Abstract
Plants possess highly sensitive mechanisms that monitor environmental stress levels for a dose-dependent fine-tuning of their growth and development. Differences in plant responses to severe and mild abiotic stresses have been recognized. Although many studies have revealed that glutathione can contribute to plant tolerance to various environmental stresses, little is known about the relationship between glutathione and mild abiotic stress, especially the effect of stress-induced altered glutathione levels on the metabolism. Here, we applied a systems biology approach to identify key pathways involved in the gene-to-metabolite networks perturbed by low glutathione content under mild abiotic stress in Arabidopsis thaliana. We used glutathione synthesis mutants (cad2-1 and pad2-1) and plants overexpressing the gene encoding γ-glutamylcysteine synthetase, the first enzyme of the glutathione biosynthetic pathway. The plants were exposed to two mild stress conditions—oxidative stress elicited by methyl viologen and stress induced by the limited availability of phosphate. We observed that the mutants and transgenic plants showed similar shoot growth as that of the wild-type plants under mild abiotic stress. We then selected the synthesis mutants and performed multi-platform metabolomics and microarray experiments to evaluate the possible effects on the overall metabolome and the transcriptome. As a common oxidative stress response, several flavonoids that we assessed showed overaccumulation, whereas the mild phosphate stress resulted in increased levels of specific kaempferol- and quercetin-glycosides. Remarkably, in addition to a significant increased level of sugar, osmolytes, and lipids as mild oxidative stress-responsive metabolites, short-chain aliphatic glucosinolates over-accumulated in the mutants, whereas the level of long-chain aliphatic glucosinolates and specific lipids decreased. Coordinated gene expressions related to glucosinolate and flavonoid biosynthesis also supported the metabolite responses in the pad2-1 mutant. Our results suggest that glutathione synthesis mutants accelerate transcriptional regulatory networks to control the biosynthetic pathways involved in glutathione-independent scavenging metabolites, and that they might reconfigure the metabolic networks in primary and secondary metabolism, including lipids, glucosinolates, and flavonoids. This work provides a basis for the elucidation of the molecular mechanisms involved in the metabolic and transcriptional regulatory networks in response to combined low glutathione content with mild oxidative and nutrient stress in A. thaliana.
Keywords: glutathione, mild abiotic stress, oxidative stress, phosphorus limiting stress, metabolomics, transcriptomics
Introduction
Plants can respond to environmental changes and enhance their ability to tolerate biotic and abiotic stresses, including oxidative and nutrient stress (Mittler, 2002, 2006; Vinocur and Altman, 2005; Schachtman and Shin, 2007; Qin et al., 2011). Because reactive oxygen species (ROS) produced via plant metabolism under various stress conditions induce oxidative stress, plants need to reprogram and reconfigure their metabolism to survive (Moller et al., 2007; Baxter et al., 2014). Differences in plant responses to severe and mild abiotic stress have been documented (Kreps et al., 2002; Skirycz et al., 2010; Claeys and Inze, 2013; Dubois et al., 2013, 2015; Claeys et al., 2014). These differences suggest that plants possess highly sensitive systems that monitor environmental stress levels for a dose-dependent fine-tuning of their growth and biological processes in response to stress.
Glutathione is one of the important molecules that protect cells against ROS and maintain intracellular redox homeostasis (Marrs, 1996; Noctor et al., 2012). It has two main stable forms—thiol (GSH) and disulfide (GSSG)—and is synthesized from its constituting amino acids via two steps catalyzed by γ-glutamylcysteine synthetase (GSH1) and glutathione synthetase (GSH2) (Dixon et al., 2002). GSH1 is localized in plastids; GSH2 is primarily localized in the cytosol, but it can also be found in the chloroplast (Wachter et al., 2005). A complete loss-of-function of GSH1 results in embryo lethality (Cairns et al., 2006), whereas loss-of-function of GSH2 results in a seedling-lethality phenotype (Pasternak et al., 2008). Some mutants with defects in the GSH1 gene (Cobbett et al., 1998; Vernoux et al., 2000; Ball et al., 2004; Parisy et al., 2007) harbor nucleotide transitions or deletions in a region of the putative catalytic domain of GSH1 (Parisy et al., 2007). Compared to that in wild-type (WT; Columbia; Col-0) plants, two GSH1 mutants—cadmium-sensitive 2-1 (cad2-1) (Cobbett et al., 1998) and phytoalexin-deficient mutant 2-1 (pad2-1)—showed lower glutathione content (40 and 22%, respectively) (Parisy et al., 2007). The former mutant was found to be cadmium sensitive (Howden et al., 1995), whereas the latter was highly susceptible to Pseudomonas syringae and Phytophthora brassicae (Cobbett et al., 1998). The importance of glutathione in plant defense has been well documented (for example, see reviews by Noctor et al., 2012; Queval and Foyer, 2012).
In addition to the above central functions of glutathione, many studies have revealed that glutathione content can contribute to the promotion of tolerance to various environmental stresses (Noctor et al., 1998; Zhu et al., 1999; Gullner et al., 2001; Gomez et al., 2004; Liedschulte et al., 2010). Among the key glutathione-associated genes in Arabidopsis thaliana, glutathione-S-transferase (GST) plays an important role in the mechanisms underlying plant responses to abiotic stresses, including drought and salt (Ji et al., 2010; Qi et al., 2010; Jha et al., 2011; Xu et al., 2015), cold (Huang et al., 2009), and cadmium (Dixit et al., 2011). Other researchers (Chen et al., 2012; Cheng et al., 2015) determined whether altered glutathione levels affect the abiotic stress tolerance in Arabidopsis and found that an endo- and exogenous increase in glutathione levels in GST-knockout Arabidopsis elicited both drought and salt stress tolerance; pad2-1 exhibited a survival rate of approximately 50% under stress conditions. In addition to the important role of phytohormones such as salicylic acid (SA), jasmonic acid (JA), and ethylene in plant defense responses, the relationship between glutathione and phytohormones has been considered to play a pivotal role during abiotic stress. Combined transcriptome and proteome analysis of pad2-1 subjected to combined osmotic and cold stress revealed that glutathione confers stress tolerance to plants via a process associated with lignin, phenylpropanoid, and ethylene biosynthesis (Kumar et al., 2015). Subsequent studies showed that glutathione induces the transcription of genes associated with ethylene biosynthesis in a WRKY33-dependent manner (Datta et al., 2015). Comparative transcriptome and proteome analyses by using ethylene-insensitive, abscisic acid (ABA), and glutathione mutants suggested a crosstalk among ethylene, ABA, and glutathione in inducing stress-responsive genes and proteins to mitigate osmotic and cold stress in Arabidopsis (Kumar et al., 2016). However, little is known about the relationship between glutathione and mild abiotic stress, especially nutrient stress. Furthermore, the effect of stress-induced altered glutathione levels on global primary and secondary metabolism has not been studied.
In this study, we applied a systems biology approach to identify key pathways involved in the gene-to-metabolite networks affected by low glutathione content under mild abiotic stress in Arabidopsis. We used GSH1 mutants (cad2-1 and pad2-1) and GSH1-overexpressing plants and exposed them to two conditions—mild oxidative stress elicited by methyl viologen (MV) and mild stress induced by the limited availability of phosphate (P-lim). We observed no severe visual phenotypes (e.g., chlorosis) in the assayed plants and found that glutathione synthesis mutants and the transgenic plants showed growth similar to that of Col-0 plants under the two mild abiotic stresses. Metabolite and transcript profiling and glutathione quantification showed that the GSH1 mutants exposed to MV-induced oxidative and P-lim stresses survived by modulating their metabolic and transcriptional networks associated with secondary metabolism.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis ecotype Columbia (Col-0) was used. The mutant cad2-1 (Cobbett et al., 1998) was used as an allelic mutant of pad2-1 (Parisy et al., 2007). Further, 35S::GSH1 transgenic plants, 7-5 and 13-6 (Cheng et al., 2015), were also used to evaluate the shoot phenotypic changes. Plants other than those exposed to MV stress and P-lim were grown in Murashige and Skoog agar medium. Sterilized seeds were stratified at 5°C for 2 days and then sown on Murashige and Skoog medium containing 1% sucrose. Oxidative stress was produced by adding 0.05 μM MV to the Murashige and Skoog medium. The low phosphorus condition was created using P-lim medium in which the phosphate concentration was 20% of that in the Murashige and Skoog medium. Seedlings of Arabidopsis Col-0 and mutants were cultivated in growth chambers at 22°C under 16-h light/8-h dark conditions for 18 days (light strength, 80 μmol⋅m-2⋅s-1 of the photosynthetic photon flux). GSH1-overexpression lines were cultivated under the same condition for 20 days.
Metabolite Profiling
Harvested aerial parts of WT and mutant plants (n = 8, biological replicates) were frozen immediately in liquid nitrogen to quench enzymatic activity. Primary and secondary metabolites were extracted according to previously established methods; we performed GC-TOF-MS analysis (Kusano et al., 2007a,b) for primary metabolites and LC-q-TOF-MS analysis for secondary metabolites (Matsuda et al., 2009; Nakabayashi et al., 2014) and lipids (Kimbara et al., 2013). The details of metabolite profiling are shown in Supplementary Document S1. Principal component analysis (PCA) was performed using SIMCA-P 12.0 (UmetricsAB1) with log10 transformation and autoscaling. For stress treatment- and genotype comparison, differentially abundant metabolites were identified using linear regression models in the LIMMA method (Smyth, 2005), which yields false discovery rate (FDR)-adjusted p-values for multiple testing problems (Benjamini and Hochberg, 1995). The significance level was set at FDR < 0.05. Our data are reported in a manner compliant with the guidelines recommended by Fernie et al. (2011), as shown in Supplementary Table S4. VENNY2 was used to generate the Venn diagram.
Glutathione Quantification
Plant samples (n = 3, biological replicates) were frozen immediately in liquid nitrogen and stored at -80°C until analysis. Each frozen sample was extracted with a 20-fold amount of solvent [methanol/water (8:2 v/v)]. Aerial parts were mashed for 6 min in a 2-mL tube by using a mixer mill (MM400; Retsch, Haan, Germany) at a frequency of 20 Hz. The mixture was centrifuged at 15,000 rpm, and then supernatant containing glutathione was taken out. The supernatant was used for the quantification of GSH and GSSG by using ultra high performance liquid chromatography (UHPLC-MS; Nexera, Shimadzu, Kyoto, Japan) coupled with a triple quadrupole mass spectrometer (TSQ Quantum Ultra Thermo; Fisher Scientific, San Jose, CA, United States). UHPLC separation was performed on an Acquity UPLC BEH C18 column (50 mm × 2.1 mm, 1.7 μm particle size; Waters) maintained at 40°C. Other LC conditions were as follows: flow rate, 0.4 mL/min; injection volume, 5 μL (GSH) and 20 μL (GSSG); solvent system, acetonitrile (0.1% formic acid):water (0.1% formic acid); and gradient program, 2:98 v/v at 0–3 min, 98:2 at 7–10 min. For mass spectrometry (MS) detection, heated-electrospray ionization was used as the ionization source in the positive mode. Other conditions were as follows: sheath gas pressure, 50 arbitrary units; ion sweep cone gas, 0 arbitrary units; vaporizer temperature, 450°C; aux gas pressure, 20 arbitrary units; and spray voltage, 3,000 V. Selected reaction monitoring (SRM) was used to quantify GSH and GSSG. SRM was conducted by scanning the product ions at m/z 162.070 obtained from the fragmentation of the parent ions at m/z 308.167 of GSH. SRM was conducted by scanning the product ions at m/z 231.010 obtained from the fragmentation of the parent ions at m/z 613.244 of GSSG. The collision energy for MS/MS was 27 V. Identities were confirmed by comparing the MS/MS spectra with authentic standards [reduced glutathione (GSH, 97.0%) and oxidized glutathione (GSSG, 98.0%)] purchased from Tokyo Chemical Industry (Tokyo, Japan). Statistical data analysis and plotting were performed using Microsoft Excel and the unpaired Welch’s t-test by using the R-function t.test().3
RNA Isolation, Microarray Hybridization, and Data Analysis
We performed analysis of mRNA as described previously (Kusano et al., 2011). Briefly, total RNA was extracted from the 18-day-old aerial part of each mutant and WT sample using the RNeasy plant mini kit (Qiagen4) according to the manufacturer’s instructions. Three independent hybridizations were performed using the Affymetrix ATH1 GeneChip microarray, according to the manufacturer’s instructions (Affymetrix).5 A single biological replicate was used for each hybridization. Preprocessing and normalization/summarization of all CEL files were performed using R, the Bioconductor (Gentleman et al., 2004), and a robust multi-chip average (RMA) (Bolstad et al., 2003; Irizarry et al., 2003). The quality of the GeneChip data was assessed using the AFFYPLM package (Bolstad et al., 2005). The annotation of each gene in the CSV file ATH1-121501.na31.annot.csv (downloaded in April 2012)6 released by Affymetrix was used. For stress treatment- and genotype comparison, differentially expressed genes were detected using linear regression models in the LIMMA method (Smyth, 2005), which provides FDR-adjusted p-values for multiple testing problems (Benjamini and Hochberg, 1995). The significance level was set at FDR < 0.05. Genevenn7 was used to generate the Venn diagram.
Functional Enrichment Analysis
The transcript profiles were visualized using MAPMAN (v3.5.1R2) (Thimm et al., 2004) to inspect coordinated changes in metabolism and other biological process. GO enrichment analysis was performed using Cytoscape (v3.2.1) App BiNGO (Maere et al., 2005). The resulting enriched GO categories were visualized using the Enrichment Map (v2.2.0) (Merico et al., 2010; Isserlin et al., 2014) and AutoAnnotate (Kucera et al., 2016).
Results
GSH1 Mutants and 35S::GSH1 Transgenic Plants Showed Similar Growth to That of WT Plants under Mild Abiotic Stress Conditions
Previously, we reported that, under no-stress condition, the shoot phenotype of the pad2-1 mutant was similar to that of WT plants (background, Col-0), but showed significant changes in the levels of primary metabolites (Fukushima et al., 2014) (Supplementary Figure S1 and Table S1). To further understand the relationship between the GSH1 mutation related to glutathione content and mild abiotic stress, we selected two GSH1 mutants, pad2-1 and cad2-1, and two GSH1-overexpressing plants, 7-5 and 13-6 (Cheng et al., 2015). We first assessed the phenotypic changes of individual WT plants and the GSH1 mutants under different concentrations of MV (Supplementary Figure S2). Our current experimental setup was based on these pilot experiments and findings from MV treatment to isolate MV-resistant mutants in the previous studies [for example, see (Fujibe et al., 2004; Sukrong et al., 2012; Egert et al., 2013)]. GSH1 mutants and GSH1-overexpressing plants were grown on untreated medium (control condition), on medium containing a low concentration (0.05 μM) of MV compared to that used in previous studies [e.g., 10 μM MV treatment in AtGenExpress (Kilian et al., 2007)], or on P-lim medium that had a phosphate concentration of 20% of that in the medium (Supplementary Figure S3). We evaluated the shoot phenotypic changes in the GSH1 mutants (Figure 1) and GSH1-overexpressing plants (Supplementary Figure S4). No plants exhibited chlorosis and/or necrosis attributable to treatment with the low MV medium (Figure 1A and Supplementary Figure S4A). MV treatment of the mutants and WT plants resulted in an approximately 50% growth reduction (Figure 1B). The fresh weight of the aerial parts of the mutants grown under control- and P-lim conditions was approximately 70% of that of the WT plants. These findings suggest that, under the no-stress condition, the GSH1 mutations were silent at the aerial parts, although the mutants were shown to have impaired lateral root density (Schnaubelt et al., 2013, 2015). Therefore, since the applied stress treatments did not result in severe growth inhibition in the assayed plants, we considered them as mild stress treatments (Figures 1A,B and Supplementary Figure S2).
FIGURE 1.
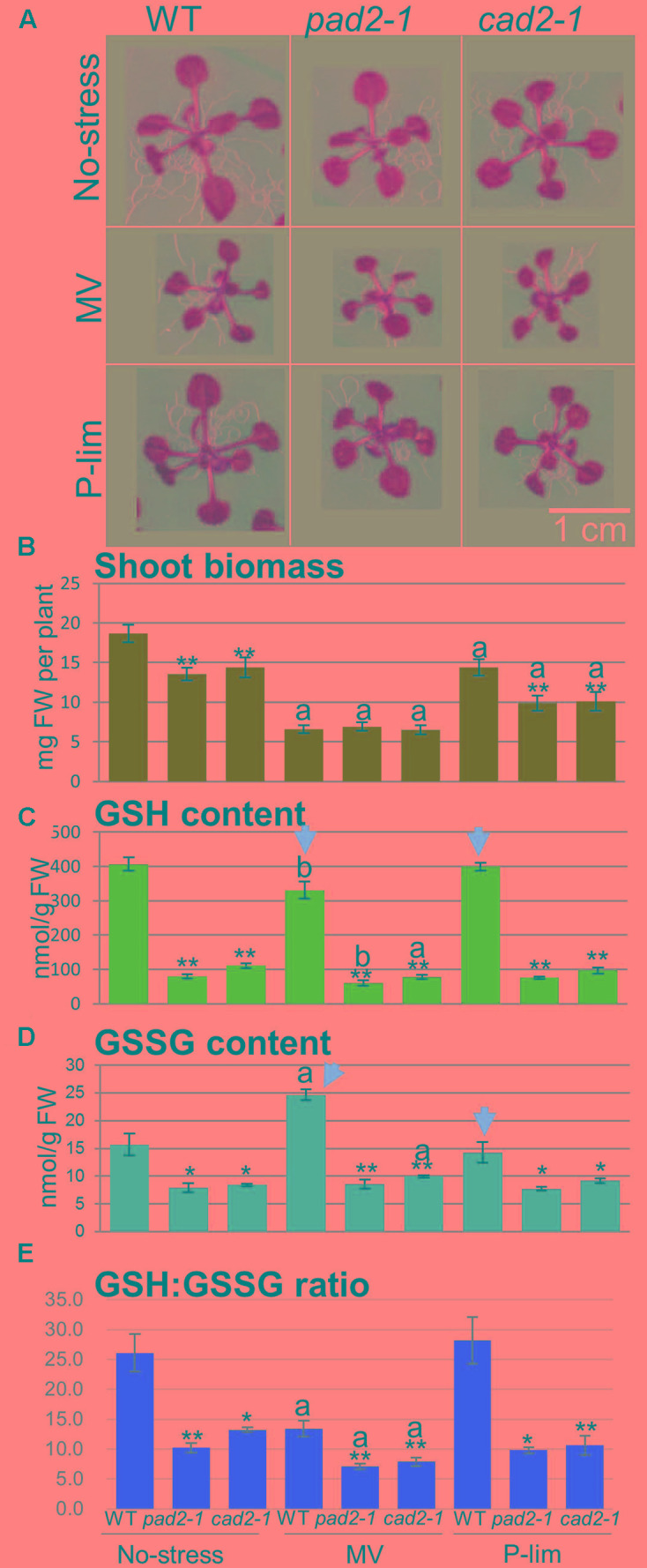
Visual phenotypes, fresh weight of shoots, and GSH and GSSG levels in WT plants and the allelic mutants of GSH1 (cad2-1 and pad2-1) grown under three different conditions. Visible phenotypic changes in 18-day-old GSH1 mutants grown on Murashige and Skoog medium (no-stress), Murashige and Skoog medium containing 0.05 μM methyl viologen (MV), and low-phosphorus (P-lim) Murashige and Skoog medium (A). Fresh weight (FW) of aerial parts of WT plants and GSH1 mutants grown under the three conditions (n = 20, biological replicates) (B). Quantification of the glutathione content in WT plants and GSH1 mutants grown under the three growth conditions. Content of the reduced form of glutathione, GSH (n = 3, biological replicates) (C). Content of the oxidized form of glutathione, GSSG (n = 3, biological replicates) (D). GSH:GSSG ratio as an indirect determinant of oxidative stress (E). Each error bar indicates the standard deviation from the mean. Asterisks represent differences from the control (significant levels were ∗∗ α = 0.01 and ∗ α = 0.05) by Welch’s t-test. The letters “a” and “b” represent significant differences compared to that under the no-stress condition (significant levels were a, α = 0.01 and b, α = 0.05) by Welch’s t-test.
The GSH and GSSG contents were lower in the mutant than in the WT plants under the no-stress condition (GSH, approximately 20%; GSSG, approximately 50%). Changes in GSH and GSSG contents differed with stress conditions (Figures 1C,D). The level of GSH was decreased significantly in WT plants subjected to MV stress, whereas that of GSSG was increased (arrows in Figures 1C,D); the GSH and GSSG contents were almost identical under the P-lim and control conditions. The two GSH1-overexpressing plants manifested no changes at the phenotypic level under the no-stress condition (Supplementary Figure S4A). The fresh weight of the shoot biomass of the GSH1 mutants, WT plants, and transgenic lines grown under the two mild stress conditions showed similar trends (Supplementary Figure S4B). Increased levels of GSH and GSSG failed to rescue the shoot biomass of the GSH1-overexpressing plants under the MV condition (Supplementary Figures S4C,D). The GSH:GSSG ratio can be regarded as an indirect determinant of oxidative stress; the GSSG level increases and the GSH:GSSG ratio decreases, when the plant cells are exposed to oxidative stress (Queval et al., 2007; Noctor et al., 2012). Under the MV and P-lim conditions, the measured GSH:GSSG ratio was lower in the two GSH1 mutants than in the WT plants (Figure 1E). The GSH:GSSG ratio was largely unaltered in the transgenic lines (Supplementary Figure S4E). On the other hand, the overexpression transgenic plants showed high GSH:GSSG ratio under P-lim conditions. Taken together, our findings suggest that, under both mild abiotic conditions, neither low nor high concentrations of glutathione elicited critical changes in the shoot growth and biomass of the examined plants. In our subsequent analysis, we focused on the GSH1 mutants to avoid pleiotropic effects from 35S-mediated overexpression.
Environmental and Genetic Perturbation in the Metabolic Pathways of the GSH1 Mutants under Mild Abiotic Stresses
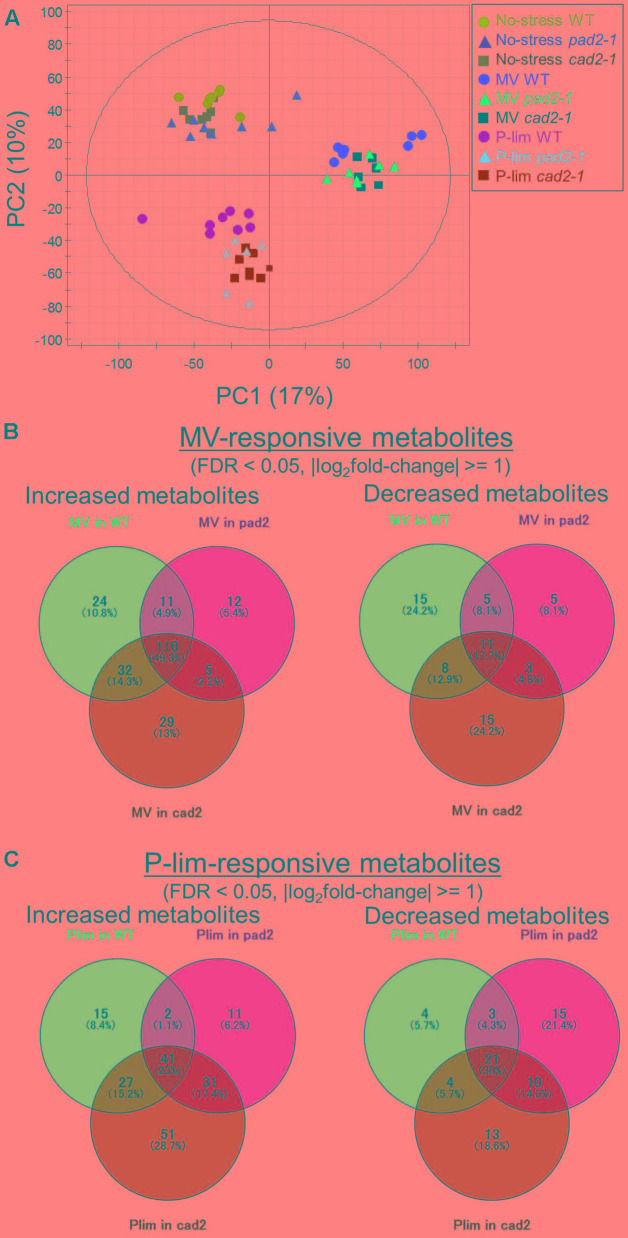
To assess the wide range of metabolic impacts attributed to genotypic differences and types of mild abiotic stresses, we performed multi-platform metabolite profiling (Supplementary Table S2). For a visual inspection of the extent of metabolomic changes under the MV and P-lim stress conditions, we subjected the metabolite profile data to PCA. On the PCA score scatter plot (Figure 2A), the samples were clustered according to the applied abiotic stress conditions, as evidenced by the separation of PC1. We observed a genotype-dependent separation in the score scatter plot in the PC1/PC3 or PC2/PC3 direction (Supplementary Figure S5). These results indicate that global perturbations due to abiotic stress strongly affected the changes in the metabolite level of the mutant and WT plants.
FIGURE 2.
Summary of metabolite profiling conducted in this study. Principal component analysis (PCA) of the metabolite profiles of WT plants and mutants, cad2-1 and pad2-1, exposed to MV and P-lim treatment; integrated data obtained from multi-platform metabolite profiling were used (n = 8, biological replicates). The PCA score scatter plot shows that the samples were clustered according to the corresponding abiotic conditions as evidenced by the separation of PC1 (A). Venn diagram of MV- (B) and P-lim-responsive (known) metabolites (C). FDR, false discovery rate.
We compared the metabolic responses in the metabolite profiles of the WT plants and GSH1 mutants grown under the no-stress condition or exposed to MV or P-lim stress. Under MV-induced oxidative stress, a total of 285 significantly changed metabolites was identified (Figure 2B), of which 223 were increased and 62 were decreased, respectively, including the commonly and exclusively accumulated metabolites. In all, 110 metabolite levels increased in the WT plant and GSH1 mutants after MV treatment, whereas 11 metabolite levels decreased (Figure 2B). For P-lim, we identified 178 increased and 70 decreased metabolites, respectively. The levels of 41 metabolites increased in the 3 genotypes under P-lim condition, whereas 21 metabolite levels decreased (Figure 2C). The number of significantly altered metabolites between GSH1 mutants and WT plants under mild abiotic stress was smaller than that of the treatment (Supplementary Figure S6), indicating consistency with the results of PCA score plots.
MV-Responsive Metabolites
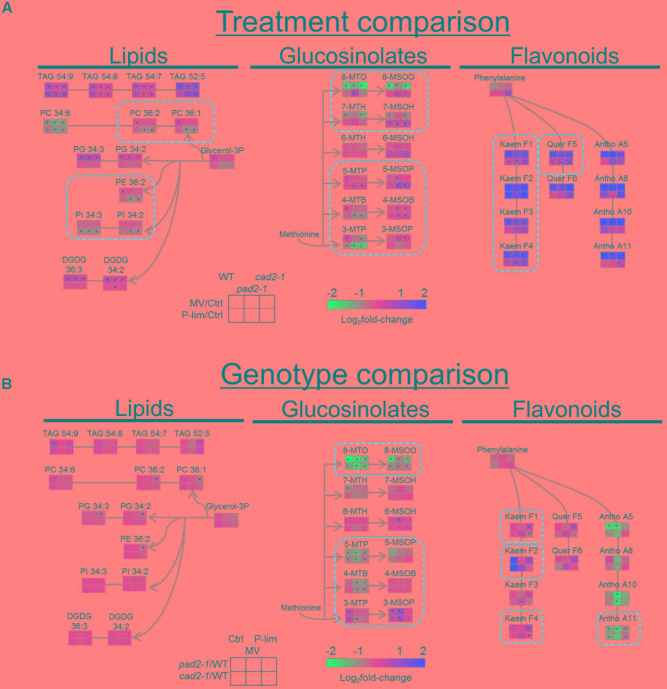
The differentially accumulated metabolites involved in primary and secondary metabolism were visualized on a simplified metabolic map (Supplementary Figure S7). Stress-specific metabolite alterations against oxidative stress were significantly increased, for example, γ-amino butyrate (GABA), 2-oxo glutarate, proline, and putrescine. MV treatment resulted in a significant increase in the level of sugar and osmolytes. Sucrose, glucose, fructose, xylose, maltose, arabinose, trehalose, raffinose, inositol, and galactinol were observed in all the samples (Supplementary Figure S7A). Notable changes were observed in metabolite levels associated with secondary metabolism in the GSH1 mutants and WT plants by MV and P-lim treatments (Figure 3). Triacylglycerols (TAG) increased under almost all conditions, whereas phosphatidylcholine (PC) 34:6 decreased under all conditions. The phospholipids, 34:2 and 34:3 phosphatidylglycerols (PG) and 34:2 and 36:3 digalactosyldiacylglycerols (DGDG), increased after MV treatment. As for glucosinolates, the level of long-chain aliphatic glucosinolates [8-MTO (8-methylthio-n-octylglucosinolate) and 8-MSOO (8-methylsulfinyl-n-octylglucosinolate)] was remarkably decreased after MV treatment. Antioxidants, including flavonols and anthocyanins except kaempferol (F2), were significantly increased across the GSH1 mutants after MV treatment.
FIGURE 3.
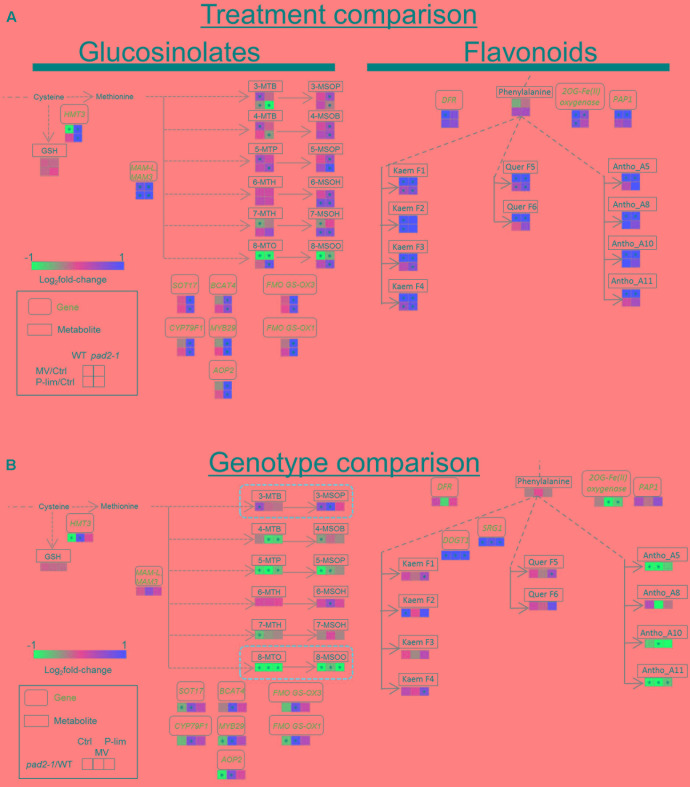
Changes in metabolite levels associated with secondary metabolism in the GSH1 mutants and WT after MV and P-lim treatments. (A) Stress treatment comparison: Log2fold-change in metabolites exposed to the two abiotic stresses. The fold-change is represented by two directed colors; red and blue indicate increase and decrease, respectively, in the metabolite level elicited by MV treatment or P-lim. (B) Genotype-dependent comparison: Red and blue indicate increase and decrease, respectively, in the metabolite levels in the mutants vs. WT plants. For abbreviations of the metabolite names, see Supplementary Table S5. n = 8, biological replicates, ∗FDR < 0.05.
Low Phosphate-Responsive Metabolites
In addition to lowered glucose-6-phosphate and fructose-6-phosphate levels under P-lim condition in both the WT and the mutants, several low phosphate-responsive meta-bolites were identified. Examples include the phospholipids, 36:1 and 36:2 PCs, 36:2 phosphatidylethanolamine (PE), and 34:2 and 34:3 phosphatidylinositol (PI), decreased in the GSH1 mutants under P-lim condition. In the glucosinolate biosynthesis, the level of aliphatic glucosinolates, 4-MSOB (4-methylsulfinyl-n-butylglucosinolate), 5-MTP (5-methylthio-n-pentylglucosinolate), 5-MSOP (5-methylsulfinyl-n-pentylglucosinolate), 7-MSOH (7-methylsulfinyl-n-heptylglucosinolate), and 8-MSOO (8-methylsulfinyl-n-octylglucosinolate), increased in the GSH1 mutants under P-lim condition (Figure 3A). In contrast, P-lim stress elicited a significant decrease in 3-MTP (3-methylthio-n-propylglucosinolate) in all genotypes/conditions. P-lim stress also resulted in moderate increases of detected flavonoids with kaempferol aglycone (F1, F2, F3, and F4) and that of quercetin (F5) in the GSH1 mutants. There was also opposite changes in dihydrouracil levels in a genotype/stress dependent manner. Other metabolites such as the monosaccharides xylose and arabinose and the disaccharide maltose, tryptophan, octadecatrienoate, fumarate, and threonine were also increased under P-lim condition (Supplementary Figure S7A).
GSH1 Mutant-Specific Changes in Metabolite Levels under Mild Abiotic Stress
A genotype-dependent comparison of the metabolite profiles of WT plants and GSH1 mutants grown under abiotic stress conditions is shown in Figure 3B. Compared to that of WT plants, the metabolite profile of each mutant allele under both stress conditions exhibited a similar trend in metabolic responses. Metabolite changes in the biosynthetic pathways of glucosinolates and anthocyanins in the pad2-1 and cad2-1 mutants were noted; the flavonol level tended to increase and the level of anthocyanins was remarkably decreased under both the stress conditions. The level of low-phosphate responsive kaempferol-type flavonols, F1 and F4, was higher in the GSH1 mutants than in the WT plants under P-lim condition. In the mutants subjected to no-stress and MV conditions, short-chain glucosinolates, 3-MTP and 3-MSOP (3-methylsulfinyl-n-propylglucosinolate), were increased and long-chain glucosinolates, 8-MTO and 8-MSOO, were decreased. The level of metabolites [e.g., 8-MSOO and A11, one of the major anthocyanin that contains three acyl groups, namely cyanidin-3-O-[2-O-(2-O-(sinapoyl)-xylosyl)-6-O-(p-O-coumaroyl)-glucoside]-5-O-[6-O-(malonyl)-glucoside (Tohge et al., 2005)] downstream of glucosinolate and anthocyanin pathways was decreased.
Genome-Wide Expression Changes in Response to Mild Abiotic Stress
Functional Enrichment Analysis
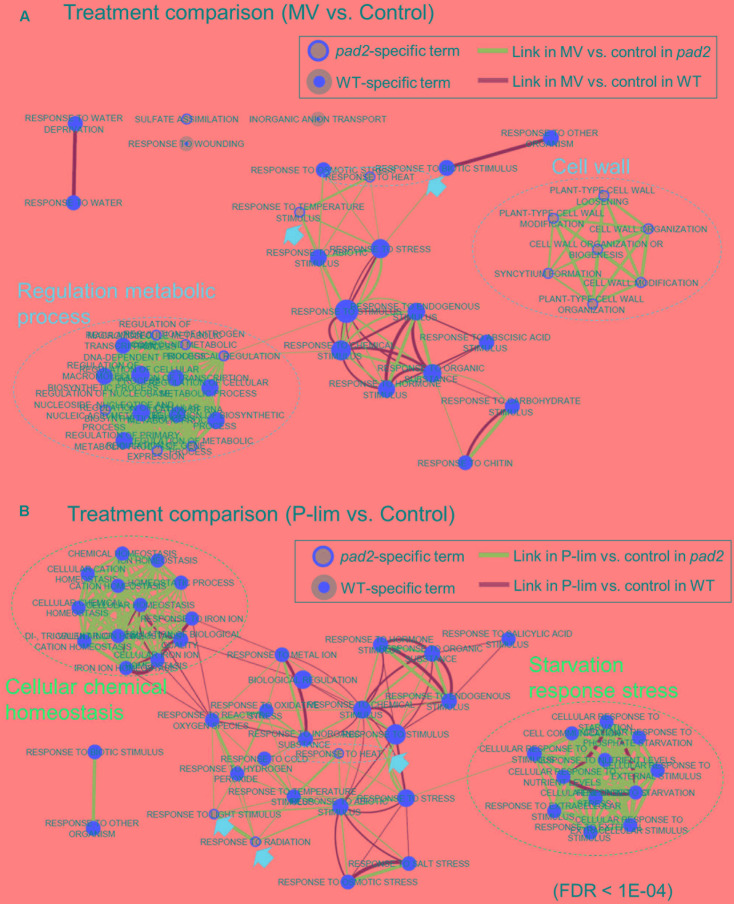
For a better understanding of the metabolic responses to the imposed stress conditions, we performed transcript profiling by using microarray analysis. In this experiment, we focused on pad2-1 as a representative GSH1 allele in transcript profiling (Supplementary Table S3). To characterize the gene expression patterns of differentially expressed genes (DEGs) in the WT and pad2-1 plants subjected to MV or P-lim stress conditions, we performed gene ontology (GO) enrichment analysis. By using Enrichment Map (Merico et al., 2010; Isserlin et al., 2014), we revealed biological processes between stress treatment and no-stress condition based on hypergeometric tests of GO terms (Figure 4). The enriched GO terms in MV-treated vs. no-stress condition for each genotype are shown in Figure 4A. We also created an enrichment map showing biological processes between pad2 and WT under two differential conditions based on hypergeometric tests of GO term enrichment analysis (Supplementary Figure S8). We identified GO terms “sulfate assimilation,” “response to heat,” and “response to temperature stimulus” as well as a cluster-comprising terms related to “cell wall” and partially related to “regulation metabolic process” as pad2-1-specific expression responses to MV treatment (Figure 4A). For the enriched GO terms in P-lim vs. no-stress, GO terms “response to heat,” “response to light stimulus,” and “response to radiation” were enriched in only pad2-1 (Figure 4B). To inspect the expression level of genes in the primary and secondary metabolism, we also performed MAPMAN (Thimm et al., 2004) analysis. For metabolism, remarkable changes were noted for many transcripts directly or indirectly involved in hormone, secondary, and cell wall metabolism (Supplementary Figures S9–S11). Coordinated induction of genes for ABA and JA metabolism was noted in the P-lim treated pad2-1. For secondary metabolism, we identified coordinated expression for glucosinolate metabolism as pad2-1-specific response to both mild abiotic stresses. In addition, there were coordinated changes in transcript levels associated with phenylpropanoids and phenolics as a pad2-1-specific response to P-lim. Cell wall modification and pectin esterases were identified as coordinated pathways in only WT plants under P-lim condition.
FIGURE 4.
Enrichment map showing biological processes between stress treatment and no-stress condition based on hypergeometric tests of gene ontology (GO) terms by using BiNGO software (Maere et al., 2005). The map shows the enriched GO terms in MV-treated vs. no-stress condition (A) and in P-lim vs. no-stress (B). Enrichment map can be used to compare differential transcriptomic responses to abiotic stress between 2 genotypes (i.e., pad2-1 vs. WT). Nodes represent GO terms, and links between nodes represent gene overlap between GO terms, resulting in a rapid identification of the major enriched functional categories. Inner circle size of each node represents the number of DEGs in “comparison 1” (e.g., MV vs. no-stress in WT) within the GO term in a biological process. Node border size represents the number of DEGs in “comparison 2” (e.g., MV vs. no-stress in pad2) within the GO term in a biological process. Color of the node and border indicate significance based on the BiNGO FDR of the GO term for “comparison 1” and “comparison 2,” respectively. The red-filled nodes highlight the major GO functional terms. Link size shows the number of DEGs that overlap between the two connected GO terms (Jaccard coefficient, the cut-off is 0.25). Green links correspond to both datasets when it is the only colored link. Green links indicate “comparison 1” and blue indicates “comparison 2.” Blue dotted circles represent summarized GO term clusters based on AutoAnnotate (Kucera et al., 2016). The maps were generated using Cytoscape (v3.2.1) Enrichment Map plugin (Merico et al., 2010; Isserlin et al., 2014).
Mild Oxidative- and Low Phosphate-responsive Genes
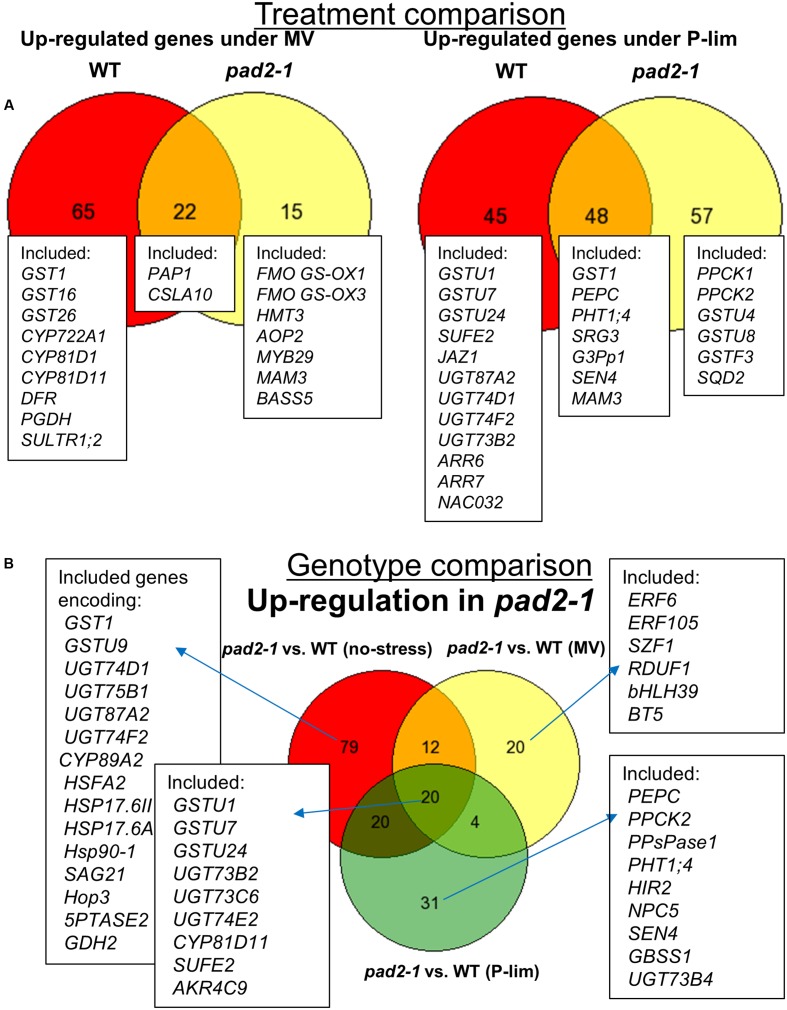
Methyl viologen treatment resulted in 87 (WT plants) and 37 (pad2-1) MV-inducible genes (Figure 5A, left). Genes down-regulated by genetic and environmental perturbations are shown in Venn diagrams that represent the classification of genes (Supplementary Figure S12). The stress-inducible genes included those encoding GST, UDP-glycosyl transferase (UGT), and cytochrome P450 (CYP), which are associated with detoxification and transcriptome responses when plants are exposed to oxidative stress (Dixon et al., 2002). In addition, genes related to flavonoids [e.g., PRODUCTION OF ANTHOCYANIN PIGMENT 1 (PAP1)] and glucosinolates [e.g., MYB DOMAIN PROTEIN 29 (MYB29)/PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 (PMG2)] were up-regulated.
FIGURE 5.
Venn diagrams representing the classification of genes based on microarrays. (A) We identified 87 MV-inducible genes in WT plants and 37 MV-inducible genes in the pad2-1 mutant. We also identified 93 low-P-inducible and 105 low-P-inducible genes in WT plants and the pad2-1 mutant, respectively. (B) Genotype-dependent up-regulated genes. Under the control condition, MV-treatment, and low P stress condition, we identified 131, 56, and 75 up-regulated genes, respectively, in pad2-1. We used genevenn (http://genevenn.sourceforge.net/vennresults.php) to draw the diagrams. Differentially expressed genes were identified using a threshold |log2 fold-change| ≥ 1 and FDR < 0.05.
P-lim condition resulted in 93 (WT) and 105 (pad2-1) low phosphorus-inducible genes (Figure 5A, right). The inducible genes included SPX1, phosphate starvation-induced genes (PS2 and PS3), phosphate transporter 1;4 (PHT1;4), and PHOSPHOCHOLINE PHOSPHATASE 1 (PEPC1), which are known to be up-regulated during phosphorus deprivation (Morcuende et al., 2007). As pad2-1-specific transcriptional responses, genes encoding phosphoenolpyruvate carboxylase kinase (PPCK1 and PPCK2), glutathione transferases (e.g., GSTU4, GSTU8, GSTF3), and lipid biosynthesis enzymes [e.g., SULFOQUINOVOSYLDIACYLGLYCEROL 2 (SQD2)] were up-regulated.
We also identified genotype-dependent altered genes. Under the no-stress condition, 131 up-regulated genes were identified in pad2-1; their number was 56 after MV treatment and 75 after exposure to low P-stress (Figure 5B). Several pad2-1-specific expression changes were noted under the different environmental conditions in genes encoding GSTs, UGTs, CYPs, heat shock proteins (HSPs), senescence-associated genes (SAGs), and transcription factors such as ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 6 (ERF6) and HEAT SHOCK TRANSCRIPTION FACTOR A2 (HSFA2).
Transcript Changes in Mild Stress-Responsive Genes and Well-Characterized Stress-Responsive Factors
Among the DEGs, we focused on “marker genes” that are highly induced by stress treatments (Claeys et al., 2014). They include marker genes elicited under oxidative stress [NAC DOMAIN-CONTAINING PROTEIN32 (NAC032) and ALDO-KETO REDUCTASE 4C9 (AKR4C9)], those induced by abscisic acid (ABA) [NINE-CIS-EPOXYCAROTENOID DIOXYGENASE3 (NCED3) and CYP707A3] and dehydration [LATE EMBRYOGENESIS ABUNDANT-LIKE5 (LEA5), also known as SAG21], mild osmotic stress markers [MYB51, ERF5, and WRKY DNA-BINDING PROTEIN33 (WRKY33)], and genes overexpressed in response to phosphate starvation [SQD1 and SQD2] (Table 1). We found that the expression of the oxidative stress markers NAC032 and AKR4C9 was remarkably up-regulated in pad2-1 grown under MV stress and control conditions. Under P-lim stress, NAC032 and AKR4C9 genes were up-regulated in WT plants; genotype comparison showed the up-regulation of the AKR4C9 gene in pad2-1 plants. Marker gene expressions associated with ABA and mild osmotic stress were largely unaltered except for LEA5/SAG21 (up-regulated in pad2-1 under control conditions) and WRKY33 (down-regulated in pad2-1 by MV treatment). With respect to stress marker responses to phosphate starvation, both SQD1 and SQD2 genes were up-regulated in pad2-1 under P-lim stress; the SQD1 gene was only up-regulated in WT plants.
Table 1.
Transcriptional changes in well-characterized stress-responsive factors in Arabidopsis.
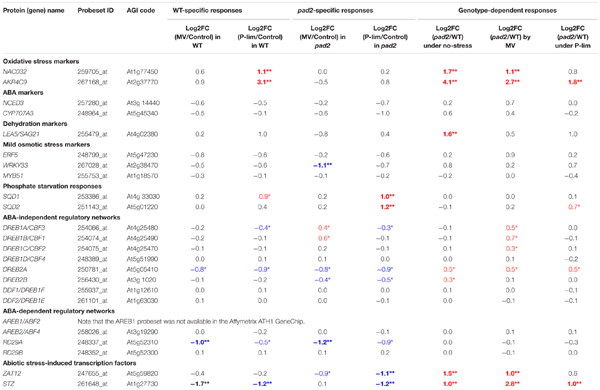 |
A single asterisk (∗) indicates significance (FDR < 0.05). Bold numbers with double asterisks (∗∗) identify a |log2 fold-change|≥ 1.0 and FDR < 0.05.
To assess the changes in other stress-responsive genes, including well-known transcription factors, in the WT plants and the GSH1 mutants exposed to abiotic stress, we investigated the general patterns of gene expression associated with DREB/CBF (dehydration-responsive-element binding protein/C-repeat-binding factor) and AREB/ABF (ABA-responsive element-binding/ABA-responsive element-binding factor) protein. They play a crucial role in the adaptation to various stresses (Yamaguchi-Shinozaki and Shinozaki, 2006). Among DREB/CBF and AREB/ABF genes, the transcript level of DREB2A was down-regulated (log2 fold-change, approximately -1) in the WT and pad2-1 plants grown under both stress conditions. The expression of DREB2A was gradually induced by H2O2 (Sakuma et al., 2006). Under the MV condition, the mutants showed slight up-regulation of DREB1A/CBF3, DREB1C/CBF2, and DREB2A. The expression of AREB2/ABF4 in the WT and pad2-1 plants was unchanged under the three growth conditions. An abiotic stress-responsive gene in Arabidopsis is Responsive to Desiccation (RD) (Yamaguchi-Shinozaki and Shinozaki, 1993). Both WT and pad2-1 plants showed down-regulation of RD29A under both stress conditions; the expression of RD29B in WT and pad2-1 plants was unaltered under the three conditions (Table 1). The expression of two genes encoding ZAT12 (Davletova et al., 2005; Vogel et al., 2005) and STZ (Sakamoto et al., 2004) was markedly higher in the MV-treated pad2-1 than in WT plants (Table 1).
Integrated Pathway Analysis Reveals Specific Changes in the Biosynthesis of Aliphatic Glucosinolates and Anthocyanins at the Metabolite and Transcript Levels in Mild Stress-Treated pad2-1 Mutants
To assess the coordinated responses of glucosinolate and flavonoid biosynthesis under mild stress conditions, we performed pathway analysis of combined metabolite and transcript profile data. The integrated metabolomic and transcriptomic responses in WT and pad2-1 plants under the two stress conditions are shown in Figure 6. As pad2-1-specific metabolite responses, the level of short-chain aliphatic methylsulphinylalkyl glucosinolates [4-MSOB, and 5-MSOP] and 6-MSOH (6-methylsulfinyl-n-hexylglucosinolate) was increased in pad2-1 under both stress conditions; the difference was a significant but moderate effect size (all the ranges from 0.39 to 0.97 in log2 fold-change) (Figure 6A, left). The level of 8-MTO and 8-MSOO was decreased by MV treatment, irrespective of the genotype (all the ranges from -1.37 to -0.56 in log2 fold-change). The level of 5-MTP, 7-MSOH, 8-MTO, and 8-MSOO was increased in pad2-1 under P-lim condition (all the ranges from 0.45 to 0.82 in log2 fold-change). Gene expressions associated with glucosinolate biosynthesis were up-regulated in pad2-1 grown under abiotic stress conditions. These included some genes encoding methylthioalkylmalate synthase (MAM) (Textor et al., 2007), MYB29 (Hirai et al., 2007; Sonderby et al., 2007; Gigolashvili et al., 2008), branched chain aminotransferase (BCAT) (Schuster et al., 2006), flavin-containing monooxygenase (FMO) (Hansen et al., 2007), and other genes involved in glucosinolate biosynthesis. Under the MV condition, the transcript levels of these genes were significantly higher in pad2-1 mutants than in WT plants, except for the MAM gene (Figure 6B, left). Compared to that in WT plants, the level of 8-MSOO was decreased in pad2-1 mutants under all conditions, whereas that of 3-MSOP was increased under no-stress and MV conditions.
FIGURE 6.
Integrated metabolomic and transcriptomic responses in the glucosinolate and flavonoid pathways of WT plants and pad2-1 mutant exposed to MV and P-lim stress. (A) Stress-treatment comparison: The log2fold-change in the metabolomic/transcriptomic levels elicited by exposure to MV or P-lim. Red indicates an increase (up-regulation) by MV or P-lim and blue indicates decrease (down-regulation) in the pad2-1 mutant compared to that in WT plants. (B) Genotype- dependent comparison: Red and blue indicate an increase and a decrease, respectively, in the metabolite and transcript levels in the mutant compared to that in WT plants. ∗FDR < 0.05. Abbreviations: Ctrl, control (no-stress) condition; HMT3, homocysteine S-methyltransferase; MAM3/MAM-L, methylthioalkylmalate 3/methylthioalkylmalate-L; SOT17, sulfotransferase 17; BCAT4, branched chain aminotransferase 4; FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3; CYP79F1, cytochrome P450 CYP79F1; MYB29, MYB DOMAIN PROTEIN 29; FMO GS-OX1, flavin-containing monooxygenase glucosinolate S-oxygenase 1; AOP2, alkenyl hydroxialkyl producing 2; DFR, dihydroflavonol 4-reductase; 2OG-Fe(II) oxygenase, 2-(oxo)glutarate and Fe(II)-dependent oxygenase; PAP1, production of anthocyanin pigments 1; SRG1, SENESCENCE-RELATED GENE 1; DOGT1, DON-GLUCOSYLTRANSFERASE 1; Kaem F1, Kaempferol- 3,7-O-di-rhamnopyranosid; Kaem F2, kaempferol-3-O-beta-glucopyranosyl-7-O-alpha-rhamnopyranoside; Kaem F3, kaempferol-3-O-alpha-L-rhamnopyranosyl (1→2)-beta-D-glucopyranoside-7-O-alpha-L-rhamnopyranoside; Kaem F4, kaempferol-3-Galactoside-6″-Rhamnoside-3′′′′-Rhamnoside; Quer F5, quercetin-3,7-O-alpha-L-dirhamnopyranoside; Quer F6, quercetin-3-O-beta-glucopyranosyl-7-O-alpha-rhamnopyranoside; Antho A5, cyanidin conjugate (A5); Antho A8, cyanidin conjugate (A8); Antho A10, cyanidin conjugate (A10); Antho A11, cyanidin conjugate (A11); 8-MTO, 8-methylthio-n-octylglucosinolate; 7-MTH, 7-methylthio-n-heptylglucosinolate; 6-MTH, 6-methylthio-n-hexylglucosinolate; 5-MTP, 5-methylthio-n-pentylglucosinolate; 4-MTB, 4-methylthio-n- butylglucosinolate; 3-MTP, 3-methylthio-n-propylglucosinolate; 8-MSOO, 8-methylsulfinyl-n-octylglucosinolate; 7-MSOH, 7-methylsulfinyl-n-heptylglucosinolate; 6-MSOH, 6-methylsulfinyl-n-hexylglucosinolate; 5-MSOP, 5-methylsulfinyl-n-pentylglucosinolate; 4-MSOB, 4-methylsulfinyl-n-butylglucosinolate; 3-MSOP, 3-methylsulfinyl-n-propylglucosinolate. See also Supplementary Table S5.
In WT and pad2-1 plants, MV treatment triggered the up-regulation of the transcript levels of dihydroflavonol reductase (DFR), 2OG-Fe(II) oxygenase, and PAP1 after a remarkable increase in flavonols and anthocyanins (Figure 6A, right). As discussed previously (Saito et al., 2008; Fukushima and Kusano, 2014; Nakabayashi and Saito, 2015), the metabolomic and transcriptomic responses in the anthocyanin pathway in Arabidopsis were well correlated under oxidative stress conditions. In contrast to mild MV-induced oxidative stress, the increased level of kaempferols mentioned above was not correlated with the transcript levels in the pathway under P-lim condition. Compared to that in WT plants, pad2-1 mutants under MV and P-lim conditions showed a significant increase in two genes encoding SENESCENCE-RELATED GENE 1 (SRG1) and DON-GLUCOSYLTRANSFERASE 1 (DOGT1) in the flavonol biosynthetic pathway; the level of flavonols was largely unaltered (Figure 6B, right). When we focused on genotype-dependent changes, we found that, in the anthocyanin synthetic pathway, a remarkable decrease in the transcript level of 2OG-Fe(II) oxygenase and the anthocyanin level in pad2-1 mutant was noted compared to that in WT plants. Our finding that the anthocyanin level was lower in the mutant was consistent with that of previous findings on GSH1-antisense transgenic plants reported by (Xiang et al., 2001).
Discussion
Impact of Low Glutathione Content and Visual Phenotypes of GSH1 Mutants and Transgenic Plants
In this study, we investigated the changes in metabolite and transcript levels that reflect regulatory networks compensating for the reduction in the glutathione levels to enable the survival of GSH1 mutants under two mild abiotic stress conditions. We chose MV because the reduction of GSSG to GSH plays an important role in maintaining and regulating the cellular redox status during oxidative stress induced by MV. GSH1 is localized in plastids (Wachter et al., 2005), and MV is a fairly specific stimulus for ROS production in the chloroplast (Noctor et al., 2015). We chose the P-lim condition because phosphorus is an important component of the phospholipid membrane in the chloroplast (Hartel et al., 2000; Andersson et al., 2003; Jouhet et al., 2004; Okazaki et al., 2013). While phosphorus is also involved in plant growth and metabolism as one of the major nutrient ions, the molecular basis of plant adaptation to deal with a low phosphorus environment has not been completely elucidated. Although the series of genes and enzymes that are associated with these plant responses and adaptations (GSH1 is an example) are known, the comprehensive molecular mechanisms of their regulation and control are less well understood. Therefore, we applied the integrated omic approach by using high-throughput molecular phenotyping to GSH1 mutants grown under mild MV and P-lim conditions.
GSH1 mutants, cad2-1 (Cobbett et al., 1998) and pad2-1 (Parisy et al., 2007), showed differences in sensitivity to cadmium and biotic stresses from that of WT plants (Bednarek et al., 2009; Clay et al., 2009; Schlaeppi et al., 2010). Generally, glutathione synthesis mutants show shorter primary roots and lower levels of lateral root density compared to that in WT plants because of glutathione depletion (Schnaubelt et al., 2015). Low glutathione levels do not largely affect the shoot phenotypes of glutathione synthesis mutants under no-stress conditions. The characterization of leaf area phenotypes by using a high-resolution phenomic approach showed that glutathione synthesis mutants, except for an allele of GSH1 mutant called regulator of APX2 1-1 (rax1-1) (Ball et al., 2004), were not as sensitive as the WT plants under 1 μM MV treatment (Schnaubelt et al., 2013). We found that the two GSH1 mutants (cad2-1 and pad2-1), GSH1-overexpressing plants [7-5 and 13-6 (Cheng et al., 2015)], and WT plants exhibited a similar level of growth reduction under the mild MV stress condition (Figure 1 and Supplementary Figure S4). This suggests that neither a low nor high content of glutathione can compensate for the reduction in the shoot biomass of Arabidopsis under MV treatment. In WT plants, the level of GSH was slightly lower in the MV than in the no-stress condition and the GSSG level was increased (Figures 1C,D). The GSH:GSSG ratio in WT plants was similar under the control and P-lim condition, suggesting that the oxidative stress status was low in both (Figure 1E). Since glutathione peroxidase catalyzes the changes from GSH to GSSG, tracing the GSH:GSSG ratio by measuring GSH and GSSG might yield a specific marker of oxidative stress, e.g., increased H2O2 metabolism (Queval et al., 2007; Noctor et al., 2012).
Effects of Combined Low Glutathione and Mild Abiotic Stress on Metabolism in Arabidopsis
Our metabolite and transcript profiling revealed distinctive metabolic and transcriptional responses against two mild abiotic stresses. Genetic perturbation of metabolic responses in the WT plants and the two allelic mutants subjected to oxidative stress by MV clearly showed a specific enhancement in metabolite production in the sugar and flavonoid metabolism (Figure 3 and Supplementary Figure S7). This type of stress might result in an altered metabolic status to compensate for the reduced ROS scavenging ability by reprogramming the sugar and flavonoid metabolism to produce specific antioxidant metabolites that consist of aglycone with sugar conjugates, i.e., flavonols and anthocyanins, as a common oxidative stress response. We observed that our P-lim condition resulted in increased levels of specific flavonol-glycosides, kaempferol 3-O-rhamnoside-7-O-rhamnoside (F1), kaempferol 3-O-[glucosyl(1 → 6)glucoside] 7-O-rhamnoside (F4), and quercetin 3-O-rhamnoside-7-O-rhamnosides (F5). Previous transcript- and flavonoid profiling demonstrated that the LATERAL ORGAN BOUNDARY DOMAIN (LBD) gene family of transcription factors (LBD37, LBD38, and LBD39) regulate the late steps of anthocyanin-specific biosynthesis and flavonol glycosides [F1 to F6, see (Tohge et al., 2005)] derived from kaempferol or quercetin were largely unaltered in the LBD-overexpression or mutant seedlings irrespective of nitrogen supply (Rubin et al., 2009). Another report also demonstrated that flavonoid overaccumulation [e.g., flavonols (F1, F2, F3, F5, and F8) and anthocyanin] was important to enhanced tolerance to oxidative- and drought stresses (Nakabayashi et al., 2014). Together these observations suggest that specific flavonols and anthocyanins play a distinct role in mitigating abiotic stress in Arabidopsis.
Changes in the GSH level under each stress condition (MV or P-lim vs. control) were almost identical in WT plants and the mutants (Figure 1 and Supplementary Figure S7). Antioxidants such as ascorbate and glutathione play a central role in plant defense under oxidative stress conditions (for example, see Foyer and Noctor, 2009). Conversely, phytochelatins, polymerized to form GSH, protect plants from heavy metal toxicity (Xiang et al., 2001; Schat et al., 2002). In general, GSH is maintained at a high concentration in the sulfur assimilation pathway (Saito, 2000, 2004). Glucosinolates and phytochelatins are downstream metabolites in the pathway. No accumulation of phytochelatins was noted in the cad2-1 mutant (Jozefczak et al., 2015). Our metabolite profiling showed that, with the exception of 8-MTO and 8-MSOO, the level of glucosinolates was higher under both stress conditions than under no-stress in pad2-1 and cad2-1 mutants (Figures 3A, 5A, left). The accumulation patterns of glucosinolates under the applied stress conditions might be explained by metabolic perturbation resulting in a decrease of phytochelatin and an increase of glucosinolate levels to compensate for the reduced glutathione level against mild oxidative stress. Glucosinolates might protect against oxidative stress in the GSH1 mutants.
As mentioned above, our results imply that coordinated responses of glucosinolate and flavonoid biosynthesis at the metabolite and transcript levels are associated with the plant response to MV-induced oxidative stress and to stress elicited by P-lim (Figure 6). We found that, in the GSH1 mutants, a short-chain aliphatic glucosinolate (3-MSOP) was over-accumulated under mild oxidative stress, and the level of long-chain aliphatic glucosinolates was decreased (Figures 3B, 6B, left). Glucosinolate biosynthesis involves the breakdown of methionine to produce short-chain glucosinolates that harbor three carbons in their moiety (3C-glucosinolates) in the first round of glucosinolate chain elongation. Subsequently, 4C- and long-chain glucosinolates are synthesized by further rounds of elongation (Field et al., 2004). These elongation reactions are mediated by genes depicted in Figure 6 (left). The increase in the 3-MSOP level together with a decrease in the level of long-chain glucosinolates might be due to the gap and could result in the production of glucosinolates exhibiting different chain lengths. By using the insect herbivore Spodoptera littoralis, Schlaeppi et al. (2008) found that the pad2-1 mutant exhibited a reduction of two 3C- and 4C-glucosinolates, 4-methylsulfinylbutylglucosinolate and indolyl-3-methylglucosinolate, and that their reduction was correlated with the reduced GSH level (approximately 20% of that in WT plants). In addition, glutathione has possible roles as an s atom donor for glucosinolates (Noctor et al., 2012). It could be a reason of the decrease of long-chain glucosinolate level in pad2-1 mutants. In the flavonoid biosynthetic pathway in pad2-1 mutants, the decrease in the anthocyanin level is likely due to a reduction in the transcript level of genes encoding 2OG-Fe(II) oxygenase and PAP1 (Figure 6B, right). Under the control condition, the accumulation patterns of GSH and anthocyanins were roughly correlated in Arabidopsis (Xiang et al., 2001). Our results suggest that the correlation between GSH content and anthocyanin level is regulated at the transcript level.
Identification of Key Pathways Associated with the Gene-to-Metabolite Networks Perturbed by Low Glutathione Content under Mild Abiotic Stress
In this study, we imposed mild stress conditions to obtain a better understanding of the fundamental basis of plant responses to abiotic stress. Changes in the GSH:GSSG ratio reflected their oxidative stress status under the assayed conditions (Figure 1). Clauw et al. (2015) who used an automated phenotyping system and genome-wide transcriptome analysis based on RNA sequencing showed that six Arabidopsis accessions exhibited common and specific response to mild drought stress (Clauw et al., 2015). Our comparative transcript profiling of the GSH1 mutants and WT plants revealed common as well as specific stress responses, particularly against mild oxidative stress induced by a low concentration of MV and against mild stress elicited by low phosphorus (Table 1, Figure 5, and Supplementary Figure S12). We also evaluated the specific changes in transcript levels in response to environmental and genetic perturbations (Figure 5). We showed that stress-inducible genes included those encoding specific GSTs, UGTs, or CYPs that were associated with a well-known detoxification and transcriptome response when plants are exposed to oxidative stress. Our transcriptome results are in agreement with previous findings obtaining by conducting expression analysis of known “severe” oxidative stress-related genes (Dubreuil-Maurizi et al., 2011). However, we were able to find the pad2-specific transcript and metabolic responses under mild oxidative stress; an example is related to glucosinolate biosynthesis. The pad2-1 mutant clearly reprograms the cellular metabolic networks and the transcriptome in response to the applied mild abiotic stress, but whether they can be reversible is unclear and needs further studies. Very severe stress causes visible symptoms in plant growth and development, for example, stunted growth, light yellow or green leaves, and rolled leaves. Such phenotypic changes generally affect metabolite profiles (for example, see Fiehn et al., 2000). In addition, we were able to see significant metabolic fluctuation even under the strictly controlled conditions (Weckwerth et al., 2004; Kusano et al., 2007a). Combined metabolite- and transcript profiling has important implications for the diagnostic characterization of the “mild” stress level and fine-tuning molecular responses in plants.
WRKY-type transcription factors play an important role in the response to biotic and abiotic stress and are also important components of a plant signaling pathway that controls developmental processes (Eulgem et al., 2000; Ulker and Somssich, 2004; Rushton et al., 2010). Interplay among three types of group I WRKY proteins, WRKY33, WRKY25, and WRKY26, is crucial for promoting heat and salt tolerance in Arabidopsis (Li et al., 2011). Arabidopsis sigma factor-binding proteins (SIB1 and SIB2) stimulate the DNA binding activity of the WRKY33 transcription factor in plant defense (Lai et al., 2011). MV treatment also induces WRKY33 expression (Zheng et al., 2006; Miller et al., 2008). Our microarray experiment showed remarkable down-regulation of WRKY33 gene expression in pad2-1 by mild MV treatment (Table 1). Cross-talk between glutathione and phytohormones is also important for abiotic stress responses in Arabidopsis. Datta and colleagues showed that glutathione regulates the transcription levels of genes involved in ethylene biosynthesis via WRKY33 (Datta et al., 2015). Ethylene is known to regulate a wide range of plant processes, including growth, ripening, and responses to environmental stress (Kieber, 1997; Guo and Ecker, 2004; Klee, 2004; Yoo et al., 2009; Wang et al., 2013). As genotype-dependent altered genes and pad2-1-specific responsive genes, we detected changes in transcript levels of genes, including GSTs, UGTs, CYPs, HSPs, and SAGs, and transcription factors such as ERF6 and HSFA2. Our findings suggest that WRKY33 protein plays a vital role either as positive or negative transcriptional regulators, perceiving direct or indirect signal transduction in different ways under mild oxidative stress, though detailed regulatory mechanisms are still unclear.
The Relationship between Known Stress Marker Genes and Low Glutathione Content
Although the glutathione biosynthetic pathway can be considered as a key component in the sulfur assimilation pathway (Saito, 2000, 2004; Kopriva, 2006; Takahashi et al., 2011), except for the gene encoding SUFE2 that was up-regulated in pad2-1 compared with that in WT plants under all conditions, we observed no significant changes in the transcript level of genes involved in this pathway. Unlike our MV treatment and P-lim condition, earlier transcriptome analysis of cadmium-treated plants showed the induction of genes associated with sulfur assimilation reduction and glutathione pathways in Arabidopsis roots (Herbette et al., 2006). During oxidative stress elicited by MV and under P-lim conditions, DREB1A, DREB1B, DREB1C, and DREB2A genes were slightly up-regulated in pad2-1, but not in WT plants (Table 1). This suggests that DREB/CBF factors contribute, at least partly, to plant survival by increasing resistance under the imposed mild stress conditions. DREB2A interacts with the Radical-Induced Cell Death1 (RCD1) protein (Jaspers et al., 2010; Vainonen et al., 2012) that is involved in programmed cell death signaling and regulates plant stress responses (Overmyer et al., 2000; Ahlfors et al., 2004). The rcd1 mutant exhibits MV and UV-B tolerance (Fujibe et al., 2004). UV-B also elicits oxidative stress, resulting in changes in photomorphogenesis, photosynthesis, membranes, and secondary metabolism (Landry et al., 1995; Mittler, 2002; Frohnmeyer and Staiger, 2003; Kusano et al., 2011). Our current understanding of the molecular mechanisms controlling programmed cell death (PCD) in plants is limited, particularly with regard to how signaling by ROS drives/regulates the changes in expression leading to PCD (Kragelund et al., 2012). These mechanisms have not yet been identified. Genes encoding ZAT12 and STZ were also up-regulated in pad2-1 under both stress conditions (Table 1). ZAT12 is related to the control of cold-responsive gene expression and to the adaptation of plants to a freezing environment (Vogel et al., 2005). STZ is related to the Cys2/His2-type zinc-finger transcriptional repressor that involves drought stress tolerance (Sakamoto et al., 2004). These findings suggest that ZAT12 and STZ are also involved in plant responses to mild oxidative and low phosphorus stress.
Conclusion
Glutathione is a critical molecule that protects plant cells against ROS and is one of the important antioxidants in abiotic stress responses in plants. Despite efforts focused on elucidating the relationship among glutathione content, biosynthesis, and abiotic/biotic stress, understanding of low glutathione-mediated plant responses to mild rather than severe oxidative and nutrient stress remains incomplete. In this study, we assessed the effects of combined low glutathione with mild oxidative and low phosphorus stress on the metabolism in Arabidopsis. This study presents integrated metabolomics and transcriptomics analysis of glutathione depletion mutants, or GSH1 mutants (cad2-1 and pad2-1), in Arabidopsis in response to mild MV-induced oxidative and low phosphate stress. The data presented here suggest that sensitivity to mild oxidative stress induced by a low concentration of MV in the mutant of the GSH1 gene is similar to that of WT plants in terms of plant shoot growth. Our broad metabolite profiling showed that several flavonoids overaccumulated as a common oxidative stress response, whereas increased levels of flavonols with specific kaempferol- and quercetin-glycosides were observed as a common mild phosphate stress response. In addition to a significant production of sugar, osmolytes, and lipids as mild oxidative stress-responsive metabolites, we identified opposite alteration between short-chain- and long-chain aliphatic glucosinolates in the GSH1 mutants. Genome-wide transcriptome analysis supports the metabolite responses by detecting coordinated gene expressions related to glucosinolate and flavonoid biosynthesis in the pad2-1 mutant. We also hypothesize that pad2-1 mutants accelerate transcriptional regulatory networks to control the biosynthetic pathways involved in glutathione-independent scavenging metabolites, and that they might reconfigure the metabolic networks in the primary and secondary metabolism of compounds, including lipids, glucosinolates, and flavonoids. The findings of our study provide a basis for the elucidation of the molecular mechanisms involved in the transcriptional regulation and reprogramming of metabolic networks in response to mild oxidative and nutrient stress in Arabidopsis.
Availability of Supporting Data
All microarray data are available in the NCBI GEO database (Barrett et al., 2013) (accession no. GSE57286).
Author Contributions
AF, MI, and MK conceived the project, analyzed the data, and interpreted the majority of the results. MI and TN participated in experiments on the Arabidopsis mutants, cad2-1 and pad2-1, and the 35S::GSH1 transgenic lines 7-5 and 13-6, and in transcript profiling by microarrays. MK, RN, MKo, YO, and KS designed and performed metabolite profiling and drafted the Materials section. MI conducted glutathione quantification. AF, MI, and MK wrote the article with contributions from other co-authors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Christopher Corbett (University of Maryland, Baltimore, MD, United States) for providing cad2-1 seeds; Dr. Tsan-Piao Lin (National Taiwan University) for providing 35S::GSH1 transgenic lines; Drs. Thomas Moritz, Pär Jonsson, and Hans Stenlund (Umeå Plant Science Centre) for their assistance with hyphenated and raw data analysis; Mr. Koji Takano (RIKEN Center for Sustainable Resource Science) for preparing the standard compounds; Ms. Akiko Watanabe-Takahashi, Ms. Naomi Hayashi, Mr. Makoto Suzuki, and Ms. Tomoko Narisawa (RIKEN) for their experimental assistance; Dr. Tetsuya Sakurai and Mr. Yutaka Yamada (RIKEN) for computational support; and Ms. Ursula Petralia for editorial assistance. The Japan Advanced Plant Research Network supported by the Japan Society for the Promotion of Science is also acknowledged for allowing use of LC-q-TOF-MS.
Funding. This work was supported in part by the Japan Science and Technology Agency (JST)-Strategic International Collaborative Research Program (SICORP) and by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. This work was also supported in part by a Grant-in-Aid for Scientific Research on Innovative Area (22119003 to MK) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by JST PRESTO Grant Number JPJPMJPR13B5, Japan.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01464/full#supplementary-material
Strategy to choose the candidate lines that showed no visible phenotypes, but had altered metabotypes compared to that of WT. See also Supplementary Table S1. (A) Workflow of the screening for the eight candidate lines from the 50 mutant lines. Visual phenotypes of the 50 lines were preliminary observed as described in (Fukushima et al., 2014). (B) PCA of metabolite profiles of the “batch 2” dataset obtained using GC-TOF-MS. Score scatter plots were shown using PC1 (13%) and PC2 (10%) in the left panel and PC1 (13%) and PC3 (8%) in the right panel, respectively.
Visible phenotypic changes in 18-day-old samples grown under different concentrations of methyl viologen (MV). We observed WT individual plants grown under Murashige and Skoog medium containing 0.01, 0.03, and 0.05 μ MV (A). We also compared WT with pad2-1 plants under different MV treatments (B).
Visible phenotypic changes in 18-day-old samples grown under Murashige and Skoog medium (control MS), MS medium containing 0.05 μM of methyl viologen (MV), and phosphorous-limiting MS (P-lim, the concentration corresponds to 20% of ordinary MS medium).
Visual phenotypes, fresh weight of shoots, and GSH and GSSG levels in WT plants and 35S::GSH1-overexpressing transgenic lines, 7-5 and 13-6, grown under three different conditions. Visible phenotypic changes in 20-day-old GSH1-overexpressing lines grown on Murashige and Skoog medium (no stress), Murashige and Skoog medium containing 0.05 μM methyl viologen (MV), and low-phosphate (P-lim) Murashige and Skoog medium (A). Fresh weight (FW) of aerial parts of WT and transgenic plants grown under the three conditions (n = 8) (B). Quantification of the glutathione content in WT and transgenic plants grown under the three growth conditions. Content of the reduced form of glutathione, GSH (n = 3, biological replicates) (C). Content of the oxidized form of glutathione, GSSG (n = 3, biological replicates) (D). GSH:GSSG ratio as an indirect determinant of oxidative stress (E). Each error bar indicates the standard deviation from the mean. Asterisks represent differences from the control (significant levels were ∗∗ α = 0.01 and ∗ α = 0.05) by Welch’s t-test. The letters “a” and “b” represent significant differences compared to the no-stress condition (significant levels were a, α = 0.01 and b, α = 0.05) by Welch’s t-test. Note that the control values in glutathione levels are considerably higher than those shown in Figure 1, because the differences can arise due to both technical (e.g., unwanted instrumental variation) and/or biological factors (e.g., quality of the Arabidopsis seeds).
Principal component analysis of the metabolite profiles of WT plants and GSH1 mutants (cad2-1 and pad2-1) exposed to MV and P-lim; integrated data obtained from multi-platform metabolite profiling were used (n = 8, biological replicates). Detected metabolites contained sugar and sugar alcohols, amino acids, organic acids, fatty acids, glucosinolates, flavonoids, and lipids. The PCA score scatter plots indicate that the samples were clustered according to the corresponding genotype-dependent separation in the PC1/PC3 or PC2/PC3 direction.
Venn diagram of identified/annotated metabolites under different conditions (control, MV, and P-lim). The threshold was set at FDR < 0.05 and |log2fold-change|≥ 1.
Metabolite changes in the GSH1 mutants and WT by MV and P-lim treatments. (A) Stress-treatment comparison: Log2fold-change in metabolites exposed to the two abiotic stresses. The fold-change is represented by two directed colors; red and blue indicate increase and decrease, respectively, in the metabolite level elicited by MV treatment or P-lim. (B) Genotype-dependent comparison: Red and blue indicate increase and decrease, respectively, in the metabolite levels in the mutants vs. WT plants. For abbreviations of the metabolite names, see Supplementary Table S5. n = 8, biological replicates, ∗FDR < 0.05.
Enrichment map showing biological processes between pad2-1 and WT under 2 differential conditions based on hypergeometric tests of gene ontology (GO) terms by using BiNGO software (Maere et al., 2005). The map indicates the enriched GO terms in pad2 vs. WT under MV (A) and under P-lim condition (B). Enrichment map can be used to inspect differential transcriptomic responses between 2 genotypes (i.e., pad2-1 vs. WT) under abiotic stress. Nodes represent GO terms and links between nodes represent gene overlap between GO terms. Inner circle size of each node represents the number of DEGs in “comparison 1” (e.g., pad2 vs. WT under no-stress condition) within the GO term in biological process. Node border size represents the number of DEGs in “comparison 2” (e.g., pad2-1 vs. WT under MV stress) within the GO term in biological process. Color of the node and border refer to the significance based on the BiNGO FDR of the GO term for “comparison 1” and “comparison 2,” respectively. The red-filled nodes highlight the major GO functional terms. Link size shows the number of DEGs that overlap between the two connected GO terms (Jaccard coefficient, the cut-off is 0.25). Green links correspond to both datasets when it is the only color link. Green links indicate “comparison 1” and blue indicates “comparison 2.” Blue dotted circles represent summarized GO term clusters based on AutoAnnotate (Kucera et al., 2016). The maps were generated using Cytoscape (v3.2.1) Enrichment Map plugin (Merico et al., 2010; Isserlin et al., 2014).
Overview of the transcript profiles associated with hormone metabolism based on MapMan. The Affymetrix ATH1 microarray of treatment (A) and genotype comparison by using MapMan software (http://mapman.gabipd.org/web/guest/mapman). The log2fold-change is shown by color: for (A) red, upregulated by stress treatment; blue, downregulated by stress treatment; for (B) red, upregulated in pad2-1; blue downregulated in pad2-1. A Wilcoxon test with Benjamini–Hochberg multiple-testing correction was used to identify MapMan BINs that were significant, coordinately (FDR < 0.05) compared to the others.
Overview of the transcript profiles associated with secondary metabolism based on MapMan. For detail explanation, see the legend to Supplementary Figure S9.
Overview of the transcript profiles associated with cell wall metabolism based on MapMan. For detail explanation, see the legend to Supplementary Figure S9.
Venn diagrams representing the classification of genes based on microarray data. (A) Down-regulated genes by MV treatment and P-lim stress. (B) Genotype-dependent down-regulated genes.
References
- Ahlfors R., Lang S., Overmyer K., Jaspers P., Brosche M., Tauriainen A., et al. (2004). Arabidopsis RADICAL-INDUCED CELL DEATH1 belongs to the WWE protein-protein interaction domain protein family and modulates abscisic acid, ethylene, and methyl jasmonate responses. Plant Cell 16 1925–1937. 10.1105/tpc.021832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M. X., Stridh M. H., Larsson K. E., Liljenberg C., Sandelius A. S. (2003). Phosphate-deficient oat replaces a major portion of the plasma membrane phospholipids with the galactolipid digalactosyldiacylglycerol. FEBS Lett. 537 128–132. 10.1016/S0014-5793(03)00109-1 [DOI] [PubMed] [Google Scholar]
- Ball L., Accotto G. P., Bechtold U., Creissen G., Funck D., Jimenez A., et al. (2004). Evidence for a direct link between glutathione biosynthesis and stress fefense gene expression in Arabidopsis. Plant Cell 16 2448–2462. 10.1105/tpc.104.022608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T., Wilhite S. E., Ledoux P., Evangelista C., Kim I. F., Tomashevsky M., et al. (2013). NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 41 D991–D995. 10.1093/nar/gks1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter A., Mittler R., Suzuki N. (2014). ROS as key players in plant stress signalling. J. Exp. Bot. 65 1229–1240. 10.1093/jxb/ert375 [DOI] [PubMed] [Google Scholar]
- Bednarek P., Pislewska-Bednarek M., Svatos A., Schneider B., Doubsky J., Mansurova M., et al. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323 101–106. 10.1126/science.1163732 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57 289–300. [Google Scholar]
- Bolstad B. M., Collin F., Brettschneider J., Simpson K., Cope L. M., Irizarry R., et al. (2005). “Quality assessment of affymetrix genechip data,” in Bioinformatics and Computational Biology Solutions Using R and Bioconductor, eds Gentleman R., Carey V., Huber W., Irizarry R., Dudoit S. (New York, NY: Springer; ). [Google Scholar]
- Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19 185–193. 10.1093/bioinformatics/19.2.185 [DOI] [PubMed] [Google Scholar]
- Cairns N. G., Pasternak M., Wachter A., Cobbett C. S., Meyer A. J. (2006). Maturation of arabidopsis seeds is dependent on glutathione biosynthesis within the embryo. Plant Physiol. 141 446–455. 10.1104/pp.106.077982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. H., Jiang H. W., Hsieh E. J., Chen H. Y., Chien C. T., Hsieh H. L., et al. (2012). Drought and salt stress tolerance of an Arabidopsis glutathione S-transferase U17 knockout mutant are attributed to the combined effect of glutathione and abscisic acid. Plant Physiol. 158 340–351. 10.1104/pp.111.181875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M. C., Ko K., Chang W. L., Kuo W. C., Chen G. H., Lin T. P. (2015). Increased glutathione contributes to stress tolerance and global translational changes in Arabidopsis. Plant J. 83 926–939. 10.1111/tpj.12940 [DOI] [PubMed] [Google Scholar]
- Claeys H., Inze D. (2013). The agony of choice: how plants balance growth and survival under water-limiting conditions. Plant Physiol. 162 1768–1779. 10.1104/pp.113.220921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys H., Van Landeghem S., Dubois M., Maleux K., Inze D. (2014). What is stress? dose-response effects in commonly used in vitro stress assays. Plant Physiol. 165 519–527. 10.1104/pp.113.234641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauw P., Coppens F., De Beuf K., Dhondt S., Van Daele T., Maleux K., et al. (2015). Leaf responses to mild drought stress in natural variants of Arabidopsis. Plant Physiol. 167 800–816. 10.1104/pp.114.254284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay N. K., Adio A. M., Denoux C., Jander G., Ausubel F. M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323 95–101. 10.1126/science.1164627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbett C. S., May M. J., Howden R., Rolls B. (1998). The glutathione-deficient, cadmium-sensitive mutant, cad2-1, of Arabidopsis thaliana is deficient in gamma-glutamylcysteine synthetase. Plant J. 16 73–78. 10.1046/j.1365-313x.1998.00262.x [DOI] [PubMed] [Google Scholar]
- Datta R., Kumar D., Sultana A., Hazra S., Bhattacharyya D., Chattopadhyay S. (2015). Glutathione regulates 1-aminocyclopropane-1-carboxylate synthase transcription via WRKY33 and 1-aminocyclopropane-1-carboxylate oxidase by modulating messenger RNA stability to induce ethylene synthesis during stress. Plant Physiol. 169 2963–2981. 10.1104/pp.15.01543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S., Schlauch K., Coutu J., Mittler R. (2005). The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol. 139 847–856. 10.1104/pp.105.068254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit P., Mukherjee P. K., Ramachandran V., Eapen S. (2011). Glutathione transferase from Trichoderma virens enhances cadmium tolerance without enhancing its accumulation in transgenic Nicotiana tabacum. PLoS ONE 6:e16360 10.1371/journal.pone.0016360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon D. P., Lapthorn A., Edwards R. (2002). Plant glutathione transferases. Genome Biol. 3:REVIEWS3004 10.1186/gb-2002-3-3-reviews3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M., Skirycz A., Claeys H., Maleux K., Dhondt S., De Bodt S., et al. (2013). Ethylene Response Factor6 acts as a central regulator of leaf growth under water-limiting conditions in Arabidopsis. Plant Physiol. 162 319–332. 10.1104/pp.113.216341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M., Van Den Broeck L., Claeys H., Van Vlierberghe K., Matsui M., Inze D. (2015). The ETHYLENE RESPONSE FACTORs ERF6 and ERF11 Antagonistically Regulate Mannitol-Induced Growth Inhibition in Arabidopsis. Plant Physiol. 169 166–179. 10.1104/pp.15.00335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil-Maurizi C., Vitecek J., Marty L., Branciard L., Frettinger P., Wendehenne D., et al. (2011). Glutathione deficiency of the Arabidopsis mutant pad2-1 affects oxidative stress-related events, defense gene expression, and the hypersensitive response. Plant Physiol. 157 2000–2012. 10.1104/pp.111.182667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egert A., Keller F., Peters S. (2013). Abiotic stress-induced accumulation of raffinose in Arabidopsis leaves is mediated by a single raffinose synthase (RS5, At5g40390). BMC Plant Biol. 13:218 10.1186/1471-2229-13-218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T., Rushton P. J., Robatzek S., Somssich I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5 199–206. 10.1016/S1360-1385(00)01600-9 [DOI] [PubMed] [Google Scholar]
- Fernie A. R., Aharoni A., Willmitzer L., Stitt M., Tohge T., Kopka J., et al. (2011). Recommendations for reporting metabolite data. Plant Cell 23 2477–2482. 10.1105/tpc.111.086272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O., Kopka J., Dormann P., Altmann T., Trethewey R. N., Willmitzer L. (2000). Metabolite profiling for plant functional genomics. Nat. Biotechnol. 18 1157–1161. 10.1038/81137 [DOI] [PubMed] [Google Scholar]
- Field B., Cardon G., Traka M., Botterman J., Vancanneyt G., Mithen R. (2004). Glucosinolate and amino acid biosynthesis in Arabidopsis. Plant Physiol. 135 828–839. 10.1104/pp.104.039347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C. H., Noctor G. (2009). Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxid. Redox. Signal. 11 861–905. 10.1089/ars.2008.2177 [DOI] [PubMed] [Google Scholar]
- Frohnmeyer H., Staiger D. (2003). Ultraviolet-B radiation-mediated responses in plants. Balancing damage and protection. Plant Physiol. 133 1420–1428. 10.1104/pp.103.030049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujibe T., Saji H., Arakawa K., Yabe N., Takeuchi Y., Yamamoto K. T. (2004). A methyl viologen-resistant mutant of Arabidopsis, which is allelic to ozone-sensitive rcd1, is tolerant to supplemental ultraviolet-B irradiation. Plant Physiol. 134 275–285. 10.1104/pp.103.033480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima A., Kusano M. (2014). A network perspective on nitrogen metabolism from model to crop plants using integrated ‘omics’ approaches. J. Exp. Bot. 65 5619–5630. 10.1093/jxb/eru322 [DOI] [PubMed] [Google Scholar]
- Fukushima A., Kusano M., Mejia R. F., Iwasa M., Kobayashi M., Hayashi N., et al. (2014). Metabolomic characterization of knockout mutants in Arabidopsis: development of a metabolite profiling database for knockout mutants in Arabidopsis. Plant Physiol. 165 948–961. 10.1104/pp.114.240986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., et al. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5 R80 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigolashvili T., Engqvist M., Yatusevich R., Muller C., Flugge U. I. (2008). HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol. 177 627–642. 10.1111/j.1469-8137.2007.02295.x [DOI] [PubMed] [Google Scholar]
- Gomez L. D., Noctor G., Knight M. R., Foyer C. H. (2004). Regulation of calcium signalling and gene expression by glutathione. J. Exp. Bot. 55 1851–1859. 10.1093/jxb/erh202 [DOI] [PubMed] [Google Scholar]
- Gullner G., Komives T., Rennenberg H. (2001). Enhanced tolerance of transgenic poplar plants overexpressing gamma-glutamylcysteine synthetase towards chloroacetanilide herbicides. J. Exp. Bot. 52 971–979. 10.1093/jexbot/52.358.971 [DOI] [PubMed] [Google Scholar]
- Guo H., Ecker J. R. (2004). The ethylene signaling pathway: new insights. Curr. Opin. Plant Biol. 7 40–49. 10.1016/j.pbi.2003.11.011 [DOI] [PubMed] [Google Scholar]
- Hansen B. G., Kliebenstein D. J., Halkier B. A. (2007). Identification of a flavin-monooxygenase as the S-oxygenating enzyme in aliphatic glucosinolate biosynthesis in Arabidopsis. Plant J. 50 902–910. 10.1111/j.1365-313X.2007.03101.x [DOI] [PubMed] [Google Scholar]
- Hartel H., Dormann P., Benning C. (2000). DGD1-independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 97 10649–10654. 10.1073/pnas.180320497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbette S., Taconnat L., Hugouvieux V., Piette L., Magniette M. L., Cuine S., et al. (2006). Genome-wide transcriptome profiling of the early cadmium response of Arabidopsis roots and shoots. Biochimie 88 1751–1765. 10.1016/j.biochi.2006.04.018 [DOI] [PubMed] [Google Scholar]
- Hirai M. Y., Sugiyama K., Sawada Y., Tohge T., Obayashi T., Suzuki A., et al. (2007). Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 104 6478–6483. 10.1073/pnas.0611629104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden R., Andersen C. R., Goldsbrough P. B., Cobbett C. S. (1995). A cadmium-sensitive, glutathione-deficient mutant of Arabidopsis thaliana. Plant Physiol. 107 1067–1073. 10.1104/pp.107.4.1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Guo T., Zheng S. C., Feng Q. L., Liang J. H., Li L. (2009). Increased cold tolerance in Arabidopsis thaliana transformed with Choristoneura fumiferana glutathione S-transferase gene. Biol. Plant. 53 183–187. 10.1007/s10535-009-0031-1 [DOI] [Google Scholar]
- Irizarry R. A., Bolstad B. M., Collin F., Cope L. M., Hobbs B., Speed T. P. (2003). Summaries of affymetrix genechip probe level data. Nucleic Acids Res. 31:e15 10.1093/nar/gng015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isserlin R., Merico D., Voisin V., Bader G. D. (2014). Enrichment Map - a Cytoscape app to visualize and explore OMICs pathway enrichment results. F1000Res 3:141 10.12688/f1000research.4536.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers P., Brosche M., Overmyer K., Kangasjarvi J. (2010). The transcription factor interacting protein RCD1 contains a novel conserved domain. Plant Signal. Behav. 5 78–80. 10.4161/psb.5.1.10293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha B., Sharma A., Mishra A. (2011). Expression of SbGSTU (tau class glutathione S-transferase) gene isolated from Salicornia brachiata in tobacco for salt tolerance. Mol. Biol. Rep. 38 4823–4832. 10.1007/s11033-010-0625-x [DOI] [PubMed] [Google Scholar]
- Ji W., Zhu Y., Li Y., Yang L., Zhao X., Cai H., et al. (2010). Over-expression of a glutathione S-transferase gene, GsGST, from wild soybean (Glycine soja) enhances drought and salt tolerance in transgenic tobacco. Biotechnol. Lett. 32 1173–1179. 10.1007/s10529-010-0269-x [DOI] [PubMed] [Google Scholar]
- Jouhet J., Marechal E., Baldan B., Bligny R., Joyard J., Block M. A. (2004). Phosphate deprivation induces transfer of DGDG galactolipid from chloroplast to mitochondria. J. Cell Biol. 167 863–874. 10.1083/jcb.200407022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jozefczak M., Bohler S., Schat H., Horemans N., Guisez Y., Remans T., et al. (2015). Both the concentration and redox state of glutathione and ascorbate influence the sensitivity of arabidopsis to cadmium. Ann. Bot. 116 601–612. 10.1093/aob/mcv075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber J. J. (1997). The ethylene response pathway in Arabidopsis. Annu. Rev. Plant Physiol. Plant. Mol. Biol. 48 277–296. 10.1146/annurev.arplant.48.1.277 [DOI] [PubMed] [Google Scholar]
- Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., et al. (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50 347–363. 10.1111/j.1365-313X.2007.03052.x [DOI] [PubMed] [Google Scholar]
- Kimbara J., Yoshida M., Ito H., Kitagawa M., Takada W., Hayashi K., et al. (2013). Inhibition of CUTIN DEFICIENT 2 causes defects in cuticle function and structure and metabolite changes in tomato fruit. Plant Cell Physiol. 54 1535–1548. 10.1093/pcp/pct100 [DOI] [PubMed] [Google Scholar]
- Klee H. J. (2004). Ethylene signal transduction. Moving beyond Arabidopsis. Plant Physiol. 135 660–667. 10.1104/pp.104.040998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S. (2006). Regulation of sulfate assimilation in Arabidopsis and beyond. Ann. Bot. 97 479–495. 10.1093/aob/mcl006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragelund B. B., Jensen M. K., Skriver K. (2012). Order by disorder in plant signaling. Trends Plant Sci. 17 625–632. 10.1016/j.tplants.2012.06.010 [DOI] [PubMed] [Google Scholar]
- Kreps J. A., Wu Y., Chang H. S., Zhu T., Wang X., Harper J. F. (2002). Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 130 2129–2141. 10.1104/pp.008532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucera M., Isserlin R., Arkhangorodsky A., Bader G. D. (2016). AutoAnnotate: a cytoscape app for summarizing networks with semantic annotations. F1000Res. 5:1717 10.12688/f1000research.9090.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D., Datta R., Hazra S., Sultana A., Mukhopadhyay R., Chattopadhyay S. (2015). Transcriptomic profiling of Arabidopsis thaliana mutant pad2.1 in response to combined cold and osmotic stress. PLoS ONE 10:e0122690 10.1371/journal.pone.0122690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D., Hazra S., Datta R., Chattopadhyay S. (2016). Transcriptome analysis of Arabidopsis mutants suggests a crosstalk between ABA, ethylene and GSH against combined cold and osmotic stress. Sci. Rep. 6:36867 10.1038/srep36867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano M., Fukushima A., Arita M., Jonsson P., Moritz T., Kobayashi M., et al. (2007a). Unbiased characterization of genotype-dependent metabolic regulations by metabolomic approach in Arabidopsis thaliana. BMC Syst. Biol. 1:53 10.1186/1752-0509-1-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano M., Fukushima A., Kobayashi M., Hayashi N., Jonsson P., Moritz T., et al. (2007b). Application of a metabolomic method combining one-dimensional and two-dimensional gas chromatography-time-of-flight/mass spectrometry to metabolic phenotyping of natural variants in rice. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 855 71–79. [DOI] [PubMed] [Google Scholar]
- Kusano M., Tohge T., Fukushima A., Kobayashi M., Hayashi N., Otsuki H., et al. (2011). Metabolomics reveals comprehensive reprogramming involving two independent metabolic responses of Arabidopsis to UV-B light. Plant J. 67 354–369. 10.1111/j.1365-313X.2011.04599.x [DOI] [PubMed] [Google Scholar]
- Lai Z., Li Y., Wang F., Cheng Y., Fan B., Yu J. Q., et al. (2011). Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 23 3824–3841. 10.1105/tpc.111.090571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry L. G., Chapple C. C., Last R. L. (1995). Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol. 109 1159–1166. 10.1104/pp.109.4.1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Fu Q., Chen L., Huang W., Yu D. (2011). Arabidopsis thaliana WRKY25 WRKY26 and WRKY33 coordinate induction of plant thermotolerance. Planta 233 1237–1252. 10.1007/s00425-011-1375-2 [DOI] [PubMed] [Google Scholar]
- Liedschulte V., Wachter A., Zhigang A., Rausch T. (2010). Exploiting plants for glutathione (GSH) production: uncoupling GSH synthesis from cellular controls results in unprecedented GSH accumulation. Plant Biotechnol. J. 8 807–820. 10.1111/j.1467-7652.2010.00510.x [DOI] [PubMed] [Google Scholar]
- Maere S., Heymans K., Kuiper M. (2005). BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21 3448–3449. 10.1093/bioinformatics/bti551 [DOI] [PubMed] [Google Scholar]
- Marrs K. A. (1996). The functions and regulation of glutathione S-transferases in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 127–158. 10.1146/annurev.arplant.47.1.127 [DOI] [PubMed] [Google Scholar]
- Matsuda F., Yonekura-Sakakibara K., Niida R., Kuromori T., Shinozaki K., Saito K. (2009). MS/MS spectral tag-based annotation of non-targeted profile of plant secondary metabolites. Plant J. 57 555–577. 10.1111/j.1365-313X.2008.03705.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico D., Isserlin R., Stueker O., Emili A., Bader G. D. (2010). Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS ONE 5:e13984 10.1371/journal.pone.0013984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Shulaev V., Mittler R. (2008). Reactive oxygen signaling and abiotic stress. Physiol. Plant. 133 481–489. 10.1111/j.1399-3054.2008.01090.x [DOI] [PubMed] [Google Scholar]
- Mittler R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7 405–410. 10.1016/S1360-1385(02)02312-9 [DOI] [PubMed] [Google Scholar]
- Mittler R. (2006). Abiotic stress, the field environment and stress combination. Trends Plant Sci. 11 15–19. 10.1016/j.tplants.2005.11.002 [DOI] [PubMed] [Google Scholar]
- Moller I. M., Jensen P. E., Hansson A. (2007). Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 58 459–481. 10.1146/annurev.arplant.58.032806.103946 [DOI] [PubMed] [Google Scholar]
- Morcuende R., Bari R., Gibon Y., Zheng W., Pant B. D., Blasing O., et al. (2007). Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ. 30 85–112. 10.1111/j.1365-3040.2006.01608.x [DOI] [PubMed] [Google Scholar]
- Nakabayashi R., Saito K. (2015). Integrated metabolomics for abiotic stress responses in plants. Curr. Opin. Plant Biol. 24 10–16. 10.1016/j.pbi.2015.01.003 [DOI] [PubMed] [Google Scholar]
- Nakabayashi R., Yonekura-Sakakibara K., Urano K., Suzuki M., Yamada Y., Nishizawa T., et al. (2014). Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 77 367–379. 10.1111/tpj.12388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G., Arisi A. C., Jouanin L., Foyer C. H. (1998). Manipulation of glutathione and amino acid biosynthesis in the chloroplast. Plant Physiol. 118 471–482. 10.1104/pp.118.2.471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G., Lelarge-Trouverie C., Mhamdi A. (2015). The metabolomics of oxidative stress. Phytochemistry 112 33–53. 10.1016/j.phytochem.2014.09.002 [DOI] [PubMed] [Google Scholar]
- Noctor G., Mhamdi A., Chaouch S., Han Y., Neukermans J., Marquez-Garcia B., et al. (2012). Glutathione in plants: an integrated overview. Plant Cell Environ. 35 454–484. 10.1111/j.1365-3040.2011.02400.x [DOI] [PubMed] [Google Scholar]
- Okazaki Y., Otsuki H., Narisawa T., Kobayashi M., Sawai S., Kamide Y., et al. (2013). A new class of plant lipid is essential for protection against phosphorus depletion. Nat. Commun. 4 1510 10.1038/ncomms2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer K., Tuominen H., Kettunen R., Betz C., Langebartels C., Sandermann H., et al. (2000). Ozone-sensitive arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell 12 1849–1862. 10.1105/tpc.12.10.1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisy V., Poinssot B., Owsianowski L., Buchala A., Glazebrook J., Mauch F. (2007). Identification of PAD2 as a gamma-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. Plant J. 49 159–172. 10.1111/j.1365-313X.2006.02938.x [DOI] [PubMed] [Google Scholar]
- Pasternak M., Lim B., Wirtz M., Hell R., Cobbett C. S., Meyer A. J. (2008). Restricting glutathione biosynthesis to the cytosol is sufficient for normal plant development. Plant J. 53 999–1012. 10.1111/j.1365-313X.2007.03389.x [DOI] [PubMed] [Google Scholar]
- Qi Y. C., Liu W. Q., Qiu L. Y., Zhang S. M., Ma L., Zhang H. (2010). Overexpression of Glutathione S-transferase gene increases salt tolerance of Arabidopsis. Russ. J. Plant Physiol. 57 233–240. 10.1134/S102144371002010X [DOI] [Google Scholar]
- Qin F., Shinozaki K., Yamaguchi-Shinozaki K. (2011). Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 52 1569–1582. 10.1093/pcp/pcr106 [DOI] [PubMed] [Google Scholar]
- Queval G., Foyer C. H. (2012). Redox regulation of photosynthetic gene expression. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367 3475–3485. 10.1098/rstb.2012.0068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queval G., Issakidis-Bourguet E., Hoeberichts F. A., Vandorpe M., Gakiere B., Vanacker H., et al. (2007). Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 52 640–657. 10.1111/j.1365-313X.2007.03263.x [DOI] [PubMed] [Google Scholar]
- Rubin G., Tohge T., Matsuda F., Saito K., Scheible W. R. (2009). Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21 3567–3584. 10.1105/tpc.109.067041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton P. J., Somssich I. E., Ringler P., Shen Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15 247–258. 10.1016/j.tplants.2010.02.006 [DOI] [PubMed] [Google Scholar]
- Saito K. (2000). Regulation of sulfate transport and synthesis of sulfur-containing amino acids. Curr. Opin. Plant Biol. 3 188–195. 10.1016/S1369-5266(00)00063-7 [DOI] [PubMed] [Google Scholar]
- Saito K. (2004). Sulfur assimilatory metabolism. The long and smelling road. Plant Physiol. 136 2443–2450. 10.1104/pp.104.046755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Hirai M. Y., Yonekura-Sakakibara K. (2008). Decoding genes with coexpression networks and metabolomics - ‘majority report by precogs’. Trends Plant Sci. 13 36–43. 10.1016/j.tplants.2007.10.006 [DOI] [PubMed] [Google Scholar]
- Sakamoto H., Maruyama K., Sakuma Y., Meshi T., Iwabuchi M., Shinozaki K., et al. (2004). Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 136 2734–2746. 10.1104/pp.104.046599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y., Maruyama K., Qin F., Osakabe Y., Shinozaki K., Yamaguchi-Shinozaki K. (2006). Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. U.S.A. 103 18822–18827. 10.1073/pnas.0605639103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman D. P., Shin R. (2007). Nutrient sensing and signaling: NPKS. Annu. Rev. Plant Biol. 58 47–69. 10.1146/annurev.arplant.58.032806.103750 [DOI] [PubMed] [Google Scholar]
- Schat H., Llugany M., Vooijs R., Hartley-Whitaker J., Bleeker P. M. (2002). The role of phytochelatins in constitutive and adaptive heavy metal tolerances in hyperaccumulator and non-hyperaccumulator metallophytes. J. Exp. Bot. 53 2381–2392. 10.1093/jxb/erf107 [DOI] [PubMed] [Google Scholar]
- Schlaeppi K., Abou-Mansour E., Buchala A., Mauch F. (2010). Disease resistance of Arabidopsis to Phytophthora brassicae is established by the sequential action of indole glucosinolates and camalexin. Plant J. 62 840–851. 10.1111/j.1365-313X.2010.04197.x [DOI] [PubMed] [Google Scholar]
- Schlaeppi K., Bodenhausen N., Buchala A., Mauch F., Reymond P. (2008). The glutathione-deficient mutant pad2-1 accumulates lower amounts of glucosinolates and is more susceptible to the insect herbivore Spodoptera littoralis. Plant J. 55 774–786. 10.1111/j.1365-313X.2008.03545.x [DOI] [PubMed] [Google Scholar]
- Schnaubelt D., Queval G., Dong Y., Diaz-Vivancos P., Makgopa M. E., Howell G., et al. (2015). Low glutathione regulates gene expression and the redox potentials of the nucleus and cytosol in Arabidopsis thaliana. Plant Cell Environ. 38 266–279. 10.1111/pce.12252 [DOI] [PubMed] [Google Scholar]
- Schnaubelt D., Schulz P., Hannah M. A., Yocgo R. E., Foyer C. H. (2013). A phenomics approach to the analysis of the influence of glutathione on leaf area and abiotic stress tolerance in Arabidopsis thaliana. Front. Plant Sci. 4:416 10.3389/fpls.2013.00416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster J., Knill T., Reichelt M., Gershenzon J., Binder S. (2006). Branched-chain aminotransferase4 is part of the chain elongation pathway in the biosynthesis of methionine-derived glucosinolates in Arabidopsis. Plant Cell 18 2664–2679. 10.1105/tpc.105.039339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A., De Bodt S., Obata T., De Clercq I., Claeys H., De Rycke R., et al. (2010). Developmental stage specificity and the role of mitochondrial metabolism in the response of Arabidopsis leaves to prolonged mild osmotic stress. Plant Physiol. 152 226–244. 10.1104/pp.109.148965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G. K. (2005). “Limma: linear models for microarray data,” in Bioinformatics and Computational Biology Solutions using R and Bioconductor, eds Gentleman R., Carey V., Dudoit S., Irizarry R., Huber W. (New York, NY: Springer; ), 397–420. 10.1007/0-387-29362-0_23 [DOI] [Google Scholar]
- Sonderby I. E., Hansen B. G., Bjarnholt N., Ticconi C., Halkier B. A., Kliebenstein D. J. (2007). A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PLoS ONE 2:e1322 10.1371/journal.pone.0001322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukrong S., Yun K. Y., Stadler P., Kumar C., Facciuolo T., Moffatt B. A., et al. (2012). Improved growth and stress tolerance in the Arabidopsis oxt1 mutant triggered by altered adenine metabolism. Mol. Plant 5 1310–1332. 10.1093/mp/sss065 [DOI] [PubMed] [Google Scholar]
- Takahashi H., Kopriva S., Giordano M., Saito K., Hell R. (2011). Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 62 157–184. 10.1146/annurev-arplant-042110-103921 [DOI] [PubMed] [Google Scholar]
- Textor S., De Kraker J. W., Hause B., Gershenzon J., Tokuhisa J. G. (2007). MAM3 catalyzes the formation of all aliphatic glucosinolate chain lengths in Arabidopsis. Plant Physiol. 144 60–71. 10.1104/pp.106.091579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O., Blasing O., Gibon Y., Nagel A., Meyer S., Kruger P., et al. (2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37 914–939. 10.1111/j.1365-313X.2004.02016.x [DOI] [PubMed] [Google Scholar]
- Tohge T., Nishiyama Y., Hirai M. Y., Yano M., Nakajima J., Awazuhara M., et al. (2005). Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42 218–235. 10.1111/j.1365-313X.2005.02371.x [DOI] [PubMed] [Google Scholar]
- Ulker B., Somssich I. E. (2004). WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 7 491–498. 10.1016/j.pbi.2004.07.012 [DOI] [PubMed] [Google Scholar]
- Vainonen J. P., Jaspers P., Wrzaczek M., Lamminmaki A., Reddy R. A., Vaahtera L., et al. (2012). RCD1-DREB2A interaction in leaf senescence and stress responses in Arabidopsis thaliana. Biochem. J. 442 573–581. 10.1042/BJ20111739 [DOI] [PubMed] [Google Scholar]
- Vernoux T., Wilson R. C., Seeley K. A., Reichheld J. P., Muroy S., Brown S., et al. (2000). The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 12 97–110. 10.1105/tpc.12.1.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinocur B., Altman A. (2005). Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr. Opin. Biotechnol. 16 123–132. 10.1016/j.copbio.2005.02.001 [DOI] [PubMed] [Google Scholar]
- Vogel J. T., Zarka D. G., Van Buskirk H. A., Fowler S. G., Thomashow M. F. (2005). Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 41 195–211. 10.1111/j.1365-313X.2004.02288.x [DOI] [PubMed] [Google Scholar]
- Wachter A., Wolf S., Steininger H., Bogs J., Rausch T. (2005). Differential targeting of GSH1 and GSH2 is achieved by multiple transcription initiation: implications for the compartmentation of glutathione biosynthesis in the Brassicaceae. Plant J. 41 15–30. 10.1111/j.1365-313X.2004.02269.x [DOI] [PubMed] [Google Scholar]
- Wang F., Cui X., Sun Y., Dong C. H. (2013). Ethylene signaling and regulation in plant growth and stress responses. Plant Cell Rep. 32 1099–1109. 10.1007/s00299-013-1421-6 [DOI] [PubMed] [Google Scholar]
- Weckwerth W., Loureiro M. E., Wenzel K., Fiehn O. (2004). Differential metabolic networks unravel the effects of silent plant phenotypes. Proc. Natl. Acad. Sci. U.S.A. 101 7809–7814. 10.1073/pnas.0303415101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C., Werner B. L., Christensen E. M., Oliver D. J. (2001). The biological functions of glutathione revisited in arabidopsis transgenic plants with altered glutathione levels. Plant Physiol. 126 564–574. 10.1104/pp.126.2.564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Xing X. J., Tian Y. S., Peng R. H., Xue Y., Zhao W., et al. (2015). Transgenic Arabidopsis plants expressing tomato glutathione S-transferase showed enhanced resistance to salt and drought stress. PLoS ONE 10:e0136960 10.1371/journal.pone.0136960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (1993). Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol. Gen. Genet. 236 331–340. 10.1007/BF00277130 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57 781–803. 10.1146/annurev.arplant.57.032905.105444 [DOI] [PubMed] [Google Scholar]
- Yoo S. D., Cho Y., Sheen J. (2009). Emerging connections in the ethylene signaling network. Trends Plant Sci. 14 270–279. 10.1016/j.tplants.2009.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Qamar S. A., Chen Z., Mengiste T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48 592–605. 10.1111/j.1365-313X.2006.02901.x [DOI] [PubMed] [Google Scholar]
- Zhu Y. L., Pilon-Smits E. A., Tarun A. S., Weber S. U., Jouanin L., Terry N. (1999). Cadmium tolerance and accumulation in Indian mustard is enhanced by overexpressing gamma-glutamylcysteine synthetase. Plant Physiol. 121 1169–1178. 10.1104/pp.121.4.1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Strategy to choose the candidate lines that showed no visible phenotypes, but had altered metabotypes compared to that of WT. See also Supplementary Table S1. (A) Workflow of the screening for the eight candidate lines from the 50 mutant lines. Visual phenotypes of the 50 lines were preliminary observed as described in (Fukushima et al., 2014). (B) PCA of metabolite profiles of the “batch 2” dataset obtained using GC-TOF-MS. Score scatter plots were shown using PC1 (13%) and PC2 (10%) in the left panel and PC1 (13%) and PC3 (8%) in the right panel, respectively.
Visible phenotypic changes in 18-day-old samples grown under different concentrations of methyl viologen (MV). We observed WT individual plants grown under Murashige and Skoog medium containing 0.01, 0.03, and 0.05 μ MV (A). We also compared WT with pad2-1 plants under different MV treatments (B).
Visible phenotypic changes in 18-day-old samples grown under Murashige and Skoog medium (control MS), MS medium containing 0.05 μM of methyl viologen (MV), and phosphorous-limiting MS (P-lim, the concentration corresponds to 20% of ordinary MS medium).
Visual phenotypes, fresh weight of shoots, and GSH and GSSG levels in WT plants and 35S::GSH1-overexpressing transgenic lines, 7-5 and 13-6, grown under three different conditions. Visible phenotypic changes in 20-day-old GSH1-overexpressing lines grown on Murashige and Skoog medium (no stress), Murashige and Skoog medium containing 0.05 μM methyl viologen (MV), and low-phosphate (P-lim) Murashige and Skoog medium (A). Fresh weight (FW) of aerial parts of WT and transgenic plants grown under the three conditions (n = 8) (B). Quantification of the glutathione content in WT and transgenic plants grown under the three growth conditions. Content of the reduced form of glutathione, GSH (n = 3, biological replicates) (C). Content of the oxidized form of glutathione, GSSG (n = 3, biological replicates) (D). GSH:GSSG ratio as an indirect determinant of oxidative stress (E). Each error bar indicates the standard deviation from the mean. Asterisks represent differences from the control (significant levels were ∗∗ α = 0.01 and ∗ α = 0.05) by Welch’s t-test. The letters “a” and “b” represent significant differences compared to the no-stress condition (significant levels were a, α = 0.01 and b, α = 0.05) by Welch’s t-test. Note that the control values in glutathione levels are considerably higher than those shown in Figure 1, because the differences can arise due to both technical (e.g., unwanted instrumental variation) and/or biological factors (e.g., quality of the Arabidopsis seeds).
Principal component analysis of the metabolite profiles of WT plants and GSH1 mutants (cad2-1 and pad2-1) exposed to MV and P-lim; integrated data obtained from multi-platform metabolite profiling were used (n = 8, biological replicates). Detected metabolites contained sugar and sugar alcohols, amino acids, organic acids, fatty acids, glucosinolates, flavonoids, and lipids. The PCA score scatter plots indicate that the samples were clustered according to the corresponding genotype-dependent separation in the PC1/PC3 or PC2/PC3 direction.
Venn diagram of identified/annotated metabolites under different conditions (control, MV, and P-lim). The threshold was set at FDR < 0.05 and |log2fold-change|≥ 1.
Metabolite changes in the GSH1 mutants and WT by MV and P-lim treatments. (A) Stress-treatment comparison: Log2fold-change in metabolites exposed to the two abiotic stresses. The fold-change is represented by two directed colors; red and blue indicate increase and decrease, respectively, in the metabolite level elicited by MV treatment or P-lim. (B) Genotype-dependent comparison: Red and blue indicate increase and decrease, respectively, in the metabolite levels in the mutants vs. WT plants. For abbreviations of the metabolite names, see Supplementary Table S5. n = 8, biological replicates, ∗FDR < 0.05.
Enrichment map showing biological processes between pad2-1 and WT under 2 differential conditions based on hypergeometric tests of gene ontology (GO) terms by using BiNGO software (Maere et al., 2005). The map indicates the enriched GO terms in pad2 vs. WT under MV (A) and under P-lim condition (B). Enrichment map can be used to inspect differential transcriptomic responses between 2 genotypes (i.e., pad2-1 vs. WT) under abiotic stress. Nodes represent GO terms and links between nodes represent gene overlap between GO terms. Inner circle size of each node represents the number of DEGs in “comparison 1” (e.g., pad2 vs. WT under no-stress condition) within the GO term in biological process. Node border size represents the number of DEGs in “comparison 2” (e.g., pad2-1 vs. WT under MV stress) within the GO term in biological process. Color of the node and border refer to the significance based on the BiNGO FDR of the GO term for “comparison 1” and “comparison 2,” respectively. The red-filled nodes highlight the major GO functional terms. Link size shows the number of DEGs that overlap between the two connected GO terms (Jaccard coefficient, the cut-off is 0.25). Green links correspond to both datasets when it is the only color link. Green links indicate “comparison 1” and blue indicates “comparison 2.” Blue dotted circles represent summarized GO term clusters based on AutoAnnotate (Kucera et al., 2016). The maps were generated using Cytoscape (v3.2.1) Enrichment Map plugin (Merico et al., 2010; Isserlin et al., 2014).
Overview of the transcript profiles associated with hormone metabolism based on MapMan. The Affymetrix ATH1 microarray of treatment (A) and genotype comparison by using MapMan software (http://mapman.gabipd.org/web/guest/mapman). The log2fold-change is shown by color: for (A) red, upregulated by stress treatment; blue, downregulated by stress treatment; for (B) red, upregulated in pad2-1; blue downregulated in pad2-1. A Wilcoxon test with Benjamini–Hochberg multiple-testing correction was used to identify MapMan BINs that were significant, coordinately (FDR < 0.05) compared to the others.
Overview of the transcript profiles associated with secondary metabolism based on MapMan. For detail explanation, see the legend to Supplementary Figure S9.
Overview of the transcript profiles associated with cell wall metabolism based on MapMan. For detail explanation, see the legend to Supplementary Figure S9.
Venn diagrams representing the classification of genes based on microarray data. (A) Down-regulated genes by MV treatment and P-lim stress. (B) Genotype-dependent down-regulated genes.