Abstract
Background and Purpose
The immune checkpoint PD-1 and its receptor B7-H1 (PD-L1) are successful therapeutic targets in cancer but less is known about other B7 family members. Here, we determined the expression level of B7-H3 protein in non-small cell lung cancer (NSCLC) and evaluated its association with tumor infiltrating lymphocytes (TILs), PD-L1, B7-H4 and major clinico-pathological characteristics is in 3 NSCLC cohorts.
Experimental design
We used multiplexed automated quantitative immunofluorescence (QIF) to assess the levels of B7-H3, PD-L1, B7-H4 and TILs in 634 NSCLC cases with validated antibodies. Associations between the marker levels, major clinico-pathological variables and survival were analyzed.
Results
Expression of B7-H3 protein was found in 80.4% (510/634) of the cases. High B7-H3 protein level (top 10 percentile) was associated with poor overall survival (p <0.05). Elevated B7-H3 was consistently associated with smoking history across the 3 cohorts, but not with sex, age, clinical stage and histology. Co-expression of B7-H3 and PD-L1 was found in 17.6% of the cases (112/634) and with B7-H4 in 10% (63/634). B7-H4 and PD-L1 were simultaneously detected only in 1.8% of NSCLCs (12/634). The expression of B7-H3 was not associated with the levels of CD3, CD8 and CD20 positive TILs.
Conclusion
B7-H3 protein is expressed in the majority of NSCLCs and is associated with smoking history. High levels of B7-H3 protein has a negative prognostic impact in lung carcinomas. Co-expression of B7-H3 with PD-L1 and B7-H4 is relatively low, suggesting a non-redundant biological role of these targets.
Keywords: immune checkpoints, biomarkers, survival, prognosis, quantitative immunofluorescence
Introduction
Immune checkpoints are the T cell regulatory mechanisms of co-stimulatory and inhibitory signals that control the amplitude and quality of immune response. The expression of inhibitory immune checkpoints can be upregulated by tumors and serve as an adaptive immune evasion mechanism(1). Activation of the programmed death 1 (PD-1) receptor by its ligand programmed death ligand 1 (PD-L1) has been recognized as a major immune inhibitory mechanism in solid tumors (2, 3). While antibodies that inhibit the PD-1/PD-L1 pathway produce durable clinical responses in various solid tumors including Non-Small Cell Lung Cancer (NSCLC) (4–7), they only benefit a fraction of patients. Efforts are now focusing on combination strategies to block additional immune suppressive signals and activate co-stimulatory receptors to increase response rates, prolong responses, and prevent acquired resistance to monotherapy regimens.
B7-H3 (CD276) is a type I transmembrane protein that belongs to the Ig superfamily and a member of the B7 immunoregulatory molecules (8). While B7-H3 mRNA is broadly expressed in several organs including human breast, bladder, liver, lung, lymphoid organs, placenta, prostate and testis (9–11), at the protein level, B7-H3 expression is low and rare (12). B7-H3 upregulation has been reported in multiple malignancies including NSCLC(13). In preclinical models both stimulatory and inhibitory properties of B7-H3 have been postulated in T cell directed cancer immunity (8, 9, 11, 12, 14). In human hepatocellular carcinoma, B7-H3 expression is linked to decreased T cell proliferation and decreased interferon-γ production(15). In murine pancreatic cancer model B7-H3 blockade resulted in an increased CD8+ T cell influx and antitumor effect(16). In NSCLC, B7-H3 protein expression has been associated with a negative impact in prognosis(17, 18). A humanized, Fc-optimized monoclonal antibody that targets B7-H3, Enoblituzumab (also referred to as MGA271) was shown to produce antitumor responses in a fraction of heavily pre-treated solid tumors and was well tolerated at dose levels in a Phase 1 study (19). Currently, clinical activity of Enoblituzumab is under investigation as a monotherapy and in combination with either CTLA-4 or PD-L1 targeting monoclonal antibodies (19, 20). The biologic significance of co-expression of B7 immunoregulatory molecules, their interaction in the tumor microenvironment, and their role in primary and acquired resistance to PD-1 axis inhibitors are unclear.
In this study, we measured the levels of B7-H3 protein both in the tumor and peritumoral stromal tissue and correlated it with clinico-pathological characteristics and outcome in in three independent lung cancer cohorts. We have also studied its association with major tumor infiltrating lymphocyte (TIL) subsets, levels of PD-L1, B7-H4 using quantitative objective methods and validated antibodies.
Materials and Methods
Patients, cohorts and tissue microarrays
Samples from 3 retrospective collections of lung cancer, two from Yale University (Cohort A and Cohort C) and one from University of Athens, Greece (Cohort B) represented in tissue microarrays (TMAs) were used. 2 of these cohorts were previously described and include total of 552 formalin-fixed, paraffin-embedded, primary NSCLC tumors samples (Cohort A: 202 and Cohort B: 350 lung carcinomas) (21, 22). A serial collected cohort of patients seen in the Yale Surgical Pathology suite, called YTMA 250, comprises a sample collection from 314 NSCLC patients that had surgical resection of their primary tumor between 2004 and 2011 also used in this study (Cohort C). Clinico-pathological information is summarized in supplemental table 1.
For the analysis of B7-H3 tumor and stroma protein expression and their correlation with overall survival, we used 2 different built TMA for the analysis using 0.6 mm tissue cores. These blocks were constructed with the same TMA map but cores obtained by nonadjacent sampling of the same tumors. To address run-to-run variability, slides were stained and analyzed on same days using the same protocol. Serial cut index slides were used for quality control and regression coefficients (R2) between independent runs for these index slides were found to be high (>0.9). Two TMA histospots were evaluated for each case and average is obtained. Cases which has less than 2% tumor tissue in a histospot has been excluded from the analysis in these cohorts. For the B7-H3 protein expression correlation with PD-L1, B7-H4 protein and TIL subsets; only cases which had staining for all markers included in the correlation studies.. All small cell lung cancer and cell controls within the TMAs excluded for the analysis.
The actual number of samples analyzed for each study is lower due to unavoidable loss of tissue, absence of tumor cells or poor quality of the staining in some spots as is commonly seen in TMA studies. Due to these selection criteria 130 cases out of 202 cases in Cohort A, 345 cases out of 350 cases in Cohort B and 279 cases out of 314 cases for Cohort C included in the final analysis. All tissue was collected in accordance with US Common Rule after approval from the Yale Human Investigation Committee (also known as the institutional review board) protocol #9505008219, which approved the patient consent forms or in some cases waiver of consent.
Antibody validation
Open-access databases (Expression Atlas European bioinformatics institute EMBL-EBI, https://www.ebi.ac.uk/gxa/home and Cell Line Atlas cell line atlas - The Human Protein Atlas, www.proteinatlas.org/cell ) were reviewed for cell line mRNA and protein expression of B7-H3 (CD276). SKBR3 breast cancer cell line was selected due to low mRNA expression. They were obtained from the American Type Culture Collection (ATCC) and maintained as recommended.
SKBR3 cells were seeded in 24-well plates in duplicates in Gibco™ RPMI Medium (ThermoFisher Scientific, USA), supplemented with 10% FBS, Penicillin-Streptomycin (10,000 U/mL) and kept at 37°C, 5% CO2. When they reached 70% confluency, they were transferred to Optimem-Low Serum Medium (ThermoFisher Scientific, USA), transfected with 500ng of B7-H3 plasmid (Provided by Cell Signaling Technologies, MA) with different Lipofectamine 2000 ThermoFisher Scientific, USA concentrations (2, 3, 4, 5 μl), following manufacturer’s instructions. Empty vector and untreated cells (no plasmid, no Lipofectamine) were also included as negative controls. After 72hr incubation, cells were collected and replated in 8-chamber polystyrene vessel tissue culture treated glass slides (Falcon, Cat. No.354108) in duplicates. Glass slides were washed twice in PBS (Life Technologies) and fixed in 4% Paraformaldehyde for 10 minutes. Then, following double wash in PBS they were permeabilized in 0.2% Triton x100-PBS for 3 minutes. After a double wash in PBS, a blocking in 2% BSA-PBS for 1 hr at room temperature primary antibodies against cytokeratin (Monoclonal mouse antihuman cytokeratin clone AE1/AE3) and B7-H3 (monoclonal rabbit antibody Cell Signaling Technologies, clone D9M2L) were added and left incubate overnight at 4°C, in light protected chamber. Then, after 2 washes of 1% tween-PBS and one in PBS 5 minutes each, secondary antibodies were added using GAM/Alexa Fluor 546 and Rabbit Envision Neat for 1hr at room temperature. After PBS-tween /PBS wash Cy5-tyramide (10μL Cy5 tyramide in 490 Amplification buffer) was added for 10 minutes. Finally, after a last PBS-tween /PBS wash mounting was performed with Prolong Gold – DAPI (Life Technologies).
MCF-7 cell lines were selected for B7-H4 antibody validation using the same cell transfection model described above. Quantitative immunofluorescence microscopy used for assessment of protein expression of both positive and negative controls in MCF7 for B7-H4 and SKBR-3 for B7-H3 transfection (supplemental figures 1 and 2).
Quantitative immunofluorescence (QIF)
B7-H3 (CST; clone D9M2L), PD-L1 (Spring Bioscience; clone SP142) and B7-H4 (CST; clone D1M8I) antibodies were used for staining by using a previously described protocol with simultaneous detection of cytokeratin and DAPI for tumor and nuclear compartment detection, respectively(22). Briefly, TMA slides were baked overnight at 60°C and then soaked in xylene twice for 20 minutes each. Slides were rehydrated in two 1-minute washes in 100% ethanol followed by one wash in 70% ethanol and finally rinsed in streaming tap water for 5 minutes. Antigen retrieval was performed in the PT module from LabVision for 20 min at 97°C in a pressure-boiling container (in sodium citrate buffer, pH 6 for B7-H3 and B7-H4 antibody and with EDTA buffer, pH8 for PD-L1 antibody). Blocking was performed with 0.3% bovine serum albumin in 0.05% tween solution for 30 minutes after antigen retrieval. Each of the B7 family antibodies (B7H3, B7H4 and PD-L1) was combined with 1:100 pan-cytokeratin antibody (clone AE1/AE3, M3515; Dako Corp., Carpinteria, CA, USA) in 0.3% BSA in TBST and incubated overnight at 4°C. The working concentrations for B7-H3 (clone D9M2L), PD-L1 (SP142) and B7-H4 (D1M8I were 0.05μl/mL, 0.154 μl/mL, and 1.305 μl/mL, respectively. Primary antibodies were followed by incubation with Alexa 546–conjugated goat anti-mouse or anti-rabbit secondary antibody (Molecular Probes, Eugene, OR, USA) diluted 1:100 in rabbit or mouse EnVision reagent (Dako) for 1 hour. Signal was amplified with Cyanine 5 (Cy5) directly conjugated to tyramide (Perkin-Elmer, Waltham, MA, USA) at 1:50 dilution was used for target antibody detection. ProLong mounting medium (ProLong Gold; Molecular Probes) with 4,6-diamidino-2-phenylindole (DAPI) was used to stain nuclei.
For B7-H3 and B7-H4 co-localization experiments we used a multiplexing protocol that uses benzoic hydrazide with hydrogen peroxide to quench HRP activation, as described previously(23). Briefly, following deparaffinization and antigen retrieval (pH 6, Citrate), slides were blocked with 0.3% BSA in Tris-buffered saline/Tween(TBS-T). Primary monoclonal antibody against B7-H3 (CST, Rabbit monoclonal, clone D9M2L), combined with anti-pancytokeratin conjugated with Alexafluor 488 (1:100, eBioscience, clone AE1/AE3 were incubated overnight at 4C. Then, slides were incubated with anti-rabbit Envision reagent (Dako), followed by Cy3 plus tyramide (Perkin Elmer; 1:100) amplification for 10 minutes. This step was followed by incubation with 1 mmol/L benzoic hydrazide with 0.15% hydrogen peroxide to quench HRP activation, two times 7 minutes each. Then, slides were incubated with B7-H4 antibody (CST, rabbit monoclonal, clone D1M8I) at 4C for overnight. On day two, slides were incubated with anti-rabbit Envision reagent (Dako) for an hour at room temperature, followed by signal amplification with Cy-5 tyramide (PerkinElmer; 1:50) for 10 minutes. ProLong mounting medium (ProLong Gold; Molecular Probes) with 4,6-diamidino-2-phenylindole (DAPI) was used to stain nuclei. Between every step, slides were washed with Tris-buffered saline/Tween x2, 5 minutes each followed by Tris-buffered saline wash for 5 minutes.
For B7-H3 and PD-L1 co-localization experiment EDTA pH8 used for antigen retrieval; Primary monoclonal antibodies against B7-H3 (1:200; CST, Rabbit monoclonal, clone D9M2L), Chicken anti-pancytokeratin with secondary antibody with Alexa Flour 488-conjugated goat anti-chicken and previously validated monoclonal mouse PD-L1 (clone 9A11, CST) antibody (24) used with a multiplexing protocol which was previously described (23).
Quantitative TILs measurement for with multiplexing CD3, CD8 and CD20 was described in detail in a previous work of our group (21, 23). In summary CD3, CD8, CD20 and cytokeratin were simultaneously stained in one tissue section using a sequential staining protocol, measurement of TIL markers considered the levels detected in the whole tumor sample (e.g. tumor and stroma). The levels of CD3, CD8 and CD20 were classified as high/low using the median score as cut-point.
The QIF measurements were performed using the AQUA® method of QIF (Genoptix Medical Laboratory) as previously described (25). The QIF score of B7-H3 antibody in the tumor and in the stroma was calculated by dividing the target B7-H3 pixel intensity in the area of the tumor and stroma compartment defined by the cytokeratin and DAPI positivity in TMA slides containing the cohort cases. Serial section slides measuring PD-L1 (SP142, Spring ), and B7-H4 (D1M8I, Cell Signaling Technologies) protein expression were used for the assessment of mutual expression rates between these 3 markers and their association with TILs.
Statistical analysis
Pearson’s correlation coefficient (R) was used to assess the reproducibility of the assay between near-serial sections of the index array. Differences between QIF signals between groups were analyzed using Fisher’s exact test two-sided. p values were considered statistically significant if <0.05. Linear regression coefficients were calculated to determine the association between continuous scores. For all Cohorts (A, B, C), B7-H3, PD-L1 and B7-H4 AQUA scores from two independent cores for each histospot were averaged and the averages were used for final analysis. A visual cut-point was determined for each cohort for QIF positivity. Survival functions were compared using Kaplan-Meier curves, and statistical significance was determined using the log-rank test. Survival analysis of continuous marker scores was performed using the X-tile software (Yale University, New Haven, CT, USA) for disease-specific survival differences (26). GraphPad Prism 7.01 software was used to generate Kaplan Meier survival analysis.
Results
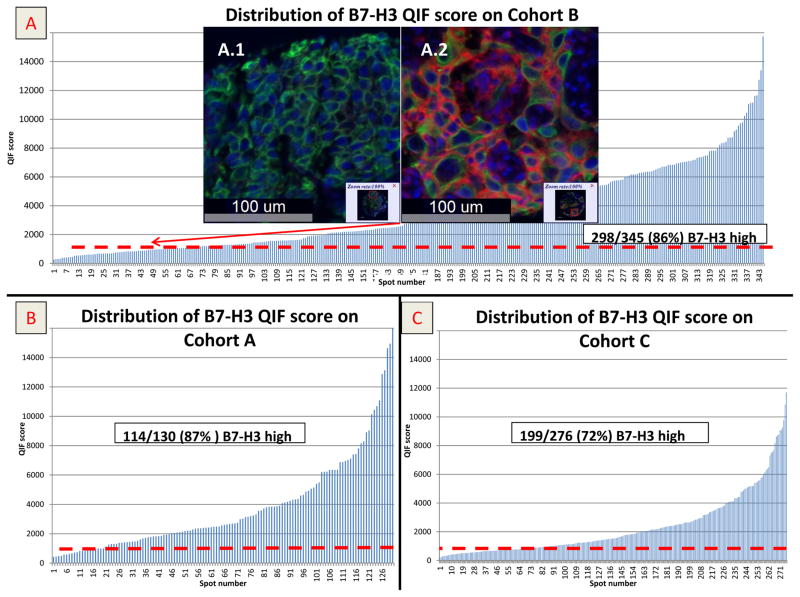
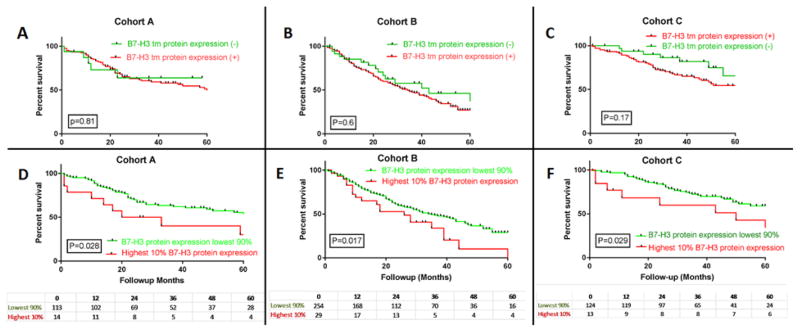
In NSCLC tissues B7-H3 protein expression showed a predominant cytoplasmic and membranous distribution in tumor cells (Figure 1). Protein expression of B7-H3 was significantly higher in the tumor compartment than in stroma and the correlation was low (linear correlation coefficient [R2] of 0.25 (Supplemental Figure 3). Since the levels of stromal B7H3 were generally low, the remainder of this work focuses on the tumor cell expression only. The levels of tumor cell B7-H3 in the lung cohorts showed a continuous distribution. A visual cut-point was determined for each cohort for QIF positivity and of the 634 cases represented in 3 cohorts B7-H3 protein expression was found in 80.4% (510/634) of cases (99/118 [83%] in cohort A, 255/291 [87%] cohort B; and 156/225 [69%] cohort C (Figure 1). Per this signal detection threshold, B7-H3 protein expression did not correlate with survival in positive and negative cases. By using X-tile, Cohort A was used as a training set to explore possible prognostic effect of B7-H3 protein expression for different expression levels. In the highest 10%, B7-H3 protein expression was found to be significantly associated with poor overall survival and this cut off was validated in 2 independent cohorts (p <0.05) (Figure 2). Elevated tumor and stromal B7-H3 expression was consistently associated with smoking history across the 3 cohorts, but not with sex, age, clinical stage and histology (Table 2). In cox regression model the negative impact of high tumor B7-H3 tumor protein expression to overall survival remained significant in 2 cohorts while there was a trend on which was not statistically significant in a third cohort.(supplemental table 2)
Figure 1.
A) QIF score distribution of tumor B7-H3 protein expression in Cohort B. Scores are expressed as arbitrary units of fluorescence and the dashed red line indicates the signal detection threshold determined by visual cut off as described in the methods; A1 and A2 are examples of TMA spots with negative and positive B7-H3 signal respectively (DAPI in blue, Cytokeratin mask in green (Cy3 channel) and B7-H3 is in red (Cy5 channel), Figure 1B and 1C display QIF score distribution of tumor B7-H3 in Cohort A and C, respectively.
Figure 2.
Kaplan Meier overall survival curve in 3 cohorts; Figure A, B and C are showing tumor B7-H3 QIF positive and negative cases (red: positive and green: negative) in Cohort A, B and C respectively. Figure D, E and F are showing highest 10% QIF B7-H3 QIF score as a cut off in Cohort A, B and C respectively (red: B7-H3 protein expression highest 10%, green: B7-H3 protein expression lowest or negative 90%).
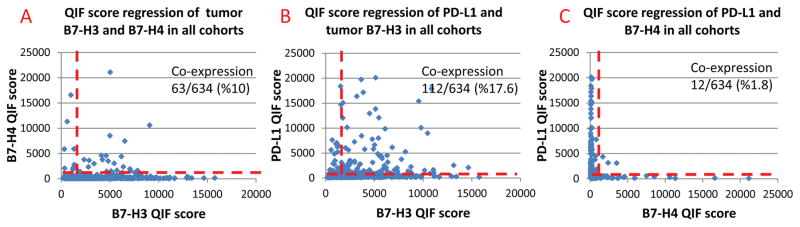
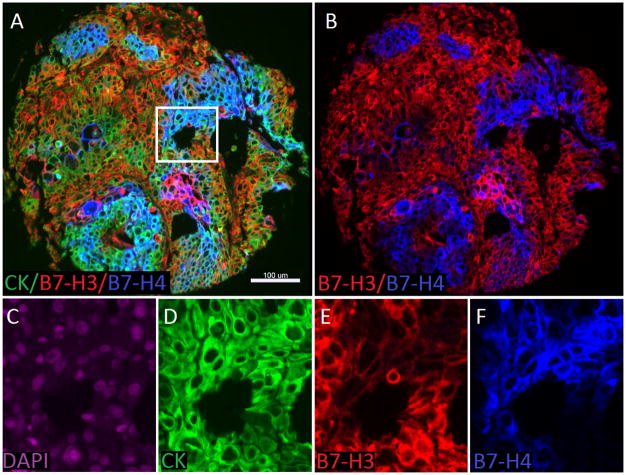
In a merged analysis of the three cohorts, B7-H3 and PD-L1 co-expression was observed in 17.6% (112/634) of all cases (Cohort A: 19/118 [16%], Cohort B 71/291 [24%], Cohort C 22/225 [9%]); B7-H3 and B7-H4 co-expression was observed in 10% (63/634) of the cases (Cohort A: 22/118 [18%], Cohort B 19/291 [6%], Cohort C 22/225 [9%]), B7-H4 and PD-L1 co-expression was observed in 1.8% (12/634) of all cases (Cohort A: 5/118 [4%], Cohort B 5/291[2%], Cohort C 2/225 [1%]). Figure 3 shows the somewhat mutually exclusive expression in the combined cohorts. In tumor samples where high protein expression is observed for multiple markers of interest (co-expression), multiplexing experiments were conducted to further assess for co-localization of these markers with-in same cell. In most of the cases with high B7-H3 and B7-H4 protein expression, tumor tissue did not show co-localization of these proteins (Figures 3 and 4). Tumors showing high levels of PD-L1 and B7-H3 showed focal co-localization (supplemental Figure 4 and 5) in most of the samples. B7-H4 and PD-L1 multiplexed co-localization assessment was not studied since these proteins are mutually exclusive in our cohorts.
Figure 3.
A-C QIF score regressions of markers the dashed red line indicates the signal detection threshold determined by visual cut off. A) QIF score regressions for co-expression rates of tumor PD-L1 and B7-H3 protein, B) QIF score regressions for co-expression rates of tumor B7-H3 and B7-H4 protein, C) QIF score regressions for co-expression rates of tumor PD-L1 and B7-H4 protein.
Figure 4.
Detection of B7-H3 and B7-H4 protein expression to assess co-localization using immunofluorescence (QIF) in lung cancer. A) Representative fluorescence images showing the simultaneous detection of B7-H3 (red channel), B7-H4 (blue channel) and cytokeratin (green channel, inserted box is highlighting the area that has been magnified in figure 2 C–F. B) Simultaneous detection of B7-H3 (red channel), B7-H4 (blue channel) C) 4′,6-diamidino-2-phenylindole (DAPI) only in purple channel, D) Cytokeratin only in green channel, E) B7-H3 only in red channel, D) B7-H4 only in blue channel. Bar=100 μm.
Neither tumor nor stromal B7-H3 expression showed any correlation with major TIL subtypes.
Discussion
Here, we show that B7-H3 protein is widely expressed in the majority of NSCLCs and is associated with smoking history. In all three cohorts overexpression of B7-H3 protein (highest 10%) was correlated with poor survival. This association indicates a prognostic effect consistent with previous observations in NSCLC cohorts (17). Co-expression or mutual exclusivity of an immune checkpoint may support understanding the mechanistic differences that affects the tumor microenvironment and tumor immune evasion. In this study, elevated levels of B7-H3 were not associated with lymphocyte infiltration including CD8 (+) lymphocyte subpopulations, suggesting that B7-H3 expression/upregulation is IFN-γ independent. Furthermore, B7-H3 and PD-L1 protein co-expression was observed in only 18% of the cases, only somewhat mutually exclusive as compared to nearly complete lack of co-expression observed for PD-L1 and B7-H4. This is suggestive of a non-redundant biological role for these targets. B7-H3 and B7-H4 co-expression was relatively low. In most of the cases high protein expression of both proteins did not show co-localization. This exclusive pattern with infrequent co-expression and co-localization is suggesting that some lung tumors may preferentially use one immune evasion mechanism/pathway.. Future studies with longitudinal samples (e.g. repeat biopsy protocols) especially in acquired resistance to immunotherapy to assess the spatiotemporal dynamics of the tumor microenvironment and assessment of co-expression rates of immune checkpoints in biopsy specimens may answer this question.
While drug development to target B7-H4 is ongoing (27), an anti B7-H3 monoclonal antibody, MGA271 (Enoblituzumab)(20), has been shown to be tolerable in a phase I study in solid tumors(19). Currently multiple phase I/II studies are ongoing to assess its safety and efficacy as a single agent and in combination with PD-1/PD-L1 or CTLA-4 axis blockers. With given different co-expression rates of B7 family immune checkpoints in NSCLC, it is useful to explore combination strategies to block additional immune suppressive signals toward the goals of increasing response rates, prolonging responses, and prevention of acquired resistance to monotherapy regimens.
There are a number of limitations to this study. The cohorts represented on our TMAs were entirely retrospectively collected samples from cases with variable follow-up, different treatments and pre-dating molecular/genomic annotation. Due to these limitations in order to avoid premature conclusions maybe related selection bias, we highlighted the findings that are consistent in all cohorts. While the mechanism of B7-H3 protein overexpression in tumors are unclear, in our analysis B7-H3 expression was not correlated to histology, tumor stage or patients age but was correlated to smoking status. The similar B7H3 protein expression in 3 cohorts with different histopathologic distribution may be associated with other factors such as the host immune system. This work was performed on TMAs which may induce under or overrepresentation of the marker levels due to tumor heterogeneity and therefore may not mirror clinical samples studied using whole tissue sections. This is a critical point, since we have found prognostic significance in only the highest 10% tumor B7-H3 tumor protein expression. In this study the proportion of lung cancer cases showing PD-L1 and B7-H4 expression in these cohorts is not identical with our previous reports. This difference is likely due to unavoidable loss of tissue or the absence or limited tumor cells in some spots in different cuts as which is commonly seen in TMA studies.
In summary, we find expression of B7-H3 is more frequent in NSCLC than any previously described immune checkpoint ligand. It’s somewhat mutually exclusive expression pattern with respect to other checkpoints is encouraging with respect to potential for complementary therapeutic approaches. We hope this study can lead to further clinical exploration of these checkpoint targeting agents in clinical trials which could play a relevant role in lung cancer clinical trial designs for combination immunotherapy strategies.
Supplementary Material
TABLE 1.
Tumor B7-H3 QIF and its correlation with clinico-pathologic features and with lymphocyte subtypes
| Characteristics | Cohort A | Cohort B | Cohort C | ||||||
|---|---|---|---|---|---|---|---|---|---|
| B7-H3 (−) | B7-H3 (+) | p | B7-H3 (−) | B7-H3 (+) | p | B7-H3 (−) | B7-H3 (+) | p | |
| Sex | |||||||||
| Male | 8 | 56 | 0.31 | 32 | 192 | 0.18 | 34 | 70 | 0.56 |
| Female | 11 | 43 | 2 | 36 | 35 | 86 | |||
| Unknown | 2 | 27 | |||||||
|
| |||||||||
| Age | |||||||||
| <70y | 11 | 59 | 1 | 30 | 173 | 0.18 | 43 | 88 | 0.46 |
| ≥70y | 8 | 40 | 4 | 53 | 26 | 68 | |||
| unknown | 2 | 29 | |||||||
|
| |||||||||
| Stage | |||||||||
| I–II | 14 | 67 | 0.78 | 19 | 139 | 1 | 59 | 124 | 0.35 |
| III–IV | 5 | 32 | 13 | 90 | 10 | 32 | |||
| unknown | 4 | 26 | |||||||
|
| |||||||||
| Histology | |||||||||
| ADC | 15 | 67 | 0.03 | 17: | 96 | 0.4 | 51 | 94 | 0.33 |
| SCC | 0 | 22 | 15 | 117 | 17 | 45 | |||
| Other | 4 | 10 | 0 | 16 | 1 | 17 | |||
| Unknown | 4 | 26 | |||||||
|
| |||||||||
| Smoker | |||||||||
| current/former | 15 | 94 | 0.03 | 24 | 192 | 0.05 | 47 | 129 | 0.02 |
| Non Smoker | 4 | 5 | 6 | 17 | 16 | 18 | |||
| unknown | 6 | 46 | 6 | 9 | |||||
|
| |||||||||
| CD3 | |||||||||
| Low | 10 | 48 | 0.8 | 21 | 124 | 0.29 | 32 | 81 | 0.47 |
| High | 9 | 51 | 15 | 131 | 37 | 75 | |||
|
| |||||||||
| CD8 | |||||||||
| Low | 9 | 51 | 0.8 | 17 | 129 | 0.72 | 32 | 81 | 0.47 |
| High | 10 | 48 | 19 | 126 | 37 | 75 | |||
|
| |||||||||
| CD20 | |||||||||
| Low | 8 | 50 | 0.61 | 22 | 124 | 0.21 | 40 | 73 | 0.14 |
| High | 11 | 49 | 14 | 132 | 29 | 83 | |||
Translational Relevance.
The biologic significance of co-expression of B7 immunoregulatory molecules, their interaction and role in primary and acquired resistance to PD-1 axis inhibitors remains unclear. By quantitatively measuring B7-H3, PD-L1 (B7-H1), B7-H4 and tumor infiltrating lymphocyte subsets in 3 large NSCLC cohorts, we showed that B7-H3 protein is expressed in the majority of NSCLCs, is associated with smoking history and when expressed in high levels is associated with a negative impact on overall survival. We also showed co-expression of B7-H3 with PD-L1 and B7-H4 is relatively low, and in most of the cases high protein expression of both proteins does not show co-localization in tumor samples. This somewhat mutually exclusive pattern suggests a non-redundant biological role of these targets which could support the use ofuse of biopsy driven personalized immunotherapy.
Acknowledgments
Grant support for this work provided by the Yale SPORE in Lung Cancer P50CA196530 (PI: Roy Herbst), and sponsored research agreements from Genoptix (PI: David Rimm) and Gilead Sciences Inc (Subcontract PI: David Rimm)
Footnotes
Relevant Conflicts of Interest: Dr. Rimm has served as a consultant/advisor for Astra Zeneca, Bristol Meyers Squib, Cell Signaling Technology, Ultivue, and Perkin Elmer and has received grant support from Genoptix and Gilead Sciences
References
- 1.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–64. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharpe AH, Wherry EJ, Ahmed R, Freeman GJ. The function of programmed cell death 1 and its ligands in regulating autoimmunity and infection. Nat Immunol. 2007;8:239–45. doi: 10.1038/ni1443. [DOI] [PubMed] [Google Scholar]
- 4.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–65. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brahmer JR, Drake CG, Wollner I, Powderly JD, Picus J, Sharfman WH, et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J Clin Oncol. 2010;28:3167–75. doi: 10.1200/JCO.2009.26.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–54. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Topalian SL, Sznol M, McDermott DF, Kluger HM, Carvajal RD, Sharfman WH, et al. Survival, durable tumor remission, and long-term safety in patients with advanced melanoma receiving nivolumab. J Clin Oncol. 2014;32:1020–30. doi: 10.1200/JCO.2013.53.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zou W, Chen L. Inhibitory B7-family molecules in the tumour microenvironment. Nat Rev Immunol. 2008;8:467–77. doi: 10.1038/nri2326. [DOI] [PubMed] [Google Scholar]
- 9.Hofmeyer KA, Ray A, Zang X. The contrasting role of B7-H3. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10277–8. doi: 10.1073/pnas.0805458105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun M, Richards S, Prasad DV, Mai XM, Rudensky A, Dong C. Characterization of mouse and human B7-H3 genes. J Immunol. 2002;168:6294–7. doi: 10.4049/jimmunol.168.12.6294. [DOI] [PubMed] [Google Scholar]
- 11.Chapoval AI, Ni J, Lau JS, Wilcox RA, Flies DB, Liu D, et al. B7-H3: a costimulatory molecule for T cell activation and IFN-gamma production. Nat Immunol. 2001;2:269–74. doi: 10.1038/85339. [DOI] [PubMed] [Google Scholar]
- 12.Yi KH, Chen L. Fine tuning the immune response through B7-H3 and B7-H4. Immunol Rev. 2009;229:145–51. doi: 10.1111/j.1600-065X.2009.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun Y, Wang Y, Zhao J, Gu M, Giscombe R, Lefvert AK, et al. B7-H3 and B7-H4 expression in non-small-cell lung cancer. Lung Cancer. 2006;53:143–51. doi: 10.1016/j.lungcan.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 14.Kreymborg K, Haak S, Murali R, Wei J, Waitz R, Gasteiger G, et al. Ablation of B7-H3 but Not B7-H4 Results in Highly Increased Tumor Burden in a Murine Model of Spontaneous Prostate Cancer. Cancer Immunol Res. 2015;3:849–54. doi: 10.1158/2326-6066.CIR-15-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun TW, Gao Q, Qiu SJ, Zhou J, Wang XY, Yi Y, et al. B7-H3 is expressed in human hepatocellular carcinoma and is associated with tumor aggressiveness and postoperative recurrence. Cancer Immunol Immunother. 2012;61:2171–82. doi: 10.1007/s00262-012-1278-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamato I, Sho M, Nomi T, Akahori T, Shimada K, Hotta K, et al. Clinical importance of B7-H3 expression in human pancreatic cancer. Br J Cancer. 2009;101:1709–16. doi: 10.1038/sj.bjc.6605375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lou Y, Diao L, Cuentas ER, Denning WL, Chen L, Fan YH, et al. Epithelial-Mesenchymal Transition Is Associated with a Distinct Tumor Microenvironment Including Elevation of Inflammatory Signals and Multiple Immune Checkpoints in Lung Adenocarcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2016;22:3630–42. doi: 10.1158/1078-0432.CCR-15-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Danilova L, Wang H, Sunshine J, Kaunitz GJ, Cottrell TR, Xu H, et al. Association of PD-1/PD-L axis expression with cytolytic activity, mutational load, and prognosis in melanoma and other solid tumors. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E7769–e77. doi: 10.1073/pnas.1607836113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Powderly J, Cote G, Flaherty K, Szmulewitz R, Ribas A, Weber JS, et al. Interim Results of an Ongoing Phase 1, Dose Escalation Study of MGA271 (Enoblituzumab), an Fc-optimized Humanized Anti-B7-H3 Monoclonal Antibody, in Patients with Advanced Solid Cancer. Society for Immunotherapy of Cancer (SITC) Annual Meeting; 2015. [Google Scholar]
- 20.Loo D, Alderson RF, Chen FZ, Huang L, Zhang W, Gorlatov S, et al. Development of an Fc-enhanced anti-B7-H3 monoclonal antibody with potent antitumor activity. Clin Cancer Res. 2012;18:3834–45. doi: 10.1158/1078-0432.CCR-12-0715. [DOI] [PubMed] [Google Scholar]
- 21.Schalper KA, Brown J, Carvajal-Hausdorf D, McLaughlin J, Velcheti V, Syrigos KN, et al. Objective measurement and clinical significance of TILs in non-small cell lung cancer. Journal of the National Cancer Institute. 2015:107. doi: 10.1093/jnci/dju435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Velcheti V, Schalper KA, Carvajal DE, Anagnostou VK, Syrigos KN, Sznol M, et al. Programmed death ligand-1 expression in non-small cell lung cancer. Lab Invest. 2014;94:107–16. doi: 10.1038/labinvest.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown JR, Wimberly H, Lannin DR, Nixon C, Rimm DL, Bossuyt V. Multiplexed Quantitative Analysis of CD3, CD8, and CD20 Predicts Response to Neoadjuvant Chemotherapy in Breast Cancer. Clin Cancer Res. 2014;20:5995–6005. doi: 10.1158/1078-0432.CCR-14-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaule P, Smithy JW, Toki M, Rehman J, Patell-Socha F, Cougot D, et al. A Quantitative Comparison of Antibodies to Programmed Cell Death 1 Ligand 1. JAMA Oncol. 2016 doi: 10.1001/jamaoncol.2016.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–7. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 26.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10:7252–9. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 27.Leong SR, Liang WC, Wu Y, Crocker L, Cheng E, Sampath D, et al. An anti-B7-H4 antibody-drug conjugate for the treatment of breast cancer. Mol Pharm. 2015;12:1717–29. doi: 10.1021/mp5007745. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.