Abstract
Overcoming tumor heterogeneity is a major challenge for personalized treatment of gastric cancer, especially for human epidermal growth factor receptor‐2 targeted therapy. Analysis of circulating tumor DNA allows a more comprehensive analysis of tumor heterogeneity than traditional biopsies in lung cancer and breast cancer, but little is known in gastric cancer. We assessed mutation profiles of ctDNA and primary tumors from 30 patients with advanced gastric cancer, then performed a comprehensive analysis of tumor mutations by multiple biopsies from five patients, and finally analyzed the concordance of HER2 amplification in ctDNA and paired tumor tissues in 70 patients. By comparing with a single tumor sample, ctDNA displayed a low concordance of mutation profile, only approximately 50% (138/275) somatic mutations were found in paired tissue samples, however, when compared with multiple biopsies, most DNA mutations in ctDNA were also shown in paired tumor tissues. ctDNA had a high concordance (91.4%, Kappa index = 0.784, P < 0.001) of HER2 amplification with tumor tissues, suggesting it might be an alternative for tissue. It implied that ctDNA‐based assessment could partially overcome the tumor heterogeneity, and might serve as a potential surrogate for HER2 analysis in gastric cancer.
Keywords: Advanced gastric cancer, ctDNA, HER2 amplification, heterogeneity, next‐generation sequencing
Although the incidence of gastric cancer (GC) has decreased in the Western countries, it remains one of the most common cancers worldwide, with the highest incidence rates in Eastern Asia. In China, GC is the second most common cancer and the second leading cause of mortality among cancers, with approximately 679 100 new cases diagnosed and approximately 498 000 deaths in 2015.1 Treatment for GC that at early stage may include surgery, chemotherapy, radiation therapy or chemoradiation. However, treatment for advanced gastric cancer (AGC) is still challenging, most of AGC may be treated but rarely can be cured.2, 3, 4 Although Trastuzumab has been demonstrated to increased overall survival in HER2‐positive AGC patients,5 there were only 13%–22% of GC patients have HER2 overexpression or amplification, and heterogeneous HER2 expression was observed in 71% of positive samples.6 Therefore, taking heterogeneous HER2 expression profile into account, it may require multiple biopsies for HER2 testing in GC. However, multiple biopsies might result in unpleasant side effects and also increase the economic burden.
One alternative to overcome these issues is using circulating tumor DNA (ctDNA) for analysis. Recent progress in ctDNA has shown that it is possible to reconstruct the tumor genome from plasma. ctDNA released from tumor carries genomic and epigenomic alternations, it may be a potential surrogate to reveal gene tumor mutational spectrum.6, 7, 8 ctDNA can be applied in detection of point mutations, copy number variation (CNV), rearrangements, microsatellite instability (MSI), loss of heterozygosity (LOH) and DNA methylation.9 Multiple studies have shown that the mutation profile detected by ctDNA has a good concordance with paired tumor tissues, with 70% in lung cancer and 39%–55% in colorectal and rectal cancer.10, 11 ctDNA analysis also facilitates the assessment of intra‐patient multiple drug resistance mechanisms caused by tumor heterogeneity in non‐small cell lung cancer (NSCLC).12 However, in gastric cancer, if the utility of ctDNA‐based assessment could overcome the intra‐tumor heterogeneity is still largely unknown.
In the current study, we used next generation sequencing (NGS) to assess ctDNA status and compared mutation profile with tumor tissues. We found that ctDNA could partially represent mutation profile of multiple tumor biopsies, with overcoming the heterogeneity of gastric cancer. Finally, we confirmed the concordance of HER2 amplification in ctDNA with IHC/DISH results of paired tumor tissues. Our results thus provided a support for the utility of ctDNA in overcoming tumor heterogeneity in gastric cancer.
Methods
Ethics statement
This study was approved by the Medical Ethics Committee of Peking University Cancer Hospital and performed according to the Declaration of Helsinki Principles. Seventy patients with pathologically confirmed AGC treated in our department at Peking University Cancer Hospital between April 2010 and October 2015 were recruited. All patients signed informed consent for their samples to be used in a future study. All clinical data and samples were received anonymously.
Patients’ cohort and study design
In this study, 70 patients with pretreated AGC (53 males and 17 females; median age, 57 years; range, 29–76 years) were enrolled. In 30 patients of them, matched plasma sample and single tumor tissue biopsy were obtained. Both sample types conducted targeted sequencing using a 483 cancer‐related genes panel, and the concordance between which was compared. To further explore whether plasma ctDNA can overcome tissue heterogeneity, another five patients with AGC each provided five tumor tissue biopsies and one paired plasma sample, the samples were also tested using NGS. To evaluate the performance of HER2 amplification in ctDNA, other 35 patients with single tumor tissue and paired plasma sample were enrolled. In total, plasma samples from 70 patients were sequenced for HER2 CNV using a 483 genes panel, and paired tumor tissue biopsies conducted HER2 test using dual‐color in situ hybridization (DISH) and immunohistochemistry (IHC) as reference. We compared the concordance of HER2 status to explore the performance of HER2 amplification detection in ctDNA. The patients’ clinical data including sex, age, primary tumor site, differentiation, Lauren classification, metastasis and therapy status were also collected from the medical records. Table 1 summarizes the patients’ characteristics. All blood samples (10 mL from each patients) and tumor tissue samples were collected simultaneously.
Table 1.
The clinic pathological characteristics of patients
| Characteristics | No. patients (%) |
|---|---|
| Sex | |
| Male | 53 (75.7%) |
| Female | 17 (24.3%) |
| Age (years) | |
| Median (range) | 57 (29‐76) |
| Primary tumor site | |
| Gastroesophageal junction | 25 (35.7%) |
| Non‐gastroesophageal junction | 45 (64.3%) |
| Differentiation† | |
| Good | 26 (37.1%) |
| Poor | 44 (62.9%) |
| Lauren classification | |
| Intestinal | 34 (48.6%) |
| Diffuse | 28 (40.0%) |
| Mixed | 8 (11.4%) |
| Liver metastasis | |
| Yes | 37 (52.9%) |
| No | 33 (47.1%) |
| Peritoneal metastasis | |
| Yes | 15 (21.4%) |
| No | 55 (78.6%) |
| Number of organs with metastasis | |
| <3 | 52 (74.3%) |
| ≥3 | 18 (25.7%) |
| Therapy status‡ | |
| Surgery | 1 (1.4%) |
| Chemotherapy | 49 (70.0%) |
| Trastuzumab therapy | 20 (28.6%) |
†Good, including high or moderate differentiation; Poor, including low differentiation, mucinous adenocarcinoma, and signet‐ring cell carcinoma. ‡Cheotherapy, patients with HER2‐negative AGC received oxapliplatin‐based rigemen; Trastuzumab,patients with HER2 positive AGC received oxapliplatin‐based regimen plus trastuzumab.
HER2 amplification detected using immunohistochemistry and dual‐color in situ hybridization
HER2 protein expression was tested using anti‐HER2/neu antibody (4B5; Roche, Basel, Switzerland) by IHC staining according to a previous report.13 HER2 amplification was determined using Ventana HER2 dual‐color ISH assay (DISH; Ventana Medical Systems, Inc., Tucson, AZ, USA). Amplification of HER2 by DISH was defined as a ratio HER2/CEP17 ≥2.2, and two independent specialists who were blinded to the study read the result.
Genomic DNA extraction, library preparation, and sequencing
Genomic DNA was extracted from tumor tissues using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). 10 mL peripheral blood samples were collected in cfDNA BCT tubes (Streck Laboratories, Omaha, NE, USA), stored at room temperature and processed within 72 h. Each tube was centrifuged at 1600 g for 10 min at 4°C. Pellets containing peripheral blood lymphocytes were stored at −20°C for future use. Aliquots of plasma were centrifuged at 16 000 g for another 10 min. ctDNA were extracted from plasma using a QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany), and genomic DNA from peripheral blood lymphocytes was extracted using the RelaxGene Blood DNA System (TianGen Biotech Co. Ltd., Beijing, China) which was used as a putative of germline mutation to filter variants in plasma. All DNA samples were stored at –80°C for future use after quantification with a Nanodrop 2000 (Thermo Fisher Scientific, Wilmington, DE, USA) or quantitative polymerase chain reaction.
A DNA library was prepared using a KAPA Hyper Prep Kit according the manufacturer's instruction (KAPA BIOSYSTEMS, Boston, MA, USA), followed by Agilent's SureSelectXT Target Enrichment System for Illumina Paired‐End Sequencing Library Protocol (Agilent Technologies, Santa Clara, CA, USA). An Agilent QPCR NGS Library Quantification kit (Agilent Technologies) was used to quantify the concentration of the DNA library, and DNA libraries with an average insert size of 150 bp (tumor tissues and lymphocytes) or 170 bp (plasma) (Fig. S1) were sequenced on an Illumina HiSeq 2000 system (Illumina, San Diego, CA, USA).
A custom designed panel, which covered 483 cancer‐related genes, were used in this study. All genes including 7011 exons (483 genes) and 94 introns from 18 genes that frequently rearranged in solid tumors (gene list in Table S1).
Sequencing quality control and alignment
The quality control was performed by filtering out the adapter sequences and low‐quality reads, and the final Q20 and Q30 of all samples were >90% and >85%, respectively. The sequencing reads were aligned to the human reference hg19 genome using the BWA software with default parameters, and duplications were marked with Picard. The alignment's results required coverage of the target region >99% and the mapping rate ≥95%. Average depth per sample required at least 200× in tissue samples and lymphocytes, while at least 1000× in plasma.
Variants calling
The workflow of gene variation calling was shown in Fig. S2. Pre‐alignment QC was performed by in‐house software for each sample. Clean reads were obtained after trimmed adaptor sequence and low mapping quality reads. Reads aligned to reference hg19 were performed with BWA. Germline mutations, somatic single nucleotide variants (SNVs) and somatic insertions and deletions (InDels) were called using Samtools, Mutect, and Strelka, respectively. Somatic mutations were determined using the following filters: (i) the minimum average sequencing depth of the target for tissue samples was at least 200× and at least 1000× for plasma DNA; (ii) the minimum number of reads carrying the mutation was ≥5; and (iii) variant allele frequency ≥0.2%. Considering the false negative of Mutect results, the mutations with the following criteria were retrieved: (i) mutation frequency of ctDNA <1%; (ii) reads in plasma and tumor tissues samples carrying mutations >10; (iii) mutations those listed in COSMIC database. All of these variations were annotated with ANNOVAR software. The CNV analysis in this study was performed using Event‐Wise Testing algorithm based on read depth of coverage according to previous report.14 To detect CNV, a neutral copy number level of each exon of each gene must be established by comparison with the matched lymphocyte sample.
Statistical analysis
Statistical analysis was performed using SAS software, version 9.2 (SAS Institute, Cary, NC, USA). The concordance of HER2 amplification between tumor tissue and plasma ctDNA was analyzed by Kappa and McNemar's tests. Values of P < 0.05 on a two‐sided test were considered statistically significant.
Results
Somatic mutations in tumor and matched plasma samples of 30 patients with AGC
Considering high heterogeneity in tumor tissue of gastric cancer and somatic genetic mutations can be detected in ctDNA, we firstly assessed the concordance of gene mutation profiles between tumor tissue DNA (tDNA) and plasma ctDNA, a total of 30 tDNA and paired ctDNA samples were both sequenced using a 483‐gene panel. We assessed the gene‐level concordance between tumor tissue and ctDNA. The four most commonly altered genes in both tumor tissue and ctDNA were TP53 (15/30, 12/30), FGF14 (3/30, 2/30), SMAD4 (3/30,2/30), PIK3CA (2/30, 3/30). There was no significant difference between tumor tissue and ctDNA on gene‐level. We then calculated the number of somatic SNVs and InDels detected in tDNA and ctDNA or in both sample type, and considered the overlap mutations between each paired sample in each sample. In a total, there were 220 and 275 somatic SNVs in tDNA and ctDNA, respectively. There were 138 overlap somatic SNVs. For the somatic InDels, it identified 13 and 10 mutations in tDNA and ctDNA and the overlap mutation was 8 (Fig. 1 and Table S2). Half of somatic SNVs (50%, 137/275) which detected in ctDNA were not found in tDNA and 37% (82/220) of somatic SNVs detected in tDNA were neither in ctDNA. Low concordance between tissue tDNA and plasma ctDNA was noted.
Figure 1.
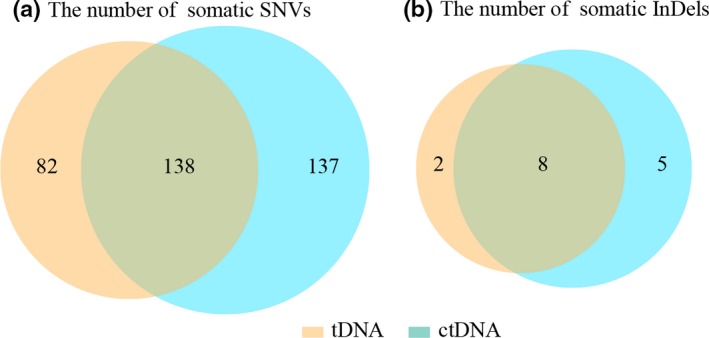
Concordance with tissue tDNA and plasma ctDNA. Thirty paired tissue and plasma samples were sequenced by a 483 genes panel; the number of somatic mutations were calculated and the overlapping mutations were also considered. (a) 220 and 275 somatic SNVs were identified in tissue tDNA and ctDNA, respectively, and the overlapping somatic SNVs were 138. (b) 10 and 15 somatic InDels were detected in tissue tDNA and plasma ctDNA, and the overlapping somatic InDels were 8.
ctDNA overcomes heterogeneity in tumor tissue samples
Since the varied concordance between ctDNA and tDNA of 30 patients, we next analyzed heterogeneity in tumor tissue and evaluated whether ctDNA could overcome heterogeneity in AGC. A total of 25 tumor tissues and five plasma samples derived from five patients (five tumor biopsies and one paired plasma sample per patient) were analyzed by NGS. Based on our results, tumor heterogeneity was identified in each patient. From these five patients, we discerned that the numbers of somatic SNVs and InDels in the plasma samples differed from those of the paired tumor tissues; however, the mutated genes identified in the plasma were all detected in one or more matched tumor tissues, which demonstrated that plasma ctDNA could partially overcome tumor heterogeneity (Table 2).
Table 2.
Number of Somatic single nucleotide variants (SNVs) and InDel in multiple tumor tissues and paired plasma
| Sample | T1 | T2 | T3 | T4 | T5 | pl | T1 shared with pl | T2 shared with pl | T3 shared with pl | T4 shared with pl | T5 shared with pl |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of Somatic SNVs | |||||||||||
| 35 | 13 | 9 | 12 | 8 | 10 | 7 | 7 | 7 | 7 | 7 | 7 |
| 36 | 1 | 6 | 0 | 1 | 0 | 2 | 1 | 2 | 0 | 1 | 0 |
| 37 | 56 | 42 | 37 | 39 | 30 | 45 | 36 | 40 | 35 | 35 | 25 |
| 38 | 11 | 10 | 12 | 10 | 10 | 1 | 1 | 1 | 1 | 1 | 1 |
| 39 | 1 | 10 | 0 | 0 | 9 | 1 | 0 | 1 | 0 | 0 | 1 |
| Number of Somatic InDel | |||||||||||
| 35 | 4 | 5 | 3 | 4 | 5 | 5 | 4 | 5 | 3 | 4 | 5 |
| 36 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 37 | 3 | 3 | 2 | 3 | 3 | 3 | 3 | 3 | 2 | 3 | 3 |
| 38 | 2 | 2 | 2 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| 39 | 1 | 3 | 1 | 2 | 4 | 1 | 1 | 1 | 1 | 1 | 1 |
T1, T2, T3, T4, T5, and pl represented tumor 1, tumor 2, tumor 3, tumor 4, tumor 5, and plasma.
Concordance of HER2 amplification between tumor tissue and plasma ctDNA
We further assessed HER2 amplification using ctDNA from all the 70 AGC patients, and compared with IHC/DISH results of HER2 expression level in paired tumor tissues, which was considered as a golden standard method in clinical‐pathology. As shown in Table 3, in this 70 patients’ cohort, it identified 20 patients with HER2 amplification in ctDNA, 16 of which were consistent with tumor tissue tested by IHC/DISH; in other 50 patients without HER2 amplification in ctDNA, two of which was positive in tumor tissue tested by IHC/DISH. However, in 52 patients without HER2 overexpression by IHC/DISH test, we identified four of which with HER2 amplified in ctDNA. The overall concordance rate between tDNA and matched plasma ctDNA was 91.43% (64/70), with a sensitivity of 88.89% (95% CI: 63.93%–98.05%), and specificity of 92.31% (95% CI: 80.60%–97.51%). The Kappa test showed no statistically significant between ctDNA and tumor tissue (Kappa index = 0.7835, P = 0.4142). Figure 2 showed HER2 amplification in ctDNA (HER2 gene sequencing coverage compared with coverage normal control) and corresponding HER2 expression detected by IHC/DISH. HER2 amplification detected using ctDNA was highly consistent with tumor tissue detected by IHC/DISH.
Table 3.
Concordance of HER2 amplification between tumor tissue and ctDNA
| HER2 overexpression in tumor tissues | HER2 CNV in ctDNA | Performance of ctDNA | ||
|---|---|---|---|---|
| Positive | Negative | Sensitivity | Specificity | |
| Positive | 16 | 2 | 88.89% | 92.31 |
| Negative | 4 | 48 | ||
| Total | 20 | 50 | ||
Kappa = 0.7835, P = 0.4142, Concordance = 91.43%.
Figure 2.
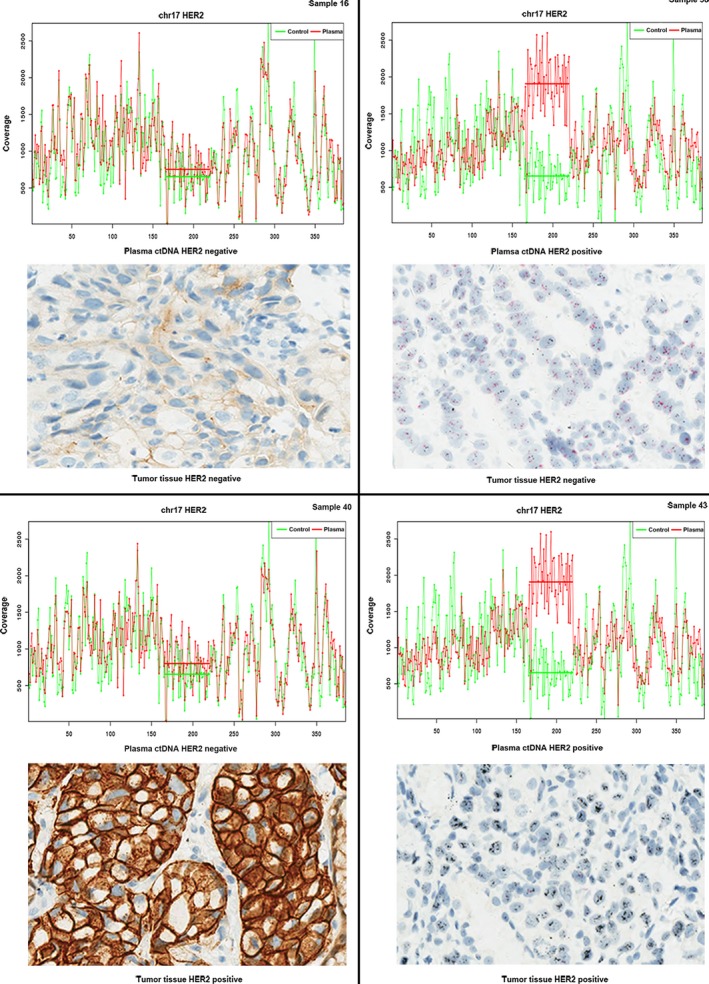
Comparison of HER2 amplification in plasma and matched primary tumor tissues. HER2 status in plasma ctDNA and matched tumor tissues were presented in four cases. Sample 16 was HER2 negtive, and sample 43 was HER2 amplified in both plasma ctDNA and matched tumor tissues. Sample 38 presented HER2 negative but HER2 amplification in plasma ctDNA, on the other hand, sample 40 was HER2 positive but HER2 amplification negative in plasma ctDNA.
Discussion
Gastric cancer patients overexpressed HER2 had a clinical benefit of targeted therapy, thus it is essential to identify HER2 status before therapy.15 Previously studies had reported that high intra‐tumor heterogeneity in AGC lead to discordant results between IHC and FISH or between endoscopic biopsy and excisional tumor specimens,16 which suggested to use multiple biopsies to confirm the HER2 status. One alternative to overcome these issues is using ctDNA for analysis. Herein, we showed ctDNA could partially overcome tissue heterogeneity and could be used for HER2 assessment for AGC patients.
In present study, we compared gene mutations to explore the concordance between single tumor biopsy and paired ctDNA sample. In 30 AGC patients, the concordance between tDNA and ctDNA was lower than which in NSCLC. For those mutations only found in tDNA but not in plasma ctDNA, it might be caused by dilution of the genomic DNA from necrotic white blood cells which released into the blood.17 Besides, the amount of ctDNA likely associated with tumor burden, status of metastasis, vascularity, cellular turnover, and response of therapy.18, 19 For mutations only detected in ctDNA, it might be due to the heterogeneity of GC.20, 21 Multiple tumor biopsies and paired plasma samples from five patients were systematically analyzed in this study. Of these five patients, different degrees of heterogeneity were observed; moreover, both intra‐ and inter‐tumoral heterogeneity was seen. It showed that single tumor biopsy could not represent entire tumor genetic profile. A previous study showed that at least four tumor biopsies were required to predict HER2 status precisely.22 In clinical practice, tumor heterogeneity is an important factor resulting in treatment failure; however, no definite strategies have been created to avoid tumor heterogeneity due to numerous risk factors on gastric cancer heterogeneity containing genetic or epigenetic factors.20 In multiple tumor biopsies, altered genes identified in the plasma were all detected in one or more matched tumor tissues, which implied that plasma ctDNA with a more comprehensive mutation profile than single tumor tissue biopsy may be a potential material to overcome tumor heterogeneity in AGC.
The feasibility of detecting HER2 amplification using ctDNA has been described in other methods except NGS. Shoda et al.23 recently provided new approach to detect HER2 amplification in ctDNA by using droplet digital PCR (ddPCR). The ddPCR measured whether amplification has occurred in a droplet, and determined CNV by assessing the number of positive droplets for HER2 and reference gene RPPH1, while the NGS determined CNV by comparing read depth of HER2 exome region between tumor sample and normal sample. The reported sensitivity of HER2 amplification detected by ddPCR in ctDNA was 73.3%, which indicated that ctDNA could be used as a noninvasive approach for evaluation of HER2 status.
HER2 gene is overexpressed or amplified in approximately 13%–22% of gastric cancers,( 24 )( 6 ) which has been found to promote tumorigenesis and is associated with poor prognosis in several human cancers.25 Accurate assessment of HER2 status is mandatory, for selecting patients who may benefit from targeted therapies with anti‐HER2 drugs such as Trastuzumab. The gold standard was based on tumor biopsy tested by IHC/FISH or other in situ hybridization methods, however, to overcome tumor heterogeneity and the inconvenience of obtaining multiple tumor biopsies for AGC, a non‐invasive yet convenient method for HER2 detection was still needed. HER2 amplification determined in plasma ctDNA was analyzed in our cohort of 70 patients by NGS, which had an overall concordance of 91.43% compared to IHC/DISH in tissue. In all 70 patients, there were four cases of HER2 positive detected in ctDNA only, only one of them accepted Trastuzumab therapy and determined a stable disease status. This patient then accepted surgery recession after two periods of targeted therapy, and the IHC/DISH results of re‐biopsied tumor tissue showed with HER2 positive. Shoda et al. indicated that an effective response to Trastuzumab was seen in patients with plasma HER2 amplification,26 but the correlation between HER2 amplification of plasma ctDNA and Trastuzumab response in this work was not analyzed due to loss of follow‐up in most Trastuzumab treated patients. That is, whether patients only detected in ctDNA with HER2 amplification could benefit from targeted therapy or not, it still needs validation in large prospective clinical trials.
The present study had several limitations. The small patient population does not allow us to draw more concrete conclusions regarding the concordance of ctDNA and tumor tissue. The mutated genes identified in the plasma were all detected in one or more matched tumor tissues only in the five patients who received multiple biopsies. Larger studies are needed to validate that plasma ctDNA could overcome tumor heterogeneity. Another limitation is that since most patients were lost to follow‐up, we had no direct evidence to show whether ctDNA detected mutation affected patient therapy and what was the response of these patients. A comparison of plasma HER2 CN status with tumor HER2 status determined using samples from multiple sites would be stronger evidence to support that ctDNA in overcoming heterogeneity.
In conclusion, here we compared the somatic mutation detected from of plasma ctDNA and paired primary tumors in patients with AGC using targeted next‐generation sequencing. Heterogeneity and homogeneity co‐existed in gastric cancer tumor tissues and paired plasma, and plasma ctDNA could partially overcome the tumor heterogeneity. Our results imply that tumor tissues and paired plasma samples could be complementary and that both should be examined in the future era of precision medicine.
Disclosure Statement
Authors Wanchun Zang, Lei Li, Guanhua Rao, Yang Yu and Zhi Jiang are employees of Novogene Bioinformatics Institute, Beijing, China. The other authors have no conflict of interest to declare.
Supporting information
Fig. S1. The distribution of insert size of DNA libraries. The distribution of insert size of DNA libraries in lymphocytes (a), tumor tissues (b), and plasma (c). (d) the average insert size of lymphocyte, tumor tissues, and plasma was 150 bp, 150 bp, and 170 bp, respectively.
Fig. S2. The work flow of gene variation calling. BWA program was used to perform alignment followed by calling germline mutations, somatic SNVs, somatic InDels, fusion genes, and CNV analysis using Samtools, Mutect, Strelka, NovoFusion, and Event‐Wise Testing algorithm, respectively. All variations were annotated with ANNOVAR.
Table S1. Gene list of 483 gene panel
Table S2. Number of SNV/InDel identified in tumor tissues and plasma ctDNA
Cancer Sci 108 (2017) 1881–1887
Funding Information
This study was funded by Beijing Municipal Science & Technology Commission Program (No. Z141107002514013), Beijing Natural Science Foundation (7161002), Beijing Municipal Administration of Hospital Clinical Medicine Development of Special Funding Support (ZYLX201701), and Beijing Municipal Senior Technical Training Plan in Health System (2015‐3‐073).
Contributor Information
Zhi Jiang, Email: jiangzhi@novogene.com.
Lin Shen, Email: lin100@medmail.com.cn.
References
- 1. Chen W, Zheng R, Baade PD et al Cancer statistics in China, 2015. CA Cancer J Clin [Internet] 2016; 66(2): 115–32. [DOI] [PubMed] [Google Scholar]
- 2. Shen L, Shan YS, Hu HM et al Management of gastric cancer in Asia: resource‐stratified guidelines. Lancet Oncol 2013; 14: e535–47. [DOI] [PubMed] [Google Scholar]
- 3. Li Y, Zhang X, Ge S et al Clinical significance of phenotyping and karyotyping of circulating tumor cells in patients with advanced gastric cancer. Oncotarget [Internet] 2014; 5: 6594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen K, Yang D, Li X et al Mutational landscape of gastric adenocarcinoma in Chinese: implications for prognosis and therapy. Proc Natl Acad Sci USA [Internet] 2015; 112: 1107–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bang Y‐J, Van Cutsem E, Feyereislova A et al Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2‐positive advanced gastric or gastro‐oesophageal junction cancer (ToGA): a phase 3, open‐label, randomised controlled trial. Lancet [Internet] 2017; 376: 687–97. [DOI] [PubMed] [Google Scholar]
- 6. Meza‐Junco J, Au HJ, Sawyer MB. Critical appraisal of trastuzumab in treatment of advanced stomach cancer. Cancer Manag Res 2011; 3: 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chan KCA, Jiang P, Zheng YWL et al Cancer genome scanning in plasma: detection of tumor‐associated copy number aberrations, single‐nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem 2013; 59: 211–24. [DOI] [PubMed] [Google Scholar]
- 8. Chan KCA, Jiang P, Chan CWM et al Noninvasive detection of cancer‐associated genome‐wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci [Internet] 2013; 110: 18761–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marzese DM, Hirose H, Hoon DSB. Diagnostic and prognostic value of circulating tumor‐related DNA in cancer patients. Expert Rev Mol Diagn [Internet] 2013; 13: 827–44. [DOI] [PubMed] [Google Scholar]
- 10. Xu S, Lou F, Wu Y et al Circulating tumor DNA identified by targeted sequencing in advanced‐stage non‐small cell lung cancer patients. Cancer Lett 2016; 370: 324–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beije N, Helmijr JC, Weerts MJA et al Somatic mutation detection using various targeted detection assays in paired samples of circulating tumor DNA, primary tumor and metastases from patients undergoing resection of colorectal liver metastases. Mol Oncol [Internet] 2016; 10: 1575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chabon JJ, Simmons AD, Lovejoy AF et al Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016; 7: 11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Spackman E, Rice S, Norman G, Suh DC, Eastwood A, Palmer S. Trastuzumab for the treatment of HER2‐positive metastatic gastric cancer: a NICE single technology appraisal. Pharmacoeconomics 2013; 31: 185–94. [DOI] [PubMed] [Google Scholar]
- 14. Yoon S, Xuan Z, Makarov V, Ye K, Sebat J. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res 2009; 19: 1586–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bang YJ, Van Cutsem E, Feyereislova A et al Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2‐positive advanced gastric or gastro‐oesophageal junction cancer (ToGA): a phase 3, open‐label, randomised controlled trial. Lancet 2010; 376: 687–97. [DOI] [PubMed] [Google Scholar]
- 16. Yang J, Luo H, Li Y et al Intratumoral heterogeneity determines discordant results of diagnostic tests for human epidermal growth factor receptor (HER) 2 in gastric cancer specimens. Cell Biochem Biophys [Internet] 2012; 62: 221–8. [DOI] [PubMed] [Google Scholar]
- 17. Schwarzenbach H, Hoon DSB, Pantel K. Cell‐free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer [Internet] 2011; 11: 426–37. [DOI] [PubMed] [Google Scholar]
- 18. Diehl F, Schmidt K, Choti M et al Circulating mutant DNA to assess tumor dynamics. Nat Med 2008; 14: 985–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kohler C, Barekati Z, Radpour R, Zhong XY. Cell‐free DNA in the circulation as a potential cancer biomarker. Anticancer Res [Internet] 2011; 31: 2623–8. [PubMed] [Google Scholar]
- 20. Hudler P. Challenges of deciphering gastric cancer heterogeneity. World J Gastroenterol 2015; 21: 10510–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wong SS, Kim KM, Ting JC et al Genomic landscape and genetic heterogeneity in gastric adenocarcinoma revealed by whole‐genome sequencing. Nat Commun [Internet] 2014; 5: 5477. [DOI] [PubMed] [Google Scholar]
- 22. Ahn S, Ahn S, Van Vrancken M et al Ideal number of biopsy tumor fragments for predicting HER2 status in gastric carcinoma resection specimens. Oncotarget [Internet] 2015; 6: 38372–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shoda K, Ichikawa D, Fujita Y et al Monitoring the HER2 copy number status in circulating tumor DNA by droplet digital PCR in patients with gastric cancer. Gastric Cancer [Internet] 2017; 20(1): 126–35. [DOI] [PubMed] [Google Scholar]
- 24. Rakha EA, Reis‐Filho JS, Ellis IO. Combinatorial biomarker expression in breast cancer. Breast Cancer Res Treat 2010; 120: 293–308. [DOI] [PubMed] [Google Scholar]
- 25. Iqbal N, Iqbal N. Human epidermal growth factor receptor 2 (HER2) in cancers: overexpression and therapeutic implications. Mol Biol Int [Internet] 2014; 2014: 852748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shoda K, Masuda K, Ichikawa D et al HER2 amplification detected in the circulating DNA of patients with gastric cancer: a retrospective pilot study. Gastric Cancer [Internet] 2014; 18: 698–710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. The distribution of insert size of DNA libraries. The distribution of insert size of DNA libraries in lymphocytes (a), tumor tissues (b), and plasma (c). (d) the average insert size of lymphocyte, tumor tissues, and plasma was 150 bp, 150 bp, and 170 bp, respectively.
Fig. S2. The work flow of gene variation calling. BWA program was used to perform alignment followed by calling germline mutations, somatic SNVs, somatic InDels, fusion genes, and CNV analysis using Samtools, Mutect, Strelka, NovoFusion, and Event‐Wise Testing algorithm, respectively. All variations were annotated with ANNOVAR.
Table S1. Gene list of 483 gene panel
Table S2. Number of SNV/InDel identified in tumor tissues and plasma ctDNA


