Abstract
Androgen deprivation therapy is initially effective for treating patients with advanced prostate cancer; however, the prostate cancer gradually becomes resistant to androgen deprivation therapy, which is termed castration‐resistant prostate cancer (CRPC). Androgen receptor splice variant 7 (AR‐V7), one of the causes of CRPC, is correlated with resistance to a new‐generation AR antagonist (enzalutamide) and poor prognosis. Heat shock protein 70 (Hsp70) inhibitor is known to decrease the levels of full‐length AR (AR‐FL), but little is known about its effects against CRPC cells expressing AR‐V7. In this study, we investigated the effect of the Hsp70 inhibitors quercetin and VER155008 in the prostate cancer cell line LNCaP95 that expresses AR‐V7, and explored the mechanism by which Hsp70 regulates AR‐FL and AR‐V7 expression. Quercetin and VER155008 decreased cell proliferation, increased the proportion of apoptotic cells, and decreased the protein levels of AR‐FL and AR‐V7. Furthermore, VER155008 decreased AR‐FL and AR‐V7 mRNA levels. Immunoprecipitation with Hsp70 antibody and mass spectrometry identified Y‐box binding protein 1 (YB‐1) as one of the molecules regulating AR‐FL and AR‐V7 at the transcription level through interaction with Hsp70. VER155008 decreased the phosphorylation of YB‐1 and its localization in the nucleus, indicating that the involvement of Hsp70 in AR regulation might be mediated through the activation and nuclear translocation of YB‐1. Collectively, these results suggest that Hsp70 inhibitors have potential anti‐tumor activity against CRPC by decreasing AR‐FL and AR‐V7 expression through YB‐1 suppression.
Keywords: Androgen receptor splice variant 7, castration‐resistant prostate cancer, heat shock protein 70, heat shock protein 70 inhibitor, Y‐box binding protein 1
Although progress has been made in the diagnosis and treatment of prostate cancer, the morbidity of prostate cancer remains high. Prostate cancer has one of the highest prevalence rates of all cancer types, with an estimated 161 360 newly diagnosed cases per year, and is the third leading cause of cancer‐related deaths, with an estimated 26 730 deaths in 2017 among men in the USA.1 The growth and progression of prostate cancer depend on androgen hormones such as dihydrotestosterone. Therefore, ADT is the standard of care and is generally initially effective for treating patients with advanced prostate cancer.2 However, most cases of prostate cancer eventually become resistant to ADT within 2–3 years. This state is termed CRPC, which is associated with a poor outcome. The mechanisms of CRPC development include AR amplification and hypersensitivity, AR splice variants, AR mutations, androgen‐independent AR activation, and alternative androgen production. Among these factors, the presence of AR splice variants has been shown to play a substantial role in the emergence of CRPC. Androgen receptor splice variant 7, one of the major splice variants expressed in human prostate cancer, is constitutively active, and its transcriptional activity is not regulated by androgens or anti‐androgens.3 The expression of AR‐V7 has been shown to accurately reflect the resistance to a novel AR antagonist (enzalutamide) and an androgen biosynthesis inhibitor (abiraterone),4 and is associated with poor cancer‐specific survival.5 Androgen receptor‐V7 lacks the C‐terminal LBD but harbors the N‐terminal transactivation domain. Because enzalutamide exerts its antitumor activity by interacting with the LBD of the androgen receptor, AR‐V7 is associated with enzalutamide resistance. Moreover, AR‐V7 is constitutively active without the ligand, and thus, also confers resistance to ligand‐depleting agents such as abiraterone.4 Therefore, AR‐V7 has been investigated as a potential biomarker and therapeutic target for CRPC.6 Indeed, several novel drugs against AR‐V7 have already been tested in clinical trials. Niclosamide inhibits AR‐V7 transcriptional activity and decreases AR‐V7 levels through a proteasome‐dependent mechanism.7 EPI‐001 interacts with the AR N‐terminal transactivation domain and attenuates AR and AR‐V transcriptional activity.8 Furthermore, it is now well established that CRPC is resistant to ADT, but that AR remains an important driver in this progression.9 Therefore, AR in addition to AR‐V7 might be important therapeutic targets in CRPC worthy of investigation.
Heat shock proteins are molecular chaperones that play important roles in the folding of misfolded proteins, and in the regulation of cellular signals and transcriptional networks.10 Heat shock proteins are overexpressed in a wide range of human cancers and are implicated in tumor cell proliferation, differentiation, invasion, metastasis, and death.11 In prostate cancer, Hsp70 and Hsp90 have cytoprotective effects, such as in the inhibition of apoptosis,12 cell cycle modulation,13 invasion and metastasis,14 and AR transcriptional activity and stability.15 Therefore, in the last decade, Hsp inhibitors have been highlighted and explored as potential targets for anticancer therapy, and clinical trials have been carried out.16
Heat shock protein 90 inhibitors have been reported as potentially effective treatments for CRPC by depressing the expression levels of AR‐FL and AR‐V7 through different mechanisms. In particular, an Hsp90 inhibitor was shown to increase the degradation rate of AR‐FL and induced AR‐V7 mRNA downregulation in vitro.17 However, it is unknown whether Hsp70 inhibitors also reduce the expression of AR‐V7, and the specific mechanism contributing to the observed effect of Hsp70 inhibitors to decrease AR‐FL has yet to be elucidated.18
LNCaP95 cells are a suitable CRPC cell line with which to explore these questions. LNCaP95 cells are derived from the LNCaP cell line, which is sensitive to ADT. LNCaP95 cells have been cultured under androgen‐stripped serum conditions for a long time, making them resistant to ADT, and they express both AR‐FL and AR‐V7. In addition to this difference in ADT sensitivity, LNCaP95 cells and the parental LNCaP cells show qualitatively similar AR transcriptional responses.19 Therefore, to expand the range of therapeutic targets and agents for prostate cancer, in this study, we investigated the effects of the Hsp70 inhibitors quercetin and VER155008 on LNCaP95 cells and the underlying mechanisms. Quercetin decreases Hsp70 expression through inhibition of heat shock factor 1, a transcription factor of Hsp70, whereas VER155008 is an ATP‐derivative inhibitor of Hsp70.
Materials and Methods
Cell line and culture conditions
The human prostate cancer cell line LNCaP95 was a generous gift from Dr. Jun O. Luo (Johns Hopkins University, Baltimore, MD, USA). The LNCaP95 cells were cultured in phenol red‐free RPMI‐1640 medium (Thermo Fisher Scientific, Waltham, MA USA) supplemented with 10% charcoal‐stripped FBS (Thermo Fisher Scientific) at 37°C in a humidified atmosphere with 5% CO2. The human prostate cancer cell line 22Rv1 was obtained from ATCC (Manassas, VA, USA) and cultured in RPMI‐1640 medium (Thermo Fisher Scientific) supplemented with 10% FBS (Thermo Fisher Scientific) and 1% penicillin/streptomycin at 37°C in a humidified atmosphere with 5% CO2.
Western blot analysis
The cells were harvested and whole‐cell lysates were prepared using lysis buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 5 mM EDTA, 1% NP‐40, and 0.65% CHAPS containing 1 mM PMSF and a protease inhibitor cocktail). Sample preparation and Western blot analysis were carried out as previously described18 using antibodies against the following proteins: Hsp70, AR (N‐20), β‐actin, c‐Jun, and Twist (Santa Cruz Biotechnology, Dallas, TX, USA); Hsp90, PSA, YB‐1, phospho‐YB‐1, and phospho‐Foxo3a (Cell Signaling Technology, Danvers, MA, USA); AR‐V7 (Precision Antibody, Columbia, MD, USA); UBE2C (BostonBiochem, Cambridge, MA, USA); FKBP5, CREB, and Sp1 (Gene Tex, CA, USA); laminB1 (Abcam, Cambridge, UK); Foxo3a (Upstate, Temecula, CA, USA); α‐tubulin (Calbiochem, San Diego, CA, USA); and HRP‐conjugated secondary antibodies (GE Healthcare, Little Chalfont, UK).
Subcellular fractionation
The nuclear and cytosolic fractions were prepared using the LysoPure Nuclear and Cytoplasmic Extractor Kit (Wako, Osaka, Japan). The cytosolic subcellular fraction was extracted according to the manufacturer's instructions. The soluble and insoluble fractions of the nuclear fraction were combined.
IncuCyte live‐cell imaging
Cell growth was monitored at 3‐h intervals using an IncuCyte live‐cell imaging system (Essen BioScience, Ann Arbor, MI, USA) as previously described.20 In brief, LNCaP95 cells were seeded in a 24‐well plate at 2.5 × 104 cells per well 48 h prior to cell monitoring. The cells were treated with 10–25 μM VER155008 (Tocris Bioscience, Bristol, UK), 10–50 μM quercetin, or DMSO (as a vehicle control) immediately following the initial image capture. For the annexin V apoptosis assay, the cells were seeded in a 96‐well plate at 1 × 104 cells per well. The cells were then simultaneously treated with the Hsp70 inhibitors and IncuCyte Annexin V Red reagent for 48 h. Annexin V‐positive cells were determined by counting of the red‐stained objects with IncuCyte image analysis software.
Quantification of mRNA
Total RNA was extracted from the cells using an RNeasy Mini Kit (Qiagen, Hilden, Germany). RNA was converted into cDNA using ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo, Osaka, Japan). Transcript expression was measured by Power SYBR Green PCR Master Mix (Thermo Fisher Scientific) using a 7500 Fast Real‐Time PCR System (Applied Biosystems, Foster City, CA, USA). The transcript levels were normalized according to the levels of HSPA8. Data were obtained from three independent biological experimental replicates with three technical replicates. Primer sequences for real‐time PCR were as follows: AR‐FL forward 5ʹ‐ACATCAAGGAACTCGATCGTATCATTGC‐3ʹ, reverse 5ʹ‐TTGGGCACTTGCACAGAGAT‐3ʹ; AR‐V7 forward 5ʹ‐CCATCTTGTCGTCTTCGGAAATGTTATGAAGC‐3ʹ, reverse 5ʹ‐TTTGAATGAGGCAAGTCAGCCTTTCT‐3ʹ; and HSPA8 forward 5ʹ‐ACCTACTCTTGTGTGGGTGTT‐3ʹ, reverse 5ʹ‐GAGATAGCTTGGAGTGGTTCG‐3ʹ.
Isolation of Hsp70 client proteins
Isolation of Hsp70‐binding proteins and identification with MS was carried out as previously described.21 For MS analyses, the cells were lysed in lysis buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 5 mM EDTA, and 1% NP‐40 containing 1 mM PMSF and a protease inhibitor cocktail). Cell lysates (500 μg protein) were precleared with inactivated NHS‐Sepharose beads (GE Healthcare) for 30 min at room temperature, and immunoprecipitated using NHS‐Sepharose beads conjugated to anti‐Hsp72 antibodies or rat IgG antibody at room temperature for 3 h, as previously described.21 The immunoprecipitates were then washed three times, and Hsp72 client proteins were eluted with 0.1 M glycine‐HCl (pH 2.0).
Sample preparation
Mass spectrometry samples were desalted and concentrated by SDS‐PAGE on a polyacrylamide gel, and the resulting gels were stained with Quick‐CBB (Wako). Samples were excised from the gel, treated with 10 mM dithiothreitol, and then with 55 mM isoamyl alcohol. In‐gel trypsin digestion (Promega, Madison, WI, USA) was then carried out, and the resulting peptides were sequentially extracted from the gel with 0.1% TFA. The samples were then desalted using StageTips with C18 Empore disk membranes (3M; St. Paul, MN, USA).
Proteomic analysis and database search
The gel‐extracted peptides were dried, dissolved in a solution containing 0.1% TFA and 2% acetonitrile, and subjected to nano‐liquid chromatography MS/MS analysis using an LTQ Orbitrap Velos Pro mass spectrometer system (Thermo Fisher Scientific) coupled with a nano‐liquid chromatography instrument (Advance LC; Michrom BioResources, Auburn, CA, USA) and an HTC‐PAL autosampler (CTC Analytics, Zwingen, Switzerland). Peptide separation was carried out with a silica capillary packed with a 3‐μM C18 L‐column (Chemicals Evaluation and Research Institute, Tokyo, Japan). Full MS spectra were obtained with Orbitrap in the mass/charge (m/z) range of 300–2000 with a resolution of 60 000 at m/z 400. The peak lists were generated using MSn.exe (Thermo Fisher Scientific) with a minimum scan/group value of 1, and were compared with the in‐house‐curated target/decoy SwissProt Release 2015_12 database (SwissProt database, 20 194 protein sequences; European Bioinformatics Institute, Cambridgeshire, UK) using the mascot algorithm (version 2.5.1; Matrix Science, Boston, MA, USA).
Criteria for protein identification
Scaffold software (version Scaffold_4.0.4; Proteome Software, Portland, OR, USA) was used to validate the MS/MS‐based peptide and protein identifications. Peptide identifications were accepted if they exceeded specific database search engine thresholds. The minimum requirement for mascot identifications was that the ion scores were greater than both the associated identity scores and 0.00. Protein identifications were accepted if they contained at least two identified peptides. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Immunofluorescence microscopy
LNCaP95 cells were seeded directly onto coverslips at 2.5 × 104 cells per well 2 days prior to fixation. Immunofluorescence staining was carried out as previously described.22 Fluorescence images of the cells were obtained with a Leica TCS‐SP5 confocal laser‐scanning microscope (Leica, Wetzlar, Germany).
Results
Inhibition of Hsp70 repressed LNCaP95 cell proliferation and survival
We initially evaluated the effect of quercetin, an inhibitor of Hsp70 expression, on the proliferation of LNCaP95 cells. The IncuCyte live‐cell imaging system showed that quercetin decreased cell proliferation in a dose‐dependent manner (Fig. 1a, Fig. S1a). To determine whether the quercetin‐induced suppression of cell proliferation was caused by chaperone activity of Hsp70, we further evaluated the effect of VER155008, an ATP‐derivative inhibitor of Hsp70, on LNCaP95 cells. VER155008 also decreased cell proliferation in a dose‐dependent manner (Fig. 1b, Fig. S1b). Quercetin and VER155008 also decreased the proliferation of 22Rv1 cells expressing higher levels of AR‐V7 (Fig. S2). As both quercetin and VER155008 notably decreased cell proliferation, we assessed their effects on the induction of apoptosis by annexin V staining. Quercetin at >25 μM and VER155008 at >10 μM significantly increased the proportion of annexin V‐positive cells (Fig. 1c,d). These results suggested that the repression of cell survival by Hsp70 inhibition was ascribed to the chaperone activity of Hsp70, and that the Hsp70 inhibitors had antitumor effects on LNCaP95 cells.
Figure 1.
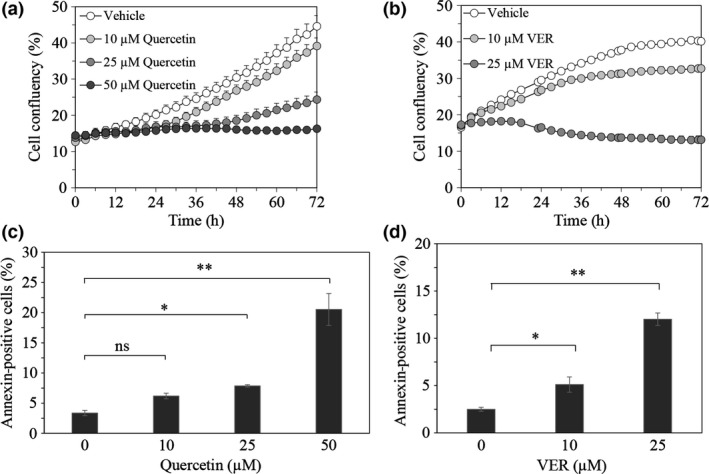
Effect of quercetin and VER155008 on survival of prostate cancer cells. Proliferation of LNCaP95 cells treated with quercetin (10 μM, 25 μM, or 50 μM) (a) or VER155008 (10 μM or 25 μM) (b) was monitored for 72 h by IncuCyte ZOOM. The percentage of annexin V‐positive cells in each field treated with quercetin (c) or VER155008 (d) was quantified at indicated concentrations for 18 h. Data represents the SEM of three independent experiments. Asterisks indicate statistical significance. *P < 0.05, **P < 0.01 versus vehicle, unpaired t‐test. ns, not significant.
Inhibition of Hsp70 decreased the expression of AR‐FL and AR‐V7 and their signals
We next evaluated the effect of quercetin and VER155008 on the expression of AR‐FL and AR‐V7. Quercetin decreased the protein expression levels of both AR‐FL and AR‐V7 in parallel with the change in Hsp70 expression in a dose‐dependent manner (Fig. 2a). This suggested a potential association between Hsp70 and ARs in LNCaP95 cells. Moreover, VER155008 decreased the expression levels of AR‐FL and AR‐V7 in a time‐dependent manner (Fig. 2b). However, the depression of AR‐V7 expression occurred earlier than that of AR‐FL.
Figure 2.
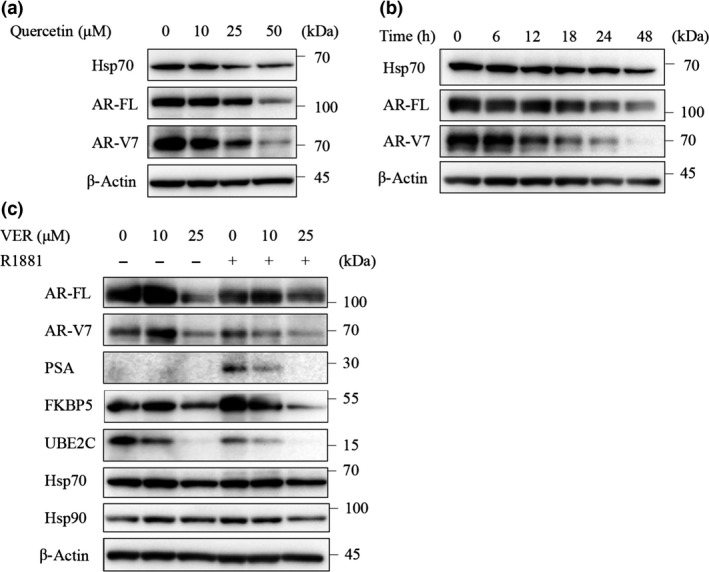
Effect of quercetin and VER155008 on full‐length androgen receptor (AR‐FL) and androgen receptor splice variant 7 (AR‐V7) protein expression. (a) LNCaP95 cells were treated with quercetin at indicated concentrations for 24 h. Protein extracts (15 μg) were subjected to Western blotting. (b) LNCaP95 cells were treated with VER155008 (25 μM) for the indicated times. Protein extracts (15 μg) were subjected to Western blotting. (c) LNCaP95 cells were treated with VER155008 at indicated concentrations for 1 h then treated with or without R1881 (1 nM) for 48 h. Protein extracts (15 μg) were subjected to Western blot analysis. FKBP5, FK506 binding protein 5; Hsp70, heat shock protein 70; PSA, prostate‐specific antigen; UBE2C, ubiquitin conjugating enzyme E2 C.
Although CRPC is resistant to ADT, ARs and their signals remain important for the survival of these cancer cells. Therefore, we also evaluated the effect of VER155008 on PSA and FKBP5 as markers of AR‐FL signaling, and on UBE2C as a marker of AR‐V7, signaling by Western blot analysis. VER155008 with R1881 decreased the PSA, FKBP5, and UBEC2 expression levels in a dose‐dependent manner. VER155008 at 25 μM decreased not only the levels of the target proteins but also the expression levels of AR‐FL and AR‐V7, whereas there was barely any change in the levels of these proteins with treatment of VER155008 at 10 μM. As VER155008 competitively blocks the binding of ATP to Hsp70, the protein levels of Hsp70 were not affected, and the expression of Hsp90, which is involved in the nuclear translocation of AR‐FL, was also not affected (Fig. 2c).
Overexpression of Hsp70 increased AR‐FL and AR‐V7 protein levels
The results described above indicated that Hsp70 inhibitors could repress the expression of ARs. Therefore, to investigate whether Hsp70 regulates the expression of ARs in LNCaP95 cells, we overexpressed Hsp70 by heat shock treatment. Heat shock‐induced Hsp70 expression increased the protein levels of AR‐FL and AR‐V7 (Fig. 3a), and these effects were negated by treatment with quercetin and VER155008. Quercetin decreased the levels of AR‐FL and AR‐V7, thereby suppressing the increase of Hsp70 (Fig. 3b), whereas VER155008 also decreased the overexpressed levels of AR‐FL and AR‐V7, but did not change the overexpressed level of Hsp70 (Fig. 3c). These results confirmed that Hsp70 regulates the expression of AR‐FL and AR‐V7.
Figure 3.
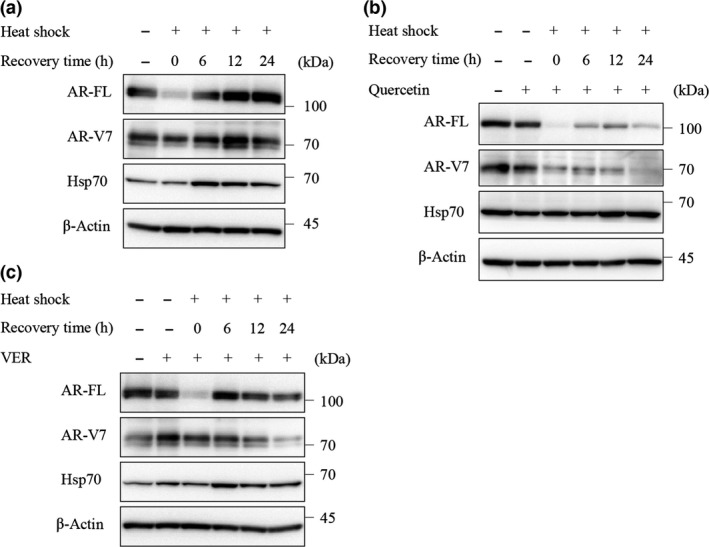
Effect of overexpressed heat shock protein 70 (Hsp70) by heat shock on full‐length androgen receptor (AR‐FL) and androgen receptor splice variant 7 (AR‐V7) protein expression. (a) LNCaP95 cells were incubated at 43°C for 1 h, then cultured at 37°C for the indicated time. Protein extracts (15 μg) were subjected to Western blot analysis. LNCaP95 cells were treated with quercetin (50 μM) (b) or VER155008 (25 μM) (c) for 1 h before heat shock (43°C, 1 h), then incubated at 37°C for the indicated time. Protein extracts (15 μg) were subjected to Western blot analysis.
Inhibition of Hsp70 decreased AR‐FL and AR‐V7 at the transcription level
Quantitative RT‐PCR was carried out to clarify whether such repression of AR‐FL and AR‐V7 expression by Hsp70 inhibitors occurs at the transcription level. Indeed, VER155008 significantly decreased the mRNA levels of AR‐FL and AR‐V7, indicating an effect at transcription (Fig. 4a). We next confirmed the change in the levels of AR‐FL transcription factors23 in LNCaP cells, the parent cells of the LNCaP95 cell line, following VER155008 treatment. VER155008 slightly decreased the levels of CREB, Sp1, and phosphorylated Foxo3a, but did not change the levels of c‐Jun and Twist (Fig. 4b). Therefore, VER155008 might decrease AR‐FL and AR‐V7 expression levels by suppressing these transcription factors.
Figure 4.
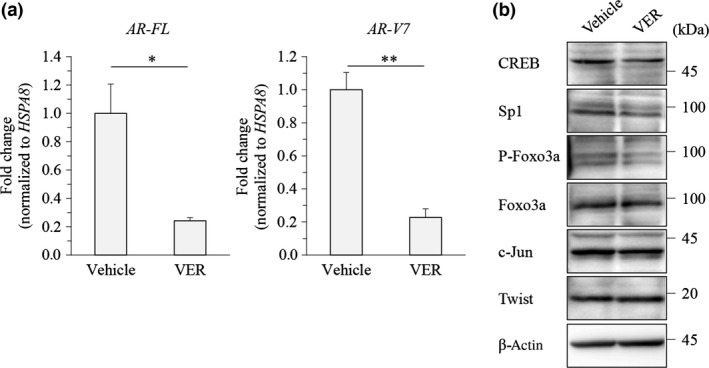
Effect of VER155008 on full‐length androgen receptor (AR‐FL) and androgen receptor splice variant 7 (AR‐V7) transcription level. (a) LNCaP95 prostate cancer cells were treated with DMSO or VER155008 (VER; 25 μM) for 24 h. The AR‐FL and AR‐V7 transcripts were examined using quantitative RT‐PCR. The mRNA levels were normalized to that of HSPA8 as internal standard, and the values of relative target expressions in vehicle‐treated cells were set to 1. Asterisks indicate statistical significance. *P < 0.05, **P < 0.01 versus vehicle by unpaired t‐test; values are SEM (n = 3). (b) LNCaP95 cells were treated with VER155008 (25 μM) for 48 h. Protein extracts (15 μg) were subjected to Western blot analysis. CREB, cAMP response element binding protein; Sp1, specificity protein 1.
Heat shock protein 70 interacts with YB‐1
Next, to identify the molecules involved in the regulation of AR‐FL and AR‐V7 under the control of Hsp70, we explored the Hsp70‐binding proteins by immunoprecipitation and MS analysis. Sixty‐two molecules were identified as candidate Hsp70‐binding proteins (Table 1, Table S1). We focused on the transcription factors among these molecules, particularly on YB‐1, as it has been reported as a transcription factor of AR‐FL in LNCaP cells,24 and its protein level is correlated with AR‐V7 expression in other CRPC cell lines, VCaP and 22Rv1.25 To verify the results of MS, we validated the binding of Hsp70 and YB‐1 by immunoprecipitation with anti‐YB‐1 antibody, which clearly revealed an interaction in LNCaP95 cells (Fig. 5a). Immunofluorescence showed that YB‐1 was mainly localized with Hsp70 in the cytoplasm (Fig. 5b). Moreover, treatment with quercetin or VER155008 for 24 h decreased the level of YB‐1 expression and remarkably repressed the levels of phosphorylated YB‐1 along with AR‐FL and AR‐V7 (Fig. 5c). VER155008 also decreased the level of YB1 along with AR‐FL and AR‐V7 in 22Rv1 cells (Fig. S3). To investigate whether Hsp70 is involved in the function of YB‐1, we assessed the localization of YB‐1 in both the cytosolic and nuclear fractions after Hsp70 blockade. YB‐1 was only decreased in the nuclear fraction, and phosphorylated YB‐1 was only detected in cytosolic fraction, which decreased with VER155008 treatment (Fig. 5d). These results suggested that the Hsp70 inhibitors decreased AR‐FL and AR‐V7 through suppression of YB‐1 expression and activity.
Table 1.
Heat shock protein 70‐binding proteins in the LNCaP95 prostate cancer cell line
| No. | Identified proteins | Accession no. | Identified proteins, kDa |
|---|---|---|---|
| 1 | Transaldolase | TALDO_HUMAN | 38 |
| 2 | Proteasome subunit α type‐7 | PSA7_HUMAN | 28 |
| 3 | Proteasome subunit α type‐1 | PSA1_HUMAN | 30 |
| 4 | Endoplasmic reticulum resident protein 29 | ERP29_HUMAN | 29 |
| 5 | Proteasome subunit β type‐1 | PSB1_HUMAN | 26 |
| 6 | Isocitrate dehydrogenase [NADP], mitochondrial | IDHP_HUMAN | 51 |
| 7 | Proteasome subunit α type‐3 | PSA3_HUMAN | 28 |
| 8 | Alpha‐actinin‐4 | ACTN4_HUMAN | 105 |
| 9 | PTB domain‐containing engulfment adapter protein 1 | GULP1_HUMAN | 34 |
| 10 | Proteasome subunit α type‐6 | PSA6_HUMAN | 27 |
| 11 | Delta‐aminolevulinic acid dehydratase | HEM2_HUMAN | 36 |
| 12 | 60S ribosomal protein L35 | RL35_HUMAN | 15 |
| 13 | DnaJ homolog subfamily A member 3, mitochondrial | DNJA3_HUMAN | 52 |
| 14 | Nuclease‐sensitive element‐binding protein 1 | YBOX1_HUMAN | 36 |
| 15 | Elongation factor 1‐δ | EF1D_HUMAN | 31 |
| 16 | Proteasome subunit β type‐4 | PSB4_HUMAN | 29 |
| 17 | Ribose‐5‐phosphate isomerase | RPIA_HUMAN | 33 |
| 18 | 40S ribosomal protein S6 | RS6_HUMAN | 29 |
| 19 | Stromal cell‐derived factor 2‐like protein 1 | SDF2L_HUMAN | 24 |
| 20 | Selenide, water dikinase 1 | SPS1_HUMAN | 43 |
| 21 | 40S ribosomal protein S25 | RS25_HUMAN | 14 |
| 22 | Phosphoglycerate mutase 1 | PGAM1_HUMAN | 29 |
| 23 | NADH dehydrogenase [ubiquinone] iron‐sulfur protein 3, mitochondrial | NDUS3_HUMAN | 30 |
| 24 | Cancer‐related nucleoside‐triphosphatase | NTPCR_HUMAN | 21 |
| 25 | Proteasome subunit α type‐2 | PSA2_HUMAN | 26 |
| 26 | 60S ribosomal protein L28 | RL28_HUMAN | 16 |
| 27 | Ketosamine‐3‐kinase | KT3K_HUMAN | 34 |
| 28 | DnaJ homolog subfamily B member 11 | DJB11_HUMAN | 41 |
| 29 | Tricarboxylate transport protein, mitochondrial | TXTP_HUMAN | 34 |
| 30 | 14‐3‐3 protein ζ/δ | 1433Z_HUMAN | 28 |
| 31 | Cleavage and polyadenylation specificity factor subunit 5 | CPSF5_HUMAN | 26 |
| 32 | Proteasome subunit β type‐3 | PSB3_HUMAN | 23 |
| 33 | Nucleotide exchange factor SIL1 | SIL1_HUMAN | 52 |
| 34 | Proteasome subunit β type‐2 | PSB2_HUMAN | 23 |
| 35 | Fructose‐bisphosphate aldolase C | ALDOC_HUMAN | 39 |
| 36 | Ferrochelatase, mitochondrial | HEMH_HUMAN | 48 |
| 37 | rRNA 2′‐O‐methyltransferase fibrillarin | FBRL_HUMAN | 34 |
| 38 | 60S ribosomal protein L36a | RL36A_HUMAN | 12 |
| 39 | 26S proteasome non‐ATPase regulatory subunit 7 | PSMD7_HUMAN | 37 |
| 40 | Proteasome subunit α type‐5 | PSA5_HUMAN | 26 |
| 41 | 40S ribosomal protein S15a | RS15A_HUMAN | 15 |
| 42 | Splicing factor 3B subunit 6 | SF3B6_HUMAN | 15 |
| 43 | Coiled‐coil‐helix‐coiled‐coil‐helix domain‐containing protein 2 | CHCH2_HUMAN | 16 |
| 44 | Sideroflexin‐1 | SFXN1_HUMAN | 36 |
| 45 | Proteasome subunit β type‐6 | PSB6_HUMAN | 25 |
| 46 | Proteasome subunit β type‐7 | PSB7_HUMAN | 30 |
| 47 | GTP‐binding nuclear protein Ran | RAN_HUMAN | 24 |
| 48 | Calmodulin | CALM_HUMAN | 17 |
| 49 | Protein disulfide‐isomerase A6 | PDIA6_HUMAN | 48 |
| 50 | NADH dehydrogenase [ubiquinone] 1 α subcomplex subunit 5 | NDUA5_HUMAN | 13 |
| 51 | Nucleoside diphosphate kinase A | NDKA_HUMAN | 17 |
| 52 | Stromal cell‐derived factor 2 | SDF2_HUMAN | 23 |
| 53 | Protein SETSIP | SETLP_HUMAN (+1) | 35 |
| 54 | 10‐kDa heat shock protein, mitochondrial | CH10_HUMAN | 11 |
| 55 | Growth factor receptor‐bound protein 10 | GRB10_HUMAN | 67 |
| 56 | 26S protease regulatory subunit 10B | PRS10_HUMAN | 44 |
| 57 | 60S ribosomal protein L22‐like 1 | RL22L_HUMAN | 15 |
| 58 | Histidine triad nucleotide‐binding protein 1 | HINT1_HUMAN | 14 |
| 59 | Malate dehydrogenase, mitochondrial | MDHM_HUMAN | 36 |
| 60 | GTP‐binding protein SAR1a | SAR1A_HUMAN (+1) | 22 |
| 61 | Protein LSM14 homolog B | LS14B_HUMAN | 42 |
| 62 | 45‐kDa calcium‐binding protein | CAB45_HUMAN | 42 |
Proteins were identified at a 95% confidence level with the mascot algorithm.
Figure 5.
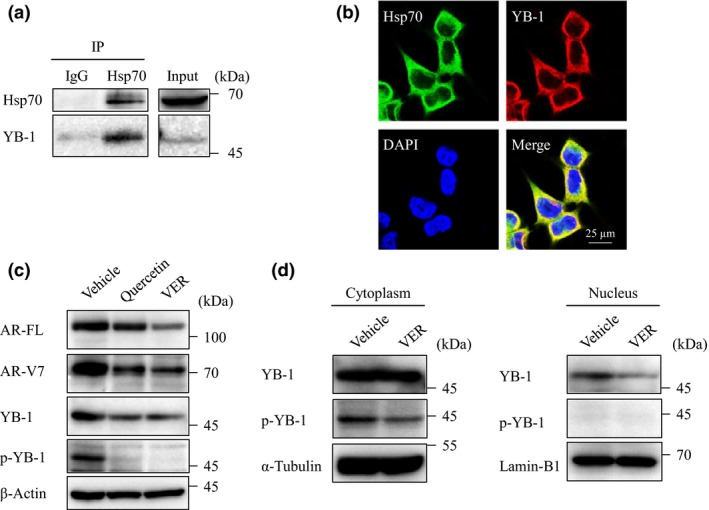
Correlation between heat shock protein 70 (Hsp70) and Y‐box binding protein 1 (YB‐1). (a) LNCaP95 cell lysates were immunoprecipitated (IP) with anti‐Hsp70 antibody and were blotted with YB‐1 and Hsp70 antibodies. (b) Localization of Hsp70 and YB‐1 was determined by double immunofluorescence staining in LNCaP95 cells. (c) LNCaP95 cells were treated with quercetin (25 μM) or VER155008 (VER; 25 μM) for 24 h. Protein extracts (15 μg) were subjected to Western blot analysis. (d) LNCaP95 cells were treated with VER155008 (25 μM) for 12 h. Cell lysates were separated into cytosolic and nuclear fractions. Protein extracts (10 μg) were subjected to Western blot analysis.
Discussion
Some of the mechanisms of resistance to ADT in prostate cancer have been elucidated. Among these mechanisms, androgen receptor splice variants have been investigated most extensively because they are associated with both prognosis and the development of drug resistance. Twelve androgen receptor splice variants that lack the LBD have been reported to date, including AR‐V7 and AR,v567es which have been identified in clinical samples and correlate with cancer‐specific survival. The majority of research has focused on AR‐V7 because of its association with resistance to novel ADT.26
In this study, we investigated the efficiency of two Hsp70 inhibitors, quercetin and VER155008, against LNCaP95 cells, a CRPC cell line expressing AR‐V7. Although quercetin has previously been shown to reduce cell proliferation and AR‐FL expression in prostate cancer,18, 27 these studies did not indicate whether the effect was attributed to Hsp70 chaperone activity. Moreover, the role of Hsp70 in regulating the expression of AR‐V7 is currently unclear. Our data revealed that the Hsp70 inhibitors had antitumor effects and decreased AR‐FL and AR‐V7 expression levels in LNCaP95 cells. Moreover, AR‐FL and AR‐V7 expression was upregulated along with overexpression of Hsp70 induced by heat‐shock treatment, and these effects were negated by treatment with the Hsp70 inhibitors. These results suggested that Hsp70 regulates AR‐FL and AR‐V7, indicating that an Hsp70 inhibitor would be an effective therapeutic agent for prostate cancer, even for CRPC expressing both AR‐FL and AR‐V7.
Given that VER155008 depressed AR‐FL and AR‐V7 expression at the transcription level, we assessed the transcription factors of AR‐FL. VER155008 slightly decreased the levels of CREB, Sp1, and phosphorylated Foxo3a, but not the levels of c‐Jun and Twist. Therefore, we further investigated the molecule that binds with Hsp70 to control AR‐FL and AR‐V7 transcription using immunoprecipitation and MS, and the results revealed YB‐1 as a strong candidate for this interaction. YB‐1 is a member of a family of proteins containing an evolutionarily ancient cold‐shock domain. This protein participates in DNA repair, pre‐mRNA transcription and splicing, mRNA packaging, and regulation of mRNA stability and translation.28 YB‐1 has also been reported as a transcription factor of AR‐FL in LNCaP cells, the parental cell line of LNCaP95.24 Moreover, YB‐1 expression levels are correlated with the level of AR‐V7 protein, and phosphorylated YB‐1 correlates with the transcription level of AR‐V7 in VCaP and 22Rv1 cells, other CRPC cell lines expressing AR‐V7.25 Based on the known functions of YB‐1, we assumed that Hsp70 regulates AR‐FL and AR‐V7 at the transcription level by interacting with and regulating YB‐1 expression and activation. In addition, the transcription level of YB‐1 was found to be upregulated over time as LNCaP cells were induced to transform to CRPC cells through long‐term ADT exposure, suggesting that it plays a role as an oncogene.29 In the present study, VER155008 reduced the YB‐1 expression level in the nuclear fraction and the phosphorylation of YB‐1, with the latter reduction being more remarkable. This suggests that Hsp70 might regulate the phosphorylation of YB‐1 and its nuclear translocation, as phosphorylation of YB‐1 at Ser102 is required for its nuclear translocation.30 Moreover, previous reports related to YB‐1 and AR‐FL also suggest that Hsp70 inhibitors decrease AR‐FL and AR‐V7 mRNA levels by depressing the nuclear localization of YB‐1.
Furthermore, AR‐V7 expression was reduced earlier than AR‐FL following VER155008 treatment, even though the half‐life of AR‐V7 is longer than that of AR‐FL.17 In addition, AR‐V7 transcription should be correlated with the AR‐FL mRNA level as AR‐V7 mRNA is generated by the splicing of AR‐FL mRNA.31 Thus, our findings suggest that there is likely another mechanism by which Hsp70 inhibitors selectively decrease AR‐V7, rather than AR‐FL, at the protein level. The mechanism might involve an influence on the stability of AR‐V7 protein by Hsp70 or an interaction of YB‐1 with the splicing of AR‐FL mRNA.
In summary, we have shown that Hsp70 inhibitors decreased cell survival, and suppressed AR‐FL and AR‐V7 expression with YB‐1 inhibition in LNCaP95 prostate cancer cells. There are currently no Hsp70 inhibitors available for clinical use because of their high renal toxicity.32 Thus, further development of clinically safe Hsp70 inhibitors is expected to provide a new line of valuable antitumor agents for treating CRPC by suppressing AR‐FL and AR‐V7.
Disclosure Statement
The authors have no conflict of interest.
Abbreviations
- ADT
androgen deprivation therapy
- AR‐FL
full‐length androgen receptor
- AR‐V7
androgen receptor splice variant 7
- CREB
cAMP response element binding protein
- CRPC
castration‐resistant prostate cancer
- FKBP5
FK506 binding protein 5
- Hsp
heat‐shock protein
- LBD
ligand‐binding domain
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- PSA
prostate‐specific antigen
- Sp1
specificity protein 1
- UBE2C
ubiquitin conjugating enzyme E2 C
- YB‐1
Y‐box‐binding protein 1
Supporting information
Fig. S1. Effects of quercetin and VER155008 on cell proliferation. Images of LNCaP95 cells treated with quercetin (10 μM, 25 μM, or 50 μM) (a) or VER155008 (10 μM or 25 μM) (b) at 48 h captured by IncuCyte ZOOM.
Fig. S2. Effects of quercetin and VER155008 on cell proliferation. Proliferation of 22Rv1 cells treated with quercetin (10 μM, 25 μM, or 50 μM) (a) or VER155008 (10 μM or 25 μM) (b) was monitored for 72 h by IncuCyte ZOOM.
Fig. S3. Effect of VER155008 on full‐length androgen receptor (AR‐FL) and androgen receptor splice variant 7 (AR‐V7), and Y‐box‐binding protein 1 (YB‐1) protein expression in 22Rv1 cells. 22Rv1 cells were treated with VER155008 (25 μM) for 48 h. Protein extracts (15 μg) were subjected to Western blot analysis.
Table S1. Protein identification with mass spectrometry. Heat shock protein 70‐binding proteins were identified at a 95% confidence level with the mascot algorithm.
Acknowledgments
The authors wish to thank members of the Central Laboratory of Osaka City University Graduate School of Medicine for technical support. This work was partly undertaken in the Cooperative Research Project Program of the Medical Institute of Bioregulation, Kyushu University.
Cancer Sci 108 (2017) 1820–1827
Funding information
None.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017; 67: 177–193. [DOI] [PubMed] [Google Scholar]
- 2. Denmeade SR, Isaacs JT. A history of prostate cancer treatment. Nat Rev Cancer 2002; 2: 389–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guo Z, Yang X, Sun F et al A novel androgen receptor splice variant is up‐regulated during prostate cancer progression and promotes androgen depletion‐resistant growth. Cancer Res 2009; 69: 2305–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Antonarakis ES, Lu C, Wang H et al AR‐V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 2014; 371: 1028–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Qu Y, Dai B, Ye D et al Constitutively active AR‐V7 plays an essential role in the development and progression of castration‐resistant prostate cancer. Sci Rep 2015; 5: 7654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Steinestel J, Luedeke M, Arndt A et al Detecting predictive androgen receptor modifications in circulating prostate cancer cells. J Clin Oncol 2015; 23: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu C, Lou W, Zhu Y et al Niclosamide inhibits androgen receptor variants expression and overcomes enzalutamide resistance in castration‐resistant prostate cancer. Clin Cancer Res 2014; 20: 3198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Andersen RJ, Mawji NR, Wang J et al Regression of castrate‐recurrent prostate cancer by a small‐molecule inhibitor of the amino‐terminus domain of the androgen receptor. Cancer Cell 2010; 17: 535–46. [DOI] [PubMed] [Google Scholar]
- 9. Chandrasekar T, Yang JC, Gao AC, Evans CP. Mechanisms of resistance in castration‐resistant prostate cancer (CRPC). Transl Androl Urol 2015; 4: 365–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Mol Cell 2010; 40: 253–66. [DOI] [PubMed] [Google Scholar]
- 11. Ciocca DR, Calderwood SK. Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones 2005; 10: 86–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gibbons NB, Watson RW, Coffey RN, Brady HP, Fitzpatrick JM. Heat‐shock proteins inhibit induction of prostate cancer cell apoptosis. Prostate 2000; 45: 58–65. [DOI] [PubMed] [Google Scholar]
- 13. Centenera MM, Gillis JL, Hanson AR et al Evidence for efficacy of new Hsp90 inhibitors revealed by ex vivo culture of human prostate tumors. Clin Cancer Res 2012; 18: 3562–70. [DOI] [PubMed] [Google Scholar]
- 14. Teng Y, Ngoka L, Mei Y, Lesoon L, Cowell JK. HSP90 and HSP70 proteins are essential for stabilization and activation of WASF3 metastasis‐promoting protein. J Biol Chem 2012; 287: 10051–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. He S, Zhang C, Shafi AA et al Potent activity of the Hsp90 inhibitor ganetespib in prostate cancer cells irrespective of androgen receptor status or variant receptor expression. Int J Oncol 2013; 42: 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer 2010; 10: 537–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ferraldeschi R, Welti J, Powers MV et al Second‐generation HSP90 inhibitor onalespib blocks mRNA splicing of androgen receptor variant 7 in prostate cancer cells. Cancer Res 2016; 76: 2731–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ferruelo A, Romero I, Cabrera PM, Arance I, Andres G, Angulo JC. Effects of resveratrol and other wine polyphenols on the proliferation, apoptosis and androgen receptor expression in LNCaP cells. Actas Urol Esp 2014; 38: 397–404. [DOI] [PubMed] [Google Scholar]
- 19. Hu R, Lu C, Mostaghel EA et al Distinct transcriptional programs mediated by the ligand‐dependent full‐length androgen receptor and its splice variants in castration‐resistant prostate cancer. Cancer Res 2012; 72: 3457–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tanaka M, Mun S, Harada A et al Hsc70 contributes to cancer cell survival by preventing Rab1A degradation under stress conditions. PLoS One 2014; 9: e96785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tanaka M, Shiota M, Nakao T et al Identification of low‐abundance proteins in serum via the isolation of HSP72 complexes. J Proteomics 2016; 136: 214–21. [DOI] [PubMed] [Google Scholar]
- 22. Tanaka M, Yamaguchi M, Shiota M et al Establishment of neutralizing rat monoclonal antibodies for fibroblast growth factor‐2. Monoclon Antib Immunodiagn Immunother 2014; 33: 261–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shiota M, Yokomizo A, Naito S. Increased androgen receptor transcription: a cause of castration‐resistant prostate cancer and a possible therapeutic target. J Mol Endocrinol 2011; 47: R25–41. [DOI] [PubMed] [Google Scholar]
- 24. Shiota M, Takeuchi A, Song Y et al Y‐box binding protein‐1 promotes castration‐resistant prostate cancer growth via androgen receptor expression. Endocr Relat Cancer 2011; 18: 505–17. [DOI] [PubMed] [Google Scholar]
- 25. Shiota M, Fujimoto N, Imada K et al Potential role for YB‐1 in castration‐resistant prostate cancer and resistance to enzalutamide through the androgen receptor V7. J Natl Cancer Inst 2016; 108. [DOI] [PubMed] [Google Scholar]
- 26. Wadosky KM, Koochekpour S. Molecular mechanisms underlying resistance to androgen deprivation therapy in prostate cancer. Oncotarget 2016; 7: 64447–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xing N, Chen Y, Mitchell SH, Young CY. Quercetin inhibits the expression and function of the androgen receptor in LNCaP prostate cancer cells. Carcinogenesis 2001; 22: 409–14. [DOI] [PubMed] [Google Scholar]
- 28. Lyabin DN, Eliseeva IA, Ovchinnikov LP. YB‐1 protein: functions and regulation. Wiley Interdiscip Rev RNA 2014; 5: 95–110. [DOI] [PubMed] [Google Scholar]
- 29. Gimenez‐Bonafe P, Fedoruk MN, Whitmore TG et al YB‐1 is upregulated during prostate cancer tumor progression and increases P‐glycoprotein activity. Prostate 2004; 59: 337–49. [DOI] [PubMed] [Google Scholar]
- 30. Evdokimova V, Ruzanov P, Anglesio MS et al Akt‐mediated YB‐1 phosphorylation activates translation of silent mRNA species. Mol Cell Biol 2006; 26: 277–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Watson PA, Chen YF, Balbas MD et al Constitutively active androgen receptor splice variants expressed in castration‐resistant prostate cancer require full‐length androgen receptor. Proc Natl Acad Sci USA 2010; 107: 16759–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Propper DJ, Braybrooke JP, Taylor DJ et al Phase I trial of the selective mitochondrial toxin MKT077 in chemo‐resistant solid tumours. Ann Oncol 1999; 10: 923–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Effects of quercetin and VER155008 on cell proliferation. Images of LNCaP95 cells treated with quercetin (10 μM, 25 μM, or 50 μM) (a) or VER155008 (10 μM or 25 μM) (b) at 48 h captured by IncuCyte ZOOM.
Fig. S2. Effects of quercetin and VER155008 on cell proliferation. Proliferation of 22Rv1 cells treated with quercetin (10 μM, 25 μM, or 50 μM) (a) or VER155008 (10 μM or 25 μM) (b) was monitored for 72 h by IncuCyte ZOOM.
Fig. S3. Effect of VER155008 on full‐length androgen receptor (AR‐FL) and androgen receptor splice variant 7 (AR‐V7), and Y‐box‐binding protein 1 (YB‐1) protein expression in 22Rv1 cells. 22Rv1 cells were treated with VER155008 (25 μM) for 48 h. Protein extracts (15 μg) were subjected to Western blot analysis.
Table S1. Protein identification with mass spectrometry. Heat shock protein 70‐binding proteins were identified at a 95% confidence level with the mascot algorithm.


