Abstract
The major driver mutations of lung cancer, EGFR mutations and EML4‐ALK fusion, are mainly detected in terminal respiratory unit (TRU)‐type lung adenocarcinomas, which typically show lepidic and/or papillary patterns, but are rarely associated with a solid or invasive mucinous morphology. In order to elucidate the key genetic events in non‐TRU‐type lung cancer, we carried out whole‐exome sequencing on 43 non‐TRU‐type lung adenocarcinomas based on morphology (17 acinar, nine solid, and two enteric adenocarcinomas, and 15 adenocarcinomas with a mucinous morphology). Our analysis identified mutations in TP53 (16/43, 37.2%), KRAS (13/43, 30.2%), and NKX2‐1/TTF‐1 (7/43; 16.3%) as the top three significantly mutated genes, while the EGFR mutation was rare (1/43, 2.3%) in this cohort. Eight NKX2‐1/TTF‐1 mutations (five frameshift, two nonsense, and one missense) were identified, with one case harboring two distinct NKX2‐1/TTF‐1 mutations (one missense and one frameshift). Functional assays with the NK2 homeobox 1 (NKX2‐1)/thyroid transcription factor 1 (TTF‐1) mutants revealed that none of them retain the activity as a transcriptional factor. Histologically, invasive mucinous adenocarcinomas accounted for most of the NKX2‐1/TTF‐1 mutations (five cases), as well as one enteric and one acinar adenocarcinoma. Immunohistochemistry showed that the cohort was largely divided into TTF‐1‐postive/hepatocyte nuclear factor 4‐α (HNF4‐α)‐negative and TTF‐1‐negative/HNF4‐α‐positive groups. NKX2‐1/TTF‐1 mutations were exclusively found in the latter, in which the gastrointestinal markers, mucin 5AC and cytokeratin 20, were frequently expressed. Bisulfite sequencing revealed that the NKX2‐1/TTF‐1 gene body was highly methylated in NKX2‐1/TTF‐1‐negative cases, including those without the NKX2‐1/TTF‐1 mutations. The genetic or epigenetic inactivation of NKX2‐1/TTF‐1 may play an essential role in the development and aberrant differentiation of non‐TRU‐type lung adenocarcinomas.
Keywords: HNF4‐α, hypermethylation, NKX2‐1 mutation, non‐TRU type, TTF‐1
Lung cancer is the leading cause of cancer death in many developed countries, including the USA and Japan,1, 2 and adenocarcinoma is the most common histological subtype of primary lung cancer. Lung adenocarcinoma appears to represent a heterogeneous group of diseases that differ in histogenesis, key genetic events, smoking status, and other clinicopathological backgrounds.3 The existence of a distinct subset of lung adenocarcinomas arising from a TRU was proposed by Yatabe et al.4, 5. The TRU‐type lung adenocarcinomas are prevalent in Asian women and non‐smokers, histologically show non‐mucinous lepidic growth or papillary components, and frequently show strong expression of transcription factor NKX2‐1, also known as TTF‐1.4, 5 EGFR mutations and ALK fusions are predominantly detected in the TRU type,5, 6 suggesting a distinct molecular pathway for carcinogenesis in this type of lung adenocarcinoma.
In contrast to the advances achieved in the research for TRU‐type lung cancer, studies on non‐TRU‐type lung adenocarcinoma are limited and the mechanisms underlying its oncogenesis have not yet been elucidated in detail. Non‐TRU‐type adenocarcinomas may consist of various histological subtypes; Yatabe et al. 4 previously reported that non‐TRU‐type adenocarcinomas are poorly differentiated tumors with a solid morphology; whereas other studies indicated that mucinous adenocarcinomas, which are NKX2‐1/TTF‐1‐negative, may also be classified as the non‐TRU type.7 Zhang et al. 8 recently reported that the major subtype among the NKX2‐1/TTF‐1‐negative group was acinar adenocarcinomas, followed in frequency by the solid and invasive mucinous subtypes. These findings suggest that solid, acinar, and mucinous patterns are the representative morphologies of non‐TRU‐type lung adenocarcinomas.
In the present study, we undertook whole exome sequencing on lung adenocarcinomas with the non‐TRU morphology. Using this histology‐driven approach, we found that mutations in the NKX2‐1/TTF‐1 gene are exclusively present in NKX2‐1/TTF‐1‐negative lung adenocarcinomas that histologically include mucinous adenocarcinomas as well as enteric and acinar adenocarcinomas. Furthermore, our results showed that the NKX2‐1/TTF‐1 gene is specifically hypermethylated in NKX2‐1/TTF‐1‐negative cases, including those without the NKX2‐1/TTF‐1 mutation.
Materials and Methods
Tumor specimens
Tumor specimens were obtained from 43 patients who underwent lung cancer surgery at Jichi Medical University Hospital (Shimotsukeshi, Japan) between August 2010 and June 2015. During this period, 490 cases of lung adenocarcinoma were surgically resected in our University Hospital. We reviewed the pathology files of these cases and selected those that met the following histological criteria: (i) acinar or solid adenocarcinomas without a lepidic/papillary component; or (ii) carcinomas with a mucinous morphology. The histological slides of the selected cases were carefully re‐examined and the eligibility of the cases was confirmed by two of the authors (T.N. and D.M.). Some cases that met the histological criteria were excluded from the analysis because of extensive necrosis or paucity of cancer cells. According to the new WHO classification,9 42 cases were adenocarcinomas that comprised 13 invasive mucinous adenocarcinomas, one mucinous adenocarcinoma in situ (JMULK‐23), 17 acinar adenocarcinomas, nine solid adenocarcinomas, and two enteric adenocarcinomas. One rare case of adenosquamous carcinoma, composed of invasive mucinous adenocarcinoma and squamous cell carcinoma (JMULK‐34), was included in this analysis. This cohort, consisting of 42 adenocarcinomas and one adenosquamous carcinoma, is described as comprising tumors with the non‐TRU‐type histology in the present study for convenience.
Our cohort contains 24 men and 19 women, ranging in age from 41 to 83 years (average, 67.5 years). Each case was re‐examined for the TNM classification and pathological stage based on the new International Association for the Study of Lung Cancer staging system.10 One case was classified as stage 0, 25 were stage I (1 stage IA1, 8 stage IA2, 6 stage IA3, and 10 stage IB), 7 were stage II (1 stage IIA and 6 stage IIB), 9 were stage III (6 stage IIIA, 2 stage IIIB, and 1 stage IIIC), and the stage of one case was undetermined because a limited surgery without lymph node dissection was carried out. Informed consent was obtained from all patients, and the study was approved by the Institutional Ethics Review Committee. Patient charts and CT scans were reviewed (by S.T.) to document the smoking status of patients and presence of the UIP pattern, respectively. Only cases in which the presence of the UIP pattern was suspected on CT scans and histologically confirmed by pathological specimens were considered to be positive for the UIP pattern.
Immunohistochemical evaluation
Formalin‐fixed, paraffin‐embedded tumor specimens were analyzed by immunohistochemistry with antibodies to NKX2‐1/TTF‐1, Napsin A, HNF4‐α, MUC5AC, MUC2, CDX2, and CK20. Tissue sections were treated with 0.3% hydrogen peroxide in methanol for 30 min to block endogenous peroxidase activity. Tissue sections for NKX2‐1/TTF‐1, HNF4‐α, MUC2, and CDX2 were then autoclaved in 10 mM citrate buffer (pH 6.0) at 120°C for 10 min, while those for Napsin A, MUC5AC, and CK20 were heated in 10 mM citrate buffer (pH 6.0) at 95°C for 10 min in a microwave oven. Sections were then preincubated with 10% normal horse serum in PBS, incubated with mouse monoclonal anti‐human NKX2‐1/TTF‐1 (M3575, clone 8G7G3/1) from Dako (Glostrup, Denmark) at a dilution of 1:100, mouse monoclonal anti‐human Napsin A (10221, clone TMU‐Ad02) from Immuno‐Biological Laboratories (Gunma, Japan) at a dilution of 1:400, mouse monoclonal anti‐human HNF4‐α (H1415; Perseus Proteomics, Tokyo, Japan) at a dilution of 1:60, mouse monoclonal anti‐human MUC5AC (NCL‐MUC‐5AC) from Leica Biosystems (Wetzlar, Germany) at a dilution of 1:100, mouse monoclonal anti‐human MUC2 (NCL‐MUC‐2) from Leica Biosystems at a dilution of 1:100, rabbit monoclonal anti‐human CDX2 (NBP1‐40553) from Novus Biologicals (Minneapolis, MN, USA) at a dilution of 1:500, and mouse monoclonal anti‐human CK20 (M7019, clone Ks20.8) from Dako at a dilution of 1:100 at 4°C overnight. All antibodies were detected with streptavidin–HRP conjugate according to the manufacturer's instructions. 3,3′‐Diaminobenzidine tetrahydrochloride was used as a chromogen, and hematoxylin was used as a light counterstain. When staining was carried out on cancer tissue sections, a positive control section was also stained, and we always confirmed that positive control cells were correctly stained. Immunohistochemical staining was evaluated independently by two pathologists (D.M. and T.Y.). Immunoreactivity was scored based on the percentage of cells that stained positively: 0, negative; 1+, <10%; 2+, 10–50%; 3+, >50%. Only foci with the adenocarcinoma component were evaluated for the adenosquamous carcinoma case. The expression of each marker antibody (NKX2‐1/TTF‐1, HNF4‐α, MUC5AC, MUC2, CDX2, and CK20) in a tumor was defined as positive when 10% of tumor cells or greater were stained (scores 2+ and 3+) and negative when <10% were stained (scores 0 and 1+).
Cell lines
HEK293T cells, human non‐small‐cell lung cancer H1299 and H2228, and 3T3 mouse fibroblast cell lines were obtained from ATCC (Manassas, VA, USA), and maintained in DMEM‐F12 supplemented with 10% FBS (both from Thermo Fisher Scientific, Waltham, MA, USA).
Whole‐exome sequencing
Exon fragments were isolated from the genomic DNA of patient samples using a SureSelect Human All Exon kit (Agilent Technologies, Santa Clara, CA, USA), and were subjected to next‐generation sequencing analyses with the HiSeq2500 platform (Illumina, San Diego, CA, USA) with the paired‐end option. We selected sequence reads with a Quality Value of <20 at each base from raw reads, and further extracted unique reads that were subsequently mapped to the reference human genome sequence (hg38) with BWA‐MEM (http://bio-bwa.sourceforge.net/), Bowtie 2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml), and NovoAlign (http://www.novocraft.com/products/novoalign/) independently. Somatic mutations were called by MuTect (http://www.broadinstitute.org/cancer/cga/mutect), SomaticIndelDetector (http://www.broadinstitute.org/cancer/cga/node/87), and VarScan (http://varscan.sourceforge.net). Mutations were discarded if: (i) they had a read depth of <20 or allele frequency for tumor of <0.1; (ii) they were supported by only one strand of the genome; and (iii) they were already present in the “1000 genomes” database (http://www.1000genomes.org) or in normal human genome alterations of our in‐house database. Gene mutations were annotated by SnpEff (http://snpeff.sourceforge.net). In order to validate HiSeq2500 data, the corresponding regions of genomic DNA prepared from tumor as well as paired normal tissue were PCR amplified, ligated to the plasmid pT7Blue‐2 (Novagen, Madison, WI, USA), and then subjected to Sanger sequencing or high‐throughput sequencing with the MiSeq system (Illumina). Significantly mutated genes were identified by MutSigCV calculations (q value <0.1).11
RNA sequencing
Complementary DNAs were prepared from tumor tissue with the use of a NEBNext Ultra Directional RNA Library Prep Kit (New England BioLabs, Ipswich, MA, USA) and subjected to HiSeq2500 sequencing for 100 bp from both ends. The paired‐end reads were aligned to the hg 19 human genome assembly with TopHat2.12 The expression level of each gene was calculated by using Cufflinks (http://cole-trapnell-lab.github.io/cufflinks/), and gene fusions were detected by deFuse (https://bitbucket.org/dranew/defuse) pipeline.
Bisulfite sequencing
Genomic DNA was modified by bisulfite conversion using the EpiTect Bisulfite kit (Qiagen, Valencia, CA, USA). The PCR primer sets targeting NKX2‐1/TTF‐1 were designed with the MethPrimer software,13 and listed in Table S1. Modified DNA was PCR amplified using AmpliTaq Gold DNA Polymerase (Thermo Fisher Scientific) with these 10 primer sets, and subjected to high‐throughput sequencing.
Isolation of full‐length NKX2‐1/TTF‐1 cDNA
Full‐length cDNA for wild‐type human NKX2‐1/TTF‐1 (NM_003317) was amplified from the cDNA of the H2228 cell line by PCR with PrimeSTAR HS DNA polymerase (Takara Bio, Shiga, Japan) using the following primers: 5′‐GAATCATGTCGATGAGTCCAAAGC‐3′ and 5′‐GGTTGTTAAGAAAAGTCGAAGCGC‐3′, and its mutant forms were generated using PrimeSTAR Max DNA polymerase (Takara Bio). The primers for mutagenesis sequences are listed in Table S2. All cDNA sequences were verified by Sanger sequencing, and ligated into the pMXS retroviral vector (kindly provided by Dr. Toshio Kitamura, Institute of Medical Science, University of Tokyo, Tokyo, Japan).
Luciferase reporter assays
H1299 cells were transfected with pMXS‐based expression plasmids for various NKX2‐1/TTF‐1 mutants, a firefly luciferase‐based MYBPH reporter plasmid,14 and the pGL4.75 plasmid for Renilla luciferase (Promega, Madison, WI, USA) using the Lipofectamine 3000 reagent (Thermo Fisher Scientific). The luciferase activities of the transfected cells were measured using the Dual‐Glo Luciferase Assay System (Promega). Data are means ± SD of values from three separate experiments. Statistical analyses were carried out with the two‐sided tests indicated. A P‐value of <0.05 was considered to be significant.
Functional analyses of novel fusion genes
Methods for functional analyses are described in Appendix S1.
Accession codes
Raw sequencing data have been deposited in the Japanese Genotype‐Phenotype Archive, which is hosted by the DNA Databank of Japan, under accession number JGAS00000000105.
Results
NKX2‐1/TTF‐1 mutations in non‐TRU‐type lung adenocarcinomas
We analyzed 43 cases with the non‐TRU‐type histology using next‐generation sequencing, and selected significantly mutated genes using MutSigCV calculations.11 Figure 1 shows the profile of the mutated genes, together with the data on EGFR mutations as well as EML4‐ALK and other fusions identified in this cohort (Table S3).
Figure 1.
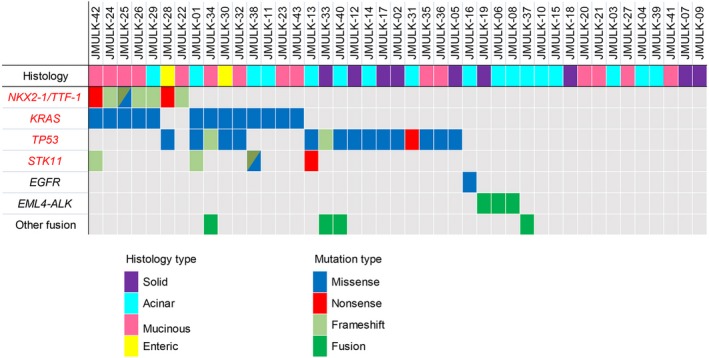
Gene mutations and fusions in the screening cohort of patients with non‐terminal respiratory unit‐type lung adenocarcinomas with the histological type. The histology status of each patient is shown in the upper panel. Four genes (red text) were listed as significantly mutated genes using MutSigCV calculations (q value <0.1). EGFR, EML4‐ALK, and other fusion candidates in this cohort are shown together in the same panel. Among 15 mucinous adenocarcinomas, JMUK‐34 was mucinous adenocarcinoma with squamous differentiation, JMUK‐23 was mucinous adenocarcinoma in situ, and the others were invasive mucinous adenocarcinomas.
Non‐synonymous mutations in the NKX2‐1/TTF‐1 gene were detected in seven cases (16.3%). The incidence of NKX2‐1/TTF‐1 mutations in this cohort was markedly higher than that (<1%) in the large cohort of lung adenocarcinomas reported by Hwang et al.15 NKX2‐1/TTF‐1 ranked among the top three significantly mutated genes, together with TP53 (16 cases, 37.2%) and KRAS (13 cases, 30.2%) in this cohort. KRAS mutations were simultaneously detected in five of seven (71.4%) NKX2‐1/TTF‐1 mutants, whereas neither EGFR mutations nor ALK fusions were identified in NKX2‐1/TTF‐1 mutant cases.
In total, eight independent NKX2‐1/TTF‐1 non‐synonymous mutations were detected: Ser305fs, Gly307fs, Gln172Glu, Ser187fs, Ala296fs, Phe168*, Ala52fs, and Gln170*. One case (JMULK‐25) harbored two mutations (one frameshift and one missense). The positions of these mutations within the NKX2‐1/TTF‐1 protein are diagrammatically shown in Figure 2. Four mutations are within the homeobox domains, whereas three are found within the C‐terminal region that plays an essential role for its transcriptional factor activity.16 Each mutation was confirmed by Sanger sequencing as shown in Figure S1 and Table S4. Hwang et al.15 identified 12 NKX2‐1/TTF‐1 mutations (three nonsense and nine frameshift mutations). Although there was no identical mutation between Hwang et al.'s study and ours, the distribution of NKX2‐1/TTF‐1 mutations appears similar in the two studies; of the total 20 mutations (12 Hwang et al.'s and eight our findings), 10 mutations were within the homeobox domain, and five mutations were within the C‐terminal region, suggesting that NKX2‐1/TTF‐1 mutations are frequent in the homeodomain.
Figure 2.
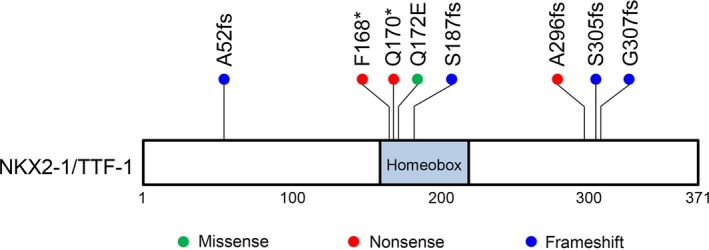
Domain organization of NK2 homeobox 1 (NKX2‐1)/thyroid transcription factor 1 (TTF‐1) showing the identified mutations. A schematic representation of human NKX2‐1/TTF‐1 showing the positions of non‐synonymous mutations detected in this cohort. Numbers refer to amino acid residues. A missense mutation is shown as a green point, nonsense as red, and frameshift as blue.
In order to test whether these mutations affect the NKX2‐1/TTF‐1 activity, we examined their transcriptional effects on the promoter of MYBPH, a previously reported NKX2‐1/TTF‐1 target gene.14 As shown in Figure 3, the MYBPH reporter activity was reduced to a basal level for all NKX2‐1/TTF‐1 mutants except Q172E (P < 0.05). This may be because only Q172E was a missense mutation, while the other mutations were null mutations (frameshift or nonsense mutations), and this suppression tendency was also observed for the Q172E mutant (P < 0.1), strongly suggesting that all NKX2‐1/TTF‐1 mutants found in this study are inactivating forms.
Figure 3.
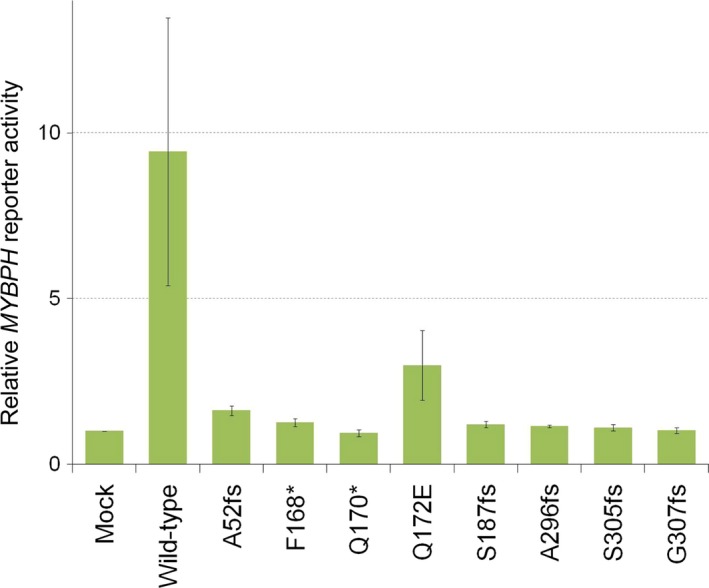
Mutant NK2 homeobox 1 (NKX2‐1)/thyroid transcription factor 1 (TTF‐1) is the inactive form. Luciferase reporter activity was measured for H1299 cells transfected with pMXS (Mock) or pMXS‐based expression plasmids for the wild‐type or A52fs, F168*, Q170*, Q172E, S187fs, A296fs, S305fs, or G307fs mutant form of NKX2‐1/TTF‐1 as well as with a MYBPH reporter plasmid and the pGL4.75 plasmid for Renilla luciferase. Data represent firefly luciferase activity normalized by Renilla luciferase activity and the amounts of the corresponding proteins, and are shown as the means ± SD of three independent experiments. Statistical analyses were carried out with the two‐sided tests indicated. A P‐value of <0.05 was considered to be significant. The MYBPH reporter activity was reduced to basal levels for all NKX2‐1/TTF‐1 mutants other than Q172E (P < 0.05), and this suppression tendency was also observed for the Q172E mutant (P = 0.06).
Immunohistochemical features, smoking status, and relationship with the UIP pattern in non‐TRU‐type adenocarcinomas
We undertook immunostaining for NKX2‐1/TTF‐1, Napsin A, and gastrointestinal markers such as MUC2, MUC5AC, HNF4‐α, CDX2, and CK20 in our cohort of 43 lung adenocarcinomas. As shown in Figure 4, NKX2‐1/TTF‐1 staining was positive in 16 cases (37.2%), 15 of which largely overlapped with those that stained positive for Napsin A (18 cases, 41.9%). These NKX2‐1/TTF‐1‐positive cases, although lacking the TRU‐type morphology, appeared to have arisen from the TRU.
Figure 4.
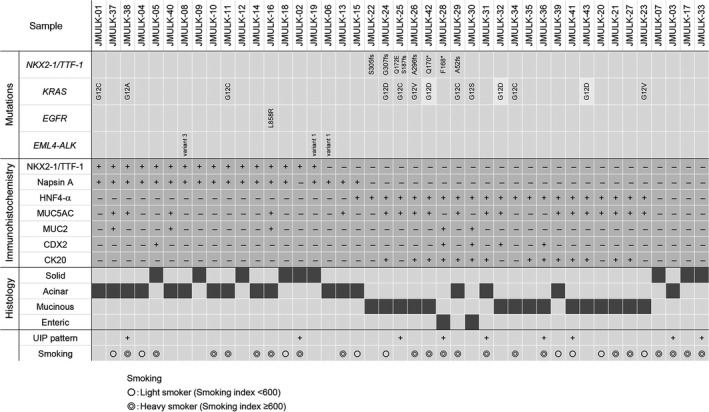
Genetic status of NKX2‐1/TTF‐1, KRAS, EGFR, and EML4‐ALK (upper panel), immunohistochemical expression of NK2 homeobox 1 (NKX2‐1)/thyroid transcription factor 1 (TTF‐1), Napsin A, hepatocyte nuclear factor 4‐α (HNF4‐α), mucin (MUC)5AC, MUC2, caudal‐type homeobox 2 (CDX2), and cytokeratin 20 (CK20) (middle panel), and histological subtypes (lower panel) of 43 cases of lung adenocarcinoma with the non‐terminal respiratory unit‐type morphology. We also added clinical information on the presence or absence of the usual interstitial pneumonitis (UIP) pattern on computed tomography and smoking history.
Regarding gastrointestinal markers, HNF4‐α was positive in 21 cases (48.8%), MUC5AC in 19 (44.2%), MUC2 in 5 (11.6%), CDX2 in 5 (11.6%), and CK20 in 14 (32.6%). Expression of NKX2‐1/TTF‐1 and HNF4‐α was mutually exclusive (Fig. 4), and TTF‐1‐postive/HNF4‐α‐negative and TTF‐1‐negative/HNF4‐α‐positive cases appeared to constitute two major groups in the cohort. Hepatocyte nuclear factor 4‐α has been reported as a marker of invasive mucinous adenocarcinomas,17, 18 and we confirmed that all invasive mucinous adenocarcinomas were positive for HNF4‐α (Fig. 4). However, two cases of enteric adenocarcinoma and four of acinar adenocarcinomas were also positive for HNF4‐α (Figs 4, 5). Hepatocyte nuclear factor 4‐α‐positive cases frequently showed expression of the gastrointestinal markers MUC5AC (14 of 21, 66.7%) and CK20 (14 of 21, 66.7%), and included most MUC5AC‐ and CDX2‐positive cases (14 of 19, 73.7%; 4 of 5, 80.0%, respectively) and all CK20‐positive cases (14 of 14, 100%) (Figs 4, 5).
Figure 5.
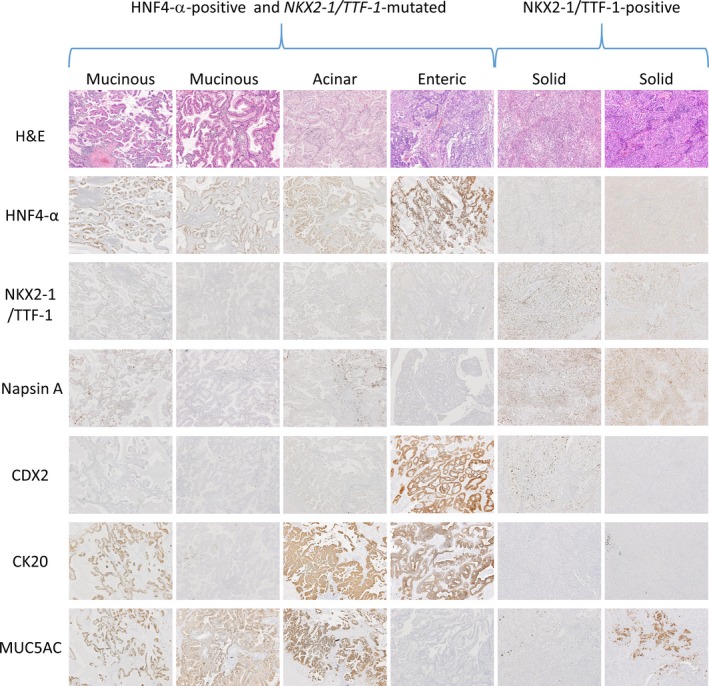
Staining of sections from six cases with the non‐terminal respiratory unit‐type morphology, including four representative cases of hepatocyte nuclear factor 4‐α (HNF4‐α)‐positive and NKX2‐1/TTF‐1‐mutated adenocarcinomas (two mucinous adenocarcinomas, one acinar adenocarcinoma, and one enteric adenocarcinoma) (left side) and two representative cases of NK2 homeobox 1 (NKX2‐1)/thyroid transcription factor 1 (TTF‐1)‐positive adenocarcinomas (two solid adenocarcinomas) (right side) for H&E, HNF4‐α, NKX2‐1/TTF‐1, Napsin A, caudal‐type homeobox 2 (CDX2), cytokeratin 20 (CK20), and mucin 5AC (MUC5AC).
Masai et al.19 recently reported that invasive mucinous adenocarcinoma was the most common histotype in adenocarcinoma with the UIP pattern. Non‐TRU‐type adenocarcinomas are often associated with a history of heavy smoking.7, 8 In our cohort, nine cases (20.9%) were associated with the UIP pattern. Thirty cases (69.8%) had a history of smoking, 21 of which (48.8% of the whole cohort) had a smoking index of 600 or more.
Mutations in the NKX2‐1/TTF‐1 gene are exclusively detected in HNF4‐α‐positive, NKX2‐1/TTF‐1‐negative adenocarcinomas
Recent findings obtained using transgenic mice suggest that the loss of NKX2‐1/TTF‐1 expression activates an intrinsic program of gastrointestinal differentiation in Kras‐driven, but not Egfr‐driven lung adenocarcinomas.20, 21 Therefore, it was of interest to compare the genetic status of NKX2‐1/TTF‐1, EGFR, KRAS, and EML4‐ALK with the NKX2‐1/TTF‐1 and HNF4‐α expression patterns and histological subtypes. As shown in Figure 4, all NKX2‐1/TTF‐1 mutated cases were NKX2‐1/TTF‐1‐negative and HNF4‐α‐positive. NKX2‐1/TTF‐1‐mutated adenocarcinomas showed various histological patterns; mucinous adenocarcinomas (n = 5), enteric adenocarcinoma (n = 1), and acinar adenocarcinoma (n = 1).
The EGFR mutation (n = 1) and EML4‐ALK fusion (n = 3) were only detected in HNF4‐α‐negative cases, whereas KRAS mutations were present in HNF4‐α‐positive cases (10 of 21, 47.6%) and NKX2‐1/TTF‐1‐positive cases (three of 16, 18.8%). Five cases, including four mucinous adenocarcinomas and one enteric adenocarcinoma, harbored NKX2‐1/TTF‐1 and KRAS mutations (Fig. 4).
Hypermethylation of the NKX2‐1/TTF‐1 gene in NKX2‐1/TTF‐1‐negative adenocarcinomas
NKX2‐1/TTF‐1 mutations were only detected in seven of 27 (25.9%) NKX2‐1/TTF‐1‐negative cases. We speculated that the loss of NKX2‐1/TTF‐1 function might be caused by not only NKX2‐1/TTF‐1 mutations, but also epigenetic silencing. In order to test this hypothesis, we undertook a methylation analysis on the NKX2‐1/TTF‐1 gene in 43 cases. The results obtained are shown in Figure 6. Significant difference in the expression level of NKX2‐1/TTF‐1 was observed between the NKX2‐1/TTF‐1 immunostaining‐negative (orange column) and ‐positive (green column) groups (P = 0.001477). Three cases (JMULK‐03, ‐42, and ‐36) were heavily methylated in almost the entire NKX2‐1/TTF‐1 gene including the 5′‐upstream region, while two cases (JMULK‐05 and ‐10) were not. The regions at which significant difference in the methylated level was observed are positioned in intron 2 and exon 3 rather than at the promoter region of NKX2‐1/TTF‐1. In seven NKX2‐1/TTF‐1 mutation‐positive cases, two were extensively methylated (JMULK‐42 and ‐26) and two (JMULK‐25 and ‐24) were not. In the case of JMULK‐22, ‐28, and ‐29, expression was considered to be decreased by epigenetic silencing as well as inactivation by the NKX2‐1/TTF‐1 inactive form.
Figure 6.
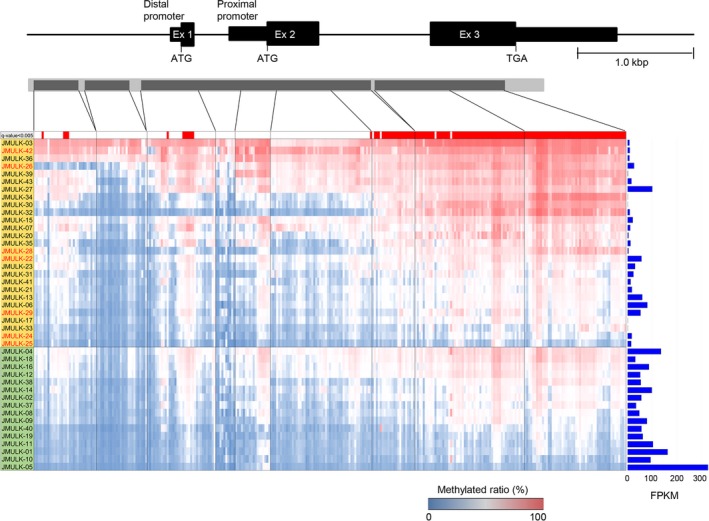
Methylation status of NKX2‐1/TTF‐1. The presence or absence of methylated cytosine was confirmed for CpG islands in almost the entire region of NK2 homeobox 1 (NKX2‐1)/thyroid transcription factor 1 (TTF‐1) including upstream of the 5′‐translated region. There are two isoforms in NKX2‐1/TTF‐1: a major form that is encoded by mRNA harboring exons 2 and 3, and a minor isoform encoded by all exons. There are proximal and distal promoters. In this heat map, the horizontal axis shows cytosine, which may be methylated among PCR‐amplified regions (dark gray bar regions within light gray bar). The region at which cytosine was converted to uracil by a bisulfite treatment is indicated by blue, and the region not converted is indicated by red. The methylation site is indicated by red, unmethylated cytosine by blue, and the methylated ratio is by a blue–red color bar in the lower panel. In NKX2‐1/TTF‐1 immunostaining negative cases (orange column) and positive cases (green column), the sums of the methylation ratio of cytosine that may be methylated at 303 sites in each sample were calculated and arranged in descending order of their values. The expression level of NKX2‐1/TTF‐1 is shown as the FPKM value calculated from RNA analysis data on the right side of the heat map. In the sample column, seven NKX2‐1/TTF‐1 mutation‐positive cases are indicated using red letters. Methylation regions where there are differences between positive and negative groups for NKX2‐1/TTF‐1 immunostaining are red‐coded in the upper part of the heat map (q value <0.005).
Identification of novel fusion genes
We identified three novel fusion genes in the present study; a fusion between the NTPCR and ACBD6 genes, a fusion between TMEM53 and ASPHD2, and a fusion between DNAJC3 and B3GLCT (Figs S2, Table S3). In order to examine the transforming potential of these fusions, we carried out a 3T3 focus formation assay, but failed to observe transformed foci in any of the three fusions (data not shown).
Discussion
In the present study, we frequently detected inactivating mutations in the NKX2‐1/TTF‐1 gene (seven of 43, 16.3%) of lung adenocarcinomas showing the non‐TRU‐type histology, namely acinar and solid adenocarcinoma, and adenocarcinoma with a mucinous morphology. While this study was underway, Hwang et al.15 reported NKX2‐1/TTF‐1 mutations in invasive lung adenocarcinoma. They analyzed a “discovery set” that consisted of 21 cases of invasive mucinous adenocarcinomas, mixed mucinous/non‐mucinous adenocarcinomas, and adenocarcinomas with mucinous features. They found NKX2‐1/TTF‐1 mutations in four of 21 cases (19.0%), comprised of three invasive mucinous adenocarcinomas and one mixed mucinous/non‐mucinous adenocarcinoma. Additional analyses of a large panel of unselected lung adenocarcinomas (n = 954) revealed NKX2‐1/TTF‐1 mutations in four invasive mucinous adenocarcinomas (0.42%). The findings reported by Hwang et al. and the present results collectively show that NKX2‐1/TTF‐1 mutations may be rare in lung adenocarcinomas in total, but are enriched in non‐TRU‐type adenocarcinomas including not only invasive mucinous adenocarcinomas, but also enteric adenocarcinomas, a rare distinct histological entity reported by Inamura et al.,22 and some acinar adenocarcinomas.
The frequency of NKX2‐1/TTF‐1 mutations of HNF4‐α‐positive cases was 33.3% (seven of 21) in our discovery set. Although we did not confirm whether our results reflected the whole picture of NKX2‐1/TTF‐1 mutations of HNF4‐α‐positive cases, this frequency was not particularly high compared with the frequency of NKX2‐1/TTF‐1 mutations of invasive mucinous adenocarcinomas (43%; three of seven) reported in Hwang's study,15 considering that HNF4‐α was reported to be a specific marker for invasive mucinous adenocarcinoma.18
NKX2‐1/TTF‐1 may act as a lineage‐specific oncogene or tumor suppressor in lung adenocarcinomas.23, 24 Therefore, it is important to investigate whether NKX2‐1/TTF‐1 mutations are activating or inactivating. We herein revealed that all NKX2‐1/TTF‐1 mutants observed are inactivating mutations, using reporter assays.
NKX2‐1/TTF‐1‐mutated cases were all negative for NKX2‐1/TTF‐1, suggesting that the NKX2‐1/TTF‐1 mutations are not only functionally inactive, but also downregulated in their expression. Because all of the seven NKX2‐1/TTF‐1‐mutated cases harbored nonsense or frameshift mutations of the NKX2‐1/TTF‐1 gene, we speculate that these mutant NKX2‐1/TTF‐1 mRNAs could be targeted for decay through the nonsense‐ or non‐stop‐mediated mRNA decay pathways.25, 26
Non‐TRU‐type lung adenocarcinomas that are typically NKX2‐1/TTF‐1‐negative account for 15–20% of lung adenocarcinomas.5, 7, 8 Due to the low incidence of NKX2‐1/TTF‐1 mutations in lung adenocarcinomas, it was of interest to investigate whether NKX2‐1/TTF‐1 is epigenetically silenced in NKX2‐1/TTF‐1 WT, non‐TRU‐type adenocarcinomas. We found that the NKX2‐1/TTF‐1 gene was hypermethylated in NKX2‐1/TTF‐1‐negative cases, including those without NKX2‐1/TTF‐1 mutations. These novel results strongly support the inactivation of NKX2‐1/TTF‐1 by genetic and/or epigenetic mechanisms occurring in a whole spectrum of non‐TRU‐type lung adenocarcinomas, including not only invasive mucinous adenocarcinomas, but also enteric and some acinar adenocarcinomas. Some NKX2‐1/TTF‐1 negative cases (for example, JMULK‐27 and ‐33) showed neither hypermethylation nor mutation of the NKX2‐1/TTF‐1 gene. In these cases, other mechanisms of epigenetic silencing, such as miRNA, and histone modification may be involved.
Previous studies identified a subset of lung adenocarcinomas that ectopically express various proportions of gastrointestinal markers such as MUC5AC, MUC2, CDX2, CK20, and HNF4‐α.7, 8, 17, 18, 22 This aberrant differentiation is often associated with the negative expression of NKX2‐1/TTF‐1 and the non‐TRU‐type histology.7, 8, 17, 18, 22 Recent studies using genetically engineered mouse models of lung cancer have shown that deletions of the NKX2‐1/TTF‐1 gene induced mucinous adenocarcinomas in Kras‐driven lung adenocarcinomas.20, 21 Based on these findings, NKX2‐1/TTF‐1 has been suggested to repress the gastric differentiation program in the lung, and the loss of NKX2‐1/TTF‐1 leads to reactivation of the intrinsic gastric differentiation program, resulting in the induction of mucinous adenocarcinomas.21 The results of the present study, in combination with previous findings, provide a molecular basis for the development of non‐TRU‐type lung adenocarcinomas. We speculate that the genetic or epigenetic inactivation of NKX2‐1/TTF‐1 may be an essential step in the early stage of the development of non‐TRU‐type lung adenocarcinomas.
The timing of NKX2‐1/TTF‐1 mutations in lung carcinogenesis remains unclear; it has not yet been established whether NKX2‐1/TTF‐1 mutations occur before or after KRAS mutations. In this setting, it is important to note that non‐TRU‐type cancer is strongly associated with a heavy smoking history and the presence of UIP pattern.7, 8, 19 An analysis of early or precursor lesions potentially occurring in these backgrounds could provide insights into these issues.
Disclosure Statement
The authors have no conflict of interest.
Abbreviations
- CDX2
caudal‐type homeobox 2
- CK20
cytokeratin 20
- CT
computed tomography
- HNF4‐α
hepatocyte nuclear factor 4‐α
- MUC
mucin
- NKX2‐1
NK2 homeobox 1
- TRU
terminal respiratory unit
- TTF‐1
thyroid transcription factor 1
- UIP
usual interstitial pneumonia
Supporting information
Appendix S1. Materials and Methods.
Fig. S1. Sequence chromatogram of NKX2‐1/TTF‐1 mutation‐positive patients.
Fig. S2. Schematic representation of three novel fusion proteins.
Fig. S3. Sequence chromatogram of three novel gene fusions.
Table S1. Primer sequence for bisulfite PCR.
Table S2. Primer sequence for NKX2‐1/TTF‐1 mutagenesis PCR.
Table S3. Summary of significantly mutated genes.
Table S4. Nucleotide substitution ratio of NKX2‐1/TTF‐1.
Acknowledgments
We would like to thank Tomoko Tamura and Miyoko Noguchi, both from Jichi Medical University, for technical assistance and secretarial work, respectively. We also thank Saori Sugaya at The University of Tokyo for technical support.
Cancer Sci 108 (2017) 1888–1896
Funding Information
This study was supported in part by JSPS KAKENHI Grant Numbers 20390103 (to T.N.), 26293080 (to T.N.), 16K08672 (to. D.M.), and 26461155 (to M.S.), Subsidies to Private Universities and MEXT‐supported Program for the Strategic Research Foundation at Private Universities from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to T.N.), grants for Leading Advanced Projects for Medical Innovation (LEAP) (to H.M.), and the Project for Cancer Research and Therapeutic Evolution (P‐CREATE) (to M.S. and M.K.), both from the Japan Agency for Medical Research and Development.
References
- 1. Statistics and Information Department . Vital Statistics, 2000. Tokyo: Ministry of Health, Labor and Welfare, 2001. [Google Scholar]
- 2. Jemal A, Siegel R, Ward E et al Cancer statistics, 2006. CA Cancer J Clin 2006; 56: 106–30. [DOI] [PubMed] [Google Scholar]
- 3. Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non‐small‐cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 2014; 14: 535–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yatabe Y, Mitsudomi T, Takahashi T. TTF‐1 expression in pulmonary adenocarcinomas. Am J Surg Pathol 2002; 26: 767–73. [DOI] [PubMed] [Google Scholar]
- 5. Yatabe Y, Kosaka T, Takahashi T, Mitsudomi T. EGFR mutation is specific for terminal respiratory unit type adenocarcinoma. Am J Surg Pathol 2005; 29: 633–9. [DOI] [PubMed] [Google Scholar]
- 6. Inamura K, Takeuchi K, Togashi Y et al EML4‐ALK lung cancers are characterized by rare other mutations, a TTF‐1 cell lineage, an acinar histology, and young onset. Mod Pathol 2009; 22: 508–15. [DOI] [PubMed] [Google Scholar]
- 7. Kim YK, Shin DH, Kim KB et al MUC5AC and MUC5B enhance the characterization of mucinous adenocarcinomas of the lung and predict poor prognosis. Histopathology 2015; 67: 520–8. [DOI] [PubMed] [Google Scholar]
- 8. Zhang Y, Wang R, Li Y et al Negative thyroid transcription factor 1 expression defines an unfavorable subgroup of lung adenocarcinomas. J Thorac Oncol 2015; 10: 1444–50. [DOI] [PubMed] [Google Scholar]
- 9. Travis WD, Brambilla E, Burke AP et al World Health Organization Classification of Tumours of the Lung, Pleura, Thymus and Heart, 4th ed Lyon: International Agency for Research on Cancer Press, 2015. [DOI] [PubMed] [Google Scholar]
- 10. Goldstraw P, Chansky K, Crowley J et al The IASLC lung cancer staging project: proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J Thorac Oncol 2016; 11: 39–51. [DOI] [PubMed] [Google Scholar]
- 11. Lawrence MS, Stojanov P, Polak P et al Mutational heterogeneity in cancer and the search for new cancer‐associated genes. Nature 2013; 499: 214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013; 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics 2002; 18: 1427–31. [DOI] [PubMed] [Google Scholar]
- 14. Hosono Y, Yamaguchi T, Mizutani E et al MYBPH, a transcriptional target of TTF‐1, inhibits ROCK1, and reduces cell motility and metastasis. EMBO J 2012; 31: 481–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hwang DH, Sholl LM, Rojas‐Rudilla V et al KRAS and NKX2‐1 mutations in invasive mucinous adenocarcinoma of the lung. J Thorac Oncol 2016; 11: 496–503. [DOI] [PubMed] [Google Scholar]
- 16. De Felice M, Damante G, Zannini M, Francis‐Lang H, Di Lauro R. Redundant domains contribute to the transcriptional activity of the thyroid transcription factor 1. J Biol Chem 1995; 270: 26649–56. [DOI] [PubMed] [Google Scholar]
- 17. Kunii R, Jiang S, Hasegawa G et al The predominant expression of hepatocyte nuclear factor 4α (HNF4α) in thyroid transcription factor‐1 (TTF‐1)‐negative pulmonary adenocarcinoma. Histopathology 2011; 58: 467–76. [DOI] [PubMed] [Google Scholar]
- 18. Sugano M, Nagasaka T, Sasaki E et al HNF4α as a marker for invasive mucinous adenocarcinoma of the lung. Am J Surg Pathol 2013; 37: 211–8. [DOI] [PubMed] [Google Scholar]
- 19. Masai K, Tsuta K, Motoi N et al Clinicopathological, immunohistochemical, and genetic features of primary lung adenocarcinoma occurring in the setting of usual interstitial pneumonia pattern. J Thorac Oncol 2016; 11: 2141–9. [DOI] [PubMed] [Google Scholar]
- 20. Maeda Y, Tsuchiya T, Hao H et al Kras(G12D) and Nk2–1 haploinsufficiency induce mucinous adenocarcinoma of the lung. J Clin Invest 2012; 122: 4388–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Snyder EL, Watanabe H, Magendantz M et al Nk2–1 represses a latent gastric differentiation program in lung adenocarcinoma. Mol Cell 2013; 50: 185–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Inamura K, Satoh Y, Okumura S et al Pulmonary adenocarcinomas with enteric differentiation: histologic and immunohistochemical characteristics compared with metastatic colorectal cancers and usual pulmonary adenocarcinomas. Am J Surg Pathol 2005; 29: 660–5. [DOI] [PubMed] [Google Scholar]
- 23. Yamaguchi T, Hosono Y, Yanagisawa K, Takahashi T. NKX2‐1/TTF‐1: an enigmatic oncogene that functions as a double‐edged sword for cancer cell survival and progression. Cancer Cell 2013; 23: 718–23. [DOI] [PubMed] [Google Scholar]
- 24. Winslow MM, Dayton TL, Verhaak RG et al Suppression of lung adenocarcinoma progression by Nk2–1. Nature 2011; 473: 101–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vasudevan S, Peltz SW, Wilusz CJ. Non‐stop decay – a new mRNA surveillance pathway. BioEssays 2002; 24: 785–8. [DOI] [PubMed] [Google Scholar]
- 26. Baker KE, Parker R. Nonsense‐mediated mRNA decay: terminating erroneous gene expression. Curr Opin Cell Biol 2004; 16: 293–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Materials and Methods.
Fig. S1. Sequence chromatogram of NKX2‐1/TTF‐1 mutation‐positive patients.
Fig. S2. Schematic representation of three novel fusion proteins.
Fig. S3. Sequence chromatogram of three novel gene fusions.
Table S1. Primer sequence for bisulfite PCR.
Table S2. Primer sequence for NKX2‐1/TTF‐1 mutagenesis PCR.
Table S3. Summary of significantly mutated genes.
Table S4. Nucleotide substitution ratio of NKX2‐1/TTF‐1.


