Abstract
Standardization of results is an important milestone in the maturation of any truly quantitative methodology. For instance, a lack of measurement agreement across imaging platforms limits multisite studies, between-study comparisons based on the literature, and inferences based on and the generalizability of results. In GABA-edited MEGA-PRESS, two key sources of differences between implementations are: differences in editing efficiency of GABA and the degree of co-editing of macromolecules (MM). In this work, GABA editing efficiency κ and MM-co-editing μ constants are determined for three widely used MEGA-PRESS implementations (on the most common MRI platforms; GE, Philips, and Siemens) by phantom experiments. Implementation-specific κ,μ-corrections were then applied to two in vivo datasets, one consisted of 8 subject scanned on the three platforms and the other one subject scanned eight times on each platform. Manufacturer-specific κ and μ values were determined as: κGE = 0.436, κSiemens = 0.366 and κPhilips = 0.394 and μGE = 0.83, μSiemens = 0.625 and μPhilips = 0.75. Applying the κ,μ-correction on the Cr-referenced data decreased the coefficient of variation (CV) of the data for both in vivo data sets (multisubjects: uncorrected CV = 13%, κ,μ-corrected CV = 5%, single subject: uncorrected CV = 23%, κ,μ-corrected CV = 13%) but had no significant effect on mean GABA levels. For the water-referenced results, CV increased in the multisubject data (uncorrected CV = 6.7%, κ,μ-corrected CV = 14%) while it decreased in the single subject data (uncorrected CV = 24%, κ,μ-corrected CV = 21%) and manufacturer was a significant source of variance in the κ,μ-corrected data. Applying a correction for editing efficiency and macromolecule contamination decreases the variance between different manufacturers for creatine-referenced data, but other sources of variance remain.
Keywords: GABA, Multi-site, Editing efficiency, Cross-platform, Macromolecular co-editing, MEGA-PRESS
1. Introduction
GABA-edited MRS is an important modality for studying inhibitory processes in normal brain function and disease [1,2]. However, a lack of standardization of MRS editing methods, in terms of acquisition parameters, reference compounds, and data analysis methods, currently makes comparing data between studies difficult. With standardized measurements and/or established post-hoc corrections, it would be possible to compare literature data quantitatively between studies, strengthening the impact and scientific potential of the data published.
One major reason why no concerted effort has been made to standardize quantification methodologies is the heterogeneity of acquisition methodologies. Excluding alternative pulse sequences such as J-PRESS [3] and MEGA-SPECIAL [4], there still remain many different implementations of the MEGA-PRESS sequence [5], the most widely used method for measuring GABA [6,7]. Therefore, differences in GABA quantification exist, even when data processing methodologies are rigidly set.
The MEGA-PRESS pulse sequence is generally implemented starting from the vendor-standard PRESS acquisition, incorporating the additional editing pulses and moving crusher gradients. The waveforms and bandwidths of the slice-selective pulses therefore depend on the vendor’s implementation of the PRESS sequence, and this results in different voxel profiles for the excitation and refocusing dimensions. The minimum-achievable duration of the first spin-echo (TE1) is also different between vendors. Additionally, the duration, shape, bandwidth and timing of editing pulses differs between implementations. These various parameters have a significant impact on the intensity and multiplet pattern of the edited 3.0 ppm GABA signal [6,8] as well as co-editing of other compounds (in particular, macromolecule resonances (MM) which also resonate at 3.0 ppm). Overall, the differences between GABA-editing sequences can be summarized by two metrics: (a) how much GABA signal is produced (i.e. the editing efficiency), and (b) how much MM signal is co-edited.
Editing efficiency can be intuitively described as “the fraction of the total possible signal that is observed in the edited experiment”, and the simplest experimental measurement that would approximate it is the integral ratio between the edited difference spectrum and the editing-on subspectrum. However, imperfections in coupling refocusing during the editing-on experiment (whether due to chemical-shift-related inhomogeneities in coupling evolution, sub-optimal editing-pulse timing or the evolution of couplings other than that refocused) make this a poor measurement of the true underlying editing efficiency. In this paper, we use glycine as a reference signal, as it is not troubled by coupling evolution and is resolved from the GABA signals. Thus, the editing efficiency (κ) of GABA was measured as the ratio of the edited signal in the difference spectrum to the signal of glycine in the same experiment, which has no coupling and no overlap with GABA (correcting for differential relaxation of the two signals). This was performed in phantoms containing GABA and glycine. If the detected 3.0 ppm GABA signal is considered as a triplet and the central line of the triplet is removed from the edited spectrum, the maximum editing efficiency would be 0.5. Losses in editing efficiency can occur due to imperfect refocusing of coupling in the ON experiment, e.g., if the editing pulses have reduced flip angle, are not separated by TE/2, or because of imperfect evolution of coupling in the OFF experiment, as, for instance, can be caused by chemical shift displacement effects [8,9].
As mentioned above, a second feature that impacts the acquired signal in vivo is the co-editing of MM [10–12]. Because co-edited MM signals overlap with GABA, the term ‘GABA+’ is commonly used to indicate the sum of GABA and MM. The GABA+ signal typically includes a contribution of ~50% from MM, depending on pulse sequence parameters such as the echo time and bandwidth of editing pulses. In this paper, μ is defined as the relative co-editing efficiency of MM signal; high values of μ (maximum = 1) indicate non-selective editing (i.e. MM is co-edited with the same editing efficiency as GABA), whereas highly selective (or deliberately MM-suppressed [4,10–12]) editing which does not co-edit MM would have a μ of 0.
The purpose of this study was to characterize three widely used [13–20] implementations of GABA-edited MRS acquisitions (typical implementations on GE, Siemens, and Philips systems), and to use the results to normalize in vivo GABA measurements across differing implementations of the MEGA-PRESS sequence. No attempt was made to implement identical pulse sequences on the three scanners. Currently-existing pulse sequences were compared without modification, other than to homogenize data acquisition parameters as much as possible. κ and μ were measured in phantoms on each platform, and it was investigated whether correcting for these two parameters improved the agreement between the three sequences for data acquired in healthy controls. This approach enables a thorough investigation as to the underlying differences between the sequences using first principles. Understanding differences between implementations is necessary to properly address variance and discrepancies. Furthermore, the approach here provides a framework to account for differences between implementations not evaluated in the current study.
2. Methods
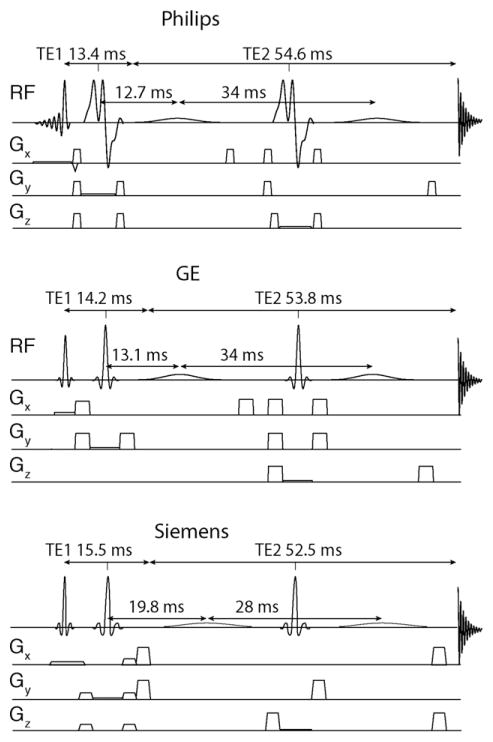
MRS data were acquired on GE Signa HDx, Siemens Tim Trio and Philips Achieva scanners all operating at 3 Tesla. Experiments were performed using either 32-channel (Philips, Siemens) or 8-channel head coils (GE). The MEGA-PRESS pulse sequence for the Siemens scanner was provided by Siemens (‘Work-in-Progress’, WIP), whereas the GE [14] and Philips [21] pulse sequences were provided by research sites. Schematic diagrams of the 3 sequences are provided in Fig. 1.
Fig. 1.
Schematic pulse sequences of the MEGA-PRESS pulse sequence as implemented on each of the 3 scanners in this study.
2.1. Phantom preparation
A phosphate-buffered phantom was prepared containing 20 mM GABA and 20 mM glycine (Gly). Gly was chosen as a reference compound because its 1H-MR spectrum contains a single resonance at 3.5 ppm [22], which will not evolve under any couplings and which does not overlap with any of the GABA resonances. This phantom was used to measure the editing efficiency of GABA (κ) on the 3 scanners. In order to obtain a relaxation-independent measurement of κ, experiments to measure the longitudinal and transverse relaxation times, T1 and T2, of Gly and GABA in the phantom were performed.
2.2. Measurement of phantom GABA and glycine T1 and T2 relaxation times
As defined, κ and μ are independent of T1 and T2 relaxation. However, for the measurement of relaxation-independent values of κ and μ in phantoms, relaxation that occurs within the experiment must be addressed. While TE impacts both T2 relaxation and J-modulation, these two processes are independent. J-coupling of a spin system and the resulting J-modulation of a signal is defined by the molecule itself and not the environment (in vivo or in vitro) or relaxation. Therefore the J-modulation will be the same in a phantom as in vivo and, consequently, κ will be the same in vitro and in vivo. Similarly, μ describes the extent to which the editing pulses invert other coupled resonances (in this case macromolecules), a process that is independent of situation, in vitro or in vivo. Thus while the total signal is a function of relaxation, J-modulation and co-editing, these individual processes are separable. In the calculation of κ and μ (as performed below), to ensure these parameters are not biased by relaxation in the experiment, we account for T1 and T2 of GABA and Gly in the phantom measurements. The result is parameter estimates (κ and μ that are independent of relaxation and thus can be applied to in vitro or in vivo data.
Using the Siemens Tim Trio, GABA-edited MEGA-PRESS spectra were acquired at a range of relaxation times (TR) ranging from 0.6 to 20 s (0.6, 0.8, 1.0, 1.2, 1.4, 1.6, 2.0, 2.4, 2.8, 3.2, 3.6, 4, 5, 6, 8, 10, 15, and 20 s) to determine T1 for GABA and Gly. Other experimental parameters included: TE = 70 ms; 64 averages of 512 datapoints; voxel size = 8 cm3; editing pulses of 15 ms duration applied at 1.9 ppm (ON) and 7.5 ppm (OFF).
These data were analyzed using in-house Matlab code, applying 8-Hz line-broadening, and integrating the editing-pulse ON spectra from 2.68 to 3.31 for GABA and 3.37 ppm to 3.77 ppm for Gly. These integrals were fit to a single-exponential saturation-recovery model with two free parameters, the fully relaxed integral and the phantom relaxation time constant T1.
Using the GE Signa HDx scanner, MEGA-PRESS GABA-edited spectra were acquired at a range of echo times (TE) ranging from 68 to 350 milliseconds (68 ms, 70 to 280 ms in steps of 10 ms, 315 ms and 350 ms) to determine the T2 of GABA and Gly. Other experimental parameters included: TR 2 s; 64 averages of 4096 datapoints; 5000 Hz spectral width; voxel size = 8 cm3; editing pulses of 14 ms duration applied at 1.9 ppm (ON) and 7.5 ppm (OFF). The phantom editing-ON spectra give GABA signals that are not (to a first approximation) modulated by J-coupling. So, the editing-ON spectra can be fit to estimate the T2 of the 3.0 ppm GABA resonance. The log of these integrals were fit to a linear model with two free parameters, the y-intercept and the inverse of the phantom relaxation time constant T2. This approach has been previously applied to measure GABA T2 [23].
2.3. Determination of editing efficiencies κ in the phantom
The editing efficiency for each scanner (κi) was calculated according to:
| (1) |
where IGABA,DIFF is the area of GABA in the averaged difference spectrum (i.e. the difference between the ON-spectrum and the OFF-spectrum, divided by two) and IGly,OFF is the integral of Gly in the time-averaged OFF spectrum. This is simply the ratio of the relaxation-corrected GABA difference signal to the relaxation-corrected glycine signal.
2.4. Determination of MM co-editing ratios μ in the phantom
Co-editing of MM was quantified as the relative editing of the 1.7 ppm MM peak, defined as μi. To measure μi, on each of the three systems the edit-ON frequency was varied from 1.9 ppm to 1.3 ppm in increments of 0.04 ppm. The resulting signal was fit with a Gaussian curve and the amplitude at an offset of −0.2 ppm (i.e., equivalent to 1.7 ppm, the shift of the MM resonance from the GABA resonance) was used to determine μi. It should be noted that the parameter μ is not the absolute editing efficiency; rather, it is the fraction of maximal (on-resonance) editing efficiency that occurs at an editing frequency offset of −0.2 ppm (i.e., μi = IGABA,ON(offset = −0.2)/IGABA,ON(offset = 0)).
2.5. In vivo experiments
GABA-edited data were acquired from a 3 × 3 × 3 cm3 voxel in the right sensorimotor cortex in 8 individuals (4 male, 4 female, 37 ± 8 y), each scanned on the three scanners and additionally in one individual (male, 26 y) who was scanned eight times on all three scanners. For the multisubject data, the same two operators performed all scanning on all scanners and both agreed upon voxel placement for each scan. For the single subject data, a third operator performed scanning on one of the platforms. This operator provided screenshots of the voxel placement for the other two operators to place the voxel when scanning on the other platforms. All participants had been scanned multiple times prior to participating in this study. Ethical approval was obtained from local IRBs, and all participants provided written informed consent. The common acquisition parameters were: TR/TE = 2 s/68 ms; 320 dynamics alternating ON-OFF every 2 averages.
The GE and Philips acquisitions acquired 4096 points, sampled at 5 kHz and used editing pulses of 14 ms, while Siemens was 2048 points at 2 kHz and 15 ms editing pulse duration. First- and second-order shimming were performed on the Philips and Siemens scanners, whereas only first-order shimming was applied on GE. Water suppression was achieved using: VAPOR [24] on Philips; a modified WET using CHESS pulses [25] on Siemens; and CHESS [26] on GE.
All in vivo data were processed using ‘Gannet 2.0’ [27], applying 3-Hz line-broadening, spectral registration for frequency and phase correction [28], zero-filling to 32 k points, and fitting the 3.0 ppm peak with a Gaussian function and linear baseline correction. Note, because the export routine used for the Siemens scanner already averaged ON and OFF subspectra, frequency and phase correction has limited impact on spectral quality for this sequence. For the other 2 sequences, individual dynamics were exported separately, which allows for more effective frequency and phase correction.
In correcting for differences in GABA and MM editing efficiency, it was assumed that the GABA signal generated by a sequence was proportional to κ, that the MM signal generated by a sequence was proportional to κ, and that the fraction of in vivo signal that is MM acquired using the Philips scanner and implementation is 50%, based on previous data acquired with and without MM suppression [10]. Based on these assumptions, the κ,μ-correction factor to account for inter-sequence differences is:
| (2) |
where i designates the sequence (GE, Siemens or Philips). This factor simplifies to 1/2κPhilips for the Philips sequence.
The estimated fraction of the in vivo signal that originates from GABA (fGABA) is given by:
| (3) |
and the estimated fraction of the in vivo signal that from MM (fMM) is given by:
| (4) |
Relative GABA levels (in institutional units) were estimated relative to the brain water signal using two approaches: 1) using the same values of κ = 0.5 [6,10] and MM correction factor of 0.45 [6] for all data; and 2) using the phantom-determined, sequence-specific κ and μ corrections (referred to as κ,μ-corrected GABA levels). Quantification of water-referenced data was therefore determined by the equation. [6]:
| (5) |
where SGABA and SH2O are the areas of the GABA and water peaks, respectively. The assumed constants are: pure water concentration, [H2O] = 55,000 mmol/dm3; the fraction of water, including the bound and free water, that is detected in the experiment (detectH2O) = 0.65 [29,30]; water T1 and T2 1.1 s and 0.095 s, respectively [31]; and GABA T1 and T2 0.80 s (intermediate between MM [32] and GABA [33]) and 0.088 s [23], respectively. These constants are assumed as representative values for the entire voxel, as tissue content was not accounted for in this experiment. GABA/Cr was also quantified, and corrections for sequence-specific κ and μ were performed according to
| (6) |
2.6. Statistics
The mean and variance of measured GABA across the subjects was calculated and compared for the sequences. Coefficients of variation (CVs) were determined for each sequence as the standard deviation across the subjects compared divided by the mean. To compare the mean GABA levels across the three sequences, two 2-way ANOVAs examined the impact of the vendor-specific sequence implementation and the application of the correction factor on the water-referenced and on the Cr-referenced data. Follow-up 1-way ANOVAs tested for differences across the means with and without the correction. Levene’s tests compared the variance (normalized by the means) across all vendors between the uncorrected and the corrected GABA levels for both the water-referenced and the Cr-referenced data.
3. Results
3.1. T1 and T2 of GABA and Gly
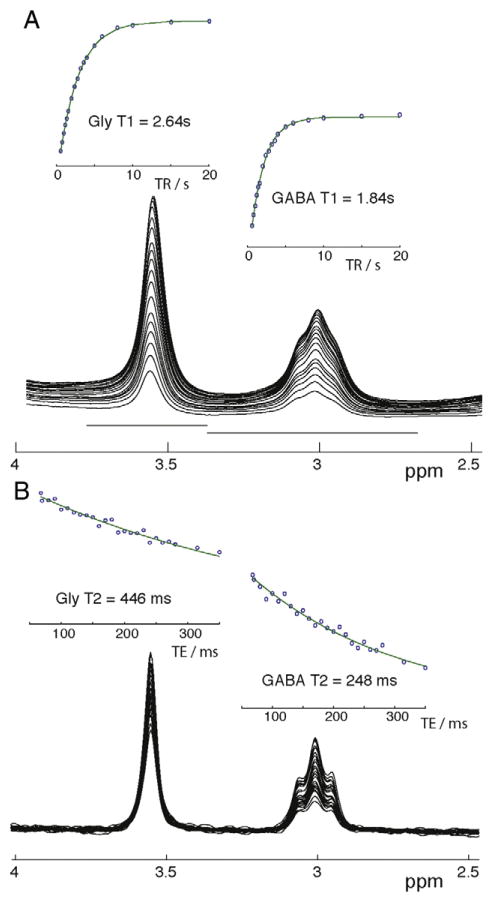
As shown in Fig. 2A, single-exponential saturation-recovery models gave good fits to the data for both GABA and Gly. The phantom longitudinal relaxation time constants were measured as 2.64 s for T1,Gly and 1.84 s for T1,GABA. As shown in Fig. 2B, single-exponential transverse decay models also gave good fits to the data for both GABA and Gly, with 446 ms for T2,Gly and 248 ms for T2,GABA. Therefore, phantom signals acquired at a TR of 2 s and a TE of 70 ms are relaxation-weighted by a factor of 0.500 for GABA and 0.456 for Gly, and the relaxation-weighting differential between GABA and Gly is 1.10.
Fig. 2.
(A) Stacked plot of the phantom MEGA-PRESS ON spectra from the TR series (TR varied from 0.6 to 20 s) and calculation of T1 for 3.5 ppm glycine peak (Gly) and 3.0 ppm GABA peak GABA. (B) Phantom spectra for TE series (TE varied from and calculation of T2 for Gly and GABA).
3.2. Determination of κ and μ
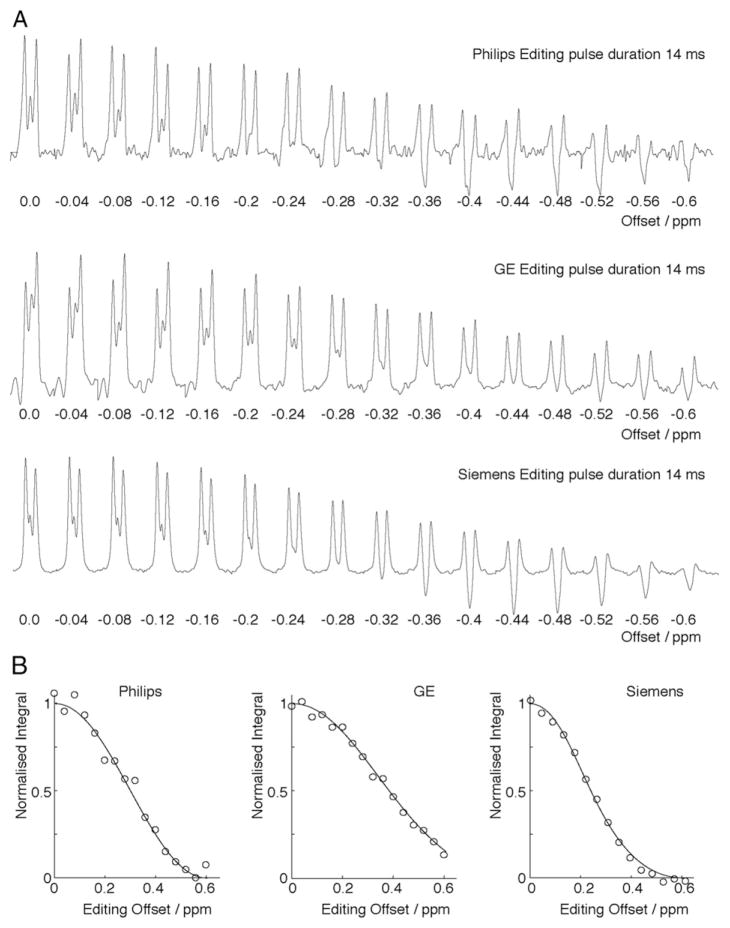
The editing efficiency, κ, was determined to be 0.43, 0.34 and 0.39 for GE, Siemens and Philips, respectively. MEGA-PRESS difference spectra acquired at a range of offsets: 0 to −0.6 ppm on each platform are shown in Fig. 3A. The area of the GABA peak is shown in Fig. 3B with a Gaussian model of best-fit. Based on the relative editing efficiency of the GABA signals at an editing offset of 0.2 ppm, the relative MM editing efficiency ratio μ was measured to be 0.83, 0.63 and 0.73, for GE, Siemens and Philips, respectively. These values correspond to an edited signal that is estimated to be 0.47:0.53, 0.54:0.46, and 0.5:0.5 ratios of fGABA:fMM for GE, Siemens and, Philips respectively.
Fig. 3.
Frequency series to determine μ. A. 3 ppm GABA signal plotted for each frequency offset in the frequency series. B. Normalized integral of the GABA peak for each of the three sequences used to determine μ.
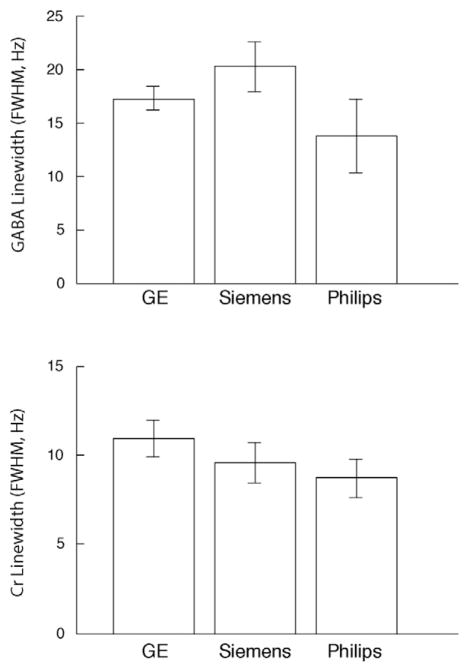
3.3. In Vivo measurements
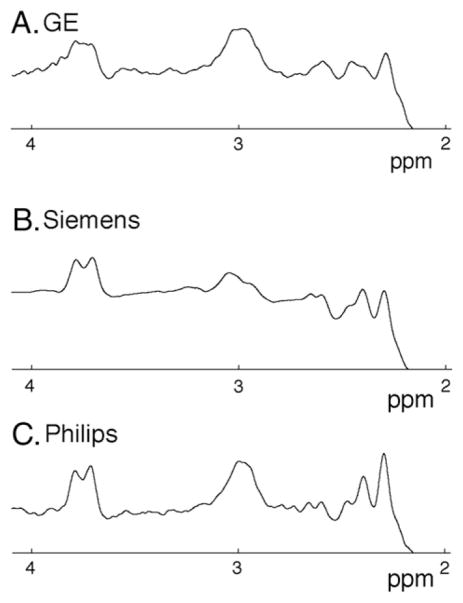
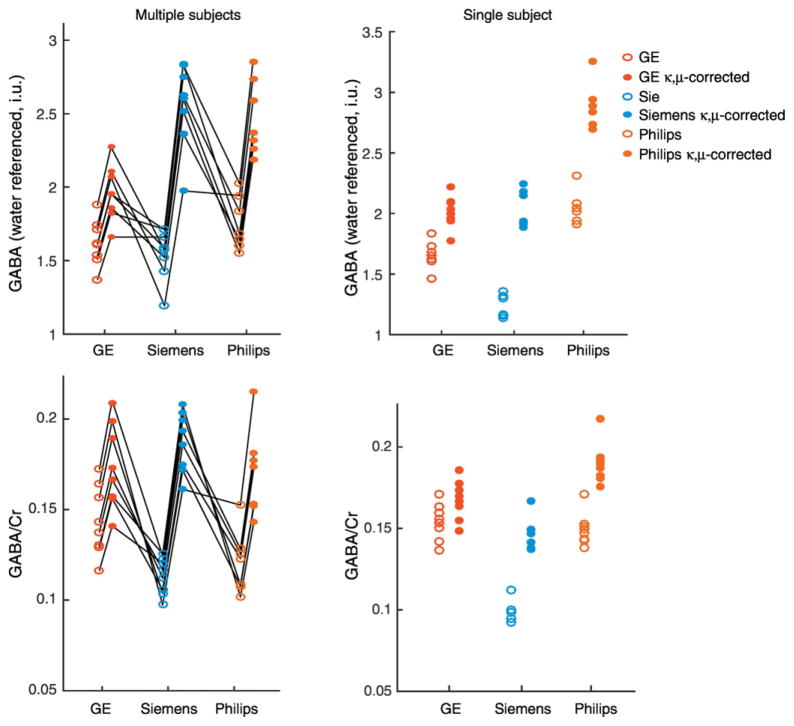
One Philips dataset from the multiple subjects and two of the Siemens data sets from the single subject with multiple repeats was removed due to poor spectral quality, as indicated by excessive subtraction artifacts that could not be resolved with retrospective frequency correction. Example in vivo spectra from one individual are shown in Fig. 4. Linewidths of both GABA and Cr were similar between vendors (Fig. 5). Using the κ and μ values from the phantom data, in vivo GABA measurements were κ,μ-corrected (Fig. 6 and Table 1). The CV of the mean across the sequences for the 8 subjects shows an increase in CV for water referenced results (uncorrected CV = 6.7%, κ,μ-corrected CV = 14%) and a decrease for Cr referenced results (uncorrected CV = 13%, κ,μ-corrected CV = 5%). The CV of the mean measures across the sequences decreases for the single subject in GABA measures referenced to water (uncorrected CV = 24%, κ,μ-corrected CV = 21%) and referenced to Cr (uncorrected CV = 23%, κ,μ-corrected CV = 13%).
Fig. 4.
Example GABA-edited spectra from (A) GE, (B) Siemens and (C) Philips for the same individual. Scan parameters were 3×3×3 cm3 voxel; 320 averages; TR/TE = 2 s/68 ms.
Fig. 5.
Bar graph summarizing the GABA and Cr linewidths (Hz) for each of the three scanners in multiple subjects (error bars indicate the standard deviation).
Fig. 6.
In vivo GABA measures on each scanner. GABA was quantified relative to the water signal and the Cr peak. Uncorrected GABA values are shown in open circles and κ,μ-corrected values are shown in closed circles. On the left, the multiple subjects (individuals connected by black lines) and on the right, a single subject scanned multiple times on each scanner.
Table 1.
Summary of in vivo GABA measurements, comparing the uncorrected to sequence-specific κ,μ corrected data.
| GABA/WATER | GABA/Cr | ||||
|---|---|---|---|---|---|
|
| |||||
| uncorrected | κ,μ-corr | uncorrected | κ,μ-corr | ||
| GE | Single Subject | 1.66 ± 0.11 | 2.01 ± 0.13 | 0.15 ± 0.01 | 0.19 ± 0.01 |
| Eight Subjects | 1.62 ± 0.16 | 1.96 ± 0.19 | 0.14 ± 0.02 | 0.17 ± 0.02 | |
| Siemens | Single Subject | 1.24 ± 0.09 | 2.05 ± 0.16 | 0.10 ± 0.01 | 0.16 ± 0.01 |
| Eight Subjects | 1.55 ± 0.17 | 2.56 ± 0.29 | 0.11 ± 0.01 | 0.19 ± 0.02 | |
| Philips | Single Subject | 2.03 ± 0.13 | 2.87 ± 0.18 | 0.15 ± 0.01 | 0.21 ± 0.01 |
| Eight Subjects | 1.75 ± 0.18 | 2.47 ± 0.26 | 0.12 ± 0.02 | 0.17 ± 0.02 | |
For the multisubject data, for the water-referenced GABA data, the 2-way ANOVA indicated that both the sequence implementation (p = 0.002) and the correction factor (p < 0.001) were significant factors of variance. For the Cr-referenced GABA data, only the correction factor (p < 0.001) is a significant factor of variance. In the follow-up 1-way ANOVAs examining the uncorrected GABA data, sequence implementation was not a significant factor of variance for water-referenced data (p = 0.09) while it was a significant factor of variance for the Cr-referenced data (p = 0.003). When examining the κ,μ-corrected data, water-referenced GABA levels were significantly different across the different sequence implementations (p < 0.001) while Cr-referenced GABA levels were not significantly difference across the different sequences (p = 0.3). For the single subject, with multiple repeats, the sequence implementation and correction factor were highly significant (p < 0.001) and all follow-up 1-way ANOVAs showed the sequence implementation was a significant factor of variance.
Levene’s tests showed that there was a significant difference in variance between the uncorrected and the corrected GABA levels for the water-referenced data (p = 0.04) and a non-significant decrease in variance for the Cr-referenced data (p = 0.4). For the single subject, there was a no difference in variance water-referenced data and a significant decrease in the Cr-referenced data (p = 0.046).
4. Discussion
Three commonly-used implementations of the MEGA-PRESS [5] sequence, as applied for the detection of GABA, were compared. The three sequences differed substantially in editing efficiency and co-editing of MM. The GE sequence showed the highest editing efficiency and the greatest co-editing of MM. The Siemens sequence produced the least amount of MM contamination. Using the editing efficiency and MM co-editing values from calibration experiments in phantoms, it was possible to improve agreement between scanners for GABA/Cr ratios in 8 healthy control subjects and for the GABA/Cr and the water-referenced GABA levels in a single subject who was scanned 8 times on each of the three platforms.
Losses in editing efficiency may arise both from the ON and OFF halves of the experiment. In the case of the ON experiment, one major factor is the extent to which the coupling evolution is perfectly refocused at the point of acquisition. This largely depends on the timing of the editing pulses – they are spaced by exactly TE/2 or 34 ms in the GE and Philips sequences, but only approximately separated by TE/2 in the Siemens sequence (27.2 ms). This results in an ON signal that is less positive than it might be, and therefore losses in the difference (ON-OFF) signal. Losses in the OFF experiment arise from spatial heterogeneity of the coupling evolution [8,9,21], and due to the fact that the four couplings between spins at 1.9 ppm and 3.0 ppm are not equal so a single TE cannot capture all four fully antiphase. This results in an OFF signal that is less negative than it might be, and therefore, losses in the difference (ON-OFF) signal. A further difference between sequences is the extent of editing of the center peak of the 3 ppm signal, which has been discussed previously [6]. All three sequences have different editing pulse profiles, each making compromises between bandwidth and off-resonance editing, leading to different editing selectivity profiles and μ-factors.
The term ‘editing efficiency’ can be defined in several ways; in this paper, it is defined as the fraction of the perfectly refocused signal acquired at the same echo time. In order to derive editing efficiencies of approximately one half, the maximum value if the 3-ppm GABA signal is considered a simple triplet, the averaged difference (ON-OFF)/2 must be integrated and compared to the ideally refocused signal. Editing efficiency may also be calculated by numerical simulations; although, this would require simulations of sufficient complexity to capture all inter-sequence differences, and will also depend on the accuracy of spin-system parameters used [34]. Therefore the experimental approach used here appears more practical.
Differences between sequences, while not negligible, are sufficiently small that a reasonable level of agreement exists between in vivo measurements prior to correction (Fig. 6). Statistical analysis of the Cr-referenced data examining the κ,μ-correction and the sequence implementation as factors showed only the correction had a significant impact. After applying the κ,μ-correction the increased agreement between the measurements is pronounced and supported by the one-way ANOVA analyses that showed sequence implementation as a significant factor of variance in the un-corrected data and that it was not a significant factor in the κ,μ-corrected data. A similar pattern is seen in the single-subject data when looking at Fig. 6, while some consistency exists prior to correction, after correction there is greater consistency amongst the GABA/Cr data. However, in this case the ANOVA analysis maintains a significant difference between the three sequence implementations. The water reference data is not as clear; the Siemens and Philips implementations appear consistent with each other after correction while the GE data appears low (Fig. 6). Interestingly, prior to correction, the Siemens and the GE data appear more consistent. The two-way ANOVA indicates both the application of the κ,μ-correction and the vendor are significant factors and one-way ANOVA confirms the sequence is a significant factor of variance after the κ,μ-correction on the water data. In the single subject data, the GE and Siemens data are very consistent after correction; however, the Philips data agrees less well.
In spite of unexplained sources of variance that remain after the κ,μ-correction, this study explicitly quantifies the editing efficiency and co-editing of macromolecules across 3 implementations of the GABA-edited MEGA-PRESS sequence. Correcting for these differences between implementations is an important initial step as to reconciling differences in GABA levels between these implementations, and the success in reducing the CVs for the both sets of the GABA/Cr data and the single subject water-referenced GABA data supports this approach. The increase in variance in the multi-subject water-referenced data after correction indicates there are other sources of variance that have yet to be identified and incorporated into the correction.
Although MRS aims to quantify metabolite levels, measurements are ‘relative’, and are usually presented as integral ratios to another signal originating from the same region of interest - either a metabolite, typically creatine (Cr), or water (often expressed in ‘institutional units’ after further relaxation correction) [6]. The benefit of the κ,μ-correction is seen quantifying relative to Cr, but not as obviously in the water-referenced results. One explanation is that the measured correction factors improve scanner-to-scanner comparability of GABA signal quantification, and the discrepancy is in the water signal or the processing of the water data. The mathematical development of the κ,μ-correction is independent of the referencing, so this correction cannot address patterns of sequence-to-sequence variance that differ between water-and Cr-referenced measures. This discrepancy must be further investigated to improve the inter-implementation corrections for reliable comparisons between platforms.
It has previously been shown that Cr referencing can result in slightly improved reproducibility of GABA measurements [35], in spite of the fact that the Cr signal has a lower SNR compared to the water signal that is presumably due to the benefit of acquiring the reference at the same time as the GABA data and challenges associated with water referencing. Resolving the ideal reference for MRS data remains an issue. While Cr-referencing is being increasingly challenged because Cr levels can change with pathology (e.g [36,37].), the same may also be true of the water signal. Water has a large signal and is easily modeled but it requires the acquisition of additional TRs and requires correction for cerebrospinal fluid water. The Cr signal is acquired with the metabolite signal however is not as large and is more complex to model. The results of the current study indicate water referencing may have additional sources of variance between implementation and we believe that progressing both referencing approaches is important.
There are relatively few studies that have attempted to reconcile MRS measures across sequences [38,39], even for unedited single-voxel spectra, and they have generally only been moderately successful. More recently, an MRSI between manufacturer comparison using the same sequence generally found the concordance of the primary singlet metabolite levels; however, it did depend on the region (i.e, temporal, occipital, frontal or parietal lobe) and metabolite ratio of interest [40]. The difference method of the MEGA-PRESS sequence may “hide” some inter-sequence differences and the discordance across sequences/manufacturers of the typical PRESS sequence suggests that accounting for all inter-sequence differences is challenging [38].
There are differences between the sequences that are not captured by the κ,μ-correction. Firstly, there are scanner-to-scanner differences in B0 field stability, depending on the power and cooling efficiency of gradient systems and the amount of passive shim iron within the magnet bore, which will impact the level of frequency drift during an MRS acquisition [41]. Although post-processing frequency and phase correction can reduce the impact of such effects, they do still have an impact on measurements – for example, they do not address changes in editing efficiency that occur as a result of the editing pulses no longer being applied on-resonance. Secondly, in addition to sequence differences in the underlying lineshape, in vivo differences such as quality of shimming (in vivo linewidth) and baseline shape (due to, e.g., quality of water suppression) can impact the modeling and quantification of signals. Thirdly, there are additional different processing steps that are included by the sequences. For example, different eddy current corrections are applied, and the Siemens RDA data format averages all the ON and OFF subspectra prior to export, limiting the impact of retrospective frequency corrections. While this limitation can be overcome by using exports in which each average is saved, this data format was not possible for the data collection in this study. Finally measurement reproducibility will influence these results but is not accounted for in the κ,μ-correction. Most recently, Shungu et al. [42] showed excellent test-retest GABA repeatability for both water-referenced and Cr-referenced data. In the literature, reproducibility of measurements is variable with test-retest studies presenting CVs from as low as 5%, as per Shungu et al., intermediate values of 8–15% as seen by Evans et al. [14] and O’Gorman et al. [43], to larger values 13–20%, as seen by Bogner et al. [35]. Test-retest reproducibility may vary between vendors and analysis methods, and by the compliance of subjects recruited, but this is beyond the scope of the current study. Consistency of voxel placement is another factor in measurement reproducibility [44]. Unfortunately we do not have access to the T1-weighted anatomical images to compare voxel placement and therefore are unable to control for the white matter and gray matter content of the voxel. A second limitation of not having the T1-anatomical images for segmentation is the assumption of the detectable water as well as T1 and T2 relaxation times used in the calculation of the GABA levels relative to water. Consistent with standards in the literature [6], we have used constants to represent the entire voxel, for example the fraction of detectable water of 0.65 was originally measured in white matter. Additionally, the current study was not designed to account for environmental factors such as stress, comfort, brightness of the room, presence of a sound system, temperature, time of day, or other identified factors that have the potential to alter GABA levels, or natural within-subject fluctuations of GABA.
In conclusion, differences in MEGA-PRESS sequence implementation result in differences in GABA editing efficiency and MM co-editing. These differences can be summarized by two parameters, κ and μ, that can be measured from phantom experiments. Correcting for these parameters results in an improvement in agreement between GABA measurements made using sequences implemented on the three main scanner manufacturer platforms though the resolution of cross-platform differences is not yet complete. In particular further investigation is warranted to understand water-referenced differences that remain.
Acknowledgments
Funding: This work was supported by NIH grants R01EB016089 and P41EB015909.
References
- 1.Puts NA, Edden RA. In vivo magnetic resonance spectroscopy of GABA: a methodological review. Prog Nucl Magn Reson. 2012;60:29–41. doi: 10.1016/j.pnmrs.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rae CD. A guide to the metabolic pathways and function of metabolites observed in human brain 1H magnetic resonance spectra. Neurochem Res. 2014;39(1):1–36. doi: 10.1007/s11064-013-1199-5. [DOI] [PubMed] [Google Scholar]
- 3.Lymer K, et al. Reproducibility of GABA measurements using 2D J-resolved magnetic resonance spectroscopy. Magn Reson Imaging. 2007;25(5):634–40. doi: 10.1016/j.mri.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Near J, et al. Efficient gamma-aminobutyric acid editing at 3T without macromolecule contamination: MEGA-SPECIAL. NMR Biomed. 2011;24(10):1277–85. doi: 10.1002/nbm.1688. [DOI] [PubMed] [Google Scholar]
- 5.Mescher M, et al. Simultaneous in vivo spectral editing and water suppression. NMR Biomed. 1998;11:266–72. doi: 10.1002/(sici)1099-1492(199810)11:6<266::aid-nbm530>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 6.Mullins PG, et al. Current practice in the use of MEGA-PRESS spectroscopy for the detection of GABA. Neuroimage. 2014;86:43–52. doi: 10.1016/j.neuroimage.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris AD, Saleh MG, Edden RA. Edited 1H magnetic resonance spectroscopy in vivo: Methods and metabolites. Magn Reson Med. 2017;77(4):1377–89. doi: 10.1002/mrm.26619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Near J, et al. J-difference editing of gamma-aminobutyric acid (GABA): simulated and experimental multiplet patterns. Magn Reson Med. 2013;70(5):1183–91. doi: 10.1002/mrm.24572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaiser LG, et al. A detailed analysis of localized J-difference GABA editing: theoretical and experimental study at 4 T. NMR Biomed. 2008;21(1):22–32. doi: 10.1002/nbm.1150. [DOI] [PubMed] [Google Scholar]
- 10.Harris AD, et al. Spectral-editing measurements of GABA in the human brain with and without macromolecule suppression. Magn Reson Med. 2014 doi: 10.1002/mrm.25549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henry P-G, et al. Brain GABA editing without macromolecule contamination. Magn Reson Med. 2001;45:517–20. doi: 10.1002/1522-2594(200103)45:3<517::aid-mrm1068>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 12.Edden RA, Puts NA, Barker PB. Macromolecule-suppressed GABA-edited magnetic resonance spectroscopy at 3T. Magn Reson Med. 2012;68(3):657–61. doi: 10.1002/mrm.24391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris AD, et al. Multi-regional investigation of the relationship between functional MRI blood oxygenation level dependent (BOLD) activation and GABA concentration. PLoS One. 2015;10(2):e0117531. doi: 10.1371/journal.pone.0117531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans CJ, McGonigle DJ, Edden RA. Diurnal stability of gamma-aminobutyric acid concentration in visual and sensorimotor cortex. J Magn Reson Imaging. 2010;31(1):204–9. doi: 10.1002/jmri.21996. [DOI] [PubMed] [Google Scholar]
- 15.Foerster BR, et al. An imbalance between excitatory and inhibitory neurotransmitters in amyotrophic lateral sclerosis revealed by use of 3-T proton magnetic resonance spectroscopy. JAMA Neurol. 2013;70(8):1009–16. doi: 10.1001/jamaneurol.2013.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edden RAE, et al. Reduced GABA concentration in attention-deficit/hyperactivity disorder. Arch Gen Psychiatry. 2012;69(7):750–3. doi: 10.1001/archgenpsychiatry.2011.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaetz W, et al. GABA estimation in the brains of children on the autism spectrum: measurement precision and regional cortical variation. Neuroimage. 2014;86:1–9. doi: 10.1016/j.neuroimage.2013.05.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cleve M, Gussew A, Reichenbach JR. In vivo detection of acute pain-induced changes of GABA+ and Glx in the human brain by using functional 1H MEGA-PRESS MR spectroscopy. Neuroimage. 2015;105:67–75. doi: 10.1016/j.neuroimage.2014.10.042. [DOI] [PubMed] [Google Scholar]
- 19.Puts NAJ, et al. Reduced GABAergic inhibition and abnormal sensory symptoms in children with Tourette syndrome. J Neurophysiol. 2015;114:808–17. doi: 10.1152/jn.00060.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oeltzschner G, et al. Low visual cortex GABA levels in hepatic encephalopathy: links to blood ammonia, critical flicker frequency, and brain osmolytes. Metab Brain Dis. 2015 doi: 10.1007/s11011-015-9729-2. [DOI] [PubMed] [Google Scholar]
- 21.Edden RA, Barker PB. Spatial effects in the detection of gamma-aminobutyric acid: improved sensitivity at high fields using inner volume saturation. Magn Reson Med. 2007;58(6):1276–82. doi: 10.1002/mrm.21383. [DOI] [PubMed] [Google Scholar]
- 22.Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000;13(3):129–53. doi: 10.1002/1099-1492(200005)13:3<129::aid-nbm619>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 23.Edden RA, et al. Measuring T2 in vivo with J-difference editing: application to GABA at 3 Tesla. J Magn Reson Imaging. 2012;35(1):229–34. doi: 10.1002/jmri.22865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tkac I, et al. In vivo 1H NMR spectroscopy of rat brain at 1ms echo time. Magn Reson Med. 1999:41. doi: 10.1002/(sici)1522-2594(199904)41:4<649::aid-mrm2>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 25.Ogg RJ, Kingsley PB, Taylor JS. WET, a T1- and B1-insensitive water-suppression method for in vivo localized 1H NMR spectroscopy. J Magn Reson B. 1994;104:1–10. doi: 10.1006/jmrb.1994.1048. [DOI] [PubMed] [Google Scholar]
- 26.Haase A, et al. 1H NMR chemical shift selective (CHESS) imaging. Phys Med Biol. 1985;30(4):341–4. doi: 10.1088/0031-9155/30/4/008. [DOI] [PubMed] [Google Scholar]
- 27.Edden RA, et al. Gannet: A batch-processing tool for the quantitative analysis of gamma-aminobutyric acid-edited MR spectroscopy spectra. J Magn Reson Imaging. 2014;40(6):1445–52. doi: 10.1002/jmri.24478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Near J, et al. Frequency and phase drift correction of magnetic resonance spectroscopy data by spectral registration in the time domain. Magn Reson Med. 2014 doi: 10.1002/mrm.25094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ernst T, Kreis R, Ross BD. Absolute quantitation of water and metabolites in the human brain. I. Compartments and water. J Magn Reson. 1993;102:1–8. Series B. [Google Scholar]
- 30.Helms G. Volume correction for edema in single-volume proton MR spectroscopy of contrast-enhancing multiple sclerosis lesions. Magn Reson Med. 2001;46(2):256–63. doi: 10.1002/mrm.1186. [DOI] [PubMed] [Google Scholar]
- 31.Wansapura JP, et al. NMR relaxation times in the human brain at 3.0 tesla. J Magn Reson Imaging. 1999;9(4):531–8. doi: 10.1002/(sici)1522-2586(199904)9:4<531::aid-jmri4>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 32.Behar KL, et al. Analysis of macromolecule resonances in 1H NMR spectra of human brain. Magn Reson Med. 1994;32(3):294–302. doi: 10.1002/mrm.1910320304. [DOI] [PubMed] [Google Scholar]
- 33.Puts NA, Barker PB, Edden RA. Measuring the longitudinal relaxation time of GABA in vivo at 3 Tesla. J Magn Reson Imaging. 2013;37(4):999–1003. doi: 10.1002/jmri.23817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kreis R, Bolliger CS. The need for updates of spin system parameters, illustrated for the case of gamma-aminobutyric acid. NMR Biomed. 2012;25(12):1401–3. doi: 10.1002/nbm.2810. [DOI] [PubMed] [Google Scholar]
- 35.Bogner W, et al. In vivo quantification of intracerebral GABA by single-voxel (1)H-MRS-How reproducible are the results? Eur J Radiol. 2010;73(3):526–31. doi: 10.1016/j.ejrad.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 36.Ongur D, et al. Creatine abnormalities in schizophrenia and bipolar disorder. Psychiatry Res. 2009;172(1):44–8. doi: 10.1016/j.pscychresns.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiduschat N, et al. Motor cortex glutathione deficit in ALS measured in vivo with the J-editing technique. Neurosci Lett. 2014;570:102–7. doi: 10.1016/j.neulet.2014.04.020. [DOI] [PubMed] [Google Scholar]
- 38.Keevil SF, et al. Absolute metabolite quantification by in vivo nmr spectroscopy: II. a multicentre trial of protocols for in vivo localised proton studies of human brain. Magn Reson Imaging. 16(9):1093–1106. doi: 10.1016/s0730-725x(98)00118-0. [DOI] [PubMed] [Google Scholar]
- 39.van de Bank BL, et al. Multi-center reproducibility of neurochemical profiles in the human brain at 7 T. NMR Biomed. 2015;28(3):306–16. doi: 10.1002/nbm.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sabati M, et al. Multivendor implementation and comparison of volumetric whole-brain echo-planar MR spectroscopic imaging. Magn Reson Med. 2014;74(5):1209–20. doi: 10.1002/mrm.25510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris AD, et al. Impact of frequency drift on gamma-aminobutyric acid-edited MR spectroscopy. Magn Reson Med. 2014;72(4):941–8. doi: 10.1002/mrm.25009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shungu DC, et al. Brain gamma-aminobutyric acid (GABA) detection in vivo with the J-editing (1) H MRS technique: a comprehensive methodological evaluation of sensitivity enhancement, macromolecule contamination and test-retest reliability. NMR Biomed. 2016;29(7):932–42. doi: 10.1002/nbm.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Gorman RL, et al. In vivo detection of GABA and glutamate with MEGA-PRESS: reproducibility and gender effects. J Magn Reson Imaging. 2011;33(5):1262–7. doi: 10.1002/jmri.22520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bai X. Voxel placement precision for GABA-edited magnetic resonance spectroscopy. O J Rad. 2017;7:35–44. doi: 10.4236/ojrad.2017.71004. [DOI] [PMC free article] [PubMed] [Google Scholar]