Abstract
Extreme ecosystems can be a source of untapped microorganisms to produce novel bioactive compounds of industrial interest. Consequently, in this work, 32 actinomycetes were isolated from 6 soil samples collected from Algerian Sahara in searching for untapped producers of novel antimicrobial compounds. All the isolates were further subjected to antimicrobial screening against pathogenic bacteria, yeast and fungi. The obtained results indicated that three of the isolates (named C, MS1 and 10) showed antimicrobial activities against most of the tested pathogenic microorganisms. Therefore, these three promising isolates, previously identified as Streptomyces by morphological, biochemical and physiological methods, were selected for their subsequent identification by the whole cell matrix assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS) analysis. Thus, the isolates C, MS1 and 10 were identified as Streptomyces violaceoruber B263 UFL, Streptomyces albus B262 UFL and Streptomyces badius B192 UFL, respectively. These results pointed out actinomycetes from Sahara soils as potential sources of novel antimicrobial compounds. Also, MALDI-TOF MS showed to be a robust technique for bacteria identification.
Key Words: Actinobacteria, antimicrobial activities, MALDI-TOF MS, Sahara soils, strain identification
Actinomycetes are ubiquitous Gram-positive aerobic bacteria which present a wide variety of morphologies and some of them, such as those belonging to the Streptomyces genus, resemble the filamentous fungi (1). The biotechnological interest of these microorganisms resides in their ability to produce different bioactive compounds. In fact, about two-thirds of natural antibiotics have been isolated from actinomycetes (2,3).
To find novel bioactive compound producers, exploration of ecosystems exposed to extreme environmental conditions is an interesting approach. Hence, research in later years is oriented towards the screening and isolation of actinomycetes from untapped habitats (4). The exploration of such habitats could even provide new taxa which, in turn, could be promising sources of novel bioactive compounds (5-8). In this sense, Algerian Sahara soils, exposed to hard climate conditions, represent particular ecosystems worthy of being explored. In addition, the Algerian Sahara soils have a significant biodiversity (9). Accordingly, the isolation of different actinomycetes strains from Algerian Sahara soils, their antimicrobial activity against pathogenic microorganisms and their characterization via conventional and molecular methods (i.e. MALDI-TOF MS) was investigated.
Materials and methods
Sampling
Six soils samples were collected from different Sahara areas in the south of Algeria, about 15 cm below the surface of the soil. All the soil samples were collected randomly, then packed in zipper bags and stored in a refrigerated container (4.C) during transportation to the laboratory.
The samples were air dried and heated aseptically to remove the undesired Gram-negative bacteria. Appropriate selective media such as yeast extract– malt extract agar medium (ISP2) and peptone yeast extract– malt extract agar medium (GLM) supplemented with actidione (5 µg/mL) and rifampicin (5 µg/mL) were used to promote actinomycetes conditions of growth and prevent fungal contamination (10).
Isolation and maintenance of actinomycetes
Ten-fold serial dilutions of soil samples were done using sterile distilled water. The soil suspensions were plated using ISP2 medium supplemented with 40 mg/mL of actidione to inhibit the development of eukaryotic microorganisms. The plates were incubated at 30 C for 7-10 days. Pure colonies were selected by observing the fine filaments around the actinomycete colonies under light microscopy and taken using a sterile inoculation loop. The isolated colonies were maintained on ISP2 agar slants at 4C for subsequent studies.
Pathogenic bacteria and yeasts
The pathogenic bacterial strains Bacillus cereus ATCC 10876, Micrococcus luteus ATCC 533, Enterococcus faecalis ATCC 29212, Lococcus aureus ATCC 43300, Staphylococcus aureus ATCC 25923, Staphylococcus epidermidis ATCC 1222, Escherichia coli ATCC 25922, Klebsiella pneumonia ATCC 43816, Pseudomonas aeruginosa ATCC 82827 and Salmonella enterica ATCC 14028 and the yeasts Candida albicans ATCC 10231 and Saccharomyces cerevisiae ATCC 4226 were obtained from the Pasteur Institute in Alger (Algeria). The strains were maintained on Petri plates containing nutrient agar at 4 C and subcultured every 2 months.
Pathogenic filamentous fungi
The pathogenic fungi Fusarium culmorum, Verticillium dahliae and Fusarium oxysporum f. sp. albedinis were obtained from the Institute of Phytopathology and Plant Protection of Messerghine in Oran (Algeria). The fungi were maintained on Petri plates containing potato dextrose agar (PDA) at 4 C and subcultured every 2 months.
Screening of the actinomycete isolates for antimicrobial activity
Primary screening was performed by the cross streak method against selected fungi and yeasts (11) and secondary screening by the agar cylinder method (12) against selected pathogenic bacteria (13). The isolates showing the highest antimicrobial activity were selected for further studies. The antimicrobial activity was evaluated by:
the cross streak method, measuring the distance of inhibition between the pathogenic bacteria and yeasts after incubation at 30 C for 24 h (Figure 1) and fungi after incubation at 25 C for 48 h (Figure 1).
Fig. 1.
Cross streak method of the isolates C, MS1 and 10 against different pathogenic bacteria, and fungi.
Pathogenic bacteria tested were 1: Bacillus cereus; 2: Micrococcus luteus; 3: Enterococcus faecalis; 4. Staphylococcus aureus ATCC 44300; 5: Staphylococcus aureus ATCC 25923; 6: Staphylococcus epidermidis; 7: Escherichia coli; 8: Klebsiella pneumonia). Pathogenic fungi were: F. c.: Fusarium culmorum; F. o. a.: Fusarium oxysporum f. sp. albedini; V. d: Verticillium dahliae.
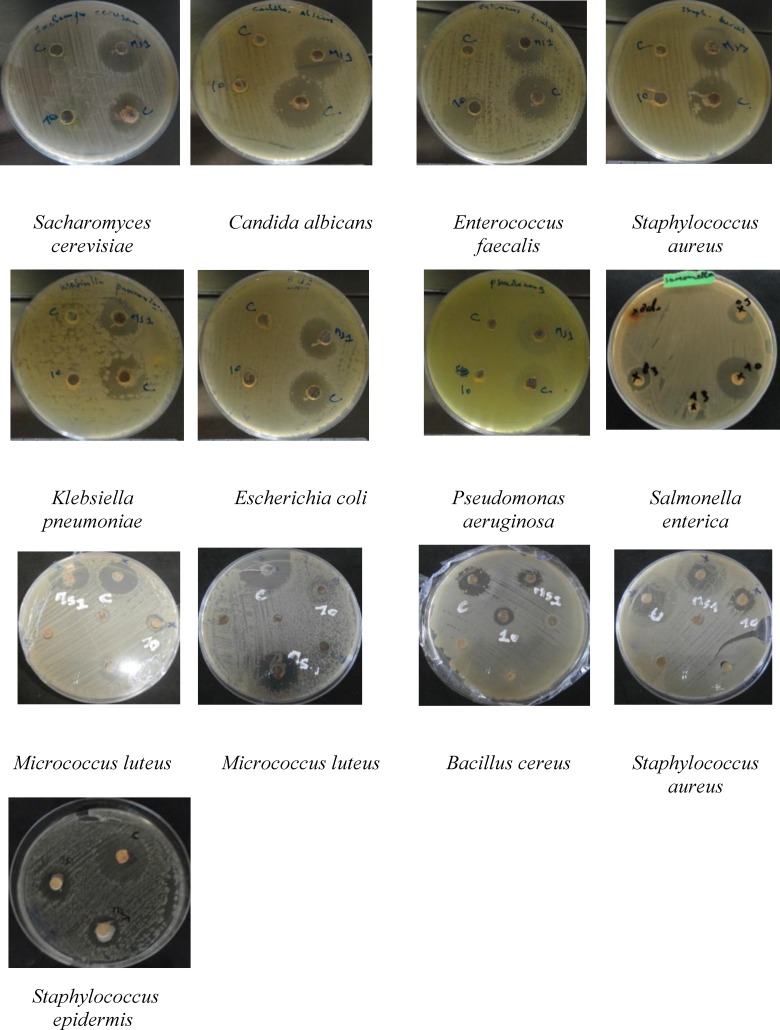
the agar cylinder method, measuring the inhibition zones around the colony of each isolate after incubation at 30 C for 24 h (bacteria and yeast) and 25 C for 48 h (fungi) (Figure 2).
Fig. 2.
Antimicrobial activity of isolates C, MS1 and 10 against different pathogenic bacteria and yeasts
Percentage of inhibition was calculated using the following formula (14).
(%)inhibition=(Rcontrol-Rtest)/Rcontrol x100
where Rtest is the colony diameter of the pathogenic fungus with actinomycete isolates on PDA plates and Rcontrol is the colony diameter of the pathogenic fungus on PDA plates.
The degree of antimicrobial activity of the isolates is classified depending on the mean diameter of the inhibition zone of inhibition. In the present case, the diameter of the zone of inhibition was divided as follows: excellent activity (≥18 mm), good activity (12-15 mm), moderate activity (10-12 mm) and weak activity (≤9 mm). Triplicate samples were performed.
Characterization of the actinomycete isolates
The isolates showing antimicrobial activity were characterized morphologically, biochemically and physiologically following the methods given in the International Streptomyces Project (ISP) (15).
Color determination was carried out using ISCC-NBS colour charts (16). The micromorphology of the strains was observed by light microscopy after incubation at 30 C for 2 weeks on petri plates containing ISP2 medium. The pigmentation of the aerial mycelium and the structure of sporophores, which are highly charac-teristic and useful in the classification of actinomy-cetes, were observed by cultivating the strains on different ISP media (i.e. yeast extract-malt extract agar (ISP2), oatmeal agar (ISP3), inorganic salts-starch agar (ISP4), glycerol-asparagine agar (ISP5), nutrient agar and Bennett medium). The arrange-ment of spores and sporulating structures was examined microscopically using the cover slip culture method by inserting a sterile cover slip at an angle of 45 C in starch casein agar medium (17,18). A loopful of each isolate was taken from a 7-day old culture, inoculated at the insertion place of the cover slip and incubated at 30 C for 7 days. The cover slip was carefully removed using a sterile forceps and placed upwards on a clean glass slide. The bacterial growth on the cover slip was fixed with a few drops of absolute methanol for 15 min, washed with tap water and then flooded with crystal violet reagent for 1 min followed by washing and blot drying. Finally, the cover slip was examined under microscope using oil immersion lens (100X). Biochemical and physiological characterization of the isolates were performed by streaking them on starch agar plates and incubating them at 30 C for 7 days. After incubation, iodine solution was poured onto the agar and examined for hydrolysis of starch by the production of a clear zone around the microbial growth. Furthermore, the isolates were streaked on gelatine agar plates and incubated at 30 C for 7 days to test for gelatine hydrolysis. After incubation, the plates were flooded with 1 mL of mercuric chloride solution and the diameters of the hydrolyzed zones around the colonies were measured. Also, the isolates were streaked on plates containing skimmed milk agar medium, incubated at 30 C for 7 days and the diameters of the hydrolyzed zones around the colonies were measured. To test sodium chloride resistance, starch casein agar was prepared in three batches and supplemented with 5%, 7% and 10% (w/v) sodium chloride. The medium was autoclaved, poured onto Petri plates and allowed for solidification. Then, the plates were streaked with the isolates and incubated at 30 C for 7 days. Visual observations were done to record the growth of the isolates. Finally, the isolates were streaked on starch casein agar plates and incubated at 25 C,30 C, 35 C, and 40 C for 7 days. The optimum temperature for maximum growth was determined through visual examination (19).
MALDI- TOF MS identification of the actino-mycetes isolates
The identification of the actinomycete isolates C, MS1 and 10 by MALDI-TOF MS was perform-ed on a Bruker Microflex system (Bruker, Germa-ny) instrument equipped with a nitrogen laser with an output wavelength of 337 nm used at a repetition rate of 60 Hz. All spectra were acquired in the linear positive mode within a range of 2-20 kDa.
A rapid, on-plate method was used for sample preparation. This method requires a small amount of bacteria which was picked up with a sterile toothpick from the bacteria colony and hand spotted onto a 96-spot polished stainless steel MALDI target plate. The spots were allowed to dry at room temperature and overlaid with 1 µL of MALDI matrix α-cyano-4-hydroxycinnamic acid (CHCA). CHCA was dissolved in a solvent (acetonitrile 50%, water 47.5% and trifluoroacetic acid 2.5%) to a final concentration of 2.5 mg/mL. When the matrix was air dried, the MALDI sample plate was inserted into the spectrometer and spectra were acquired under high vacuum conditions.
MALDI-Biotyper 3.1 software, library version V4.0.0.1 (5.627 MSPs) (Bruker Daltonik GmbH, Bremen, Germany) was used for the identification of each bacteria. This software allows discovering bacteria’s identity by its own unique molecular composition, revealing a characteristic peak pattern, even for reliable differentiation of species: the individual fingerprint. The molecular fingerprint is used for pattern matching. Sophisticated recalibra-tion and statistical algorithms allow robust and accurate identification. Matching scores supported by color codes are used for ranking the results. MALDI Biotyper integrates a ready to-use reference library comprising thousands of individual species and is growing continuously. The identification of unknown bacteria was performed by comparing their spectral fingerprints with those existing in the database (composed for 5627 entries). A matching score based on identified masses and their intensity correlation was generated and used for ranking the results.
Biotyper 3.1 software (Bruker), returned the top 10 identification matches along with confidence scores ranging from 0.0 to 3.0. Estimated values of 2.3 or higher were considered high-confidence scores and indicate that of genus and species identifications is reliable (secure species), score values between 2.0 and 2.29 show that the genus is reliable and the species is probable. Score values between 1.7 and 1.99 were considered intermediate confidence and indicate that the identification of genus was probable. Score values lower than 1.7 were considered “not reliable” evincing that spectra acquisition was insufficient or no peak protein was detected, and further analysis is required for this sample.
Production, extraction, and detection of antimic-robial compounds
Each isolate of actinomycetes was cultivated in 500-mL Erlenmeyer flasks containing 100 mL of ISP2 medium (1% malt extract, 0.4% yeast extract, 0.4% glucose, pH 7.2). The flasks were incubated at 30 C for 5 days on an orbital shaker (250 rpm). After that, the culture broth was centrifuged for 20 min at 8000g to remove the mycelium. The supernatant was divided into 4 equal volumes (60 mL each) and extracted with 60 mL of an organic solvent. Four different organic solvents ranging from non-polar to polar ones were screened for effectiveness, including n-hexane, dichloromethane, n-butanol and ethyl acetate. The organic phases of strains C and MS1 and the aqueous phase of strain 10 were evaporated to dryness using a Rotavapor (Laborota 4000). To select the best extraction solvents, according to their quantity and antimicrobial activity, the activities of each crude extract of the selected isolates (i.e. C, MS1 and 10) were examined. Briefly, each crude extract was defatted with 1 mL of methanol and subjected to biological assay (disc of 6 mm in diameter, Pasteur Institute) against Micrococcus luteus (60 µL per disc) (data not shown). The solvents which gave the highest inhibition diameter were then used for the extraction of the active substances (20).
To test the antimicrobial activities of each crude extract, the agar well-diffusion method on Muller Hinton medium (MHA) was performed. For this, a volume of 25 µL of the crude extract of each strain (i.e. MS1, C and 10) was carefully dispended into each well, allowed to diffuse for 2 h at 4 C and incubated at 37 C for 24 h.
After incubation, the zone of inhibition (in mm) around each well was recorded.
Results
Actinomycetes isolation
Among the 32 actinomycetes isolated from Sahara soils, 13 isolates showed antibacterial activities against at least one of the pathogenic bacteria by the cross streak method as primary screening and the agar cylinder method as secondary screening. The results revealed that isolates C, MS1 and 10 exhibited broad spectrum activities against pathogenic bacteria (Gram-positive and Gram-negative), especially against M. luteus, S. aureus and S. epidermidis. Anti-yeast activity was also recorded against S. cerevisiae and C. albicans, whereas the antifungal activity was moderate (Table 1).
Table 1.
Antimicrobial activity of strains C, MS1 and 10
| Test organisms | Inhibition zone (mm) | ||
|---|---|---|---|
| Gram-positive bacteria | C | MS1 | 10 |
| 1. Bacillus cereus 2. Micrococcus luteus 3. Enterococcus faecalis 4. Staphylococcus aureus ATCC 44300 5. Staphylococcus aureus ATCC 25923 6 .Staphylococcus epidermidis |
21 28 20 20 21 22 |
14 35 20 19 22 18 |
15 22 13 21 20 13 |
| Gram-negative bacteria | |||
| 7. Escherichia coli 8. Klebsiella pneumoniae 9. Pseudomonas aeruginosa 10. Salmonella enteric |
19 20 18 11 |
19 20 18 15 |
22 22 20 21 |
| Yeasts | |||
| 11. Candida albicans 12. Saccharomyces cerevisiae |
18 17 |
20 20 |
16 11 |
| Filamentous fungi (% of growth inhibition) | |||
| 13. Fusarium culmorum 14. Verticillium dahliae 15. Fusarium oxysporum f. sp. albedinis |
36 34 39 |
38 35 35 |
32 40 37 |
The values are the mean of triplicate samples with a standard deviation less than 10%
Morphological, biochemical and physiological characterization of the isolates
Colonies of strains C, MS1 and 10 grew well on most of the organic media used and were convex and smooth. The aerial mycelium was grey for strains C and MS1 and yellowish grey for strain 10. The substrate mycelium was pale yellow for strain MS1, blue violet for strain C and brilliant orange yellow for strain 10 (http://people.csail.mit.edu/ jaffer/Color/Dictionaries). Abundant dark green diffusible pigments were formed only on ISP2 and Bennett media for the strain 10. The cultural characteristics of the isolates are given in details in Table 2.
Table 2.
Culture characteristics of strains C, MS1 and 10 on different media
|
Medium
|
Growth
|
Spore colour
|
Vegetative mycelium
|
Soluble pigment
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | MS1 | 10 | C | MS1 | 10 | C | MS1 | 10 | C | MS1 | 10 | |
| ISP2 | +++ | +++ | +++ | Grey | Grey | Yellowish grey | Pale yellow | Blue violet | Brilliant orange yellow | - | - | +Dark green |
| ISP3 | + | + | + | Grey white | Grey white | Yellowish Gray | Pale yellow | Blue violet | Brilliant orange yellow | - | - | - |
| ISP4 | ++ | ++ | ++ | White | White | Yellowish Gray | Pale yellow | Blue violet | Brilliant orange yellow | - | - | - |
| ISP5 | + | + | + | Grey | Grey | Yellowish Gray | Pale yellow | Blue violet | Brilliant orange yellow | - | - | - |
| Nutrient agar | + | + | + | Grey white | Grey white | Yellowish Gray | Pale yellow | Blue violet | Brilliant orange yellow | - | - | - |
| Benett medium | +++ | +++ | +++ | Grey white | Grey white | Yellowish Gray | Pale yellow | Blue violet | Brilliant orange yellow | - | - | +Dark green |
ISP: International Streptomyces Project; ISP2: yeast extract—malt extract agar medium; ISP3: oatmeal agar medium; ISP4: inorganic salts—starch agar medium; ISP5: glycerol—asparagine agar medium.
The isolates were able to hydrolyze a great number of compounds such as casein, arabinose, fructose, galactose, glucose, mannitol and xylose. They were resistant to sodium azide (0.05 g/L), crystal violet (0.05 mg/mL) and several antibiotics such as ampicillin (20 mg/L), kanamycin (25 mg/L) and tetracycline (30 mg/L). The optimum growth temperature of most isolates was between 25 and 30 C, growth being inhibited at temperatures above 40 C (Table 3).
Table 3.
Physiological and biochemical properties of strains C, MS1 and 10
|
Isolates
|
|||
|---|---|---|---|
| Property | C | MS1 | 10 |
| Melanin formation (ISP6 and ISP7) Starch hydrolysis (tryptone soya agar medium) Casein hydrolysis (casein agar medium) Urease production (nitrate peptone broth medium) Gelatine hydrolysis (nutrient gelatine medium) Soluble pigment production (ISP media) H2S production (triple sugar iron agar medium) pH range of growth (ISP4) 6 - 9 Temperature range of growth (ISP4) 25-45 C Antibiotic resistance Ampicillin (20 mg/L) Kanamycin (25 mg/L) Tetracycline (30 mg/L) Chloramphenicol (25 mg/L) Chlortetracycline hydroxychloride (30 mg/L) NaCl tolerance NaCl (5% (ISP4) NaCl 7% (ISP4) NaCl 10% (ISP4) Sporophore morphology Straight Spiral Flexous Retinaculum apertum Growth on inhibitory compounds Phenol 0.1% (ISP4) Lysozyme 0.005% (ISP4) Sodium azide 0.01% (ISP4) Crystal violet 0.05% (ISP4) |
- + + + + + + - - + + + - R S R R S - + + + - - - + + - + - - - - - |
+ + + + + + + - - + + + - R S R S S - + + + - - - + + - - - - - - - |
+ + + + + + + + - + + Green + + + + - R S R S S - + + + - - - + + - - - - - ND - |
ISP: International Streptomyces Project; ISP4: inorganic salts-starch agar medium; ISP6: peptone-yeast extract-iron agar medium; ISP7: tyrosine agar medium. +: Growth; -: no growth; ND: not determined. R: Resistant, S: Sensible.
Identification of the isolates by MALDI-TOF MS analysis
The three isolates C, MS1 and 10 were identified by MALDI-TOF MS as Streptomyces violaceoruber (NCBI code 1935, score 1.912), Streptomyces albus (NCBI code 1888; score 1.261) and Streptomycete badius (NCBI code 1941; score 1.514), respectively. The first one presented high score and the third one was acceptable. Nevertheless, the second one did not present a high score by MALDI-TOF MS analysis, but with the additional information given by the morphological, physiological, biochemical and cultural characteristics tests, we concluded that it was very likely that strain.
Discussion
The increased emergence of multidrug resistant organisms makes the treatment of numerous infectious diseases difficult. Hence, the development of novel effective drugs against the abovementioned organisms is needed. For this, the exploration of untapped and extreme habitats can lead to the isolation of novel microorganisms to produce novel bioactive compounds, recently several researchers have shown the potential of extreme habitats as reservoirs of promising antimicrobial compounds producers (4,7,21, 22-26).
Taking into account the results exposed above, the isolation of microorganisms with promising antimicrobial activities from Algerian Sahara soils as a model of an extreme ecosystem was pursued. Among the 32 actinomycetes isolated from Algerian Sahara soils, 3 of them (named as C, MS1 and 10) exhibited broad spectrum antimicrobial activities against different pathogenic bacteria, yeasts and even fungi. These C, MS1 and 10 isolates were identified by combining the results obtained via conventional and molecular methods, as Streptomyces violaceoruber, Streptomyces albus and Streptomycete badius, respectively. This is not surprising since Streptomyces is the most common genus in the actinomycetes order (27). Our results are in agreement with those reported by Kumar et al. (28). As a consquence, they found that Streptomyces was the predominant genus in the actinomycete strains isolated from soil samples of the Uttarakhand state (India).
The isolated strains (i.e. C, MS1 and 10) are potential producers of bioactive compounds as shown in the antimicrobial activities of their crude extracts against different pathogenic microorganisms (Table 4; Figure 3). These results are in agreement with those reported by different researchers. As a result, Arumugam et al. (24) found that the most common soil bacteria actinobacteria isolated from soil samples of a mangrove forest in India exhibited anti-microbial activity and antibiotic production. Moreover, Elbendary et al. (29) reported antimicrobial activity form actinobateria isolated from farm soils in Egypt. Besidess, S. albus G was reported to produce an antibiotic compound (30), S. violoaceoruber VLK-4 , isolated from soil samples in India, was shown to produces antimicrobial compound (31) and a bioactive compound was isolated from the culture broth of a S. badius strain isolated from Egyptian soil (32).
Table 4.
Zone of inhibition (mm) of crude extracts produced by strains C, MS1 and 10 on ISP2 (International Streptomyces Project yeast extract—malt extract agar) medium using the well diffusion method.
| Test organisms | Inhibition zone (mm) | ||
|---|---|---|---|
| Gram-positive bacteria | Strain C | Strain MS1 | Strain 10 |
|
Bacillus cereus
Micrococcus luteus Enterococcus faecalis Staphylococcus aureus Staphylococcus epidermidis |
14 15 13 14 12 |
11 18 --- 12 --- |
--- --- 30 35 35 |
| Gram-negative bacteria | |||
|
Pseudomonas fluorescens
Escherichia coli Klebsiella pneumoniae |
12 14 15 |
15 10 --- |
36 34 14 |
| Yeasts | |||
|
Candida albicans
Saccharomyces cerevisiae |
17 13 |
12 --- |
31 12 |
The values are the mean of triplicate samples with a standard deviation less than 10%.
---: no inhibition
Fig. 3.
Antimicrobial activity of the crude extract of the isolates C, MS1 and 10 against different pathogenic bacteria and yeasts.
Strain 10 exhibited an antimicrobial activity higher than strains C and MS1, especially against P. fluorescens (36 mm) ), S. aureus (35 mm), S. epidermidis (35 mm), E. coli (34 mm) and C. albicans (31 mm) (Table 4; Figure 3). These differences can be attributed to their different chemical structures, disintegration during the extraction process (21) and environmental factors (temperature and pH of the crude extract).
It is worthy to mention that the crude extracts from the isolates showed antimicrobial activity against Gram-negative bacteria since, in general, they are more resistant to antimicrobial compounds than the Gram-positive bacteria (29). Lee et al. (33) also reported inhibitory activity against Gram-negative bacteria for actinomycetes isolated from soil samples of the Tanjung Lumpur mangrove forest in Malaysia. On the contrary, Rabia-Boukhalfa et al. (25) detected no activity against Gram-negative bacteria by a halotolerant actino-bacterium, belonging to the genus Nocardiopis, isolated from a salt lake soil sample in the Algerian Sahara.
The obtained results point out actinomycetes from Algerian Sahara soils as potential sources of novel antimicrobial compounds. Future research will be required to identify the produced antimicrobial compounds which will involve their purification and the use of different chemical analysis such as HPLC-MS, FTIR and NMR techniques. On the other hand, MALDI-TOF MS has shown to be a fast, reliable and highly robust technique for bacteria identification.
Acknowledgment
The authors thank Ane Aguirre Emazabel (Osakidetza’s technician) for her help in MALDI-TOF MS characterization.
Conflict of interest
The authors declared no conflict of interest.
References
- 1.Ronald M. Principles of microbiology. New York: WCB McGrill-Hill; 1997. [Google Scholar]
- 2.Buckingham J. Dictionary of Natural Products, Supplement 4. Taylor & Francis; 1997. [Google Scholar]
- 3.Newman DJ, Cragg GM, Snader KM. Natural products as sources of new drugs over the period 1981-2002. J Nat Prod. 2003;66:1022–37. doi: 10.1021/np030096l. [DOI] [PubMed] [Google Scholar]
- 4.Singh LS, Sharma H, Talukdar NC. Production of potent antimicrobial agent by actinomycete, Streptomyces sannanensis strain SU118 isolated from phoomdi in Loktak Lake of Manipur, India. BMC Microbiol. 2014;14:278. doi: 10.1186/s12866-014-0278-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aguilar A. Extremophile research in the European Union: from fundamental aspects to industrial expectations. FEMS Microbiology Reviews. 1996;18:89–92. [Google Scholar]
- 6.Bouras N, Mathieu F, Sabaou N, et al. Effect of amino acids containing sulfur on dithiolopyrrolone antibiotic productions by Saccharothrix algeriensis NRRL B-24137. J Appl Microbiol. 2006;100:390–7. doi: 10.1111/j.1365-2672.2005.02762.x. [DOI] [PubMed] [Google Scholar]
- 7.Boubetra D, Zitouni A, Bouras N, et al. Saccharothrix hoggarensis sp nov, an actinomycete isolated from Saharan soil. Int J Syst Evol Microbiol. 2013;63:549–53. doi: 10.1099/ijs.0.039099-0. [DOI] [PubMed] [Google Scholar]
- 8.Meklat A, Bouras N, Zitouni A, et al. Actinopolyspora saharensis sp nov, a novel halophilic actinomycete isolated from a Saharan soil of Algeria. Antonie Van Leeuwenhoek. 2013;103:771–6. doi: 10.1007/s10482-012-9859-z. [DOI] [PubMed] [Google Scholar]
- 9.Sabaou N, Boudjella H, Bennadji A. Les sols des oasis du Sahara algérien, source d’actinomycètes rares producteurs d’antibiotiques. Sécheresse. 1998;9:147–53. [Google Scholar]
- 10.Cuesta G, Garcia-de-la-Fuente R, Abad M, et al. Isolation and identification of actinomycetes from a compost-amended soil with potential as biocontrol agents. J Environ Manage. 2012;95(Suppl):S280–4. doi: 10.1016/j.jenvman.2010.11.023. [DOI] [PubMed] [Google Scholar]
- 11.Lemos ML, Toranzo AE, Barja JL. Antibiotic activity of epiphytic bacteria isolated from intertidal seaweeds. Microb Ecol. 1985;11:149–63. doi: 10.1007/BF02010487. [DOI] [PubMed] [Google Scholar]
- 12.Audrey W. Disk Diffusion Test and Gradient Methodologies. In: Schwalbe R, Steele-Moore L, Goodwin AC, editors. Antimicrobial Susceptibility Testing Protocols. Boca Raton: CRC Press Taylor and Francis Group; 2007. pp. 53–72. [Google Scholar]
- 13.Taechowisan T, Lu C, Shen Y, et al. Secondary metabolites from endophytic Streptomyces aureofaciens CMUAc130 and their antifungal activity. Microbiology. 2005;151:1691–5. doi: 10.1099/mic.0.27758-0. [DOI] [PubMed] [Google Scholar]
- 14.Wang SL, Hsiao WJ, Chang WT. Purification and characterization of an antimicrobial chitinase extracellularly produced by Monascus purpureus CCRC31499 in a shrimp and crab shell powder medium. J Agric Food Chem. 2002;50:2249–55. doi: 10.1021/jf011076x. [DOI] [PubMed] [Google Scholar]
- 15.Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species1. International Journal of Systematic and Evolutionary Microbiology. 1966;16:313–40. [Google Scholar]
- 16.Kelly LK. Central notations for the revised iscc-nbs color-name blocks. J Res Natl Bur Stand. 1958;61:427–31. [Google Scholar]
- 17.Williams STaC T. Actinomycetes. In: Booth C, editor. Methods in Microbiology. London: Academic Press; 1971. [Google Scholar]
- 18.Kuester E, Williams ST. Selection of Media for Isolation of Streptomycetes. Nature. 1964;202:928–9. doi: 10.1038/202928a0. [DOI] [PubMed] [Google Scholar]
- 19.Muiru WM, Mutitu EW, Mukunya DM. Identification of selected actinomycetes isolates and characterization of their antibiotic metabolites. J Biol Sci. 2008;8:1021–6. [Google Scholar]
- 20.Mertz FP, Yao RC. Streptosporangium carneum sp nov isolated from soil. Int J Syst Bacteriol. 1990;40:247–53. doi: 10.1099/00207713-40-1-28. [DOI] [PubMed] [Google Scholar]
- 21.Gebreyohannes G, Moges F, Sahile S, et al. Isolation and characterization of potential antibiotic producing actinomycetes from water and sediments of Lake Tana, Ethiopia. Asian Pac J Trop Biomed. 2013;3:426–35. doi: 10.1016/S2221-1691(13)60092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rashad FM, Fathy HM, El-Zayat AS, et al. Isolation and characterization of multifunctional Streptomyces species with antimicrobial, nematicidal and phytohormone activities from marine environments in Egypt. Microbiol Res. 2015;175:34–47. doi: 10.1016/j.micres.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Saker R, Meklat A, Bouras N, et al. Diversity and antagonistic properties of culturable halophilic actinobacteria in soils of two arid regions of septentrional Sahara: M’zab and Zibans. Annals of Microbiology. 2015;65:2241–53. [Google Scholar]
- 24.Arumugam T, Senthil Kumar P, Kameshwar R, et al. Screening of novel actinobacteria and characterization of the potential isolates from mangrove sediment of south coastal India. Microb Pathog. 2017;107:225–33. doi: 10.1016/j.micpath.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 25.Hadj Rabia-Boukhalfa Y, Eveno Y, Karama S, et al. Isolation, purification and chemical characterization of a new angucyclinone compound produced by a new halotolerant Nocardiopsis sp HR-4 strain. World J Microbiol Biotechnol. 2017;33:126. doi: 10.1007/s11274-017-2292-8. [DOI] [PubMed] [Google Scholar]
- 26.Shaik M, Girija Sankar G, Iswarya M, et al. Isolation and characterization of bioactive metabolites producing marine Streptomyces parvulus strain sankarensis-A10. Journal of Genetic Engineering and Biotechnology. 2017;15:87–94. doi: 10.1016/j.jgeb.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taddei A, Rodriguez MJ, Marquez-Vilchez E, et al. Isolation and identification of Streptomyces spp from Venezuelan soils: morphological and biochemical studies I. Microbiol Res. 2006;161:222–31. doi: 10.1016/j.micres.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 28.Kumar V, Bisht GS, Gusain O. Terrestrial actinomycetes from diverse locations of Uttarakhnad, India: Isolation and screening for their antibacterial activity. Iran J Microbiol. 2013;5:299–308. [PMC free article] [PubMed] [Google Scholar]
- 29.Silhavy TJ, Kahne D, Walker S. The Bacterial Cell Envelope. Cold Spring Harbor Perspectives in Biology. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Majer J, Chater KF. Streptomyces albus G produces an antibiotic complex identical to paulomycins A and B. J Gen Microbiol. 1987;133:2503–7. doi: 10.1099/00221287-133-9-2503. [DOI] [PubMed] [Google Scholar]
- 31.Naragani K, Munaganti RK, Sirigiri CK. Antimicrobial Potential of Streptomyces violaceoruber VLK-4 Isolated from South Coast of Andhra Pradesh, India. Int J Pharm Sci Rev Res. 2014;25:125–29. [Google Scholar]
- 32.El Sayed OH, Asker MMS, Swelim MA, et al. Production of hydroxy marilone C as a bioactive compound from Streptomyces badius. Journal of Genetic Engineering and Biotechnology. 2016;14:161–8. doi: 10.1016/j.jgeb.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee LH, Zainal N, Azman AS, et al. Diversity and antimicrobial activities of actinobacteria isolated from tropical mangrove sediments in Malaysia. ScientificWorldJournal. 2014;2014:698178. doi: 10.1155/2014/698178. [DOI] [PMC free article] [PubMed] [Google Scholar]