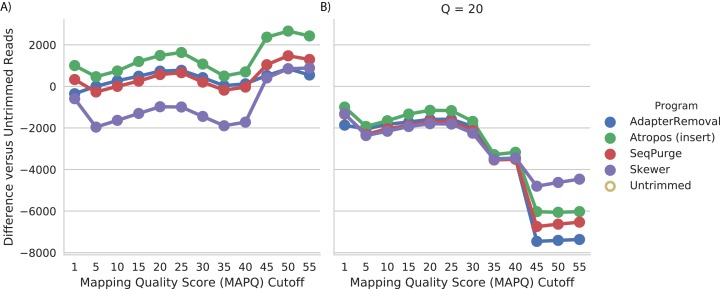
Figure 4. Atropos trimming best improves mapping of real WGBS sequencing reads.
Reads were adapter-trimmed with all four programs (A) without additional quality trimming (Q = 0) and (B) with quality trimming at a threshold of Q = 20. We mapped both untrimmed and trimmed reads to the genome. For each MAPQ cutoff M ∈ {0,5,…,60} on the x-axis, the number of trimmed reads with MAPQ >= M less the number of untrimmed reads with MAPQ >= M is shown on the y-axis for each program.