Abstract
Background and Aims
Duodenal collections of pancreatic fluid can be used as a source of mutations and other markers of pancreatic ductal neoplasia, but admixing pancreatic juice with duodenal contents lowers the concentrations of mutations. Collecting pancreatic fluid directly from the ampulla could yield a purer sample of pancreatic fluid.
Methods
We used an endoscopic distal cap attachment to “cap” the ampulla and collect secretin-stimulated pancreatic fluid samples for 5 minutes from 81 patients undergoing pancreatic evaluation as part of the Cancer of the Pancreas Screening studies. We compared mutation concentrations (KRAS and GNAS) measured by droplet-digital PCR (ddPCR) in “cap-collected juice” samples to those found in juice samples obtained from 77 patients collected by aspiration from the duodenal lumen without capping the ampulla.
Results
Among all subjects, mutation concentrations were higher in pancreatic juice samples collected using the endoscopic cap method (median 0.028%, interquartile range 0–0.077) compared to the non-cap-collected (0.019%, 0–0.044) (P = 0.055). Among pancreatic juice samples with detectable mutations, mutation concentrations were higher in the cap-collected juice samples than in those collected without the cap (0.055%, interquartile range [IQR] 0.026–0.092 vs. 0.032%, IQR 0.020–0.066, P =0.031).
Conclusions
Collecting pancreatic juice directly from the ampulla using an endoscopic distal cap yields higher concentrations of pancreatic fluid mutations.
Keywords: pancreatic fluid, pancreatic juice, secretin, pancreatic screening, KRAS, GNAS, mutation, droplet digital PCR, endoscopic distal cap
INTRODUCTION
Pancreatic cancer is expected to be the 2nd leading cause of cancer-related deaths by the year 2030 in the United States.1 Pancreatic screening and surveillance is being performed in an attempt to identify precancerous lesions and early pancreatic cancer among asymptomatic subjects judged to be at sufficiently increased risk of developing pancreatic cancer based on either their pancreatic cancer family history, an inheritance of mutations in pancreatic cancer susceptibility genes or the presence of pancreatic cysts.2–10 EUS and MRI/MRCP are accurate tests for detecting the presence of pancreatic cysts in this setting,3, 4, 9 but imaging tests are unable to detect microscopic pancreatic intraepithelial neoplasia (PanIN). Most pancreatic cancers are thought to arise from PanIN and patients with a familial susceptibility to pancreatic cancer often harbor widespread PanIN, including PanIN-3.11 Most of the pancreatic cystic lesions identified in patients undergoing pancreatic screening are thought to be intraductal papillary mucinous neoplasms (IPMNs).4, 12 PanIN and IPMN lesions can harbor the driver mutations of pancreatic ductal adenocarcinoma. For example, over 90% of PanIN-1 lesions harbor KRAS mutations,13, 14 and most IPMNs harbor GNAS and KRAS mutations.12, 15 The limitation of current pancreatic imaging is also evident in the experience patients diagnosed with pancreatic cancer who had had a non-suspicious EUS within the prior ~6 to 18 months.16 Because pancreatic imaging tests cannot reliably identify PanIN, or grade IPMN and can miss small cancers, secretin-stimulated pancreatic fluid samples is being evaluated to determine how these samples might help in the evaluation of patients undergoing screening and surveillance.3, 4, 9 Studies using these samples revealed that GNAS mutations detected in pancreatic fluid samples collected from the duodenum are highly correlated with the presence of IPMN, that KRAS mutations are commonly detected in pancreatic fluid samples of patients who are undergoing screening for their family history of pancreatic cancer and that mutation concentrations as well as mutations that are known to arise in high-grade dysplasia and invasive cancer such as in SMAD4 are helpful in distinguishing the presence of pancreatic ductal adenocarcinoma.12, 14, 16, 17 Since secretin-stimulated pancreatic fluid samples are typically collected from the duodenal lumen, they are admixed with abundant wild-type DNA in duodenal fluid effectively reducing mutation concentrations.18 The very low concentrations of mutations typically found in these duodenal collections requires that more sophisticated molecular tools be employed for their detection.16
Collecting purer samples of pancreatic fluid with less mixing of duodenal fluid contents could help pancreatic cancer early detection efforts. In this study, we compared the mutation concentrations in pancreatic juice samples collected using our standard method (from the duodenal lumen) to those obtained using a new endoscopic technique which we term “cap-assisted” pancreatic juice collection which involves “capping” the major duodenal papilla with a cap attached to the tip of a standard upper endoscope and aspirating samples from the papilla. We hypothesized that the new method would reduce contamination by duodenal contents.
MATERIALS AND METHODS
Patients and specimens
Pancreatic fluid samples and clinical information were obtained from participants enrolled in the Cancer of the Pancreas Screening (http://clinicaltrials.gov, NCT00714701 and NCT02000089).4, 9 For this study, we evaluated 158 subjects enrolled in the CAPS studies at Johns Hopkins Hospital (JHH, Baltimore, MD) who had undergone EUS and had sufficient pancreatic fluid samples for analysis. The “cap juice” collections were performed between 2014 and 2016 at a time when we were also collecting pancreatic juice samples using our standard protocol for the CAPS study. Subjects were arbitrarily assigned to undergo one of the pancreatic juice collection methods. Once we had completed the enrollment of patients who had undergone a “cap juice” collection, we matched each of these cases with another patient who had undergone a non-cap collection with the goal that the case mix be similar between the two groups. Most of the subjects enrolled were individuals (i) undergoing pancreatic screening for their familial/inherited risk of pancreatic cancer, (ii) undergoing evaluation of their “sporadic pancreatic cyst(s)” or (iii) with a diagnosis of pancreatic cancer.19
Pancreatic fluid collection
Pancreatic fluid samples were collected from all participants after infusion of intravenous human synthetic secretin (0.2 μg/kg over 1 min). The standard pancreatic juice collection approach involves collecting pancreatic fluid samples (~10 mls) from the duodenal lumen as it is secreted from the papilla by suctioning of fluid through the echoendoscopic channel over 5 minutes without the use of a catheter.9 The tip of the echoendoscope was placed next to the major papilla by endoscopic guidance. Secretin was provided by ChiRhoClin Inc (Burtonsville, MD).
The endoscopic cap collections were performed by attaching the distal cap (12.4 mm disposable distal attachment, Olympus, Tokyo, Japan) onto the tip of an upper endoscope passed into the duodenum after the EUS. The cap was placed over the major duodenal papilla by shortening the endoscope to place the distal cap edge “en face” on the mucosal surface (Movie 1). Secretin stimulated duodenal fluid samples were collected from the papilla covered by the cap by intermittent gentle suctioning of fluid through the endoscopic channel over 5 minutes. The ampulla was not visualized using the forward viewing endoscope in 5 patients so capping was not undertaken. Pancreatic fluid from these cases was aspirated from the duodenal lumen just like the non-cap cases. There were no cases with ampullary pathology in either group.
KRAS and GNAS mutation concentrations were considered the best biomarkers to use to assess the relative purity of pancreatic juice samples and their potential diagnostic utility as they are the most common mutations found in PanINs and IPMNs.
Pancreatic juice samples were stored at −80°C before use. DNA was extracted from 450 μL of pancreatic fluid using the DNeasy Blood & Tissue Kit (Qiagen, Germantown, MD) and final volume was 45 μL(1 μL sample was from 10 μL of pancreatic fluid).
All elements of this study were approved by the Johns Hopkins institutional review board and written informed consent was provided from all patients.
Droplet digital PCR methods
The mutational status of KRAS codon 12 and GNAS codon 201 were investigated with droplet-digital PCR (ddPCR™) technology (Bio-Rad Laboratories, Hercules, CA) using previously reported methods. Primers (Integrated DNA Technologies, Coralville, IA) and probes (Custom TaqMan® Probes, Thermo Fisher Scientific, Waltham, MA) were designed for KRAS (G12D, G12V and G12R) and GNAS (R201C and R201H) which cover the mutational status in KRAS and GNAS over 90% of total mutation in pancreatic neoplasms.13, 20 The MGB probes were labeled with either FAM or VIC at 5′ end and a non-fluorescent quencher (NFQ) at the 3′ end. Primers and probes sequences are available on Supplementary Table S1. Each ddPCR mix consisted of 10 μL of 2× ddPCR™ Supermix for probes (No dUTP) (Bio-Rad Laboratories), 900 nM of forward and reverse primers, 250 nM of probes (FAM for mutant and VIC for wild type) and 4 μL of DNA (5,000~25,000 copies) into a total volume of 20 μL. The PCR mix along with 70 μL Droplet Generation Oil (Bio-Rad Laboratories)were loaded into each well of the 8 channel cartridge (Bio-Rad Laboratories). The QX200™ droplet generator (Bio-Rad Laboratories) created an average of 18,000 droplets per well. Droplets were transferred to a 96-well PCR plate (Eppendorf, Hauppauge, NY), the plate was heat-sealed (PX1™ PCR Plate Sealer, Bio-Rad Laboratories) with a foil heat seal (Bio-Rad Laboratories), and placed into a thermal cycler (Verity®, Thermo Fisher Scientific). PCR cycling conditions for all assays except for GNAS R201C were as follows: 95°C for 10 minutes, 39 cycles of 94°C for 30 seconds and 63°C for 60seconds, and two final steps at 98°C for 10 minutes and 4°C hold to enhance dye stabilization.21 For GNAS R201C, the annealing and extension temperature was 61°C, and the other conditions were same as above. The droplets were subsequently read automatically by QX200™ droplet reader (Bio-Rad Laboratories), and the result was analyzed with the QuantaSoft™ software (version 1.7.4, Bio-Rad Laboratories). Each assay was conducted with no template, wild type and mutant type control wells.
The number of DNA copies is calculated by the software was converted into the number per 40 μL of pancreatic juice. The mutation concentration and the number of DNA copies were reported as median with interquartile range (IQR).
Determination of the threshold and evaluation of experiments for ddPCR
The threshold for distinguishing positive from negative droplets was determined using multiple positive and negative control samples. We then evaluated the accuracy and limit of blank (LOB) for the assays. The fluorescent value thresholds for FAM (mutant DNA) were as follows: KRAS G12D 4500, G12V 5000, G12R 5000 and GNAS R201C 4500 and R201H 5000 (Fig. S1). The fluorescent value thresholds for VIC (wild type DNA ) was 3500 for all assays.
To determine the precision of the ddPCR assays, we analyzed serial dilutions using cell line DNA samples from PANC-1, CFPAC-1 and HPNE cell lines (American Type Culture Collection, Manassas, VA) which harbored KRAS G12D, G12V and KRAS wild type codon 12, respectively. For the other mutations, we used DNA extracted from primary resected IPMN and pancreatic cancer tissue samples. The mutational status of these tumor DNA samples was determined by next-generation sequencing. Serial dilutions of reference DNA samples (10% to 0.01%) were prepared by mixing mutant and wild-type DNAs. and mutation concentration was measured 5 times. For the KRAS wild-type assay, 65 KRAS-wild-type DNA samples were analyzed containing a range of concentrations (5000, 10000, 15000 and 20000 copies/well). We determined the LOB as above the 95 percentile of the false-positive drop distribution, and the mutational status was judged to be negative (mutation concentration = 0) if the number of the positive drops at or below this value. For all assays, the concentration measured using ddPCR was highly correlated with the expected concentration (Fig. S2).
The 95 percentile upper limit of the false-positive drop distribution for these ddPCR assays was determined. Thus, we determined the threshold for calling a sample as having a mutation was if the number of positive droplets was above one, zero, zero, one and one positive droplet for KRAS G12D, G12V and G12R and GNASR201C and R201H, respectively.
Statistical analysis
Qualitative variables were compared by χ2 tests, and quantitative variables were compared by Mann–Whitney U test. Statistical analysis was performed using GraphPad Prism 6 (GraphPad Software, San Diego, CA). P <0.05 was considered statistically significant.
RESULTS
Patient characteristics
Of the 158 subjects included in this study, 97 underwent pancreatic screening because of their family history of pancreatic cancer,19 47 for evaluation of pancreatic cysts in the absence of a family history of pancreatic cancer and 14 with a diagnosis of pancreatic ductal adenocarcinoma. The “cap” assisted pancreatic juice collection was performed in 81 subjects; for these cases, the ampulla was visualized and the cap was placed over the ampulla for pancreatic fluid collection. We compared the pancreatic fluid mutation concentrations in the 81 subjects who had a pancreatic fluid collected from the ampulla with the cap (the “cap” group) to 77 subjects who had pancreatic fluid collected from the duodenal lumen (the “non-cap” group) including the five subjects who underwent an unsuccessful cap juice collection. There were no significant differences in gender, age, indication, EUS findings, cyst number, cyst size, and color and volume of pancreatic fluid collected between the cap and the non-cap groups. A summary of the patient characteristics is provided in Table 1. There were no complications related to the endoscopic cap collections.
Table 1.
Patient Characteristics
| Non-cap group (n = 77) | Cap group (n = 81) | P value | |
|---|---|---|---|
| Gender (Male/Female) | 32/45 | 36/45 | 0.714 |
| Age (median, IQR) | 62 (53–69) | 64 (57–70) | 0.128 |
| Diagnosis | |||
| PDAC | 7 | 7 | 0.990 |
| Cysts with family history | 33 | 31 | |
| Cysts without family history | 19 | 23 | |
| Other cysts | 3 | 2 | |
| No cyst with family history | 15 | 18 | |
| Number of cyst (median, IQR) | 2 (1–4) | 2 (1–4) | 0.211 |
| Cyst size (mm) (median, IQR) | 6.0 (4.4–12.5) | 9.8 (4.6–15.0) | 0.382 |
| Pancreatic fluid volume (mL) (median, IQR) | 10 (7–10) | 9 (5–10) | 0.170 |
IQR, innterquartile range; PDAC, pancreatic ductal adenocarcinoma, cysts (probable IPMNs), other cysts (SCNs)
Detection of KRAS/GNAS mutation in pancreatic fluid from duodenum lumen
KRAS mutations were detected in 88 (55.7%) of the 156 pancreatic juice samples including G12D in 46 (29.1%), G12V in 44 (27.8%) and G12R in 45 (28.5%) samples, and GNAS mutations were detected in 48 (30.4%) juice samples (R201C in 30 [19.0%] and R201H in 26 [16.5%] patients). When comparing median KRAS mutation concentration, patients with PDAC have significantly higher KRAS mutation concentration than those without PDAC (0.036% [0.020–0.088] vs. 0.010% [0–0.032], P = 0.003). Forty-eight (49.5%) of 97 high-risk patients had a detectable KRAS mutation in their pancreatic juice, including 34 (53.1%) of 64 with and 14 (42.4%) of 33 without cysts suspected to be IPMN, a prevalence very similar to what we reported previously in this population.17 Patients with KRAS mutations in their pancreatic juice were older on average than those without a KRAS mutation (P = 0.056). Thirty-six of 106 patients (34.0%) with cysts suspected to be IPMN had GNAS mutation s detected in their pancreatic juice.
Mutation detection in cap-assisted vs. non-cap-assisted pancreatic juice collections
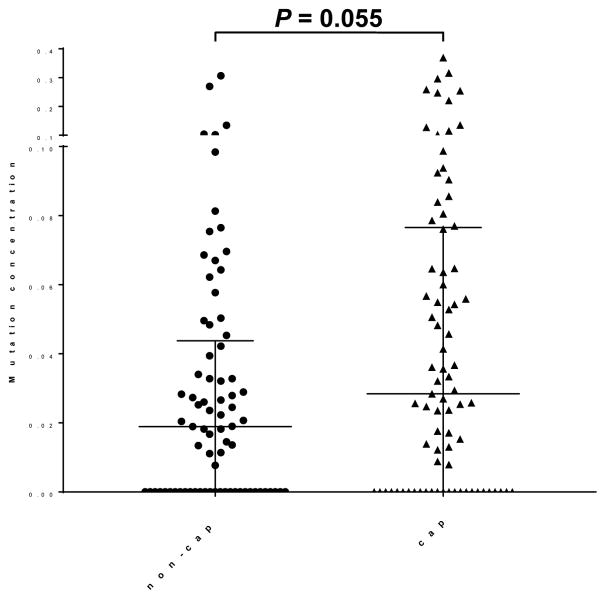
The median total mutation concentration of pancreatic juice was higher in the cap group (0.028% [0–0.077]) than in non-cap group (0.019% [0–0.044])(P = 0.055) (Fig. 1). Since patients without mutations will not have mutations detected by either collection method, we also compared mutation concentrations among those patients that had detectable mutations. In these cases, mutation concentrations were significantly higher in samples collected using the endoscopic cap method than in those collected without the cap (0.055% [0.026–0.092] vs. 0.032% [0.020–0.066], P = 0.031). The higher mutation concentration in the cap samples was reflected in the higher average wild-type DNA concentration in the non-cap juice samples (24540 [15760–56166] vs. 20832 [11896–41334] [copies/40 μL sample], P = 0.109) possibly reflecting greater contamination of wild-type DNA from duodenal fluid,18 without any difference in the average number of mutant DNA copies per sample (1.16 [0–2.70] vs. 0.68 [0–3.34] [copies/40 μL sample], respectively, P = 0.457).
Figure 1.
Pancreatic juice mutation concentrations by collection method.
DISCUSSION
In this study we describe our initial experience using cap-assisted endoscopy to collect pancreatic juice as it is being secreted from the ampulla. We find significantly higher mutation concentrations in cap-collected pancreatic juice samples. While duodenal collections of pancreatic juice can be easily collected during an endoscopic evaluation of the pancreas and these samples contain markers of pancreatic ductal neoplasia, marker concentrations in these samples are typically low (concentrations ~0.1%) and improvements in pancreatic juice sample collection would improve the detection of mutations and other markers of pancreatic neoplasia. Since pancreatic juice mutation concentrations collected from the pancreatic duct are ~10-fold higher than in samples collected from the duodenum,18 minimizing contamination from duodenal contents could be helpful. The results of this study support the hypothesis that improving how pancreatic fluid is collected can lead to a higher concentration of mutations in pancreatic juice samples collected from the duodenum.
Using an endoscopic cap to collect pancreatic juice is simple and non-invasive, but it was not always possible to visualize the papilla with forward viewing endoscope with a distal cap. The cap also does not create a complete seal around the papilla and as a result, aspiration of fluid still includes duodenal fluid contents. Further improvements in pancreatic juice collection could yield even purer samples of pancreatic fluid which would make it easier to identify genetic alterations using next-generation sequencing. For example, a custom cap that created a better seal around the papilla might provide purer juice samples. Fitting such a cap on the tip of a duodenoscope tip would make it easier to create a seal over the papilla.
In conclusion, we showed that our new method “cap” in collecting pancreatic fluid from the duodenal lumen improved rare mutation detection. Collecting high quality pancreatic fluid samples with the method combined with investigating rare mutation detection with ultrasensitive ddPCR technology may contribute to development in the genetic analysis for early detection of pancreatic neoplasms.
Supplementary Material
Movie 1. The “cap” method to collect secretin-stimulated pancreatic fluid samples.
Figure S1. Droplet digital PCR results of representative each target. Pink lines showed the thresholds of fluorescent value of FAM (vertical line, mutant type) and VIC (horizontal line, wild type).
Figure S2. Serial dilutions of positive controls were measured for each targets. Individual data point showed the mean value and error bar showed the standard deviation.
Table S1. Sequences of primers and probes for ddPCR
Acknowledgments
Grant Support: This work was supported by Susan Wojcicki and Dennis Troper, NIH grants (CA62924 and R01CA176828), the Pancreatic Cancer Action Network, and the Rolfe Pancreatic Cancer Foundation. MG is the Sol Goldman Professor of Pancreatic Cancer Research
Footnotes
Disclosures: There are no conflicts of interest for any of the authors. Recombinant secretin was generously provided for this study by ChiRhoClin, Inc. ChiRhoClin, Inc did not have any part in the study design, data analysis, interpretation or writing of this manuscript.
References
- 1.Rahib L, Smith BD, Aizenberg R, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–21. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 2.Brentnall TA, Bronner MP, Byrd DR, et al. Early diagnosis and treatment of pancreatic dysplasia in patients with a family history of pancreatic cancer. Ann Intern Med. 1999;131:247–55. doi: 10.7326/0003-4819-131-4-199908170-00003. [DOI] [PubMed] [Google Scholar]
- 3.Canto MI, Goggins M, Yeo CJ, et al. Screening for pancreatic neoplasia in high-risk individuals: an EUS-based approach. Clin Gastroenterol Hepatol. 2004;2:606–21. doi: 10.1016/s1542-3565(04)00244-7. [DOI] [PubMed] [Google Scholar]
- 4.Canto MI, Goggins M, Hruban RH, et al. Screening for early pancreatic neoplasia in high-risk individuals: a prospective controlled study. Clin Gastroenterol Hepatol. 2006;4:766–81. doi: 10.1016/j.cgh.2006.02.005. quiz 665. [DOI] [PubMed] [Google Scholar]
- 5.Poley JW, Kluijt I, Gouma DJ, et al. The yield of first-time endoscopic ultrasonography in screening individuals at a high risk of developing pancreatic cancer. Am J Gastroenterol. 2009;104:2175–81. doi: 10.1038/ajg.2009.276. [DOI] [PubMed] [Google Scholar]
- 6.Langer P, Kann PH, Fendrich V, et al. Five years of prospective screening of high-risk individuals from families with familial pancreatic cancer. Gut. 2009;58:1410–8. doi: 10.1136/gut.2008.171611. [DOI] [PubMed] [Google Scholar]
- 7.Verna EC, Hwang C, Stevens PD, et al. Pancreatic cancer screening in a prospective cohort of high-risk patients: a comprehensive strategy of imaging and genetics. Clin Cancer Res. 2010;16:5028–37. doi: 10.1158/1078-0432.CCR-09-3209. [DOI] [PubMed] [Google Scholar]
- 8.Ludwig E, Olson SH, Bayuga S, et al. Feasibility and yield of screening in relatives from familial pancreatic cancer families. Am J Gastroenterol. 2011;106:946–54. doi: 10.1038/ajg.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canto MI, Hruban RH, Fishman EK, et al. Frequent detection of pancreatic lesions in asymptomatic high-risk individuals. Gastroenterology. 2012;142:796–804. doi: 10.1053/j.gastro.2012.01.005. quiz e14–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Sukhni W, Borgida A, Rothenmund H, et al. Screening for pancreatic cancer in a high-risk cohort: an eight-year experience. J Gastrointest Surg. 2012;16:771–83. doi: 10.1007/s11605-011-1781-6. [DOI] [PubMed] [Google Scholar]
- 11.Brune K, Abe T, Canto M, et al. Multifocal neoplastic precursor lesions associated with lobular atrophy of the pancreas in patients having a strong family history of pancreatic cancer. Am J Surg Pathol. 2006;30:1067–76. [PMC free article] [PubMed] [Google Scholar]
- 12.Kanda M, Knight S, Topazian M, et al. Mutant GNAS detected in duodenal collections of secretin-stimulated pancreatic juice indicates the presence or emergence of pancreatic cysts. Gut. 2013;62:1024–33. doi: 10.1136/gutjnl-2012-302823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanda M, Matthaei H, Wu J, et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology. 2012;142:730–733. e9. doi: 10.1053/j.gastro.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanda M, Sadakari Y, Borges M, et al. Mutant TP53 in duodenal samples of pancreatic juice from patients with pancreatic cancer or high -grade dysplasia. Clin Gastroenterol Hepatol. 2013;11:719–30. e5. doi: 10.1016/j.cgh.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J, Matthaei H, Maitra A, et al. Recurrent GNAS Mutations Define an Unexpected Pathway for Pancreatic Cyst Development. Sci Transl Med. 2011;3:92ra66. doi: 10.1126/scitranslmed.3002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu J, Sadakari Y, Shindo K, et al. Digital next-generation sequencing identifies low-abundance mutations in pancreatic juice samples collected from the duodenum of patients with pancreatic cancer and intraductal papillary mucinous neoplasms. Gut. 2016 doi: 10.1136/gutjnl-2015-311166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eshleman JR, Norris AL, Sadakari Y, et al. KRAS and guanine nucleotide-binding protein mutations in pancreatic juice collected from the duodenum of patients at high risk for neoplasia undergoing endoscopic ultrasound. Clin Gastroenterol Hepatol. 2015;13:963–9. e4. doi: 10.1016/j.cgh.2014.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sadakari Y, Kanda M, Maitani K, et al. Mutant KRAS and GNAS DNA Concentrations in Secretin-Stimulated Pancreatic Fluid Collected from the Pancreatic Duct and the Duodenal Lumen. Clin Transl Gastroenterol. 2014;5:e62. doi: 10.1038/ctg.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Canto MI, Harinck F, Hruban RH, et al. International Cancer of the Pancreas Screening (CAPS) Consortium summit on the management of patients with increased risk for familial pancreatic cancer. Gut. 2013;62:339–47. doi: 10.1136/gutjnl-2012-303108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amato E, Molin MD, Mafficini A, et al. Targeted next-generation sequencing of cancer genes dissects the molecular profiles of intraductal papillary neoplasms of the pancreas. J Pathol. 2014;233:217–27. doi: 10.1002/path.4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDermott GP, Do D, Litterst CM, et al. Multiplexed target detection using DNA-binding dye chemistry in droplet digital PCR. Anal Chem. 2013;85:11619–27. doi: 10.1021/ac403061n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie 1. The “cap” method to collect secretin-stimulated pancreatic fluid samples.
Figure S1. Droplet digital PCR results of representative each target. Pink lines showed the thresholds of fluorescent value of FAM (vertical line, mutant type) and VIC (horizontal line, wild type).
Figure S2. Serial dilutions of positive controls were measured for each targets. Individual data point showed the mean value and error bar showed the standard deviation.
Table S1. Sequences of primers and probes for ddPCR