Abstract
Bathymodiolus thermophilus, a mytilid mussel inhabiting the deep-sea hydrothermal vents of the East Pacific Rise, lives in symbiosis with chemosynthetic Gammaproteobacteria within its gills. The intracellular symbiont population synthesizes nutrients for the bivalve host using the reduced sulfur compounds emanating from the vents as energy source. As the symbiont is uncultured, comprehensive and detailed insights into its metabolism and its interactions with the host can only be obtained from culture-independent approaches such as genomics and proteomics. In this study, we report the first draft genome sequence of the sulfur-oxidizing symbiont of B. thermophilus, here tentatively named Candidatus Thioglobus thermophilus. The draft genome (3.1 Mb) harbors 3045 protein-coding genes. It revealed pathways for the use of sulfide and thiosulfate as energy sources and encodes the Calvin-Benson-Bassham cycle for CO2 fixation. Enzymes required for the synthesis of the tricarboxylic acid cycle intermediates oxaloacetate and succinate were absent, suggesting that these intermediates may be substituted by metabolites from external sources. We also detected a repertoire of genes associated with cell surface adhesion, bacteriotoxicity and phage immunity, which may perform symbiosis-specific roles in the B. thermophilus symbiosis.
Keywords: Uncultured endosymbiont, Hydrothermal vents, Marine invertebrate symbiosis, Thiotrophy, Autotrophy
Introduction
Chemoautotrophic bacteria form the base of the food-chain in deep sea hydrothermal vent ecosystems [1, 2]. Many of these chemoautotrophs live in highly integrated symbiotic associations with invertebrate hosts, such as mussels, clams and tube worms, which enables megafaunal communities to thrive in the otherwise uninhabitable vent ecosystem [3–5].
The mussel Bathymodiolus thermophilus, for example, a bivalve belonging to the family Mytilidae, densely populates the hydrothermal vent fields of the Galapagos Rift and of the East Pacific Rise between the latitudes 13 °N and 21 °S [6]. Although the animal’s food groove and digestive tract are reduced [6], B. thermophilus appears to be able to ingest and assimilate suspended particles by filter feeding [7].
The major part of the bivalve’s nutrition, however, is derived from its chemosynthetic symbionts [4, 8]. The sulfur-oxidizing bacteria live within specialized gill cells, so called bacteriocytes [9]. Provided with a steady supply of reduced sulfur from the vents, these symbionts synthesize organic compounds and thus feed their host [10, 11].
Investigations on the symbiont’s physiology have hitherto been limited by the inaccessibility of mussel samples and failure to culture the symbionts in vitro. Underlying metabolic pathways that facilitate the putative inter-exchange of nutrients between the symbiotic partners therefore remain unexplored. However, culture-independent methods, such as direct genomic, transcriptomic or proteomic analyses of symbiont-containing tissue or of enriched symbiont fractions have provided useful physiological information about various uncultured marine symbionts in the past [12–17]. In this study we used symbiont-enriched preparations from B. thermophilus gill tissue to assemble the first draft genome of the B. thermophilus symbiont in order to gain preliminary insights into its metabolic potential.
Organism information
Classification and features
B. thermophilus symbiont cells are coccoid or rod-shaped (Fig. 1). In electron micrographs, they typically appear as roundish forms, whose central region is light or transparent (looking “empty”), while the outermost regions of the cytoplasm are darker and more structured (Fig. 1 and [9]). Like most sulfur-oxidizing (thiotrophic) bivalve symbionts [4, 18], the B. thermophilus symbiont has a Gram-negative cell wall. With a diameter of 0.3–0.5 μm, B. thermophilus symbiont cells are of similar size as thiotrophic symbionts from other Bathymodiolus host species [19–22], and notably smaller than sulfur-oxidizing symbionts from other invertebrate hosts [4, 23]. In the host tissue, the symbionts are usually enveloped in large vacuoles. Groups of up to 20 symbionts within a single host vacuole have previously been reported by Fisher and colleagues [9]. Imaging of purified symbiont fractions from homogenized B. thermophilus gill tissue revealed, besides a large number of free symbiont cells, some intact vacuoles encompassing multiple symbionts (Fig. 1).
Fig. 1.
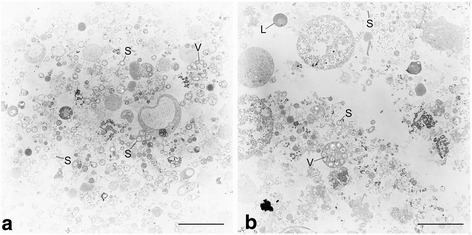
Transmission electron micrographs of Candidatus Thioglobus thermophilus. B. thermophilus gill tissue was homogenized in a glass tissue grinder and subjected to crude density gradient centrifugation using Histodenz™ gradient medium. Subsamples were taken from two visible bands and fixed for electron microscopy (a and b). Both subsamples contained numerous free symbiont cells (S) as well as some intact host vacuoles (V) containing several symbiont cells, besides various other cellular components and host tissue debris. L: Lipid drop or mucus. Scale bar: 5 μm. Electron microscopy method details: samples were fixed in a) 1% glutaraldehyde, 2% paraformaldehyde in IBS (imidazole-buffered saline; 0.49 M NaCl, 30 mM MgSO4*7H2O, 11 mM CaCl2*2H2O, 3 mM KCl, 50 mM imidazole) and b) in 2.5% glutaraldehyde, 1.25% paraformaldehyde in IBS. After embedding in low-gelling agarose and postfixation in 1% osmium tetroxide in cacodylate buffer (0.1 M cacodylate; pH 7.0), samples were dehydrated in a graded ethanol series (30 to 100%) and embedded in a mixture of Epon and Spurr (1:2). Sections were cut on an ultramicrotome (Reichert Ultracut, Leica UK Ltd., Milton Keynes, UK), stained with 4% aqueous uranyl acetate for 5 min followed by lead citrate for 1 min and analyzed with a transmission electron microscope LEO 906 (Zeiss, Oberkochen, Germany)
B. thermophilus symbionts reside intracellularly in bacteriocytes in their host’s gill tissue. Unlike some other Bathymodiolus species, such as B. azoricus that maintains a dual symbiosis with both sulfur-oxidizing and methane-oxidizing bacteria [24], B. thermophilus hosts only one type of bacterial endosymbionts. Based on 16S rRNA gene similarity [25], this sulfur-oxidizing symbiont population in B. thermophilus belongs to a single phylotype.
The B. thermophilus symbiont is a member of the 10.1601/nm.2068 (NCBI taxonomy ID 2360). It is closely related to symbionts of other Bathymodiolus species, and more distantly related to symbionts of other invertebrate hosts and to free-living 10.1601/nm.2068 from various marine habitats [26]. The B. thermophilus symbiont falls in a well-supported clade consisting of symbionts of other mytilid and vesicomyid bivalves and free-living gammaproteobacterial clones from marine vents and other submarine volcanic sites as shown in Fig. 2. Its closest relatives are the ‘Bathymodiolus aff. Thermophilus thioautotrophic gill symbiont’ from 32 °N EPR (NCBI taxonomy ID 363574; 99.85% similarity on the 16S rRNA level) and the Bathymodiolus brooksi symbiont from the Gulf of Mexico (NCBI taxonomy ID 377144; 99.53% similarity). According to our analysis, the B. thermophilus symbiont is only remotely similar to the sulfur-oxidizing symbionts of deep-sea vestimentiferan tube worms (90% 16S rRNA similarity) and of shallow water lucinid clams (87–90% similarity, see Fig. 2).
Fig. 2.
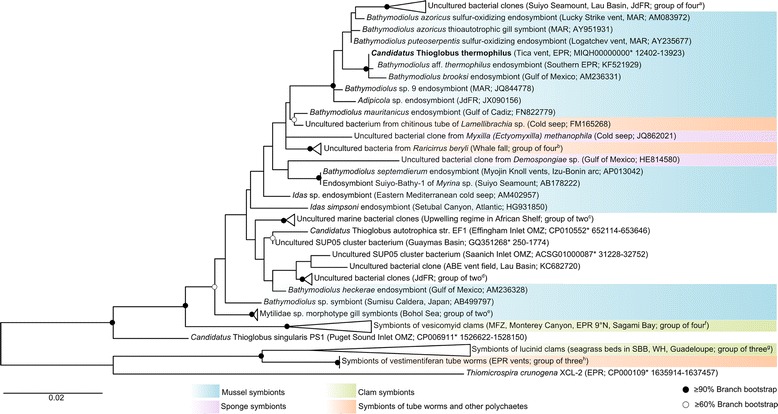
Phylogenetic tree of Candidatus Thioglobus thermophilus and related free-living and host-associated sulfur oxidizers. Ca. T. thermophilus, the thiotrophic symbiont of Bathymodiolus thermophilus, is displayed in bold. The tree was inferred from closely related 16S rRNA gene sequences obtained from the SILVA database using the SILVA Incremental Aligner (SINA) [51] and was estimated with the 16S rRNA sequence of 46 bacteria. The final alignment covered 1138 nucleotides. Sequence alignment and phylogenetic analysis were performed using the MEGA7 software tool [52]. The phylogenetic tree was constructed using the Maximum Likelihood method based on the Tamura-Nei model implemented in MEGA7 [53]. Branch bootstrap support values were calculated using 1000 replicates and are displayed as circles (black: ≥ 90%, white: ≥ 60%). For the sake of clarity some organisms were merged into groups (wedges): auncultured clones (KC682721, KC682765, JQ678401, AB193934); bwhale fall symbionts (HE814589, HE814588, HE814591 HE814585); cuncultured clones (FM246509, FM246513); duncultured clones (JQ678344, JQ678392); eMytilidae symbionts (AM503921, AM503923); fVesicomyidae symbionts (EU403432, EU403431, CP000488* 1081274–1,082,807, AP009247* 948400–949,934); gLucinidae symbionts (X84979, M99448, M90415); htube worm symbionts (NZ_AFOC01000137* 503–2033, DQ660821, NZ_AFZB01000059* 4132–5662). The lucinid clam symbionts, the vestimentiferan tube worm symbionts, and the free-living Thiomicrospira crunogena XCL-2 were included as outgroup. Branches that are not highlighted by colors represent free-living relatives. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. *these NCBI accession numbers refer to whole genome submissions and not to individually submitted 16S rRNA gene sequences (start and stop positions of the 16S rRNA gene are given after the asterisk). JdFR: Juan de Fuca Ridge, EPR: East Pacific Rise, MAR: Mid-Atlantic Ridge, OMZ: oxygen minimum zone, MFZ: Mendocino Fracture Zone, SBB: Santa Barbara Basin, WH: Woods Hole
The B. thermophilus symbiont’s closest cultured relative is the free-living Candidatus Thioglobus autotrophica, whose genome was recently sequenced [27]. The metabolic properties of both bacteria appear to be highly similar, as predicted from their genomes. Like the B. thermophilus symbiont genome presented in this study, the Ca. T. autotrophica genome encodes an incomplete TCA cycle. The high degree of 16S rRNA gene sequence similarity between Ca. T. autotrophica and the B. thermophilus symbiont (95%), suggests that both belong to the same genus. We therefore propose the tentative name Candidatus Thioglobus thermophilus for the thiotrophic B. thermophilus symbiont.
A summary of key features of Ca. T. thermophilus is given in Table 1.
Table 1.
Classification and general features of Candidatus Thioglobus thermophilus, the Bathymodiolus thermophilus gill endosymbiont, according to the MIGS recommendations [42]
| MIGS ID | Property | Term | Evidence code a |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [43] | |
| Phylum Proteobacteria | TAS [44] | ||
| Class Gammaproteobacteria | TAS [45] | ||
| Order Unclassified | NAS | ||
| Family Unclassified, S-oxidizing symbionts | TAS [46] | ||
| Genus Candidatus Thioglobus | TAS [25] | ||
| Species Candidatus Thioglobus thermophilus | TAS [25] | ||
| (Type) strain NA | NAS | ||
| Gram stain | Negative | TAS [18] | |
| Cell shape | Coccoid or short rods | TAS [9], IDA | |
| Motility | Non-motile inside the host; a putative free-living stage of the symbiont is likely motile, but no direct evidence exists |
NAS | |
| Sporulation | Unknown | ||
| Temperature range | 4 °C - 14 °C (shows psychrophilic growth characteristics) | TAS [10] | |
| Optimum temperature | Unknown | ||
| pH range; Optimum | 6.9–8.0; unknown | TAS [47] | |
| Carbon source | CO2 (autotroph) | TAS [10, 11, 48, 49] | |
| Energy source | H2S and S2O3 2− (chemotroph) | TAS [10, 49] | |
| Terminal electron acceptor | O2, NO3 2− | TAS [11] | |
| MIGS-6 | Habitat | Intracellular endosymbiont of marine bivalve inhabiting hydrothermal vents | IDA |
| MIGS-6.3 | Salinity | 35.42 PSU | IDA |
| MIGS − 22 | Oxygen | Aerobic (facultative) | TAS [11] |
| MIGS-15 | Biotic relationship | Symbiont | TAS [10, 11] |
| MIGS-14 | Pathogenicity | Non-pathogenic | NAS |
| MIGS-4 | Geographic location | East Pacific Rise (EPR) 9°N | IDA |
| MIGS-5 | Sample collection | January 2014 | IDA |
| MIGS-4.1 | Latitude | 9° 50.39′ N | IDA |
| MIGS-4.2 | Longitude | 104° 17.49′ W | IDA |
| MIGS-4.3 | Altitude | -2511 m | IDA |
aEvidence codes - IDA Inferred from Direct Assay, TAS Traceable Author Statement (i.e., a direct report exists in the literature), NAS Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [50]
Genome sequencing information
Genome project history
The genome of Candidatus Thioglobus thermophilus was sequenced to get a comprehensive insight into the metabolic potential of the bacterium. This project is part of a larger effort to compare the symbiont genomes from various Bathymodiolus species across different vent habitats in order to understand the possible effects of vent geochemistry in shaping host-symbiont evolution in Bathymodiolus. Sequencing and assembly of the symbiont genome were conducted at the Göttingen Genomics Laboratory (University of Göttingen, Germany) and at the Max-Planck-Institute of Marine Microbiology (Bremen, Germany), respectively. The sequences have been deposited in GenBank under the accession number MIQH00000000. A summary of the project information is shown in Table 2.
Table 2.
Project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Draft genome |
| MIGS-28 | Libraries used | Illumina 112 bp paired-end library (Nextera) |
| MIGS-29 | Sequencing platforms | Genome analyzer II x |
| MIGS-31.2 | Fold coverage | 86× |
| MIGS-30 | Assemblers | SPAdes v. 3.1.1 |
| MIGS-32 | Gene calling method | GeneMarkS+ (NCBI PGAP) |
| Locus Tag | BGC33 | |
| GenBank ID | o MIQH00000000 | |
| GenBank date of release | 11/16/2016 | |
| GOLD ID | - | |
| BioProject | PRJNA339702 | |
| MIGS-13 | Source material identifier | - |
| Project relevance | Vent ecosystems, Chemosynthetic symbioses, Environmental microbiology |
Growth conditions and genomic DNA preparation
Symbionts for genome sequencing were isolated from one single B. thermophilus host individual, which was collected during the R/V Atlantis cruise AT26–10 in January 2014. The mussel was collected from a diffuse-flow vent at the Tica vent field on the East Pacific Rise at 9° 50.39′ N, 104° 17.49′ W by the remotely operated vehicle (ROV) Jason. After recovery, the animal was dissected on board the research vessel and gill tissue was removed and homogenized in 1× PBS buffer (Dulbecco’s Phosphate Buffered Saline, Sigma-Aldrich order no. D5773). The resulting homogenate was diluted with 1× PBS (ratio 1:3) and subjected to multiple centrifugation steps (differential pelleting): In a first centrifugation step (500 × g, 5 min, 4 °C in a tabletop centrifuge using a swing-out rotor), crude host tissue debris and host cell nuclei were removed from the homogenate. The supernatant was centrifuged again (step 2) as described above to pellet residual host nuclei. The new supernatant was now centrifuged at maximum speed (step 3), i.e. at 15,000 × g for 20 min at 4 °C using a fixed-angle rotor. The resulting pellet contained enriched bacterial cells and was immediately frozen at −80 °C until genomic DNA preparation.
Genomic DNA was isolated from the purified bacteria using the MasterPure DNA Purification Kit (Epicentre) as recommended by the manufacturer.
Genome sequencing and assembly
Sequencing of the B. thermophilus symbiont genome was performed at the Göttingen Genomics Laboratory using the Illumina Genome Analyzer II x. A Nextera shotgun library was generated for a 112 bp paired-end sequencing run. Sequencing resulted in 7,569,934 paired-end reads. Adaptors were removed from the reads, quality-trimmed (Q = 2) with BBDuk and error-corrected with BBnorm (V35, sourceforge.net/projects/bbmap). The resulting reads were assembled with IDBA-UD [28]. To bin the symbiont genome from the metagenome assembly, we used gbtools [29] based on GC content and sequencing coverage. The corrected reads were mapped against the symbiont genome bin with BBmap and reassembled with SPAdes v. 3.1.1 [30]. This assembly resulted in 1341 contigs longer than 200 bp (1281 scaffolds). The completeness and contamination of the genome was estimated with CheckM [31]. The CheckM test showed 96.98% completeness of the genome with 11.32% contamination and 81.40% strain heterogeneity.
Genome annotation
All scaffolds were annotated using NCBI’s prokaryotic genome annotation pipeline (https://www.ncbi.nlm.nih.gov/genome/annotation_prok/), which uses the gene caller GeneMarkS+ together with a similarity-based gene detection approach [32, 33]. Predicted proteins were assigned Clusters of Orthologous Groups numbers and Protein Families domains by querying their sequences against the COG Database and the Pfam database, respectively, at NCBI (ftp://ftp.ncbi.nih.gov/pub/mmdb/cdd/little_endian/). Querying was done using the rpsblast application of the BLAST + −2.4.0 package with an E-value cutoff of 1 × 10−5 and 1 × 10−4, respectively, for COG and Pfam. To manually assign COG categories to the COG numbers returned by rpsblast, the COG category database was downloaded from the COG FTP server (ftp://ftp.ncbi.nih.gov/pub/COG/COG2014/data). For prediction of signal peptides the SignalP 4.1 Server [34], PECAS [35] and Phobius [36] were used. Transmembrane helices and CRISPR loci (CRISPR arrays) were predicted with TMHMM Server v. 2.0 [37] and the CRISPRFinder tool [38], respectively.
Genome properties
The properties of this genome are summarized in Table 3. The draft genome of the sulfur-oxidizing B. thermophilus symbiont contained 3,088,407 bp in 1281 scaffolds >200 bp. The average GC content was 37.7%. A total of 3097 genes were predicted, of which 3045 (98.3%) are predicted protein-encoding genes. The remaining 1.5% and 0.2%, respectively, consisted of RNA genes and pseudo genes. Of the protein-encoding genes, 54.5% and 65.2% were affiliated to COG- and Pfam-based functions, respectively. For an overview of predicted COG categories see Table 4.
Table 3.
Genome statistics
| Attribute | Value | % |
|---|---|---|
| Genome size (bp) a | 3,088,407 | 100 |
| DNA coding (bp) | 2,621,999 | 84.9 |
| DNA G + C (bp) | 1,164,329 | 37.7 |
| DNA scaffolds | 1281 | 100 |
| Total genes | 3097 | 100 |
| Protein-coding genes | 3045 | 98.3 |
| RNA genes | 46 | 1.5 |
| Pseudo genes | 6 | 0.2 |
| Genes in internal clusters | - | - |
| Genes with function prediction b | 2051 | 67.4 |
| Genes assigned to COGs | 1659 | 54.5 |
| Genes with Pfam domains | 1984 | 65.2 |
| Genes with signal peptides c | 337 | 11.1 |
| Genes with transmembrane helices | 626 | 20.6 |
| CRISPR repeats | 10 |
aAll 1281 scaffolds >200 bp. 478 of these (37.3%) are scaffolds >1000 bp, comprising 2,726,561 bp (88.3% of all base pairs)
bGenes with function prediction are all 3045 protein-coding genes minus those 994 genes annotated as “hypothetical proteins” that have no COG category or fall into the COG categories “unknown function” or “general function prediction only” and that have no Pfam domain or a Pfam “domain of unknown function”
cIncludes genes for which a signal peptide was predicted with at least two of the three tools used. Percentages of genes with function prediction, COGs, Pfam domains, signal peptides and transmembrane helices were calculated against a total of 3045 protein-coding genes
Table 4.
Number of genes associated with general COG functional categories
| Code | Value | % age | Description |
|---|---|---|---|
| J | 179 | 5.88 | Translation, ribosomal structure and biogenesis |
| A | 1 | 0.03 | RNA processing and modification |
| K | 50 | 1.64 | Transcription |
| L | 126 | 4.14 | Replication, recombination and repair |
| B | 0 | 0.00 | Chromatin structure and dynamics |
| D | 20 | 0.66 | Cell cycle control, cell division, chromosome partitioning |
| V | 81 | 2.66 | Defense mechanisms |
| T | 40 | 1.31 | Signal transduction mechanisms |
| M | 105 | 3.45 | Cell wall/membrane biogenesis |
| N | 6 | 0.20 | Cell motility |
| U | 47 | 1.54 | Intracellular trafficking and secretion |
| O | 99 | 3.25 | Posttranslational modification, protein turnover, chaperones |
| C | 115 | 3.78 | Energy production and conversion |
| G | 34 | 1.12 | Carbohydrate transport and metabolism |
| E | 117 | 3.84 | Amino acid transport and metabolism |
| F | 46 | 1.51 | Nucleotide transport and metabolism |
| H | 104 | 3.42 | Coenzyme transport and metabolism |
| I | 46 | 1.51 | Lipid transport and metabolism |
| P | 61 | 2.00 | Inorganic ion transport and metabolism |
| Q | 98 | 3.22 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 175 | 5.75 | General function prediction only |
| X | 32 | 1.05 | Mobilome: prophages, transposons |
| W | 9 | 0.30 | Extracellular structures |
| S | 68 | 2.23 | Function unknown |
| - | 1386 | 45.52 | Not in COGs |
The percentage is based on a total of 3045 protein-coding genes
Insights from the genome sequence
Sulfur-oxidizing symbionts of Bathymodiolus species are assumed to be horizontally transmitted, i.e., they supposedly enter their bivalve hosts from a free-living bacterial population in the environment, rather than being transferred from one mussel generation to the next [39]. The idea of a putative free-living stage of the symbiont in the hydrothermal vent environment is in accordance with our genome analysis: Unlike some insect symbionts, which are obligatorily dependent on their hosts and have a diminished genome [40], the B. thermophilus symbiont genome (3.1 Mb in size, see below) is not reduced. With the exception of the tricarboxylic acid cycle, which lacks three enzymes (see below), all necessary pathways for a host-independent life-style appear to be complete in the B. thermophilus symbiont’s genome.
Energy generation
The B. thermophilus symbiont uses reduced sulfur compounds such as sulfide and thiosulfate as its major energy sources [10]. As predicted from the genome sequence, sulfide and thiosulfate are oxidized to sulfate via the rDSR-APS-Sat pathway and the Sox multienzyme-complex, respectively. Oxygen and nitrate are used as final electron acceptors. Complete gene sets for these pathways are present in the symbiont genome.
CO2 fixation and carbon metabolism
The B. thermophilus symbiont genome furthermore encodes a modified version of the CO2-fixing Calvin-Benson-Bassham cycle: while the genes for sedoheptulose-7-phosphatase and fructose-1,6-bisphosphatase are missing, a pyrophosphate-dependent 6-phosphofructokinase is encoded, which potentially replaces the two other functions (as also described for the endosymbionts of Calyptogena magnifica [12], Riftia pachyptila [13] and Olavius algarvensis [16]). The B. thermophilus symbiont’s TCA cycle is incomplete, as the enzyme 2-oxoglutarate dehydrogenase is missing. Moreover, homologs of the enzymes malate dehydrogenase and succinate dehydrogenase are also lacking, similar to what was reported for the thiotrophic B. azoricus symbiont [17].
Nitrogen metabolism
The B. thermophilus symbiont possesses genes for assimilatory nitrate reduction, i.e. for nitrogen uptake from nitrate. Its genome also encodes the Nar complex, a membrane-bound respiratory nitrate reductase necessary for respiratory reduction of nitrate, indicating that nitrate can be used as an alternative electron acceptor besides oxygen. Several membrane transporters for the uptake of nitrate, nitrite and ammonia are also encoded.
Immunity and cell surface interactions
Of the 3045 protein-coding genes, 10.74% are predicted to contain Pfam domains related to bacterial cell surface adhesion, such as bacterial Ig-like domain proteins and cadherins, and to putative toxins, such as pore-forming RTX and MARTX cytotoxins. Another 2.17% of the protein-coding genes were associated with immunity against phages (CRISPR-Cas, restriction modification system and the Abi toxin-antitoxin system). This elaborate presence of genes associated with pathogenicity and phage defense, typical of pathogens and bacteriophages, was also observed in the related thiotrophic B. azoricus symbiont [17, 41]. This particular feature of Bathymodiolus symbionts is surprising since the bacteria a) reside in shielded intracellular niches, b) are beneficial symbionts for their host, and c) are not related to any known pathogen [26, 41]. Moreover, approximately 1.71% of the protein-coding B. thermophilus genes belonged to several classes of pathogenic and digestive peptidases. Membrane transporters of type I and type II secretion systems, which transport toxins and folded exoproteins such as peptidases, are also encoded. Although their exact roles have not been determined as yet, we postulate that these pathogenicity-related genes may be involved in protecting the symbionts against pathogens or phages or even perform symbiosis-specific functions, such as symbiont attachment to the host or defense against the host’s immune system, as suggested previously [41].
Conclusions
Sequencing of the uncultured B. thermophilus symbiont’s genome allowed preliminary insights into its genomic characteristics and metabolic potential. Candidatus Thioglobus thermophilus appears to solely rely on sulfide and thiosulfate as energy sources, as genes for the oxidation of other reduced compounds were absent from its genome. The absence of three genes encoding essential TCA cycle enzymes, which was recently also reported for the thiotrophic B. azoricus symbiont [17], may suggest that these genes are consistently missing in Bathymodiolus symbionts. The unusual presence of a repertoire of genes associated with cell adhesion, toxin production and phage immunity in the non-pathogenic B. thermophilus symbiont may point to a symbiosis-specific beneficial role of these functions other than pathogen defense.
Acknowledgements
We are indebted to the captain and the crews of the R/V Atlantis and ROV Jason for their expert help in obtaining the samples. Thanks to Julia Polzin for collecting symbiont samples used for TEM imaging and Annette Meuche for technical assistance during electron microscopy. We also thank Nikolaus Leisch for his help in interpreting transmission electron micrographs. We appreciate Nicole Dubilier’s advice with regard to symbiont naming as well as her help in providing us with the genome assembling facility and expertise.
Funding
This study was supported by the EU-funded Marie Curie Initial Training Network “Symbiomics” (project no. 264774). RP was supported by a fellowship of the Institute of Marine Biotechnology, Greifswald. MK was supported by a NSERC Banting Postdoctoral Fellowship. LS was supported by a DAAD scholarship. SMS was supported by US National Science Foundation grant OCE-1136727, which also funded the research cruise.
Abbreviations
- APS
Adenylyl-sulfate reductase
- EPR
East Pacific Rise
- rDSR
Reverse dissimilatory sulfite reductase
- Sat
ATP sulfurylase
- Sox
Sulfur oxidation
- TCA cycle
Tricarboxylic acid cycle
Authors’ contributions
RP submitted the genome, performed genome and phylogenetic analysis and drafted the manuscript. RP and HF collected mussel samples and purified the symbiont fractions. SMS was the Chief Scientist on the cruise and coordinated the sample collection. MK and StM developed the symbiont enrichment procedure. RS performed the electron microscopy. AT and RD conducted DNA isolation and genome sequencing. LS, MK and SEH assembled the genome, LS binned the genome and conducted quality control tests. LS and SEH helped with phylogenetic analyses. TS and StM supervised and coordinated the entire project. All authors reviewed and approved the final version of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Karl DM, Wirsen CO, Jannasch HW. Deep-sea primary production at the galapagos hydrothermal vents. Science. 1980;207(4437):1345–1347. doi: 10.1126/science.207.4437.1345. [DOI] [Google Scholar]
- 2.Felbeck H, Somero GN. Primary production in deep-sea hydrothermal vent organisms: roles of sulfide-oxidizing bacteria. Trends Biochem Sci. 1982;7(6):201–204. doi: 10.1016/0968-0004(82)90088-3. [DOI] [Google Scholar]
- 3.Felbeck H. Chemoautotrophic potential of the hydrothermal vent tube worm, Riftia pachyptila Jones (Vestimentifera) Science. 1981;213(4505):336–338. doi: 10.1126/science.213.4505.336. [DOI] [PubMed] [Google Scholar]
- 4.Cavanaugh CM. Symbiotic chemoautotrophic bacteria in marine invertebrates from sulfide-rich habitats. Nature. 1983;302(5903):58–61. doi: 10.1038/302058a0. [DOI] [Google Scholar]
- 5.Van Dover CL. The ecology of deep-sea hydrothermal vents. 1. New Jersey: Princeton University Press; 2000. [Google Scholar]
- 6.Kenk VC, Wilson BR. A new mussel (Bivalvia , Mytilidae) from hydrothermal vents in the galapagos rift-zone. Malacologia. 1985;26(1–2):253–271. [Google Scholar]
- 7.Page HM, Fiala-Medioni A, Fisher CR, Childress JJ. Experimental evidence for filter-feeding by the hydrothermal vent mussel Bathymodiolus thermophilus. Deep Sea Res (I Oceanogr Res Pap) 1991;38(12):1455–1461. doi: 10.1016/0198-0149(91)90084-S. [DOI] [Google Scholar]
- 8.Raulfs EC, Macko SA, Van Dover CL. Tissue and symbiont condition of mussels (Bathymodiolus thermophilus) exposed to varying levels of hydrothermal activity. J Mar Biol Assoc UK. 2004;84(1):229–234. doi: 10.1017/S0025315404009087h. [DOI] [Google Scholar]
- 9.Fisher CR, Childress JJ, Oremland RS, Bidigare RR. The importance of methane and thiosulfate in the metabolism of the bacterial symbionts of two deep-sea mussels. Mar Biol. 1987;96(1):59–71. doi: 10.1007/BF00394838. [DOI] [Google Scholar]
- 10.Nelson DC, Hagen KD, Edwards DB. The gill symbiont of the hydrothermal vent mussel Bathymodiolus thermophilus is a psychrophilic, chemoautotrophic, sulfur bacterium. Mar Biol. 1995;121(3):487–495. doi: 10.1007/BF00349457. [DOI] [Google Scholar]
- 11.Belkin S, Nelson DC, Jannasch HW. Symbiotic assimilation of CO2 in 2 hydrothermal vent animals, the mussel Bathymodiolus thermophilus and the tube worm Riftia pachyptila. Biol Bull. 1986;170(1):110–121. doi: 10.2307/1541384. [DOI] [Google Scholar]
- 12.Newton ILG, Woyke T, Auchtung TA, Dilly GF, Dutton RJ, Fisher MC, Fontanez KM, Lau E, Stewart FJ, Richardson PM, Barry KW, Saunders E, Detter JC, Wu D, Eisen JA, Cavanaugh CM. The Calyptogena magnifica chemoautotrophic symbiont genome. Science. 2007;315(5814):998–1000. doi: 10.1126/science.1138438. [DOI] [PubMed] [Google Scholar]
- 13.Markert S, Gardebrecht A, Felbeck H, Sievert SM, Klose J, Becher D, Albrecht D, Thurmer A, Daniel R, Kleiner M, Hecker M, Schweder T. Status quo in physiological proteomics of the uncultured Riftia pachyptila endosymbiont. Proteomics. 2011;11(15):3106–3117. doi: 10.1002/pmic.201100059. [DOI] [PubMed] [Google Scholar]
- 14.Gardebrecht A, Markert S, Sievert SM, Felbeck H, Thürmer A, Albrecht D, Wollherr A, Kabisch J, Le Bris N, Lehmann R, Daniel R, Liesegang H, Hecker M, Schweder T. Physiological homogeneity among the endosymbionts of Riftia pachyptila and Tevnia jerichonana revealed by proteogenomics. ISME J. 2012;6(4):766–776. doi: 10.1038/ismej.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bettencourt R, Pinheiro M, Egas C, Gomes P, Afonso M, Shank T, Santos RS. High-throughput sequencing and analysis of the gill tissue transcriptome from the deep-sea hydrothermal vent mussel Bathymodiolus azoricus. BMC Genomics. 2010;11:559. doi: 10.1186/1471-2164-11-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleiner M, Wentrup C, Lott C, Teeling H, Wetzel S, Young J, Chang YJ, Shah M, VerBerkmoes NC, Zarzycki J, Fuchs G, Markert S, Hempel K, Voigt B, Becher D, Liebeke M, Lalk M, Albrecht D, Hecker M, Schweder T, Dubilier N. Metaproteomics of a gutless marine worm and its symbiotic microbial community reveal unusual pathways for carbon and energy use. Proc Natl Acad Sci U S A. 2012;109(19):E1173–E1182. doi: 10.1073/pnas.1121198109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ponnudurai R, Kleiner M, Sayavedra L, Petersen JM, Moche M, Otto A, Becher D, Takeuchi T, Satoh N, Dubilier N, Schweder T, Markert S. Metabolic and physiological interdependencies in the Bathymodiolus azoricus symbiosis. ISME J. 2017;11(2):463–477. doi: 10.1038/ismej.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Pennec M, Hily A. Anatomie, structure et ultrastructure de la branchie d’un Mytilidae des sites hydrothermaux du Pacifique oriental. Oceanol Acta. 1984;7(4):517–523. [Google Scholar]
- 19.Fiala-Médioni A, Métivier C, Herry A, Pennec M. Ultrastructure of the gill of the hydrothermal-vent mytilid Bathymodiolus sp. Mar Biol. 1986;92(1):65–72. doi: 10.1007/BF00392747. [DOI] [Google Scholar]
- 20.Dubilier N, Windoffer R, Giere O. Ultrastructure and stable carbon isotope composition of the hydrothermal vent mussels Bathymodiolus brevior and B. sp. affinis brevior from the North Fiji Basin, western Pacific. Mar Ecol Prog Ser. 1998;165:187–193. doi: 10.3354/meps165187. [DOI] [Google Scholar]
- 21.Duperron S, Nadalig T, Caprais J-C, Sibuet M, Fiala-Médioni A, Amann R, Dubilier N. Dual symbiosis in a Bathymodiolus sp. mussel from a methane seep on the Gabon continental margin (Southeast Atlantic): 16S rRNA phylogeny and distribution of the symbionts in gills. Appl Environ Microb. 2005;71(4):1694–1700. doi: 10.1128/AEM.71.4.1694-1700.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kádár E, Bettencourt R, Costa V, Santos RS, Lobo-da-Cunha A, Dando P. Experimentally induced endosymbiont loss and re-acquirement in the hydrothermal vent bivalve Bathymodiolus azoricus. J Exp Mar Biol Ecol. 2005;318(1):99–110. doi: 10.1016/j.jembe.2004.12.025. [DOI] [Google Scholar]
- 23.Bright M, Sorgo A. Ultrastructural reinvestigation of the trophosome in adults of Riftia pachyptila (Annelida, Siboglinidae) Invertebr Biol. 2003;122(4):347–368. doi: 10.1111/j.1744-7410.2003.tb00099.x. [DOI] [Google Scholar]
- 24.Duperron S, Bergin C, Zielinski F, Blazejak A, Pernthaler A, McKiness ZP, DeChaine E, Cavanaugh CM, Dubilier N. A dual symbiosis shared by two mussel species, Bathymodiolus azoricus and Bathymodiolus puteoserpentis (Bivalvia: Mytilidae), from hydrothermal vents along the northern Mid-Atlantic Ridge. Environ Microbiol. 2006;8(8):1441–1447. doi: 10.1111/j.1462-2920.2006.01038.x. [DOI] [PubMed] [Google Scholar]
- 25.Distel DL, Lane DJ, Olsen GJ, Giovannoni SJ, Pace B, Pace NR, Stahl DA, Felbeck H. Sulfur-oxidizing bacterial endosymbionts: analysis of phylogeny and specificity by 16S rRNA sequences. J Bacteriol. 1988;170(6):2506–2510. doi: 10.1128/jb.170.6.2506-2510.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petersen JM, Wentrup C, Verna C, Knittel K, Dubilier N. Origins and evolutionary flexibility of chemosynthetic symbionts from deep-sea animals. Biol Bull. 2012;223(1):123–137. doi: 10.1086/BBLv223n1p123. [DOI] [PubMed] [Google Scholar]
- 27.Shah V, Morris RM. Genome Sequence of “Candidatus Thioglobus autotrophica” strain EF1, a chemoautotroph from the SUP05 clade of marine Gammaproteobacteria. Genome Announc. 2015;3(5):e01156–e01115. doi: 10.1128/genomeA.01156-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng Y, Leung HC, Yiu S-M, Chin FY. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28(11):1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 29.Seah BK, Gruber-Vodicka HR. gbtools: interactive visualization of metagenome bins in R. Front Microbiol. 2015;6. doi:10.3389/fmicb.2015.01451. [DOI] [PMC free article] [PubMed]
- 30.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25(7):1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29(12):2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44(14):6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 35.Cortazar AR, Oguiza JA, Aransay AM, Lavín JL. PECAS: prokaryotic and eukaryotic classical analysis of secretome. Amino Acids. 2015;47(12):2659–2663. doi: 10.1007/s00726-015-2058-2. [DOI] [PubMed] [Google Scholar]
- 36.Käll L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res. 2007;35(suppl 2):W429–WW32. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 38.Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35(Web Server issue):W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Won YJ, Hallam SJ, O'Mullan GD, Pan IL, Buck KR, Vrijenhoek RC. Environmental acquisition of thiotrophic endosymbionts by deep-sea mussels of the genus Bathymodiolus. Appl Environ Microbiol. 2003;69(11):6785–6792. doi: 10.1128/AEM.69.11.6785-6792.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gil R, Sabater-Muñoz B, Latorre A, Silva FJ, Moya A. Extreme genome reduction in Buchnera spp.: toward the minimal genome needed for symbiotic life. Proc Natl Acad Sci U S A. 2002;99(7):4454–4458. doi: 10.1073/pnas.062067299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sayavedra L, Kleiner M, Ponnudurai RP, Wetzel S, Pelletier E, Barbe V, Satoh N, Shoguchi E, Fink D, Breusing C, Reusch TB, Rosenstiel P, Schilhabel MB, Becher D, Schweder T, Markert S, Dubilier N, Petersen JM. Abundant toxin-related genes in the genomes of beneficial symbionts from deep-sea hydrothermal vent mussels. elife. 2015;4:e07966. doi: 10.7554/eLife.07966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, Ashburner M, Axelrod N, Baldauf S, Ballard S, Boore J, Cochrane G, Cole J, Dawyndt P, De Vos P, de Pamphilis C, Edwards R, Faruque N, Feldman R, Gilbert J, Gilna P, Glockner FO, Goldstein P, Guralnick R, Haft D, Hancock D, Hermjakob H, Hertz-Fowler C, Hugenholtz P, Joint I, Kagan L, Kane M, Kennedy J, Kowalchuk G, Kottmann R, Kolker E, Kravitz S, Kyrpides N, Leebens-Mack J, Lewis SE, Li K, Lister AL, Lord P, Maltsev N, Markowitz V, Martiny J, Methe B, Mizrachi I, Moxon R, Nelson K, Parkhill J, Proctor L, White O, Sansone SA, Spiers A, Stevens R, Swift P, Taylor C, Tateno Y, Tett A, Turner S, Ussery D, Vaughan B, Ward N, Whetzel T, Gil IS, Wilson G, Wipat A. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26(5):541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garrity G, Bell J, Phylum LT. XIV. Proteobacteria phyl. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey’s manual of systematic bacteriology. 2nd. New York: Springer; 2005. p. 1. [Google Scholar]
- 45.Garrity G, Bell J, Lilburn T. Class III. Gammaproteobacteria class. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey’s manual of systematic bacteriology. 2nd. New York: Springer; 2005. p. 1. [Google Scholar]
- 46.Rua CPJ, Thompson F. The Unclassified Genera of Gammaproteobacteria: Alkalimonas, Arenicella, Chromatocurvus, Congregibacter, Gallaecimonas, Halioglobus, Marinicella, Methylohalomonas, Methylonatrum, Orbus, Plasticicumulans, Porticoccus, Sedimenticola, Simiduia, Solimonas. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The prokaryotes: Gammaproteobacteria. 4th. Berlin: Springer; 2014. pp. 749–768. [Google Scholar]
- 47.Nedoncelle K, Lartaud F, Pereira LC, Yücel M, Thurnherr A, Mullineaux L, Le Bris N. Bathymodiolus growth dynamics in relation to environmental fluctuations in vent habitats. Deep Sea Res (I Oceanogr Res Pap) 2015;106:183–193. doi: 10.1016/j.dsr.2015.10.003. [DOI] [Google Scholar]
- 48.Felbeck H, Childress JJ, Somero GN. Calvin-Benson cycle and sulphide oxidation enzymes in animals from sulphide-rich habitats. Nature 1981;293(5830):291-93.
- 49.Fisher CR, Childress JJ, Arp AJ, Brooks JM, Distel D, Favuzzi JA, Felbeck H, Hessler R, Johnson KS, Kennicutt MC, Macko SA, Newton A, Powell MA, Somero GN, Soto T. Microhabitat Variation in the Hydrothermal Vent Mussel, Bathymodiolus thermophilus, at the Rose Garden Vent on the Galapagos Rift. Deep Sea Res (I Oceanogr Res Pap) 1988;35(10–11):1769–1791. doi: 10.1016/0198-0149(88)90049-0. [DOI] [Google Scholar]
- 50.The Gene Ontology C. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28(14):1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]


