts were plotted for each residue (Wishart Database as a reference).
Structural information of the Cx43CT, Cx40CT, and Cx45CT domains has advanced our understanding of gap junction regulation via protein-protein interactions, phosphorylation induced structural changes, and channel regulation. However, to date, no high resolution structural information exists for the Cx37CT. Here, we report the sequence-specific assignments for the Cx37CT domain. These assignments will be useful for mapping the binding sites for potential molecular partners, including inter- and intra-molecular connexin domains.
Methods and experiments
The recombinant Mus musculus Cx37CT (residues C233-V333) was subcloned into the pGEX-KT and pET-14b bacterial expression vectors according to standard protocol. Expression and purification of the 15N-13C-labeled Cx37CT was achieved following a previously published protocol (Kieken et al., 2010). Briefly, the pGEX-KT vector containing the Cx37CT was transformed in E. coli BL21(DE3) bacterial strain and grown in M9 minimum medium supplemented with 15N-ammonium chloride and 13C-glucose. After induction with IPTG (1 mM) at a cell density of 0.6 (OD at 600 nm), cells were grown for an additional 4 hours, pelleted, and lysed in 1× PBS lysis buffer using an EmulsiFlex-C3 homogenizer. After incubating the GST-tag Cx37CT with glutathione resin, non-specific binding proteins were removed by sequential washes with 2× PBS and 1× PBS in presence of 2 mM DTT, at pH 6.0. The GST-tag was subsequently cleaved overnight at 4°C by incubation with thrombin (7 units per liter of culture). The Cx37CT was then subjected to cation exchange chromatography for further purification (HiTrap SP HP column; Elution with an increasing gradient of sodium chloride in 1× PBS at pH 7.0 in presence of 2 mM DTT) using an ÄKTA FPLC. Fractions containing the Cx37CT were pooled and dialyzed against 1× PBS, 2 mM DTT, at pH 6.0 overnight before concentrating the protein using a 3 kDa Amicon® Ultra centrifugal filter. Final NMR sample contained 650 μM of 15N-13C-isotopically labeled Cx37CT protein in 1× PBS, 4% D2O, 2 mM DTT, at pH 5.8.
NMR data were acquired at 7°C using a Bruker Avance-III HD 600 MHz spectrometer outfitted with a z-axis PFG “inverse” triple-resonance cold probe. HNCACB, CBCA(CO)NH, HNCO, and HN(CA)CO 3D experiments were collected to carry out the backbone sequential assignment while the 3D 15N-TOCSYHSQC, 15N-NOESYHSQC, 13C-HCCH-TOCSY, and 13C- NOESYHSQC experiments were used to assign the chemical shifts of the side chain atoms. All spectra were processed using Bruker Topspin 3.2, and analyzed with NMRView (Johnson, 2004) after transfer through NMRPipe (Delaglio et al., 1995).
To aid in the unequivocal assignment of some of the amino acid chemical shifts, the E. coli DL39 strain (auxotroph for Phe, Tyr, Asp, Leu, Ile, and Val; (LeMaster and Richards, 1988)) was transformed with a pET-14b vector containing the Cx37CT. Selective amino acid labeling was achieved by growing the cells in a non-labeled M9 minimum media supplemented with each of the auxotroph amino acids (150 mg/L of Phe, 90 mg/L of Tyr, 400 mg/L of Asp, 200 mg/L of Leu, 200 mg/L of Ile, and 200 mg/L of Val; (Vourtsis et al., 2014)) until reaching an optical density of 0.6 at 600 nm, and supplementing with either 15N-Tyr, 15N-Phe, or 15N-Val, as well as the other auxotroph unlabeled amino acids when adding IPTG (same amount as described above). Cells were grown for an additional 12 hours at room temperature, pelleted, and lysed in 1× PBS buffer using an EmulsiFlex-C3 homogenizer. Unexpectedly, the Cx37CT protein expressed by the DL39 cells was found in the inclusion bodies. Purification was achieved following a protocol similar to that described in (Kellezi et al., 2008). Briefly, the inclusion bodies containing the 6× His-tag Cx37CT were solubilized overnight at 4 °C in a buffer containing 1× PBS (pH 8.0), 6 M urea, 1% Triton X-100, and 2 mM β-mercaptoethanol, and loaded onto a HisTrap HP column for purification by affinity chromatography. After loading, buffer was exchanged to eliminate the Triton X-100 before eluting with increasing amount of imidazole. Fractions containing the Cx37CT were pooled and dialyzed against 1× PBS, 2 mM DTT, at pH 6.0. After concentration through a 3 kDa Amicon® Ultra centrifugal filter, the final NMR samples contained 50 μM of the 15N-specifically labeled amino acid Cx37CT protein in 1× PBS, 4% D2O, 2 mM DTT, at pH 5.8. 2D 15N-HSQC experiments were collected to identify the specific cross peaks.
Assignments and data deposition
Data collection of the Cx37CT domain was performed in 1× PBS (pH 5.8), as previously collected for the Cx43, Cx40, and Cx45CT domains (Bouvier et al., 2007; Kopanic and Sorgen, 2013; Sorgen et al., 2004a). Assignments were made for all of the 1H, 15N, and 13C backbone resonances and 92% of the side chain resonances (BioMag ResBank database, accession number 26921). Figure 1 shows the 1H-15N HSQC spectrum and assignments for the Cx37CT domain. The low chemical shift dispersion in the 1H-15N HSQC spectrum indicates the Cx37CT is predominately unfolded. Chemical shift index predictions of secondary structure (Figure 2; (Wishart and Sykes, 1994)) and the lack of medium- and long-range NOE connectivities observed in the 1H-15N NOESY-HSQC and 1H-13C-NOESY spectra (data not shown) are consistent with the prediction of an intrinsically disordered structure for the Cx37CT. Also in the 1H-15N HSQC, N-terminal residues S254-Y259 displayed a second weaker peak and residues E261-Y266 had significant line broadening. These residues are flanked by prolines (P250, P258, P260, and P268), suggesting proline cis-trans isomerization. Consistent with this observation, the online program CISPEPpred (sunflower.kuicr.kyoto-u.ac.jp/~sjn/cispep) predicted this Cx37CT region can undergo cis-trans proline isomerization. This observation is similar, both in the location on the CT and the number of prolines involved, to what was observed for the Cx45CT domain (Kopanic and Sorgen, 2013). A potential novel role for cis-trans proline isomerization in the regulation of gap junction intercellular communication may be to modulate protein-protein interactions (69). Assignment of the Cx37CT domain will be useful for characterizing the effects of phosphorylation and protein-protein interactions on the Cx37CT structure; this information can provide novel mechanistic insight into the regulation of Cx37 gap junction intercellular communication.
Figure 1.
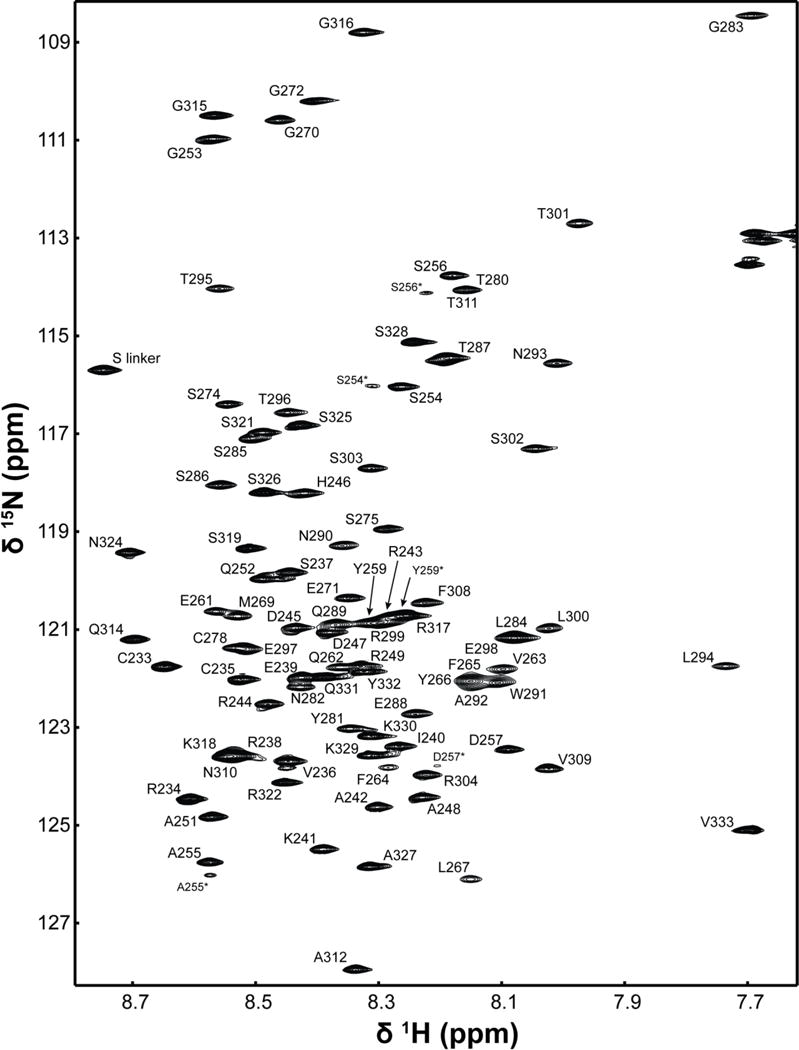
1H-15N HSQC spectrum of the Cx37 carboxyl terminal domain in 1× PBS (pH 5.8). Assignments for the backbone amide groups are labeled with numbering corresponding to the full-length mouse Cx37 protein. Asterisks (*) indicate residues potentially affected by cis-trans proline isomerization.
Acknowledgments
This work is funded by the United States Public Health Service Grants, GM072631, HL131712, CA036727, and GM103427. We would like to thank and acknowledge Ed Ezell, manager of the Nuclear Magnetic Resonance Laboratory at the University of Nebraska Medical Center, for his assistance with collection of the NMR data.
Footnotes
This work was supported by United States Public Health Service Grants GM072631, HL131712, CA036727, and GM103427.
References
- Anumonwo JM, Taffet SM, Gu H, Chanson M, Moreno AP, Delmar M. The carboxyl terminal domain regulates the unitary conductance and voltage dependence of connexin40 gap junction channels. Circ Res. 2001;88:666–673. doi: 10.1161/hh0701.088833. [DOI] [PubMed] [Google Scholar]
- Bouvier D, Kieken F, Kellezi A, Sorgen PL. Structural changes in the carboxyl terminus of the gap junction protein connexin 40 caused by the interaction with c-Src and zonula occludens-1. Cell Commun Adhes. 2008;15:107–118. doi: 10.1080/15419060802014347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier D, Kieken F, Sorgen PL. (1)H, (13)C, and (15)N backbone resonance assignments of the carboxyl terminal domain of Connexin40. Biomol NMR Assign. 2007;1:155–157. doi: 10.1007/s12104-007-9044-x. [DOI] [PubMed] [Google Scholar]
- Brisset AC, Isakson BE, Kwak BR. Connexins in vascular physiology and pathology. Antioxid Redox Signal. 2009;11:267–282. doi: 10.1089/ars.2008.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- Gilleron J, Carette D, Chevallier D, Segretain D, Pointis G. Molecular connexin partner remodeling orchestrates connexin traffic: from physiology to pathophysiology. Crit Rev Biochem Mol Biol. 2012;47:407–423. doi: 10.3109/10409238.2012.683482. [DOI] [PubMed] [Google Scholar]
- Haefliger JA, Nicod P, Meda P. Contribution of connexins to the function of the vascular wall. Cardiovasc Res. 2004;62:345–356. doi: 10.1016/j.cardiores.2003.11.015. [DOI] [PubMed] [Google Scholar]
- Harris AL. Emerging issues of connexin channels: biophysics fills the gap. Q Rev Biophys. 2001;34:325–472. doi: 10.1017/s0033583501003705. [DOI] [PubMed] [Google Scholar]
- Herve J, Bourmeyster N, Sarrouilhe D, Duffy H. Gap junctional complexes: From partners to functions. Progress in Biophysics and Molecular Biology. 2007;94:29–65. doi: 10.1016/j.pbiomolbio.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- Kellezi A, Grosely R, Kieken F, Borgstahl GE, Sorgen PL. Purification and reconstitution of the connexin43 carboxyl terminus attached to the 4th transmembrane domain in detergent micelles. Protein Expr Purif. 2008;59:215–222. doi: 10.1016/j.pep.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieken F, Spagnol G, Su V, Lau AF, Sorgen PL. NMR structure note: UBA domain of CIP75. J Biomol NMR. 2010;46:245–250. doi: 10.1007/s10858-010-9397-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopanic JL, Sorgen PL. Chemical shift assignments of the connexin45 carboxyl terminal domain: monomer and dimer conformations. Biomol NMR Assign. 2013;7:293–297. doi: 10.1007/s12104-012-9431-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird DW. The gap junction proteome and its relationship to disease. Trends Cell Biol. 2010;20:92–101. doi: 10.1016/j.tcb.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Lampe PD, Lau AF. The effects of connexin phosphorylation on gap junctional communication. Int J Biochem Cell Biol. 2004;36:1171–1186. doi: 10.1016/S1357-2725(03)00264-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMaster DM, Richards FM. NMR sequential assignment of Escherichia coli thioredoxin utilizing random fractional deuteriation. Biochemistry. 1988;27:142–150. doi: 10.1021/bi00401a022. [DOI] [PubMed] [Google Scholar]
- Morel S. Multiple roles of connexins in atherosclerosis- and restenosis-induced vascular remodelling. J Vasc Res. 2014;51:149–161. doi: 10.1159/000362122. [DOI] [PubMed] [Google Scholar]
- Moreno AP, Chanson M, Elenes S, Anumonwo J, Scerri I, Gu H, Taffet SM, Delmar M. Role of the carboxyl terminal of connexin43 in transjunctional fast voltage gating. Circ Res. 2002;90:450–457. doi: 10.1161/hh0402.105667. [DOI] [PubMed] [Google Scholar]
- Morley GE, Taffet SM, Delmar M. Intramolecular interactions mediate pH regulation of connexin43 channels. Biophys J. 1996;70:1294–1302. doi: 10.1016/S0006-3495(96)79686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson TK, Sorgen PL, Burt JM. Carboxy terminus and pore-forming domain properties specific to Cx37 are necessary for Cx37-mediated suppression of insulinoma cell proliferation. Am J Physiol Cell Physiol. 2013;305:C1246–1256. doi: 10.1152/ajpcell.00159.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revilla A, Castro C, Barrio LC. Molecular dissection of transjunctional voltage dependence in the connexin-32 and connexin-43 junctions. Biophys J. 1999;77:1374–1383. doi: 10.1016/S0006-3495(99)76986-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorgen PL, Duffy HS, Sahoo P, Coombs W, Delmar M, Spray DC. Structural changes in the carboxyl terminus of the gap junction protein connexin43 indicates signaling between binding domains for c-Src and zonula occludens-1. J Biol Chem. 2004a;279:54695–54701. doi: 10.1074/jbc.M409552200. [DOI] [PubMed] [Google Scholar]
- Sorgen PL, Duffy HS, Spray DC, Delmar M. pH-dependent dimerization of the carboxyl terminal domain of Cx43. Biophys J. 2004b;87:574–581. doi: 10.1529/biophysj.103.039230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauch K, Kieken F, Sorgen P. Characterization of the structure and intermolecular interactions between the connexin 32 carboxyl-terminal domain and the protein partners synapse-associated protein 97 and calmodulin. J Biol Chem. 2012;287:27771–27788. doi: 10.1074/jbc.M112.382572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenin AF, Kowal TJ, Fong JT, Kells RM, Fisher CG, Falk MM. Proteins and mechanisms regulating gap-junction assembly, internalization, and degradation. Physiology (Bethesda) 2013;28:93–116. doi: 10.1152/physiol.00038.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourtsis DJ, Chasapis CT, Pairas G, Bentrop D, Spyroulias GA. NMR conformational properties of an Anthrax Lethal Factor domain studied by multiple amino acid-selective labeling. Biochem Biophys Res Commun. 2014;450:335–340. doi: 10.1016/j.bbrc.2014.05.123. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]


