Dear Editor
Acetylcholinesterase (AChE) hydrolyzes the neurotransmitter acetylcholine at cholinergic synapses in the central nervous system (Toutant, 1989). Inhibition of the enzyme in insects could lead to the death of insects rapidly; thus AChE has been a molecular target for developing insecticides.
Organophosphates and carbamates represent two major classes of AChE inhibitors used as insecticides in agriculture and public health. Mosquitoes carry two AChE genes, ace-1 and ace-2, encoding AChE-1 and AChE-2 proteins, respectively (Weill et al., 2002; Radic & Taylor, 2006; Bourguet et al., 1996). In Anopheles gambiae and most other mosquitoes it appears that AChE-1 is primarily responsible for the nervous system cholinesterase activity (Weill et al., 2002). Accordingly, public health concerns have directed most interest toward the AChE-1 protein.
Mosquitoes transmit a number of diseases in animals and humans, including dengue, zika virus infection and malaria that affect millions of people each year (Enserink, 2008; Wilder-Smith et al., 2016). Controlling the disease-transmitting mosquito, An. gambiae, has proven to be a successful strategy to reduce malaria transmission (Killeen et al., 2007, Enserink, 2008). However, the emergence of resistant strains against AChE-targeting pesticides poses a significant challenge to mosquito control-based public health strategies (Ramphul et al., 2009; Djogbenou et al., 2009). One challenge in developing new AChE-targeting pesticides is minimizing mammalian toxicity. A crystal structure of AgAChE could be very useful to develop safe anticholinesterase insecticides that feature high selectivity for inhibition of AgAChE over human AChE (hAChE). Here, we reported a crystal structure of AgAChE of the malaria mosquito.
The AgAChE catalytic domain (CD) was expressed using Pichia cells. Purified recombinant AgAChE CD was concentrated to 5 mg/mL protein. The crystals were grown by a hanging-drop vapor diffusion method. The parameters of the crystals and data collections were listed in Table S1 (supplemental information). The structure was determined by the molecular replacement method using a mouse AChE structure (PDB code, 2ha2), and refined to 3.40 Å resolution. The detailed method for protein expression, purification, crystallization and data processing were described in the supplemental information. The final model contains two protein molecules in an asymmetric unit and yields a crystallographic R value of 17.6% and an Rfree value of 21.7%. No residues are in disallowed regions of the Ramachandran plot as defined with MolProbity (Chen et al., 2010) (Table S1 in the supplemental information). The biological molecules are two alternative dimers based on analysis using protein interfaces, surfaces and assemblies service, PISA (Krissinel & Henrick, 2007). Here we named dimers 1 and 2. Dimer 1 (Fig. 1A) is a new form of AChE dimers that has not been previously reported in any AChE structures and dimer 2 (Fig. 1B) is same as the previously reported AChE dimer form, called 4-helix bundle (FHB) (Dvir et al., 2010) as seen in structures of mouse AChE, Torpedo californica AChE (TcAChE) and human hAChE.
Figure 1.
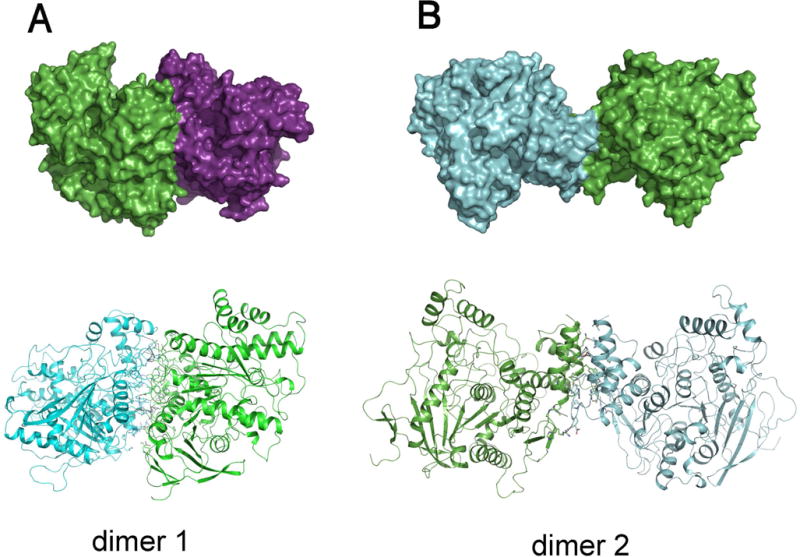
Dimer 1 (A) is the newly reported dimer herein, and dimer 2 (B) is the FHB dimer seen also in other AChE structures. Surface presentations of possible biological molecules are shown in upper panels and cartoon presentations of the structure are illustrated in the bottom panels.
In dimer 1, molecule interactions involve more than 10 salt bridges formed by R207, R210, R338, R341, D171, D342, D377, D461, and E462 (Fig. 2A), more than 10 other hydrogen bonds and many hydrophobic interactions that involve L204, F331, L335, A373, L374, L378, V455 and A458 . The total surface area buried at dimer 1 interfaces, calculated using PISA (2692.6 Å2), is bigger than those at dimer 2 interfaces (1940.9 Å2). To investigate why dimer 1 is only found in the AgAChE, we superposed AgAChE CD structure and four other AChE structures and the results demonstrated that other enzymes lack more than two third of residues required to form salt bridges (Fig. 2B), which is likely the main reason why AgAChEs from other species do not form dimer 1. The finding of a new dimer form is interesting; however, we cannot make any conclusions if this new dimeric form is biologically significant because 120 residues of the N-terminal part are missing in the crystallized enzyme and this N-terminal part could interact with the interface involved in dimer 1 formation.
Figure 2.
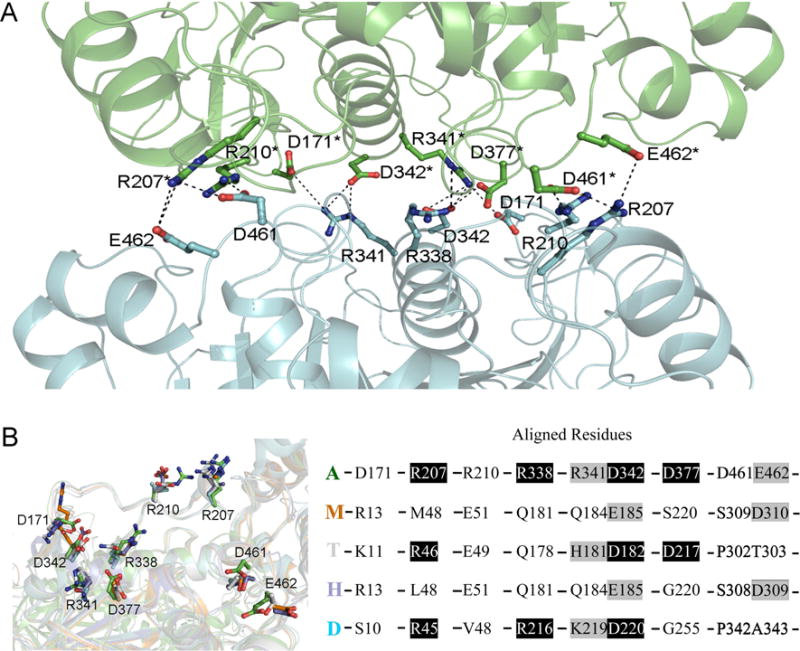
(A) Salt bridges in the intermolecular interface of dimer 1 in structure are shown with dashed lines, the involved residues are shown in sticks, and other part of the dimer is shown in cartoon presentation. (B) Superposition of AgAChE chain A (A, green), hAChE (H, 4ey4, blue), mouse AChE (M, 2ha2, brown), TcAChE (T, 2wg2, grey), and Drosophila AChE (D, 1qo9, cyan). The left panel shows the residues (in sticks) of the four other structures structurally aligned with the residues involved in salt bridge formation in AgAChE CD dimer 1; the right panel shows the residues in AgAChE CD involved in salt bridge formation and the aligned residues from other AChEs (black background: same residues, grey background: similar residues).
Each monomer of AgAChE has three intramolecular disulfide bonds, between C228 and C255, C414 and C427, C562 and C683, which are conserved across AChE structures. We used the program Dali (Holm et al., 2006) to compare the CD of AgAChE with those of other species in the PDB. rmsd values ranged from 1.2 Å (TcAChE 2wg2) to 1.5 Å (mouse AChE, 2ha2) to 1.6 Å (hAChE, 4ey4) to 1.7 Å (AgAChE homology model, 2azg) to 1.9 Å (Drosophila AChE, 1qo9). The common fold cores (the typical α/β hydrolase fold) of AChE structures from different species are very well superposed. However some significant conformational changes in other regions are seen among AChE structures from different species (Fig. 3A). The conformations of active site residues are significantly different from the previously deposited homology model (Pang, 2006) (Fig. 3B). However, the active site residue conformations of AgAChE are similar to those in TcAChE except a few residues, including C447, Y489 and D602, showing noticeable differences from their corresponding sites with Y288, F330 and Y442, respectively in TcAChE (Fig. 3C).
Figure 3.
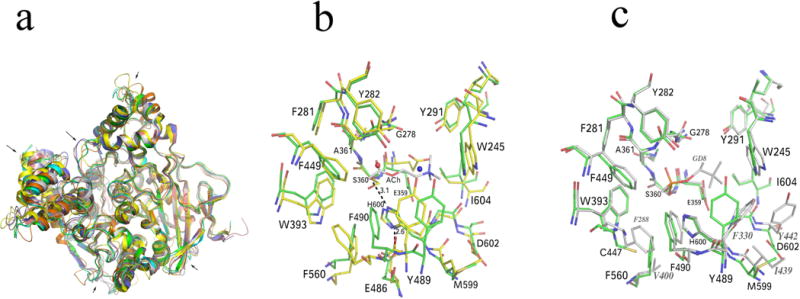
(A) Superposition of a previously deposited AgAChE homology model (pdb code: 2azg, yellow), hAChE (4ey4, blue), mouse AChE (2ha2, brown), TcAChE (2wg2, grey), Drosophila AChE (1qo9, orange) and AgAChE CD chain A (green). The main chain conformational differences are pointed by arrows. (B) Comparison of the previously deposited AgAChE homology model and AgAChE CD structure (chain A). The blue and red spheres represent an ammonium ion and a water molecule, respectively. (C) Comparison of TcAChE and AgAChE CD structure (chain A). The side chain conformational differences in the region close to the active center (6 Å around the putative ACh binding site) are shown. Residues showing a significant conformational difference with AgAChE equivalent residues in TcAChE structure are labeled with grey italic fonts.
AgAChE is a glycoprotein. After diligently considering the preferred glycans of yeast glycosylation system and observed Fo-Fc and 2Fo-Fc electro density maps, we modeled 2 N-acetyl glucosamine molecules that link residue N220 of the structure. The glygosylation site is different from that reported previously, but the linkage is similar (Engdahl et al., 2015).
Supplementary Material
Table S1 Data collection and refinement statistics of AChE structures.
Figure S1 Main chain RMSD values between AgAChE and Drosophila AChE (panel A, pdb code: 1qo9), and between AgAChE and hAChE (panel B, pdb code: 4ey4). The RMSD plots were generated using “Superpose molecules” function of the CCP4 program.
Figure S2 The electron density map (2Fo-Fc) was shown around the active center. Key residues were labeled in the map. The map was contoured at 2.0 Sigma and generated using Coot program (The University of York).
Acknowledgments
This work was carried out in part at the National Synchrotron Light Source, Brookhaven National Laboratory. We thank the NIH (AI082581) and the National Natural Science Foundation of China (31472186) for financial support.
Footnotes
The atomic coordinate and structure factor (PDB codes: 5X61) has been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org).
References
- Bourguet D, Raymond M, Fournier D, Malcolm CA, Toutant JP, Arpagaus M. Existence of two acetylcholinesterases in the mosquito Culex pipiens (Diptera:Culicidae) Journal of Neurochemistry. 1996;67:2115–2123. doi: 10.1046/j.1471-4159.1996.67052115.x. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D: Biological Crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djogbenou L, Labbe P, Chandre F, Pasteur N, Weill M. Ace-1 duplication in Anopheles gambiae: a challenge for malaria control. Malaria Journal. 2009;8:70. doi: 10.1186/1475-2875-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvir H, Silman I, Harel M, Rosenberry TL, Sussman JL. Acetylcholinesterase: from 3D structure to function. Chemico-Biological Interactions. 2010;187:10–22. doi: 10.1016/j.cbi.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engdahl C, Knutsson S, Fredriksson SA, Linusson A, Bucht G, Ekstrom F. Acetylcholinesterases from the disease vectors Aedes aegypti and Anopheles gambiae: functional characterization and comparisons with vertebrate Orthologues. PLoS ONE. 2015;10:e0138598. doi: 10.1371/journal.pone.0138598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enserink M. Epidemiology. Lower malaria numbers reflect better estimates and a glimmer of hope. Science. 2008;321:1620. doi: 10.1126/science.321.5896.1620b. [DOI] [PubMed] [Google Scholar]
- Holm L, Kaariainen S, Wilton C, Plewczynski D. Using Dali for structural comparison of proteins. Current Protocols in Bioinformatics. 2006 doi: 10.1002/0471250953.bi0505s14. Chapter 5, Unit 55. [DOI] [PubMed] [Google Scholar]
- Killeen GF, Smith TA, Ferguson HM, Mshinda H, Abdulla S, Lengeler C, Kachur SP. Preventing childhood malaria in Africa by protecting adults from mosquitoes with insecticide-treated nets. PLoS Medicine. 2007;4:e229. doi: 10.1371/journal.pmed.0040229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. Journal of Molecular Biology. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Pang YP. Novel acetylcholinesterase target site for malaria mosquito control. PLoS ONE. 2006;1:e58. doi: 10.1371/journal.pone.0000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radic Z, Taylor P. Structure and Function of Cholinesterases. In: Guptas RC, editor. Toxicology of Organophosphate and Carbamate Compounds. Elsevier; Amsterdam: 2006. pp. 161–186. [Google Scholar]
- Ramphul U, Boase T, Bass C, Okedi LM, Donnelly MJ, Muller P. Insecticide resistance and its association with target-site mutations in natural populations of Anopheles gambiae from eastern Uganda. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2009;103:1121–1126. doi: 10.1016/j.trstmh.2009.02.014. [DOI] [PubMed] [Google Scholar]
- Toutant JP. Insect acetylcholinesterase: catalytic properties, tissue distribution and molecular forms. Progress In Neurobiology. 1989;32:423–446. doi: 10.1016/0301-0082(89)90031-2. [DOI] [PubMed] [Google Scholar]
- Weill M, Fort P, Berthomieu A, Dubois MP, Pasteur N, Raymond M. A novel acetylcholinesterase gene in mosquitoes codes for the insecticide target and is non-homologous to the ace gene in Drosophila. Proceedings Biological Sciences. 2002;269:2007–2016. doi: 10.1098/rspb.2002.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder-Smith A, Gubler DJ, Weaver SC, Monath TP, Heymann DL, Scott TW. Epidemic arboviral diseases: priorities for research and public health. The Lancet Infectious Diseases. 2016 doi: 10.1016/S1473-3099(16)30518-7. Epub 2016 Dec 20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Data collection and refinement statistics of AChE structures.
Figure S1 Main chain RMSD values between AgAChE and Drosophila AChE (panel A, pdb code: 1qo9), and between AgAChE and hAChE (panel B, pdb code: 4ey4). The RMSD plots were generated using “Superpose molecules” function of the CCP4 program.
Figure S2 The electron density map (2Fo-Fc) was shown around the active center. Key residues were labeled in the map. The map was contoured at 2.0 Sigma and generated using Coot program (The University of York).


