Abstract
Tissue repair is fundamental to our survival as tissues are challenged by recurrent damage. During mammalian skin repair, cells respond by migrating and proliferating to close the wound. However, the coordination of cellular repair behaviors and their effects on homeostatic functions in a live mammal remains unclear. Here we capture the spatiotemporal dynamics of individual epithelial behaviors by imaging wound re-epithelialization in live mice. Differentiated cells migrate while the rate of differentiation changes depending on local rate of migration and tissue architecture. Cells depart from a highly proliferative zone by directionally dividing towards the wound while collectively migrating. This regional co-existence of proliferation and migration leads to local expansion and elongation of the repairing epithelium. Finally, proliferation functions to pattern and restrict the recruitment of undamaged cells. This study elucidates the interplay of cellular repair behaviors and consequent changes in homeostatic behaviors that support tissue-scale organization of wound re-epithelialization.
Keywords: Skin, Tissue repair, Intravital microscopy, Epithelial behaviors, Regeneration
Introduction
In the skin, improper wound repair can be lethal, leaving an organism without its normal water-tight barrier and vulnerable to environmental insults. While skin repair begins with an inflammatory response and clot formation, re-epithelialization is essential to seal the wound permanently over the long term1–3. Central to skin re-epithelialization is the mammalian adult epidermis, which is organized in layers composed of progressively differentiated suprabasal cells (known as the spinous, granular and cornified layers4 – see Supplementary Fig. 1a–e) that are continuously shed from the top surface and homeostatically replenished by underlying proliferative basal cells4,5. Understanding how tissue homeostatic functions can be maintained while orchestrating a wound repair response is a key question in mammalian epithelial and stem cell biology6,7.
Previous work has shown that during the repair process epidermal basal cells retain homeostatic behaviors such as proliferation and differentiation but acquire new behaviors such as migration2,8–11. Additionally, the epithelial tissue around the wound becomes patterned to form a migratory front in close proximity to the wound, surrounded distally by a concentric ring of proliferating cells12–19. While these studies elucidate critical cellular behaviors utilized during mammalian re-epithelialization, we still lack an understanding of first, how cellular repair behaviors are dynamically orchestrated at the single cell level as well as the tissue level in a live mammal20 and second, how the epithelium integrates the homeostatic program that maintains the skin barrier with the demands of rapidly creating new tissue to cover the wound. A major difficulty in addressing this question is the inability in a live mammal to resolve individual cell dynamics that will result in tissue-scale changes underlying wound repair. To tackle this problem, we have established an intravital imaging approach to study wound repair in real time and show, by combining lineage tracing tools with genetic and drug perturbations, that 1) differentiated cell migration as well as oriented cell divisions contribute to wound re-epithelialization, 2) homeostatic stratification rate is influenced by migration rates and distance from the wound, 3) interplay between areas of migration and proliferation leads to distinctive regional transformations of the epithelium close to the wound and 4) proliferation patterns and spatially restricts the repairing epithelium.
Migration of differentiated cells contributes to repair
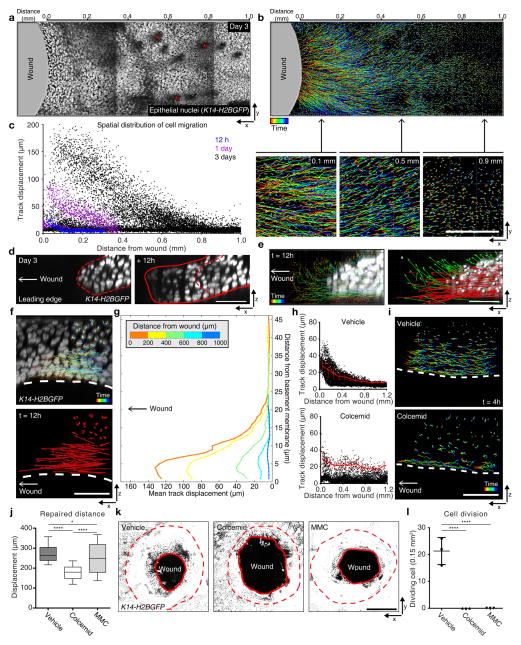
To investigate mechanisms of epithelial repair, we established a platform to study by time-lapse microscopy the repair of 1 mm wide wounds by tracking fluorescently marked epithelial cell nuclei (fusion protein of histone H2B with green fluorescent protein (GFP) driven by the keratin 14 promoter (K14-H2BGFP21; Supplementary Fig. 1f, g). We began by interrogating migration as a distinctive epithelial behavior that emerges during wound re-epithelialization. Automated quantitative tracking and analysis of cell trajectories during wound repair show that migration begins slowly around 12 hours post wound induction (PWI). Further analysis of migration tracks shows that the total displacement of nuclei per unit time, which we will refer to as the migration rate, follows a gradient distribution at 1 day PWI. At this point in time the migration rate of epithelial cells is greatest closer to the wound and progressively diminishes with distance from the wound. This gradient distribution, expanded to cover a larger area of the epithelium, is maintained through peak activity 3 days PWI (Fig. 1a–c and Supplementary Video 1). Track analyses captured a large range of migration rates (Fig. 1c). We reasoned that this spread may reflect heterogeneity in the migration rates of cells within the basal layer as suprabasal layers are thought to not migrate during re-epithelialization12,13. Instead, analyses of individual epidermal layers showed that both suprabasal and basal cell layers migrate towards the wound. Specifically, closer to the wound, we captured suprabasal cells and basal cells rapidly advancing towards the wound at the leading edge (~80 μm total displacement within 12 hours of imaging). Additionally, these cells at the leading edge display an upward movement not previously described (Fig. 1d, e green arrows; Supplementary Video 2). Similarly, analysis of the tissue behind the leading edge showed that both basal and suprabasal cells migrate during re-epithelialization with displacement highest in the basal layer and gradually decreasing in higher, more differentiated layers (Fig. 1f, g and Supplementary Video 3). Interrogation of suprabasal membrane dynamics showed lamellipodial movements similar to those occurring in basal cells, suggesting that suprabasal cells are actively migrating (Supplementary Video 4). Together our analyses show that the repair process is associated with a spatially organized migration throughout undifferentiated and differentiated cell layers in a gradient fashion along two distinct directions: along the plane of the epithelium (x-y plane) within an individual cell layer and along the apical-basal axis (x-z plane) across different cell layers.
Figure 1. Migration of differentiated layers of skin epithelium contributes to re-epithelialization.
(a) Time-lapse image of skin epithelial nuclei 3 days PWI using K14-H2BGFP mice (red asterisks, hair canals). (b) Imaris track analysis of epithelial nuclei 3 days PWI. Colors project time (blue, beginning; red, end). Scale bar, 1 mm. (c) Total displacement of individual cell tracks over 12 hours plotted as a function of distance from the wound; Imaging performed at 12 hours (blue), 1 day (purple) and 3 days (black) PWI (n=3 mice respectively). (d) Still images of the leading edge 3 day PWI and 12 hours later using K14-H2BGFP. Scale bar, 50 μm. (e) Cell tracks and displacement vectors of the leading edge (parallel (<5°), red arrow; upward (≥5°), green arrow). Scale bar, 50 μm. (f) x-z view of cell tracks located behind the leading edge. Scale bar, 50 μm. (g) Mean displacement of cell tracks that initiated within 200 μm segments of tissue plotted as a function of both distance from the wound and the basement membrane (n= 2 mice, t=12 hours). (h) Representative quantification of the migration tracks 3 days PWI. Every 100 μm from the initial wound, the displacement of migrating cell tracks are averaged (red). Mean ± S.D. (Vehicle, n=14232 tracks from 2 mice; Colcemid, n=5523 tracks from 2 mice). (i) x-z view of cell tracks after vehicle and Colcemid treatments. Scale bar, 50 μm. (j) Box plot of epidermal wound closure by determining the leading edge distance from the initial wound site. Mean ± S.D. Horizontal lines are the median values, the edges of the boxes are from 25 to 75 percentiles, the whiskers extend from Min to Max. * p<0.05 and **** p<0.0001, unpaired Student’s t-test (n=180 points pooled from n=3 mice). (k) Whole wound image 3 days PWI from Vehicle, Colcemid and MMC-treated mice. z-stacks of epidermis is projected (~50 μm from granular layer). Dotted line, initial wound boundary; solid line, edge of the repaired epithelium. Scale bar, 500 μm. (l) Cell division rate 3 days PWI from Vehicle, Colcemid and MMC-treated mice. Dividing cells, 1 mm from the wound, are quantified per 0.15mm2. Mean ± S.D. **** p<0.0001, unpaired Student’s t-test (t=4 hours; n=3 mice). Images shown in a, b, d, e, f, i and k are representative of 3 mice.
In order to interrogate the impact of basal and suprabasal cell migration on wound re-epithelialization, we sought to inhibit their migration by perturbing cytoskeletal elements. Inhibition of microtubule polymerization (via drugs such as Colcemid) has been shown to inhibit migration as well as cell divisions in vitro22,23. Drug treatments may target dermis in addition to epidermis. Thus, while possible observed defects may be the result of both direct and indirect effects of the drug, they still provide useful insights on the biology supporting epithelial repair in vivo. Unexpectedly, Colcemid-treated skin showed that epithelial migration is largely abolished in the suprabasal layers while basal cells continued to migrate in vivo (Fig. 1h, i). Strikingly, Colcemid-treated animals were also dramatically affected in their ability to close the wound when compared to vehicle-treated animals (Fig. 1j, k). To understand to what extent impaired cell divisions contribute to the wound re-epithelialization impairment observed in the Colcemid-treated mice, we evaluated the consequences of specifically inhibiting cell divisions (using Mitomycin C or MMC). MMC-treated mice re-epithelialize more efficiently than Colcemid-treated mice and more similarly to vehicle-treated controls supporting that the lack of wound re-epithelization in the Colcemid-treated mice is a result of the identified suprabasal migratory defects (Fig. 1j–l). All together these results highlight a migratory behavior of epidermal differentiated layers and support a model for suprabasal cell migration to functionally contribute to wound closure during mammalian re-epithelialization.
Migration impacts differentiation rates
While migration allows cells to build new epithelial tissue towards the wound, ongoing stratification depletes this pool of cells to generate the terminally differentiated layers needed to maintain homeostatic barrier function24. Additionally, wound-induced stratification produces a thicker epidermis with more cell layers when compared to homeostatic stratification2,15 (Supplementary Fig. 1a–e). While epithelial differentiation has been shown to still occur during wound-induced stratification25,26, it remains unknown how cells balance migration and terminal differentiation to produce a properly stratified epidermis during repair.
A challenge in addressing this question is that migration rates change as a function of wound distance as well as the length of time after wounding. Therefore, this requires a method to not only label the wound epidermis with single-cell precision but also track cells over hours and even days. To this end, we developed a transgenic mouse that expresses a photo-activatable fluorescent reporter (K14-H2BPAmCherry) that provides spatiotemporal control of labeling, to track migration and differentiation of the same population of cells over time27,28 (Supplementary Fig. 2a–d).
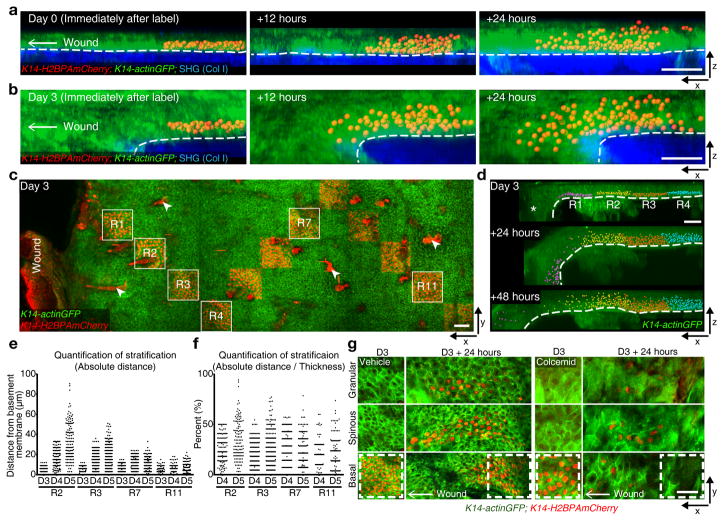
To first determine the rate of differentiation during re-epithelialization, we began by tracking geometrically defined groups of basal cells. We have previously demonstrated that basal cells labelled and tracked a day later uniformly differentiate vertically, while largely retaining the same two dimensional basal area28 (Supplementary Fig. 2d). Thus, we reasoned that if differentiation rates remain the same throughout the repairing epithelium, the acquired migratory behavior of epithelial cells would influence the resulting shape of the population leading to a shifted organization (trapezoidal) with the heaviest upward contribution at the back of the population furthest from the wound. Labelling basal cells immediately after wounding and revisiting 1 day PWI showed a slimmer migratory front of mostly basal cells with a thicker tail distribution of both basal and differentiated cells consistent with migration rate being faster than differentiation rates (Fig. 2a and Supplementary Fig. 2e). In contrast, labelling and tracking of cells 3 days PWI shows an inverted distribution with a thicker front of cells extending from basal to differentiated layers (Fig. 2b and Supplementary Figs. 2f–h). Notably, cell displacement between layers was observed to occur exclusively in an upward direction at both timepoints examined, despite previous models proposing downward cell contribution from differentiated layers to the basal one (Supplementary Fig. 3). This data indicates that while during the first day PWI migration seems to predominate over upward differentiation, by 3 days PWI cell differentiation and migration are both occurring, allowing a rapid progression of tissue thickening towards the wound.
Figure 2. Migration enhance differentiation.
(a, b) x-z view of labeled cells rendered with Imaris software as an x-z projection and their sequential changes in position from 0 day (a) and 3 days (b) PWI (representative images from 3 mice). Labeled cells (K14-H2BPAmCherry), red; epidermis (K14-actinGFP), green; dermis (SHG), blue. Scale bar, 50 μm. (c) Staggered frame tile labeling of H2BPAmCherry cells in the basal layer at varying distance from the wound using K14-H2BPAmCherry;K14-actinGFP (representative images from 3 mice). Arrowheads indicated strong red auto-fluorescence from the hair shafts. Scale bar, 50 μm. (d) Imaris visualization of labeled cells and their sequential changes in regions 1–4 (R1, purple; R2, yellow; R3, red; R4, blue; representative images from 3 mice). Asterisk indicates a fold in the epidermis. Scale bar, 50 μm. (e) Representative quantification of absolute upward cell stratification at different distances from the wound. y-axis represents the absolute value of distances of labeled cells from the basement membrane (representative graph derived from n=1823 labeled cells from n=2 mice). (f) Representative quantification of relative upward cell stratification, normalized to epidermal thickness, at different distances from the wound. y-axis represents distance of labeled cells from the basement membrane represented as a percentage of epidermal thickness (representative graph derived from n=1327 labeled cells from n=2 mice). (g) Basal cell labeling at 3 days PWI and 24 hours later in Vehicle and Colcemid-treated wounds using K14-H2BPAmCherry;K14-actinGFP (representative images from 3 mice). Scale bar, 50 μm.
Interestingly, quantifications of differentiation rates of groups of cells labeled in a spatially staggered pattern throughout the tissue showed that cells move upwards through the differentiated layers at a higher rate in regions closer to the wound, where cell migration is at its highest, compared to more distant regions (Fig. 2c–e). However, these rates appear to remain proportional to the local epidermal thickness over time (Fig. 2f). Therefore, these results suggest that cell migration and differentiation rates adjust as a function of both wound time and proximity in order to meet local tissue demands.
Given that rates of migration and differentiation seem to change together during re-epithelialization, we asked whether individual cells could contribute to repair by both migrating and differentiating. To address this question, we combined the K14-H2BPAmCherry mouse model with Colcemid treatment where basal cells can still migrate but cannot divide. This allowed us to specifically track migrating basal cells and ask whether they also differentiate. Tracking basal cells in Colcemid-treated animals showed that basal cells advance towards the wound by migrating as they progressively differentiate (Fig. 2g). Since the vast majority of stratifying cells also appear to migrate toward the wound, we next asked if wound-induced stratification is dependent on cell migration. To address this question, we sought to utilize a model where migration would be impaired in both basal and suprabasal layers. Conditional epithelial loss of function of the Rho GTPase Rac1 has been previously shown to have re-epithelialization defects as well as migratory defects in cultured keratinocytes29. Time-lapse imaging of epithelial conditional Rac1−/− mice (K14-CreER;Rac1−/−;K14-H2BGFP) shows that migration of skin epidermal cells is in fact impaired when compared to Rac1+/− littermates during re-epithelialization in vivo (Supplementary Fig. 4a). However strikingly, while we observe that cells from the basal layers can progress upwards through differentiated layers, we find that wound-induced stratification is inhibited in the Rac1−/− model when compared to control littermates (Supplementary Fig. 4b–d). All together these results suggest that migration rates impact epidermal differentiation to promote wound-induced stratification and regulate cell movement to build the progressively thicker epidermis closer to the wound.
Migration regulates proliferation
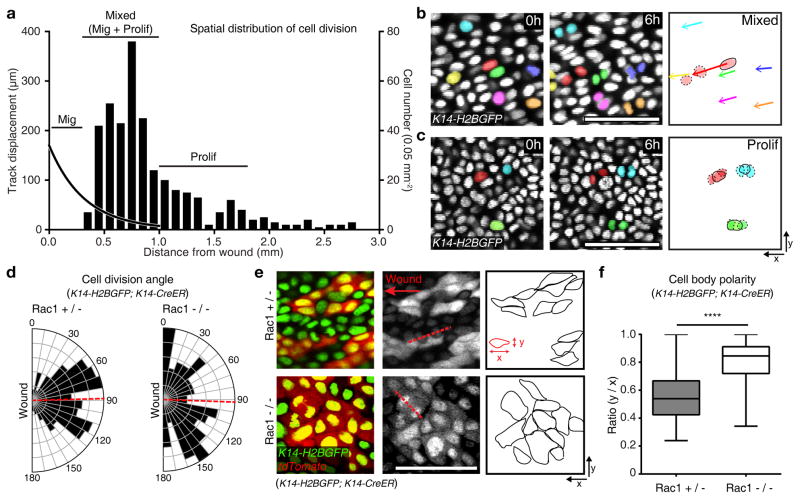
As epithelial cells migrate and stratify towards the wound, proliferation replenishes the tissue with new cells. Specifically, previous studies have identified a highly proliferative zone right behind the migratory front during re-epithelialization12–15,30. However, it is still unclear how cells transition from the proliferating zone to the migratory front. To begin to address this question, we systemically quantified cell divisions occurring at different distances from the wound during time-lapse imaging. Analysis of a time-course after wounding showed that similar to what we observed for migration, cell division events had become more frequent by 3 days PWI. Consistent with previous findings12–15,30, these divisions were organized into a clear zone of proliferation that was set back from the front of the epithelium. Unexpectedly however, the proliferation zone was wider than expected and appeared to overlap with the migration zone (Fig. 3a). Thus the repairing epithelium is organized in three territories: a migration only front (migration zone), an intermediate mixed region where both proliferation and migration occur (mixed zone), and a proliferation region further away (proliferation zone) (Fig. 3a). We therefore hypothesized that upon cell divisions, migration could be used in the mixed region to begin to displace one daughter cell towards the wound with the other remaining behind. Surprisingly, individual cells in this zone were observed performing both proliferation and migration behaviors. (Fig. 3b, c and Supplementary Videos 5, 6).
Figure 3. Migration controls cell division.
(a) Representative quantification of cell divisions relative to the distance from the wound boundary using K14-H2BGFP (representative graph derived from n=409 cells from n=3 mice). The left hand axis and curved black trend-line represents the cell track displacement trend observed over 12 hours of imaging. The right hand y axis and column plot indicates the cell divisions based on nuclei counts per 0.05 mm2. The plot is annotated with the identified regions of migration, mixed (migration and proliferation) or proliferation zones. (b, c) Representative cell divisions and migration in the mixed (b) and the proliferation zone (c) (n=3 mice). Scale bar, 50 μm. (d) Quantification of cell division axes in the Rac1+/− and Rac1−/− (Rac1+/− n=197 cells pooled from 3 mice and Rac1−/− n=99 cells pooled from 3 mice). Cell division axes were measured with respect to the wound edge. A red dashed line indicates average of angles (88.48° in Rac1+/− and 92.05° in Rac1−/−). (e) Cell shape changes in Rac1+/− and Rac1−/− mice per 0.7 mm2 over a 4 hours period. Recombined cells are indicated by the presence of tdTomato expression and outlined in the third panel (representative images from n=3 mice). (f) Box plot quantifying cell body polarity in the Rac1+/− and Rac1−/−. The polarity is measured by ratio of y/x values of 150 randomly picked tdTomato positive cells in Rac1+/− and Rac1−/− epidermis respectively 3 days PWI. Horizontal lines are the median values, the edges of the boxes are from 25 to 75 percentiles, the whiskers extend from Min to Max. Mean ± S.D. **** p<0.0001, unpaired Student’s t-test (n=150 cells pooled from n=3 mice). Scale bar, 50 μm.
This observed tight spatial and temporal connection between migration and proliferation behaviors made us wonder whether there is an interplay between migration and proliferation. We began by asking whether orientation of cell divisions is aligned with the direction of cell migration. To address this question, we first systemically quantified the rate and orientation of cell divisions and found that they largely occur within the basal layer (with parallel representing 89% and oblique 7.09 %), with a small percentage occurring in the suprabasal layer (3.91 %; Supplementary Fig. 5), consistent with previous analysis on tissue sections31,32. Interestingly, quantification of cell division angles in the plane of the tissue showed that cell divisions are largely oriented towards the wound (Fig. 3d left). Quantification of cell body orientation also demonstrated that the majority of cells are elongated towards the wound following the same orientation documented for migration (Fig. 3e top, 3f left).
To test whether migration impacts the directionality of cell divisions, we turned to a mouse model lacking migration. Rac1−/− mutant mice showed an overall reduction of cell divisions (50% reduction in proliferation when compared to control littermates) as well as loss of cell orientation towards the wound (Fig. 3e bottom; 3f right) indicating a role for Rac for both processes. Strikingly however, quantifications of the occurring proliferation events within the epithelial Rac1−/− mutant mice showed lack of directionality in cell division angles (Fig. 3d right). These data support a model where migration and cell orientation regulate the directionality of cell divisions for their delivery to the wound site33.
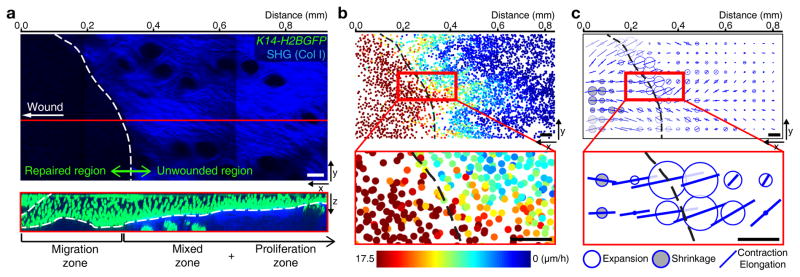
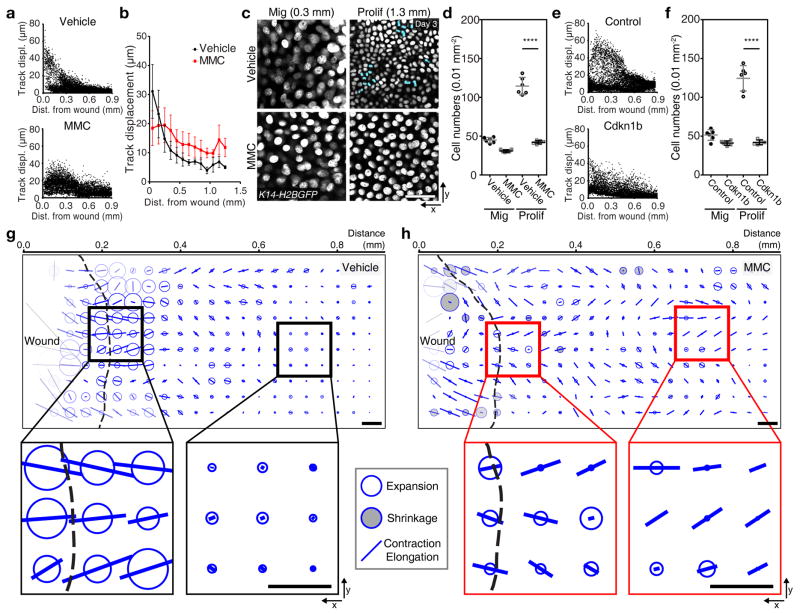
These data demonstrate a spatial interplay between proliferation and migration patterns and prompted us to interrogate its impact at a tissue-scale on the advancement of the repairing epithelium towards the wound. To this end, we used the migration profiles we had extracted at 3 days PWI to build tissue-scale maps of displacements of basal nuclei. For wild-type response at 3 days PWI, this map clearly highlights the gradient of nuclear displacements across the three zones over 12 hours (Fig. 1g) suggesting that tissue deformations may occur. To determine local tissue deformation in the basal layer of the tissue, we used a recently published tissue-scale quantitative method34 (Supplementary Fig. 6a–c and Supplementary Video 7). Strikingly, while the broad migration region translated into an overall contraction-elongation of the repairing epithelium (oriented bars), the region where migration and proliferation co-exist coincided with a peak of tissue expansion (white disks) corresponding to the border of where the wound was initially generated (Fig. 4a–c). All together these data show how the interplay of migration and proliferation that we have observed defines a distinctive area of local tissue expansion that is sandwiched between elongating regions of migration in the front and regions of high proliferation in the back.
Figure 4. Tissue expansion occurs at a distinctive area near wound.
(a) Different regions pattern the unwounded and repaired epithelium. Projection of the SHG in the dermis (top panel). White dotted line surrounds initial wounded region, which is negative for SHG signal. x-z view of red line (bottom panel) indicating that the left region without SHG (repaired epithelium) is the migration zone and the right region with SHG (unwounded epithelium) is the mixed and the proliferation zones (representative images from 3 mice). Scale bar, 50 μm. (b) Nuclei in the basal layer from K14-H2BGFP mice are represented as dots of different colors reflecting their displacement over 4 hours (representative images from 2 mice). Black dotted line surrounds initial wounded region. Scale bar, 50 μm. (c) Tissue deformation of the basal layer (representative images from 2 mice). White balls represent tissue expansion, black balls represent tissue shrinkage, and bars represent contraction/elongation rate and direction. Black dotted line surrounds initial wounded region. Scale bar, 50 μm.
Proliferation limits the repairing epithelial area
Thus far we have shown that models altering migration (Colcemid and Rac1−/−) have far worse consequences on repair than models that impair proliferation (MMC). To understand how the tissue compensates for lack of proliferation, we interrogated how migration is affected in two complementary models that inhibit proliferation at different stages of cell cycle: a drug based (MMC) model to acutely block proliferation as well as a genetic model of Cdkn1b (or p27) over-expression that we use for blocking proliferation for a longer term. In both cases, cells closer to the wound showed a decreased migration rate and cell migration appeared more uniformly spread over a larger area of the repairing epithelium (Fig. 5a, b, e). Additionally, we observed a region of low cell density that spread out over a large region of the repairing epithelium suggesting that cells compensate by increasing their size (Fig. 5c, d, f). This suggests that the proliferation zone functions not only to supply new cells but also to restrict the re-epithelialization territory by containing the range of repair behaviors.
Figure 5. Inhibition of proliferation changes repairing area.
(a) Representative quantification of cell tracks in the vehicle and the MMC-treated wound using K14-H2BGFP (Vehicle, n=8155 tracks from n=3 mice; MMC, n=9177 from n=3 mice). (b) Plot of cell tracks vs. distance from the initial wound boundary. Migrating cell tracks are averaged every 100 μm from the initial wound. Mean ± S.D. (Vehicle, n=8155 tracks from n=3 mice; MMC, n=9177 from n=3 mice). (c) Inhibition of cell division after MMC treatment. Proliferating cells are pseudo-colored in blue (representative images from 3 mice). Scale bar, 50 μm. (d) Representative quantification of cell density after MMC treatment. Mean ± S.D. **** p<0.0001, unpaired Student’s t-test (n=6 squares each sample pooled from 3 mice). (e) Representative quantification of migration tracks in the control and the Cdkn1b overexpressed wound using K14-rtTA;tetO-Cdkn1b;K14-H2BGFP (Control, n=7106 tracks from n=3 mice; MMC, n=9151 from n=3 mice). (f) Representative quantification of the cell density after Cdkn1b overexpression per 0.01 mm2. Mean ± S.D. **** p<0.0001, unpaired Student’s t-test (n=6 squares each sample pooled from 3 mice). (g, h) Tissue deformation after inhibition of the proliferation by MMC treatment. The tissue deformation analysis is performed in the Vehicle (g) and the MMC (h)-treated K14-H2BGFP mice (representative analysis from 2 mice). Scale bar, 50 μm.
To understand how this distribution of behaviors impacts repair at the tissue level, we determined tissue deformation and showed that tissue expansion and contraction-elongation rates were decreased near the wound and that contractions occurred more distally, occupying the further regions of the repairing epithelium (Fig. 5g, h and Supplementary Fig. 6d, e). Thus, while re-epithelialization can still occur (Fig. 1j) these analyses uncover that it does so at the cost of expanding the repairing epithelial region. Together these experiments show a role for the proliferation zone to function both as a source for new cells but also to limit the area of the unwounded epithelium utilized for re-epithelialization.
Discussion
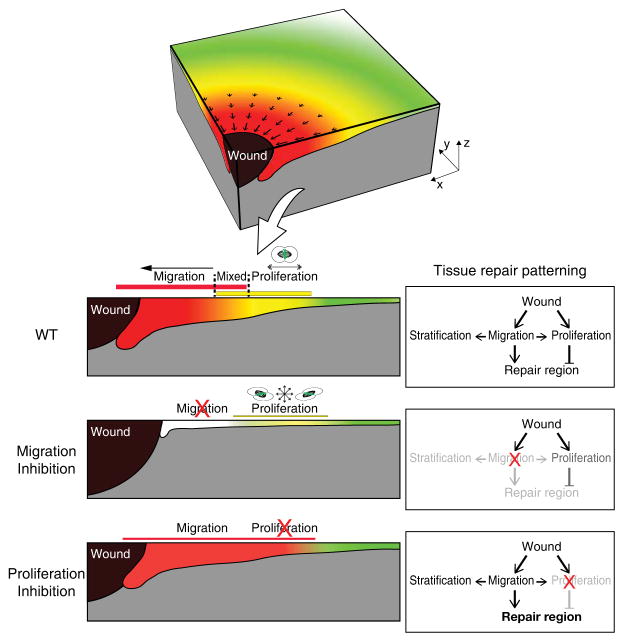
Using intravital imaging, we have captured, for the first time in a live mammal, how a tissue orchestrates the cellular mechanisms necessary to re-epithelialize a wound while maintaining the basic functions of tissue homeostasis (Fig. 6). Previous findings had defined that in response to wounding the skin epithelium organizes in an actively proliferating ring and a migratory front35–42. However, understanding how newly produced cells transition to migrate to the wound while at the same time maintaining a stratified epithelium through differentiation in a live mammal has been difficult to be directly investigated. Our results show that cells transition out of the proliferation ring through directional division as they actively migrate towards the wound. Migration of both differentiated and undifferentiated cells progressively increases when approaching the wound site impacting in turn the rate of stratification of the repairing epithelium. These interconnected behaviors result into tissue-scale changes setting an overall pattern within the repairing epithelium of contraction-elongations driven by cell migration and restrained by the proliferation zone, as well as a localized zone of tissue expansion where migration and proliferation co-exist. Taken together, our results identify how coordination of cellular behaviors is orchestrated to support wound re-epithelialization and provide insight into how the epidermis balances cellular behaviors for effective wound re-epithelialization and homeostasis.
Figure 6. Tissue-scale coordination of cellular behaviors during wound repair.
During wound repair the epithelium organizes directional migration and proliferation in overlapping regions moving towards the wound. Migration delivers cells to the wound region and also enhances stratification. Proliferation replenishes cells as well and limits the size of the repair region. Upon inhibition of migration, stratification is dramatically decreased and cell divisions lose their directionality. Moreover, wound repair is delayed compared to a wild type re-epithelialization. In contrast, upon inhibition of proliferation the tissue compensates by increasing the overall size of the repair region. Thickness of bars represents intensity of the behaviors (migration, red; proliferation, yellow).
Methods
Mice
K14-H2BGFP21 and K14-actinGFP43 mice were obtained from Elaine Fuchs (Rockefeller University). TetO-Cre44 mice were obtained from Katerina Politi (Yale). K14-rtTA45, tetO-Cdkn1b46, Rac1−/−28, Rosa-stop-tdTomato47, Rosa-mTomato-stop-mGFP (mTmG)48 and K14-CreER49 mice were obtained from Jackson Laboratories. K14-H2BPAmCherry mice were generated by the Yale Transgenic Facility28. For the photo-activation experiment, K14-H2BPAmCherry mice were mated with K14-actinGFP mice to visualize the structure of the epidermis (K14-H2BPAmCherry;K14-actinGFP). For the migration inhibition experiment, K14-CreER;Rac1−/−;tdTomato;K14-H2BGFP were generated. To block the migration of epithelial cells, three intraperitoneal injections of tamoxifen (100 mg/kg in corn oil) at postnatal day 10, 12 and 14. Inhibition of migration was monitored by time-lapse imaging via two-photon laser scanning microscopy described below. K14-CreER;Rac1+/−;tdTomato;K14-H2BGFP mice were used as controls. For the proliferation inhibition experiment, K14-rtTA;tetO-Cdkn1b;tetO-Cre;Rosa-stop-tdTomato;K14-H2BGFP were generated. To block the proliferation of epithelial cells, these mice were given doxycycicline (1 mg/ml) in potable water with 1 % Sucrose starting 1 day before the punch biopsy. Doxycycline treatment was sustained until imaging was performed. tdTomato signal was used to monitor Cre activation. Inhibition of proliferation was monitored by time-lapse imaging via two-photon laser scanning microscopy described below. K14-rtTA;tetO-Cre;Rosa-stop-tdTomato;K14-H2BGFP mice were used as controls. For cell membrane labeling experiments, Cre expression was induced with a single intraperitoneal injection of tamoxifen (1 mg/kg in corn oil) at postnatal day 21 in K14-CreER;mTmG mice. Mice from experimental and control groups were randomly selected of either gender for live imaging experiments. No blinding was done. All procedures involving animal subjects were performed under the approval of the Institutional Animal Care and Use Committee (IACUC) of the Yale School of Medicine.
Antibodies and reagents
Antibodies and reagents. Mitomycin C (MMC) was purchased from Research Products International Corp (M92010), Colcemid from Sigma (D7385-5MG), rabbit polyclonal anti-keratin 10 from Biolegend/Covance (PRB-159P, used at 1:1000), rabbit polyclonal anti-keratin 5 from Biolegend/Covance (PRB-160P, used at 1:1000), donkey anti-rabbit Rhodamine Red-X from Jackson Lab (711-295-152, used at 1:100) and donkey anti-rabbit Alexa 647 from ThermoFisher (A-31573, used at 1:200).
Wounding
Three-week-old mice were anesthetized by intraperitoneal injection of ketamine and xylazine cocktail mix (100 mg/kg and 10 mg/kg, respectively in phosphate-buffered saline). Once a surgical plane of anesthesia was verified by the absence of a physical and physiologic response to a noxious stimulus, a punch biopsy was performed using a 1 mm diameter punch biopsy tool (Miltex, USA). The punch biopsy tool was used to make a circular full thickness wound on the dorsal side of mouse ear. Wounds did not penetrate the ear and remained above the cartilage. The ear is chosen as a convenient region for imaging and for its similarity with respect to epithelial functions to other regions of mouse skin50,51. Dermal components such as cartilage are however different from back skin.
In vivo imaging
Mice were anesthetized using an isofluorane chamber and anesthesia was maintained throughout the course of the experiment with vaporized isofluorane delivered by a nose cone as previously described52,53. Image stacks were acquired with a LaVision TriM Scope II (LaVision Biotec, Germany) laser scanning microscope equipped with a tunable Two-photon Chameleon Ultra (Coherent, USA) Ti:Sapphire laser. To acquire serial optical sections, a laser beam (940 nm for H2BGFP, actinGFP and mTmG; 1040 nm for H2BPAmCherry and tdTomato) was focused through a 20X or 40X water immersion lens (N.A. 1.0 and 1.1 respectively; Zeiss, USA) and scanned with a field of view of 0.5 × 0.5 mm2 or 0.25 × 0.25 mm2 respectively at 600 Hz. Z-stacks were acquired in 1–3 μm steps to image a total depth of 200 μm of tissue52. To visualize large areas, 3–36 tiles of optical fields were imaged using a motorized stage to automatically acquire sequential fields of view as previously described54. For time-lapse imaging, serial optical sections were obtained in a range of 1 to 12 minute intervals depending on the experimental setup. The duration of time-lapse imaging ranged from 4–12 hours.
Photo-activation
Photo-activation in K14-H2BPAmCherry mice was carried out with the same optical equipment used for image acquisition. An 810 nm laser beam at 5 % laser power was used for 1 minute to scan the target area (100 × 100 mm2) and activate the PAmCherry protein.
Topical drug treatment
To inhibit cell proliferation in the epidermis, Mitomycin C (MMC)55 was delivered topically by applying it to the wound and the surrounding ear skin. MMC was dissolved in a 15 mg/ml stock solution in dimethyl sulfoxide (DMSO) and later the stock solution was diluted 100 times in 100 % petroleum jelly (Vaseline; final concentration is 150 μg/ml). 100 mg of the mixture of the MMC and the petroleum jelly was spread evenly on the wounded area at 1 and 2 days PWI. A mixture of 100 % DMSO in petroleum jelly (1:100) was used as a vehicle control. Colcemid was used to block microtubule polymerization56,57. Colcemid was dissolved to 25 mg/ml stock solution in the DMSO and delivered as described for the MMC treatment.
Image Analysis
Raw image stacks were imported into Fiji (NIH, USA) or Imaris software (Bitplane/Perkin Elmer) for further analysis. Imaris software was used to track cells and obtain xyz coordinates from individual tracked cells over time. All cell tracks were individually examined to confirm that they reported the behavior of a single cell. Only cells that could be tracked for the full duration of a movie were included in the final analysis. For the wound, a surface was manually defined in Imaris by the absence of cells within the wound area at the beginning of each movie. Imaris’ distance transformation module from their XTensions library was then utilized to determine the starting distance of each track from the wound’s surface. Prism software (Graphpad) was used to graph the data and to calculate non-linear trend-line (exponential growth equation, weight by 1/Y2) for a WT movie (Fig. 3a). To quantify migration in vehicle and drug-treated mice, migrating cells were isolated by track straightness (track length/track displacement) > 0.3 and binned every 100 μm from the wound. The bin calculated means were used to yield trend-lines (Fig. 1h and 5b). For the tracking of photolabeled H2BPAmCherry cells, an artificial surface was created using second-harmonic generation (SHG) from fibrillar collagens and the interface with the K14-actinGFP positive epithelial cells. This interface roughly defines the basement membrane. From this basement membrane, H2BPAmCherry signal was identified using Imaris’ spot analysis, and used to determine the minimum distance from each spot to the basement membrane surface using the distance transformation module in Imaris XTensions. Day 3 labeling (where only basal layer cells are labeled) was then utilized at the later time points to determine cell permanence within the basal layer or their displacement into suprabasal layers. Provided images and supplementary videos are typically presented as a maximal projection of 3 to 6 μm optical sections, unless otherwise specified. For visualizing individual labeled cells expressing the H2BPAmCherry reporter, the brightness and contrast were adjusted accordingly for the green (GFP) and red (H2BPAmCherry) channels and composite serial image sequences were assembled as previously described58. The tiled images were stitched by a grid/collection stitching plugin in Fiji. Migrating cell tracking analysis and 3D reconstitution of photo-activated cells were performed in Imaris software.
Immunostaining
Frozen skin sections were fixed in 4 % paraformaldehyde and used for histological analysis as previously described59,60. Immunofluorescence was performed by incubating sections at 4 °C overnight with primary antibodies: rabbit anti-keratin 10 (1:1000, Covance) and rabbit anti-keratin 5 (1:1000, Covance). One hour incubation at room temperature was then performed with secondary antibodies: donkey anti-rabbit Rhodamine Red-X (1:100, Jackson Lab) and donkey anti-rabbit Alexa 647 (1:200, Alexa Fluor). Two photon microscope or a Zeiss Axioimager was used to collect images.
Quantitative characterization of tissue deformation
The tracking of cell nuclei over time made the measurement of local tissue deformation possible. Based on the profile of the average displacement toward the wound and distance to the basement membrane, we first estimated that nuclei belonging to the most basal layer of cells were in the range of [0 – 10] μm distance to the basement membrane. For each time point, we therefore selected the “basal nuclei” based on this threshold.
Then we aimed to characterize the deformation of the basal layer of cells based on nuclei displacements. To achieve this, we used a quantitative approach that was recently developed and validated, and which is based on the evolution of links connecting neighboring cell centroids34. Therefore, we first determined neighbor relationships between nuclei, which then enabled us to define the pairs of neighbor nuclei that will be connected with a link. To this end, in each image, we used the nuclei positions as generators to build a Voronoi tessellation that established neighbor relationships between nuclei and between their corresponding Voronoi regions. In each image, we defined the links joining the centers of neighboring Voronoi regions. Then, based on the tracking of nuclei, we could record the evolution of those links over time (Supplementary Fig. 6a–c).
To characterize tissue deformation across the tissue surrounding the wound and its spatial variations, we defined sub-regions in the tissue of about 80 × 80 μm2 containing approximately 50 to 200 cells. The sub-region size was chosen so that it was large enough to contain many nuclei and links, ensuring accurate statistical measurements, but also small enough so that tissue deformations could be estimated at various locations across the tissue and that deformations within each sub-region remain rather homogeneous. The nuclei initially assigned to each sub-region were kept in the same sub-region deforming over time (Lagrangian approach). Then, we determined the local tissue deformation in each sub-region, the calculation of which was solely based on the change of geometry of links that were conserved in this region between two images.
Note that, if the contributions of cell behaviors to tissue deformation indeed depends on the actual cell network and requires the knowledge of actual cell geometries and topologies, this is not the case for tissue deformation that only involves the variation of conserved link geometry in the sub-region. Also, it is not sensitive to the type of tessellation chosen, as long as each sub-region contains enough conserved links between two successive images34.
Statistics and reproducibility
Data are expressed either as absolute numbers or percentages ± S.D. An unpaired Student’s t-test was used to analyze data sets with two groups and * p<0.05 to **** p<0.0001 indicate a significant difference. Statistical calculations were performed using the Prism software package (GraphPad, USA). No statistical method was used to predetermine sample size. Panels showing representative images are representative of at least two independent experiments and up to five, as indicated in the figure legends.
Code availability
Cell nuclei were tracked over time using Imaris. The Voronoï tessellation used to determine nuclei neighbor relationships was done using Matlab function “voronoi”. The network made up by Voronoï region centroids and its time evolution were used as inputs to calculate local tissue deformation rate G, as presented in Guirao et al. 34. Matlab codes to calculate G from nuclei displacements are available upon request.
Data availability
All data that support the conclusions are available from the authors on request. Source data for Fig. 1c, 1g, 1h, 2e, 2f, 3a, 4b, 4c, 5a, 5b, 5e, 5g, 5h and Supplementary Fig. 4a, 4c, 6d, 6e have been provided as Supplementary Table 7.
Supplementary Material
Acknowledgments
We thank Elaine Fuchs (Rockefeller University) for the K14-H2BGFP, K14-actinGFP, K14-rtTA mice; Katerina Politi (Yale) for the TetO-Cre mice; Vladislav V. Verkhusha (Albert Einstein) for the PAmCherry construct; we thank the Marine Biological Laboratory (MBL) for their support while writing this manuscript. Scott Williams (UNC), Markus Schober (NYU) and Minhwa Lee for critical feedback. Sabine Werner, Richard Clark and Paul Martin for their constructive criticism of this work. V.G. is supported by the National Institute of Arthritis and Musculoskeletal and Skin Disease (NIAMS), NIH, grants no. 5R01AR063663-04 and 1R01AR067755-01A1 and by a Mallinckrodt Scholar Award. V.G. is a New York Stem Cell Foundation Robertson Investigator and a HHMI Scholar. S.P. was supported by the James Hudson Brown-Alexander Brown Coxe Postdoctoral Fellowship and is currently supported by the CT Stem Cell Grant 14-SCA-YALE-05. D.G.G. and A.M.H. are supported by the Yale Rheumatic Disease Research Core Center (P30 AR053495-08). K.R.M. was and S.B. is supported by the NIH Predoctoral Program in Cellular and Molecular Biology, grant no. T32GM007223. K.R.M. is currently supported by the NSF Graduate Research Fellow. E.D.M. is supported by the National Institute of Health, grant no. T32 GM007499. P.R. was a New York Stem Cell Foundation-Druckenmiller Fellow and was supported by the CT Stem Cell Grant 13-SCA-YALE-20. K.C. is supported by a Canadian Institutes of Health Research Postdoctoral Fellowship. Y.B. is supported by CNRS, INSERM, Institut Curie, ERC Advanced (TiMoprh, 340784), ARC (SL220130607097), ANR Labex DEEP (11-LBX-0044, ANR-10-IDEX-0001-02) and PSL grants.
Footnotes
Author Contributions
S.P. and V.G. designed experiments and wrote the manuscript. S.P. performed the experiments and analyzed the data. D.G.G. and A.M.H. performed 3D imaging analysis and quantifications. J.D.B. and E.D.M. performed immunofluorescence. B.G. and Y.B. performed tissue transformation. Y.B., K.R.M., J.D.B, A.M.H, D.G.G., K.C. and S.B. assisted with critical feedback on the research and manuscript. P.R. generated the K14-H2BPAmCherry mouse line.
References
- 1.Eming S, Martin P, Tomic-Canic M. Wound repair and regeneration: mechanisms, signaling, and translation. Science translational medicine. 2014;6:265sr6. doi: 10.1126/scitranslmed.3009337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gurtner G, Werner S, Barrandon Y, Longaker M. Wound repair and regeneration. Nature. 2008;453:314–321. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- 3.Clark RA, Ghosh K, Tonnesen MG. Tissue engineering for cutaneous wounds. The Journal of investigative dermatology. 2007;127:1018–29. doi: 10.1038/sj.jid.5700715. [DOI] [PubMed] [Google Scholar]
- 4.Simpson CL, Patel DM, Green KJ. Deconstructing the skin: cytoarchitectural determinants of epidermal morphogenesis. Nat Rev Mol Cell Biol. 2011;12:565–80. doi: 10.1038/nrm3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuchs E, Raghavan S. Getting under the skin of epidermal morphogenesis. Nat Rev Genet. 2002;3:199–209. doi: 10.1038/nrg758. [DOI] [PubMed] [Google Scholar]
- 6.Xin T, Greco V, Myung P. Hardwiring Stem Cell Communication through Tissue Structure. Cell. 2016;164:1212–25. doi: 10.1016/j.cell.2016.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mesa KR, Rompolas P, Greco V. The Dynamic Duo: Niche/Stem Cell Interdependency. Stem Cell Reports. 2015;4:961–6. doi: 10.1016/j.stemcr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kandyba EE, Hodgins MB, Martin PE. A murine living skin equivalent amenable to live-cell imaging: analysis of the roles of connexins in the epidermis. J Invest Dermatol. 2008;128:1039–49. doi: 10.1038/sj.jid.5701125. [DOI] [PubMed] [Google Scholar]
- 9.Liang CCC, Park AY, Guan JLL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2:329–33. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 10.Hertle MD, Kubler MD, Leigh IM, Watt FM. Aberrant integrin expression during epidermal wound healing and in psoriatic epidermis. J Clin Invest. 1992;89:1892–901. doi: 10.1172/JCI115794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nunan R, et al. Ephrin-Bs Drive Junctional Downregulation and Actin Stress Fiber Disassembly to Enable Wound Re-epithelialization. Cell Rep. 2015;13:1380–95. doi: 10.1016/j.celrep.2015.09.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Safferling K, et al. Wound healing revised: a novel reepithelialization mechanism revealed by in vitro and in silico models. The Journal of cell biology. 2013;203:691–709. doi: 10.1083/jcb.201212020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bereiter-Hahn J. Epidermal cell migration and wound repair. 1986:443–471. doi: 10.1007/978-3-662-00989-5_23. [DOI] [Google Scholar]
- 14.Christophers E. Some observations on stratum corneum. Current medical research and opinion. 1981 [PubMed] [Google Scholar]
- 15.Coulombe PA, Takahashi K. The Cellular Mechanisms of Keratinocyte Migration into a Skin Wound Site: An Open Question with Important Implications. CELL VISION. 1996;3:217. [Google Scholar]
- 16.Danjo Y, Gipson IK. Specific transduction of the leading edge cells of migrating epithelia demonstrates that they are replaced during healing. Exp Eye Res. 2002;74:199–204. doi: 10.1006/exer.2001.1115. [DOI] [PubMed] [Google Scholar]
- 17.Zhao M, Song B, Pu J, Forrester JV, McCaig CD. Direct visualization of a stratified epithelium reveals that wounds heal by unified sliding of cell sheets. FASEB J. 2003;17:397–406. doi: 10.1096/fj.02-0610com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Usui ML, et al. Morphological evidence for the role of suprabasal keratinocytes in wound reepithelialization. Wound Repair Regen. 2005;13:468–79. doi: 10.1111/j.1067-1927.2005.00067.x. [DOI] [PubMed] [Google Scholar]
- 19.Radice GP. The spreading of epithelial cells during wound closure in Xenopus larvae. Dev Biol. 1980;76:26–46. doi: 10.1016/0012-1606(80)90360-7. [DOI] [PubMed] [Google Scholar]
- 20.Brown S, Greco V. Stem cells in the wild: understanding the World of stem cells through intravital imaging. Cell Stem Cell. 2014;15:683–6. doi: 10.1016/j.stem.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tumbar T, et al. Defining the epithelial stem cell niche in skin. Science (New York, NY ) 2004;303:359–63. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang H, Ganguly A, Cabral F. Inhibition of cell migration and cell division correlates with distinct effects of microtubule inhibiting drugs. J Biol Chem. 2010;285:32242–50. doi: 10.1074/jbc.M110.160820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ganguly A, Yang H, Sharma R, Patel KD, Cabral F. The role of microtubules and their dynamics in cell migration. J Biol Chem. 2012;287:43359–69. doi: 10.1074/jbc.M112.423905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu YCC, Li L, Fuchs E. Emerging interactions between skin stem cells and their niches. Nat Med. 2014;20:847–56. doi: 10.1038/nm.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Odland G, Ross R. HUMAN WOUND REPAIR I. Epidermal Regeneration. The Journal of Cell Biology. 1968;39:135–51. doi: 10.1083/jcb.39.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matoltsy AG, Viziam CB. Further observations on epithelialization of small wounds: an autoradiographic study of incorporation and distribution of 3H-thymidine in the epithelium covering skin wounds. The Journal of investigative dermatology. 1970;55:20–5. doi: 10.1111/1523-1747.ep12290488. [DOI] [PubMed] [Google Scholar]
- 27.Subach FV, et al. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nature methods. 2009;6:153–9. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rompolas P, et al. Spatiotemporal coordination of stem cell commitment during epidermal homeostasis. Science (New York, NY ) 2016;352:1471–4. doi: 10.1126/science.aaf7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tscharntke M, et al. Impaired epidermal wound healing in vivo upon inhibition or deletion of Rac1. J Cell Sci. 2007;120:1480–90. doi: 10.1242/jcs.03426. [DOI] [PubMed] [Google Scholar]
- 30.Werner S, et al. The function of KGF in morphogenesis of epithelium and reepithelialization of wounds. Science. 1994;266:819–22. doi: 10.1126/science.7973639. [DOI] [PubMed] [Google Scholar]
- 31.Repertinger SK, et al. EGFR enhances early healing after cutaneous incisional wounding. J Invest Dermatol. 2004;123:982–9. doi: 10.1111/j.0022-202X.2004.23478.x. [DOI] [PubMed] [Google Scholar]
- 32.Patel GK, Wilson CH, Harding KG, Finlay AY, Bowden PE. Numerous keratinocyte subtypes involved in wound re-epithelialization. J Invest Dermatol. 2006;126:497–502. doi: 10.1038/sj.jid.5700101. [DOI] [PubMed] [Google Scholar]
- 33.Das T, et al. A molecular mechanotransduction pathway regulates collective migration of epithelial cells. Nature Cell Biology. 2015;17:276–87. doi: 10.1038/ncb3115. [DOI] [PubMed] [Google Scholar]
- 34.Guirao B, et al. Unified quantitative characterization of epithelial tissue development. eLife. 2015;4 doi: 10.7554/eLife.08519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw TJ, Martin P. Wound repair at a glance. J Cell Sci. 2009;122:3209–13. doi: 10.1242/jcs.031187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaw TJ, Martin P. Wound repair: a showcase for cell plasticity and migration. Curr Opin Cell Biol. 2016;42:29–37. doi: 10.1016/j.ceb.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 37.Margadant C, et al. Integrin alpha3beta1 inhibits directional migration and wound re-epithelialization in the skin. J Cell Sci. 2009;122:278–88. doi: 10.1242/jcs.029108. [DOI] [PubMed] [Google Scholar]
- 38.Razzell W, Wood W, Martin P. Recapitulation of morphogenetic cell shape changes enables wound re-epithelialisation. Development. 2014;141:1814–20. doi: 10.1242/dev.107045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michalik L, et al. Impaired skin wound healing in peroxisome proliferator-activated receptor (PPAR)alpha and PPARbeta mutant mice. J Cell Biol. 2001;154:799–814. doi: 10.1083/jcb.200011148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jameson J, et al. A role for skin gammadelta T cells in wound repair. Science. 2002;296:747–9. doi: 10.1126/science.1069639. [DOI] [PubMed] [Google Scholar]
- 41.Keyes BE, et al. Impaired Epidermal to Dendritic T Cell Signaling Slows Wound Repair in Aged Skin. Cell. 2016;167:1323–1338.e14. doi: 10.1016/j.cell.2016.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chernyavsky AI, Arredondo J, Wess J, Karlsson E, Grando SA. Novel signaling pathways mediating reciprocal control of keratinocyte migration and wound epithelialization through M3 and M4 muscarinic receptors. J Cell Biol. 2004;166:261–72. doi: 10.1083/jcb.200401034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vaezi A, Bauer C, Vasioukhin V, Fuchs E. Actin cable dynamics and Rho/Rock orchestrate a polarized cytoskeletal architecture in the early steps of assembling a stratified epithelium. Developmental cell. 2002;3:367–81. doi: 10.1016/s1534-5807(02)00259-9. [DOI] [PubMed] [Google Scholar]
- 44.Song X, et al. ERBB3-independent activation of the PI3K pathway in EGFR-mutant lung adenocarcinomas. Cancer research. 2015;75:1035–45. doi: 10.1158/0008-5472.CAN-13-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie W, Chow LT, Paterson AJ, Chin E, Kudlow JE. Conditional expression of the ErbB2 oncogene elicits reversible hyperplasia in stratified epithelia and up-regulation of TGFalpha expression in transgenic mice. Oncogene. 1999;18:3593–607. doi: 10.1038/sj.onc.1202673. [DOI] [PubMed] [Google Scholar]
- 46.Pruitt SC, Freeland A, Rusiniak ME, Kunnev D, Cady GK. Cdkn1b overexpression in adult mice alters the balance between genome and tissue ageing. Nature communications. 2013;4:2626. doi: 10.1038/ncomms3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience. 2010;13:133–40. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis (New York, NY : 2000) 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 49.Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:8551–6. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barker JH, et al. The hairless mouse ear: an in vivo model for studying wound neovascularization. Wound Repair Regen. 1994;2:138–43. doi: 10.1046/j.1524-475X.1994.20208.x. [DOI] [PubMed] [Google Scholar]
- 51.Auffray C, et al. Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science. 2007;317:666–70. doi: 10.1126/science.1142883. [DOI] [PubMed] [Google Scholar]
- 52.Rompolas P, Mesa KR, Greco V. Spatial organization within a niche as a determinant of stem-cell fate. Nature. 2013;502:513–8. doi: 10.1038/nature12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rompolas P, et al. Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature. 2012;487:496–9. doi: 10.1038/nature11218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pineda CM, et al. Intravital imaging of hair follicle regeneration in the mouse. Nat Protoc. 2015;10:1116–30. doi: 10.1038/nprot.2015.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomasz M. Mitomycin C: small, fast and deadly (but very selective) Chemistry & biology. 1995;2:575–9. doi: 10.1016/1074-5521(95)90120-5. [DOI] [PubMed] [Google Scholar]
- 56.Rieder CL, Palazzo RE. Colcemid and the mitotic cycle. Journal of cell science. 1992;102( Pt 3):387–92. doi: 10.1242/jcs.102.3.387. [DOI] [PubMed] [Google Scholar]
- 57.Dunlap MK, Donaldson DJ. Inability of colchicine to inhibit newt epidermal cell migration or prevent concanavalin A-mediated inhibition of migration. Studies in vivo. Experimental cell research. 1978;116:15–9. doi: 10.1016/0014-4827(78)90059-9. [DOI] [PubMed] [Google Scholar]
- 58.Mesa KR, et al. Niche-induced cell death and epithelial phagocytosis regulate hair follicle stem cell pool. Nature. 2015;522:94–7. doi: 10.1038/nature14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zito G, et al. Spontaneous tumour regression in keratoacanthomas is driven by Wnt/retinoic acid signalling cross-talk. Nature communications. 2014;5:3543. doi: 10.1038/ncomms4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deschene ER, et al. β-Catenin activation regulates tissue growth non-cell autonomously in the hair stem cell niche. Science. 2014;343:1353–6. doi: 10.1126/science.1248373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that support the conclusions are available from the authors on request. Source data for Fig. 1c, 1g, 1h, 2e, 2f, 3a, 4b, 4c, 5a, 5b, 5e, 5g, 5h and Supplementary Fig. 4a, 4c, 6d, 6e have been provided as Supplementary Table 7.