Abstract
Histone lysine demethylases facilitate the activity of oncogenic transcription factors including possibly MYC. Here we show that multiple histone demethylases influence the viability and poor prognosis of neuroblastoma cells where MYC is often overexpressed. We also identified the approved small molecule antifungal agent ciclopirox as a novel pan-histone demethylase inhibitor. Ciclopirox targeted several histone demethylases including KDM4B implicated in MYC function. Accordingly, ciclopirox inhibited Myc signaling in parallel with mitochondrial oxidative phosphorylation, resulting in suppression of neuroblastoma cell viability and inhibition of tumor growth associated with an induction of differentiation. Our findings provide new insights into epigenetic regulation of MYC function and suggest a novel pharmacologic basis to target histone demethylases as an indirect MYC targeting approach for cancer therapy.
Keywords: Histone demethylase, Neuroblastoma, Ciclopirox
Introduction
Neuroblastoma, a malignancy of the sympathetic nervous system, is the most common extracranial solid tumor of childhood and accounts for 15% of all cancer-related deaths in children. MYCN amplification is an important biologic variable that is a critical factor in defining high risk disease; about 20% of neuroblastoma patients have MYCN amplification (MNA) (1). N-Myc has been clearly shown to be an oncogenic driver of neuroblastoma in transgenic mouse and zebrafish models (2, 3). Cancer genomic sequencing has revealed few genetic mutations in neuroblastoma (4), suggesting that epigenetic factors might be involved in tumorigenesis and disease progression, in addition to N-Myc activity. Myc is one of the most deregulated oncoprotein in various cancers. Unfortunately, directly targeting transcription factors such as Myc is technically challenging. Therefore, novel approaches need to be developed to block the Myc pathway for cancer treatment
Histone methylation is an epigenetic mark that is dynamically regulated by histone methyltransferases and demethylases. The most well studied histone demethylases are histone lysine demethylases (KDM) that are composed of two families. One family is the JmjC domain-containing KDMs that are α-ketoglutarate and Fe(II)-dependent while the other family is flavin adenine dinucleotide (FAD)-dependent KDMs (5, 6). KDMs play important roles in regulation of gene expression, cell cycle and genomic integrity (7). Deregulation of KDMs occurs in many diseases including cancer (8). For example, genetic alterations of KDMs, such as mutation of KDM6A, occur in many types of malignancies (8). In addition, oncogenic transcription factors frequently hijack KDMs to facilitate their transcriptional activity or to inhibit tumor suppressor expression. KDM1 has been shown to enhance c-Myc activity (9) and to complex with the EWS/FLI-1 oncoprotein to suppress TGFB1 transcription (10), while KDM5B has been shown to help c-Myc repress CDKN1A expression (11). KDM4B interacts with the estrogen receptor in breast cancer (12), while KDM4C cooperates with JAK2 to regulate c-Myc expression in a subset of lymphomas (13).
We have recently shown that KDM4B interacts with N-Myc to maintain low levels of the repressive transcription mark, H3K9me3/me2, in neuroblastoma, resulting in overexpression of genes that promote tumor progression (14). Therefore, targeting histone demethylases may block the activities of oncogenic transcription factors and activate tumor suppressive pathways, thereby achieving therapeutic efficacy. However, despite increased understanding of the importance of KDMs in cancer, successful, specific KDM inhibitors for cancer therapeutics have not yet been developed. In this study, we evaluated all known histone demethylases in the regulation of neuroblastoma and discovered a new pan-KDM inhibitor, ciclopirox, and assessed its antitumor activity.
Materials and Methods
Cell Culture, Reagents, Neurosphere Formation, Cell Viability Assay and Cell Proliferation Index
Human 293T, HS68, BE(2)-C, SK-N-AS, IMR-32, SK-N-SH, and U2OS cell lines were purchased from the American Type Culture Collection (ATCC) between 2012 to 2014. NB1, NB7, NB8, NB15, NB-1691 and NB1643 cell lines were established at St. Jude Children’s Research Hospital and obtained from Peter Houghton and Jill Lathi in 2003 and 2012, respectively. Nagai was a gift from Stephen Morris (Insight Genetics) in 2009. Cell lines were propagated, expanded, and frozen immediately into numerous aliquots after arrival. The cells revived from the frozen stock were used within 10 to 15 passages and not exceeding a period of 6 months. When we collected the data for the cell lines, which was done 2–4 years ago, we did not perform STR assay until Aug 2016 when we started performing STR and mycoplasma monthly. All cell lines were characterized by Short Tandem Repeat (STR) analysis by using Promega PowerPlex 16 HS System. PCR based method was used for detection of Mycoplasma with LookOut Mycoplasma PCR Detection Kit (Sigma) and JumpStart Taq DNA Polymerase (Sigma). Neuroblastoma cell lines were cultured in RPMI medium while 293T and HS68 cells were cultured in DMEM medium, supplemented with 10% FCS (HyClone, Thermo), 1% L-glutamine (MediaTech), 1% penicillin–streptomycin (Lonza). HepaRG cell line was purchased from ThermoFisher and cultured in Williams’ Medium E with HepaRG Supplement. Neurospheres were cultured under conditions described previously (15). Briefly, BE(2)-C and SK-N-AS were plated on an ultra-low attachment dish at 1×105 cells/well in a 6-well plate. Human recombinant basic FGF and EGF (Peprotech, Rocky Hill, NJ, USA) were added to the culture medium every 3 days. Cell viability was measured using Presto blue (Invitrogen) in a 96-well plate according to the manufacturer’s instructions. Cell proliferation index was determined in a log-phase proliferation period by dividing the viable cell number at 48h by the viable cell number at 24h. Ciclopirox and Holo-transferrin were purchased from Sigma. GST-KDM4B protein was purchased from BPS Bioscience (San Diego, CA). Tb-anti-Histone H3K9me2 Antibody and Alexa Fluor 488 streptavidin were purchased from Invitrogen (Carlsbad, CA). H3K9me3-biotin [Histone H3 (1 – 21)-K9(Me3)- GGK(Biotin)] was obtained from AnaSpec (Fremont, CA)
siRNA Knockdown
The custom siRNA library (4 siRNAs pooled together for each gene, see Supplementary Table 1) that targets genes encoding JmjC domain containing proteins and FAD-dependent KDM was purchased from Dharmacon. Reverse transfection of 20 nM siRNA in 96-well plate using RNAiMAX (Invitrogen) was performed. We used non-targeting control #2 (NT2) as the negative control. Three days after siRNA transfection, cell viability was assessed using PrestoBlue (Invitrogen). The absorbance reading for each gene was normalized to the control siRNA. The normalized value was then subtracted by 1, and was plotted using Prism program (target vs control). If the value was less than −0.2 or greater than 0.2, we concluded the cell viability as reduced or increased, respectively. For validation of pooled siRNAs, we chose two individual oligos against the selected targets. See oligo sequences in the Supplemental section. For shRNA knockdown of KDM4B we used sh814 which had the best knockdown efficiency(14).
Colony formation assay
After siRNA transfection, 17,000 BE(2)-C cells were plated in each well in a 6-well plate in 2 ml of RPMI 1640 medium in triplicates. After 6 days, colonies were fixed in 4% formaldehyde for 20 min after being washed once with PBS. 0.1% Crystal violet was added onto fixed cells and shaken for 1 hour. The stained cells were washed with ddH2O and scanned. The number of single colony (>20 cells/colony) per view was counted under microscopy. Three different views per well for all triplicates were randomly chosen and colony numbers were averaged.
Western Blotting and Immunofluorescence
Western blot was performed as described in (14). See antibodies in the Supplemental.
For immunofluorescence detection of histone methyl marks, cells transfected with HA-KDM4B were treated with DMSO or 5 µM ciclopirox for 24 hours. Cells were washed three times with PBS and fixed in 4.0% paraformaldehyde in PBS for 15 min at room temperature. Cells were permeabilized for 30 minutes using Triton X-100 (0.3%) in the presence of 10% goat serum. Primary antibodies with a dilution of 1:1,000 in PBS with 0.3% Triton X-100 and 3% goat serum were added to cells for overnight incubation. After three consecutive 5-minute washes with PBS, cells were incubated with secondary antibodies for 1 hour before being washed with PBS and mounted in a mounting medium (DAKO) containing 4,6-diamidino-2-phenylindole dihydrochloride). Imaging of the cells was carried out using a Nikon immunofluorescence microscope. The intensity of immunofluorescence was quantified by using ImageJ software.
RNA and miRNA Extraction and RT-PCR
RNA was extracted using the RNeasy Mini Kit from Qiagen while miRNA was extracted using the miVana kit from Life Technologies. RT-PCR was performed using an Applied Biosystems 7500 Real-Time PCR system. The results were analyzed using ΔΔCT methods. Fold-differences calculated using the ΔΔCT method are expressed as a range by incorporating the standard deviation of the ΔΔCT value into the fold- difference calculation according to the manufacture’s PCR Guide instructions (Applied Biosystem). The PCR primer sequences were listed in Supplementary Table 2.
Affymetrix Microarray Analysis
RNA was extracted from SK-N-AS, NB-1691 and BE(2)-C cells after 48-hours of treatment with 2.5 µM ciclopirox. After quality control with the Agilent RNA analyzer, RNA was subjected to hybridization using an Affymetrix HT HG-U133+ PM 16-Array Plate. Differential gene expression was analyzed using the GenePattern program while gene signature was analyzed using the Gene Set Enrichment Analysis (GSEA) program. 3402 gene sets (chemical and genetic perturbations) were downloaded from MSigDB. After including the MYCN signature, 3403 gene sets were used for GSEA comparison. The permutation parameter for GSEA analysis was 1000. The collapsed dataset to gene symbol parameter was “true”. The permutation type was “gene set”.
TR-FRET assay
In the individual wells of black 384-well low volume assay plates, titrations of ciclopirox, 100 µM of ciclopirox (set as 100% inhibition), or DMSO (set as 0% inhibition) were incubated with H3K9(me3)-biotin peptide (1 µM) and GST-KDM4B protein (250 nM) in 15 µl assay buffer with 50 mM Tris-HCl (pH 8.0), 50 mM KCl, 10 mM MgCl2, 1 mM α-ketoglutarate, 80 µM FeSO4, 2 mM ascorbic acid, 0.01% BSA at room temperature for 30 minutes. The final DMSO concentration was 1% for all assay wells. 5 µl/well of detection mixture of Tb-anti-Histone H3K9me2 Antibody (8 nM) and Alexa Fluor 488 streptavidin (80 nM) in the same assay buffer was then dispensed into individual assay wells. After 15 minute incubation, the TR-FRET signals (fluorescence emission ratio of 10,000 × 520 nm/490 nm) for each well in individual assay plates were collected with a PHERAstar FS (BMG Labtech, Durham, NC) using a 340-nm excitation filter, 100-µs delay time, and 200-µs integration time. The activities of ciclopirox at the indicated concentrations were normalized to that of DMSO wells (negative control, 0% inhibition) and that of 100 µM of ciclopirox (positive control, 100% inhibition) and fit into sigmoidal dose-response curves to derive IC50 values with GraphPad Prism 7.00 (GraphPad Software, Inc., La Jolla, CA).
High Content Method for H3K9me3 quantification
See details in Supplemental section.
Molecular Docking
The KDM4B crystal structure was obtained from the RCSB PDB repository (PDB accession code: 4LXL). The protein structure was prepared for docking using the Protein Preparation Wizard of Maestro. Water molecules having less than two hydrogen bonds to non-water molecules were deleted. This retained a water molecule complexed with Nickel (II). The protein structure was protonated at pH7.4. Finally, the structure was restrained minimized using the OPLS_2005 forcefield with default parameters. The cognate ligand pyridine-2,4-dicarboxylic acid formed the center of the docking box. A metal-coordination restraint was added at the Nickel center so that docked ligands could participate in metal-ligand interactions. The tri-methylated histone 3 lysine 9 portion of the complex was retained at the time of grid generation. The CPX structure was prepared using the Ligand Preparation module of Maestro which generated a deprotonated state as the dominant state. This state was docked into the KDM4B active site using the single precision scoring mode in Glide, with the metal coordination restraint in “on” mode. This generated a mono-dentate chelated CPX. In keeping with the octahedral complex seen in mono-dentate chelation (cf.PDB structure with accession code 5F37) in similar active sites, a water molecule was introduced at the position of the carboxylate oxygen of pyridine-2,4-dicarboxylic acid from PDB structure 4lxl. The entire structure was then subjected to 100 iterations of OPLS_2005 force-field based energy minimization to finally obtain an octahedral complex with two coordinated waters.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed according to the manufacturer’s protocol (Magna EZ-CHIP). Briefly, BE(2)-C cells were cross-linked with 1% paraformaldehyde for 10 minutes and quenched for 5 minutes with 125 mmol/L of glycine. After sonication, cell lysates were spun down and 100 µL of supernatant was diluted to 500 µl for immunopreciptation. H3K9me3 antibodies (Millipore, 07–523) were used for immunopreciptation of the cross-linked DNA-protein complexes. After serial washing, DNA-protein cross-links were reversed and DNA was extracted for PCR.
Oxygen Consumption Assay
Respiration was measured in intact cells using the Seahorse XF24 analyzer(16). After 48-hour treatment with CPX or siRNA knockdown, cells were seeded in plates coated with poly-L-lysine. After 5 hours, the cells were loaded into the machine to determine the oxygen consumption rate (OCR). Respiration was measured sequentially after addition of oligomycin (0.5µM), FCCP (1µM) and rotenone (0.5µM). After each injection OCR was measured for 3 min, the medium mixed and again measured for 3 min.
Database mining
The Versteeg neuroblastoma dataset (GSE16476) at R2: Genomics Analysis and Visualization Platform (http://hgserver1.amc.nl/cgi-bin/r2/main.cgi) was used to correlate expression of MYCN and KDMs and Kaplan-Meier survival analysis.
Animal Experiments
All murine experiments were done in accordance with a protocol approved by the Institutional Animal Care and Use Committee of St. Jude Children's Research Hospital. The disseminated disease model was established as described(17). 0.1 ml of the cell suspension (1 × 106 cells) of NB-1691luc cells were injected into the lateral tail vein of SCID mice with a 25-G needle. One week later pumps containing CPX were subcutaneously implanted in the dorsum of tumor bearing mice. The pumps (model 1004) were purchased from Alzet, which delivers 0.11µl/hour for 4 weeks and holds 100µl in total. CPX pumps were prepared as follows: 18 mg of CPX were dissolved in 225 µl of 95% EtOH by vortexing. 750 µl of DMSO were added and vortexed. Then 525 µl of sterile H2O were added and vortexed to have a final concentration of 12mg/ml. The CPX solution was filled in pumps using sterile forceps inside a hood. The control mice received an alzet pump with vehicle. Tumor growth was monitored using an IVIS Imaging System 100 Series (Xenogen Corporation, Alameda, Calif). Mice received a 150-mg/kg intraperitoneal injection of D-Luciferin (Xenogen). Five minutes after substrate injection the animals were imaged. Acquired images were analyzed using Living Image Software version 2.50 (Xenogen). Subcutaneous xenografts were established in male CB-17 severe combined immunodeficient mice (Taconic, Hudson, NY), Tumor measurements were done weekly using handheld calipers, and volumes calculated as width π/6 xd3 where d is the mean of two diameters taken at right angles. Subcutaneous xenografts were treated with 20mg/kg of CPX via oral gavage twice daily.
Statistics
Each experiment was carried out at least in triplicate. To determine statistical significance, the unpaired, two-tailed Student’s t test was calculated using the t test calculator available on GraphPad Prism 7.0 software. Survival analysis was done by the Kaplan–Meier method, and survival curves were compared by the log-rank test. A p value of less than 0.05 was considered statistically significant.
Microarray deposition
The microarray data have been submitted in GEO repository with accession numbers of GSE45969 and GSE45970.
Results
Identification of histone demethylases regulating neuroblastoma cell viability
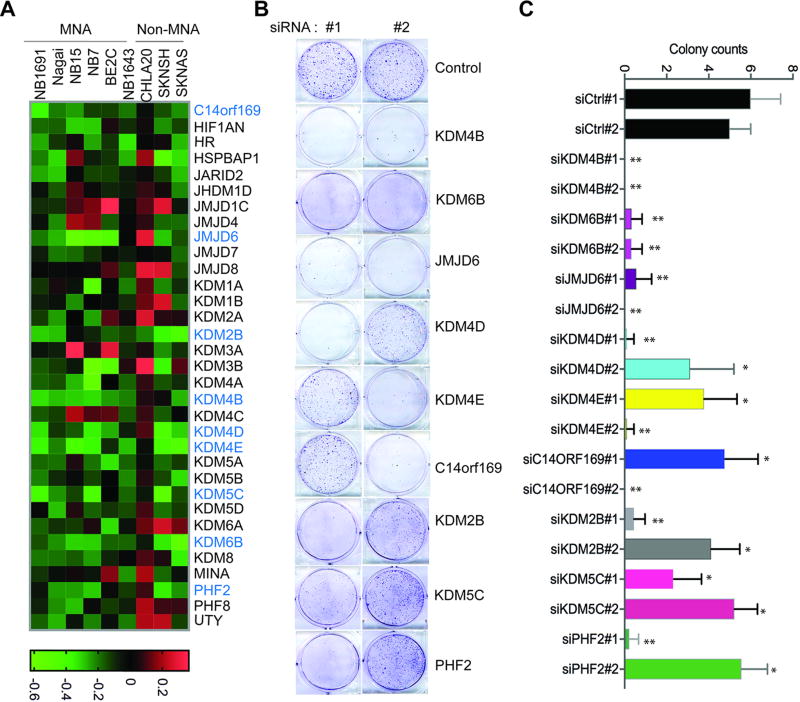
To assess the potential role of histone demethylases in the regulation of neuroblastoma cell proliferation, we used a focused siRNA pool library (Supplemental Table 1; including 32 genes encoding JmjC domain proteins with 17 of them having histone lysine or arginine demethylase activity; and 2 genes encoding FAD-dependent demethylase KDM1A and KDM1B), for screening in 9 neuroblastoma cell lines, six of which have MYCN amplification (MNA). Cell viability was assessed by PrestoBlue assay three days after introduction of the siRNAs. After filtering out targets that affected fewer than 4 cell lines (cell viability value <-0.2), 9 histone demethylases (C14orf169, JMJD6, KDM2B, KDM4B, KDM4D, KDM4E, KDM5C, KDM6B and PHF2) and 2 genes encoding JmjC domain containing non-histone demethylase proteins (HR and HSPBAP1) were found to contribute to the maintenance of neuroblastoma cell viability (Figure 1A and Supplemental Figure 1). Interestingly, three members of the KDM4 subfamily (KDM4B, KDM4D and KDM4E but not KDM4A and KDM4C), which are responsible for removing the transcription repressive mark H3K9me3/me2, were involved in limiting viability of most of the cell lines (Figure 1A, Supplemental Figure 1). Knockdown of other histone lysine demethylase subfamily members including C14orf169, KDM2B, KDM5C, KDM6B, PHF2 and histone arginine demethylase JMJD6 also affected the viability of multiple cell lines (Figure 1A). Two non-histone demethylase genes HR and HSPBAP1 were also important for neuroblastoma cell viability.
Figure 1. Identification of histone demethylases in the regulation of neuroblastoma cell viability.
(A) Heat map shows cell viability assessment after siRNA knockdown of histone demethylases and genes encoding JmjC domain containing non-histone demethylase proteins. The viability threshold is set to (+0.2, −0.2). If the value is less than −0.2 we conclude that the gene reduces cell viability. If the value is greater than 0.2 we conclude that the gene increases cell viability. The genes highlighted in blue are genes that inhibits the cell viability of at least 4 of 9 cell lines. MNA indicates MYCN amplification.
(B) Crystal violet staining of cells after 4-days of depletion of indicated histone demethylases in BE2(C).
(C) Colony number quantified by averaging counts from 5 different views under microscope. All the data are shown as mean ± S.D.; **P<0.01, *P<0.05.
To validate our screening results, we randomly took two individual oligos in the original pool and tested them separately in BE(2)-C cells, a frequently studied MYCN amplified cell line (Figure 1B, 1C). Here we focused on histone demethylases, and thus excluded HR and HSPBAP1, but tested 9 histone demethylases (Figure 1B, 1C). Consistent with our screening data, depletion of KDM4B, KDM4D, JMJD6 and KDM6B greatly reduced the cell colonies of BE(2)-C (Figure 1B, 1C). And depletion of other histone demethylases also led to a phenotype similar to the original screening in BE(2)-C cells (Figure 1B, 1C and Supplemental Figure 1). Taken together, these data confirmed that histone demethylases are important for maintaining neuroblastoma cell viability.
Identification of ciclopirox as a pharmacologic inhibitor of histone demethylases
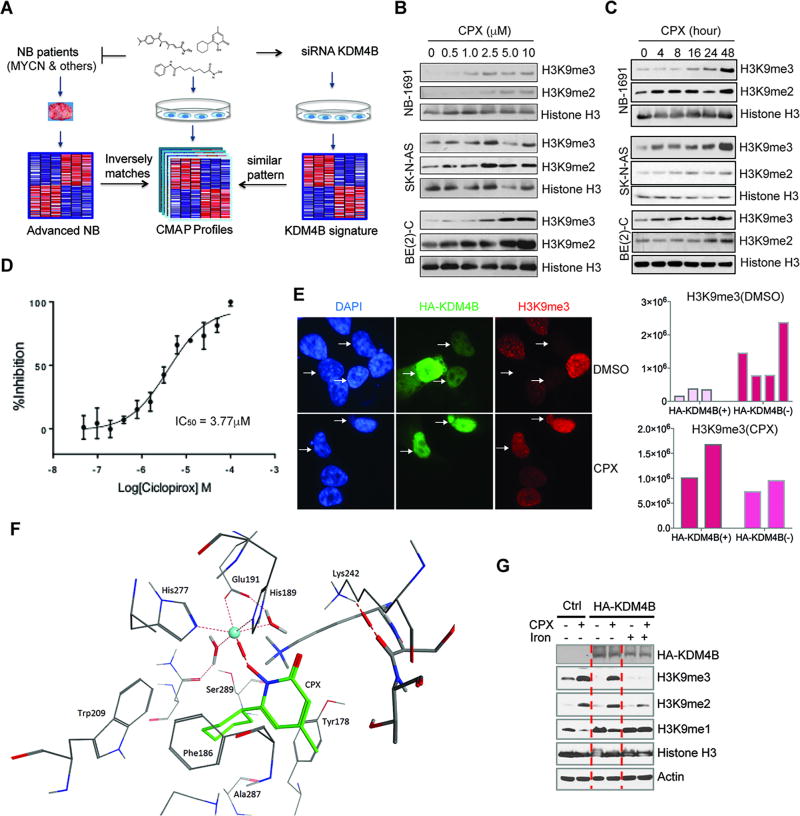
The KDM dependency of Myc-driven neuroblastoma cells as shown in Figure 1 indicates that identification and development of KDM inhibitors may serve as a new therapeutic strategy to target neuroblastoma. We recently demonstrated that KDM4B plays an important role in the regulation of N-Myc function and N-Myc-driven tumor growth (14), consistent with the screening results in this study. Since specific and potent inhibitors of KDM4B are currently not available, we used a chemical genetic approach to identify inhibitors that target KDM4B or its associated signaling pathway, since we had previously studied the global gene expression profile of KDM4B. We hypothesized that the genetic and epigenetic alterations in tumors would give rise to a transcriptome that could be pharmacologically reversed. The connectivity Map (CMAP) is a collection of genome-wide transcriptional expression data from cultured human cancer cells treated with bioactive small molecules, and these chemical genetic profiles and pattern-matching algorithms enable the discovery of functional connections between drugs, genes and diseases (18). We used CMAP to search for drugs that produce a similar transcriptome to that of KDM4B depletion, with the premise that these drugs would be able to target KDM4B function or KDM4B–related signaling pathways (Figure 2A). We also used clinical neuroblastoma gene expression data to identify drugs that produce transcriptome profiles inversely matching the signature of advanced stage neuroblastoma, since MYCN is frequently amplified in advanced stage patients. Using this strategy, we found that the KDM4B gene expression profile was enriched with CMAP profiles induced by compounds including several protein synthesis inhibitors, PI3K and mTOR inhibitors, and others such as dopamine blocking agents (Supplemental Figure 2A). Interestingly, the histone deacetylase (HDAC) inhibitors and an off-patent drug, ciclopirox (CPX), an antifungal agent for seborrheic dermatitis that has been used in a clinical trial via oral delivery for acute myeloid leukemia (19), were identified in both analyses, based on an in vitro KDM4B knockdown signature and an in vivo advanced stage neuroblastoma signature (Figure 2A, Supplemental Figure 2, A and B). It is noteworthy that CPX ranked #1 when comparing with the high-stage primary neuroblastoma profiles, which suggests MYCN signaling is more likely proxy to the CPX profile. Thus, these data suggest that these compounds affect the pathways shared by KDM4B and Myc. We chose to focus on CPX, first testing whether CPX inhibited KDM4B function.
Figure 2. Identification of ciclopirox as a pharmacological inhibitor of KDM.
(A) The rationale for identification of compounds that may target KDM4B.
(B–C) Neuroblastoma cells were treated with CPX for 48 hours with indicated concentrations (B) or treated with 2.5 µM of CPX for different lengths of time (C). Histone methyl marks in different neuroblastoma cell lines were assessed using immunoblotting.
(D) Ciclopirox inhibited KDM4B-mediated demethylation of the H3K9Mme3-biotin peptide in a dose-response manner in a biochemical TR-FRET functional assay (IC50 of 3.77 ± 0.17 µM).
(E) 8 hours after transfection of HA-tagged KDM4B in SK-N-AS cells (top), 5.0 µM of CPX was added to the medium for 24-hour treatment (bottom). Immunofluorescence was used to assess global H3K9me3 marks (63×). The intensity of H3K9me3 for each single cell in 2 groups was quantified by using ImageJ (right).
(F) Molecular docking of CPX into the JmjC domain of KDM4B. The molecule forms an octahedral complex at the nickel center via its deprotonated hydroxyl group. The oxygen from the keto group of CPX is close to Lys242 and may interact with it directly or via a water bridge (not shown). The cyclohexyl moiety was nestled in a hydrophobic pocket formed by Phe186, Tyr 178, Trp209, Ala287 and the aliphatic chain of Lys207. The blue sphere indicates nickel ion.
(G) 293T cells were transfected with HA-KDM4B. 8 hours after transfection, cells were treated with 5.0 µM of CPX and/or 500µg/ml of holo-transferrin that carries iron into cells, for 24 hours. Western blotting was used to assess the indicated markers.
First, we examined the CPX effect on histone methyl marks in neuroblastoma cells. We found that CPX treatment did cause global induction of histone H3K9 methyl marks in three different neuroblastoma cell lines in a dose- and time-dependent manner (Figure 2, B and C). To test whether CPX directly affected the enzymatic activity, we used a time-resolved fluorescence energy transfer (TR-FRET) assay to assess the KDM4B-mediated demethylation of an H3K9me3-biotin peptide. In this assay, ciclopirox inhibited KDM4B in a dose-response manner, with an IC50 of 3.77 ± 0.17 µM (Figure 2D). We then tested whether CPX was able to block the activity of overexpressed KDM4B in cells and found that CPX at 5 µM suppressed KDM4B activity in over 90% of cells by assessing H3K9me3 using immunofluorescence (Figure 2E and Supplemental Figure 2C). Correspondingly, elevated H3K9me1 levels due to conversion of H3K9me3/me2 to H3K9me1 by KDM4B, were significantly reversed by CPX (Supplemental Figure 2C). Similarly, the H3K9me3 levels that were inversely correlated with KDM4B overexpression in U2OS cells were increased by CPX, further demonstrating that CPX blocks KDM4B function (Supplemental Figure 2D). Next, we used the crystal structure of KDM4B (4LXL) as a template to dock CPX into the JmjC domain. The docking free energies are quite favorable. An octahedral complex at the active site was obtained (Figure 2F). The deprotonated hydroxyl group of CPX formed a mono-dentate interaction with the metal ion. The oxygen from the keto group of CPX was available for interaction with Lys242 either directly or via a water bridge. The cyclohexyl moiety of CPX was nestled in a hydrophobic pocket formed by Phe186, Tyr 178, Trp209, Ala287 and the aliphatic chain of Lys207.
Since histone demethylases require iron for activity and CPX has been shown to chelate iron (20), we tested whether iron was able to rescue the effect of CPX. We overexpressed KDM4B in 293T cells because of their greater transfection efficiency than neuroblastoma cells. The exogenous KDM4B markedly reduced global H3K9me3 levels and increased H3K9me1 levels (Figure 2G). 5 µM of CPX treatment efficiently blocked KDM4B activity. Iron supplementation was able to block the CPX effect (Figure 2G), suggesting that CPX inhibits KDM4B by chelating iron from its catalytic domain. Although CPX shares a similar active moiety to the HDAC inhibitor vorinostat that chelates Zinc (Supplementary Figure 2E), we found that CPX did not induce acetylation of histone H3 or alpha-tubulin (Supplementary Figure 2F), demonstrating that CPX is not an HDAC inhibitor.
To assess the specificity of CPX to KDMs, we further tested other histone methyl marks and found that CPX also increased methylation levels of H3K27, H3K4 and others (Supplemental Figure 3, A and B), suggesting CPX might be a pan-KDM inhibitor. To validate this, we overexpressed KDM4C (H3K9me3/me2), PHF8 (H3K9me2/me1), KDM6A/UTX (H3K27me3/me2), KDM5A (H3K4me3/me2), and C14orf169 (also named NO66) in 293T cells (Supplemental Figure 3, C–G). We found that CPX inhibited KDM4C, PHF8 and KDM6A activity and as in the previous case, the addition of iron at least partially rescued the effect (Supplemental Figure 3, C–E). Interestingly, NO66 did not exert histone demethylase activity in 293T cells (Supplemental Figure 3F) although it is purported to be an H3K4me3 and H3K36me3/me2 demethylase (21). Thus, CPX appears to be a pan-KDM inhibitor. Nevertheless, the discovery that multiple KDM subfamily members are involved in regulation of neuroblastoma cell proliferation or viability indicates that a pan-KDM inhibitor may be expected to suppress tumor growth better than a specific KDM inhibitor.
CPX suppresses neuroblastoma cells but not normal cells
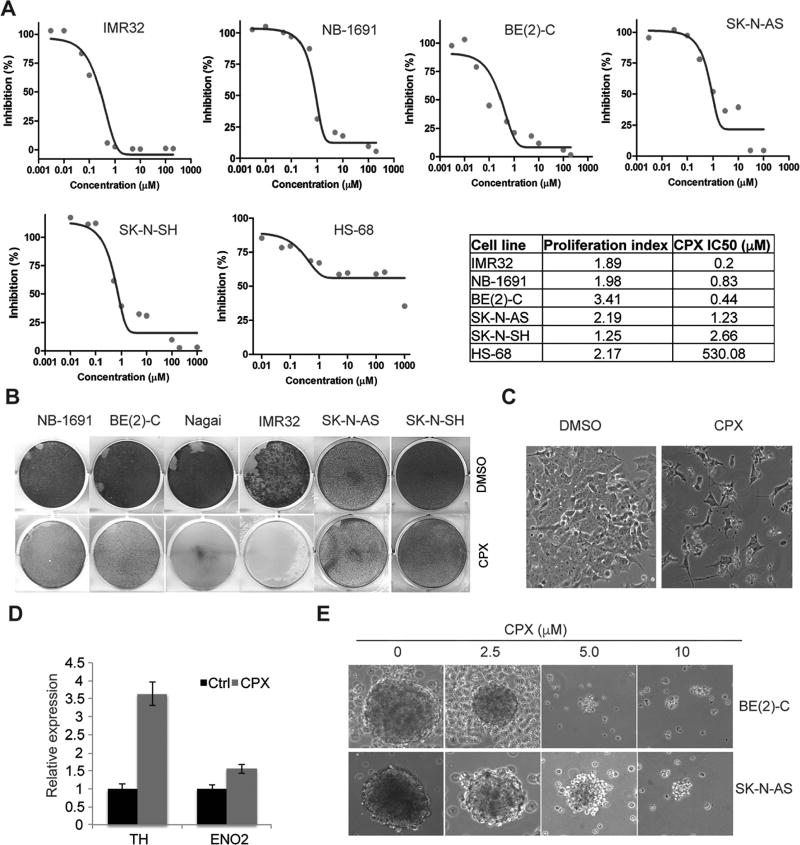
Since we have identified CPX as a novel inhibitor to KDMs that are essential to neuroblastoma cell survival (Figure 1 and 2), we next assessed its effect on cell viability. An Alamar blue assay showed that CPX potently suppressed the viability of neuroblastoma cells in vitro but not the normal human fibroblast cell line HS68 (Figure 3A). Neuroblastoma cells were at least 200-fold more sensitive to CPX than HS68 cells (Figure 3A). To assess the possibility that the drug response may be associated with a decrease in the proliferation rate we examined the proliferation index of each cell line. We found that HS68 cells proliferated even faster than IMR32, NB-1691 and SK-N-SH cells (Figure 3A), ruling out the possibility that HS68 was resistant to CPX due to its slow proliferation rate. Thus, these data suggest CPX may selectively kill cancer cells but not normal cells. This is consistent with a recent Phase I clinical trial in AML in that once-daily administration of 40mg/m2 oral ciclopirox olamine for five days was well tolerated in all patients without dose-limiting toxicity(22). A crystal violet staining assay showed that CPX potently inhibited neuroblastoma cell proliferation and survival in five MYCN amplified cell lines while the non-MYCN cell lines SK-N-AS and SK-N-SH were more resistant (Figure 3B). CPX treatment also caused cell morphology changes such as neurite outgrowth (Figure 3C), a characteristic typical of neuroblastoma differentiation. Real-time PCR showed that CPX induced expression of differentiation markers such as neuron-specific enolase 2 (ENO2) and tyrosine hydroxylase (TH) (Figure 3D). To test whether CPX was able to target potential tumor-initiating cells, we treated neurospheres derived from BE(2)-C and SK-N-AS cells. CPX treatment also inhibited the neurosphere formation in a dose-dependent manner (Figure 3E).
Figure 3. CPX selectively inhibits cancer cells in comparison with normal cells.
(A) Cells were treated with ciclopirox for 96 hours. Curves showing that ciclopirox suppresses viability of neuroblastoma cells but not HS68 cells, a normal human fibroblast cell line. The IC50 of CPX to each cell line was summarized in the table. The proliferation index for each cell line was determined and summarized in the table.
(B) Cells were stained by crystal violet after 5 µM of ciclopirox treatment for 4 days.
(C) Morphology of BE(2)-C cells following treatment with 2.5 µM of ciclopirox. Note the neurite outgrowth in treated cells.
(D) RT-PCR showing that ciclopirox induces differentiation marker gene expression in BE(2)-C cells. This experiment was carried out in triplicate.
(E) Ciclopirox suppresses formation of neurospheres derived from BE(2)-C (top) and SK-N-AS (bottom) cells.
CPX inhibits the Myc signaling pathway
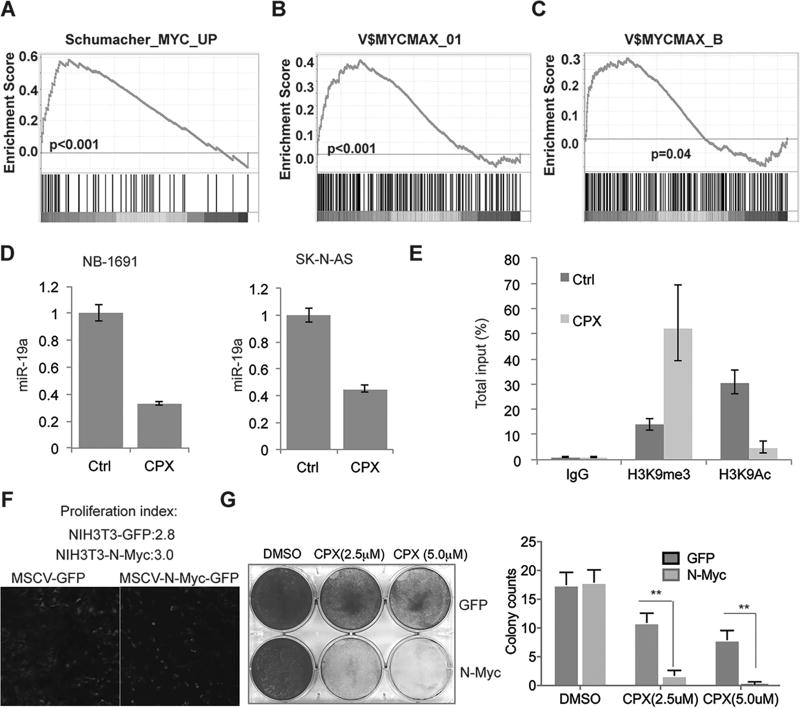
KDMs play an important role in regulation of gene transcription. To investigate the CPX effect on the transcriptome at a global level, we characterized the gene expression profile after 48-hour treatment of tumor cells with 2.5 µM of CPX, a time and dose when cell viability was not yet significantly affected. We first compared the gene expression pattern of CPX with JIB-04, a compound that has recently been found to target histone demethylases including KDM4B (23). Gene Set Enrichment Analysis (GSEA) showed that the JIB-04 signature was significantly enriched with genes affected by CPX (Supplemental Figure 3H), supporting the fact that CPX, like JIB-04, targets KDMs. Importantly, GSEA revealed that the Myc pathway was targeted by CPX (Figure 4A, Supplemental Figure 4A). Among oncogenic signatures in MSigDB that are targeted by CPX, the Myc oncogenic signature also ranked #1 (Supplemental Figure 4B). Motif analysis showed that the genes downregulated by CPX were enriched with Myc/Max binding sites (Figure 4, B and C). Nevertheless, we found that the genes affected by CPX bear potential binding sites for other factors such as upstream stimulating factors (USF) and zinc finger protein ZF5 (Supplemental Figure 4C), and therefore other regulators may also be involved in the CPX induced effect. We observed that the Myc target miR-17–92 was downregulated by CPX and performed RT-PCR to test the expression of miR-19a, the most important miRNA of the miR-17–92 cluster in oncogenesis (24, 25). Indeed, CPX inhibited expression of miR-19a (Figure 4D), indicating that CPX suppresses Myc-mediated miR-17–92 cluster expression. ChIP-PCR showed that CPX treatment caused an increase in H3K9me3 and a decrease in acetyl H3K9 (a transcriptionally active marker) on the MIR17HG E box region (Figure 4E), indicating an epigenetic change at this locus. To further understand the mechanistic association of Myc and KDM, we analyzed the ENCODE data which has available ChIP-seq data for c-Myc, KDM4A and H3K9me3. Interestingly, in human embryonic stem cells, the genomic occupancy pattern of KDM4A and c-Myc resembled and was mutually exclusive to H3K9me3 (Supplemental Figure 4D), suggesting that KDM4A cooperates with c-Myc in human embryonic stem cells, supporting the hypothesis that targeting histone demethylase by CPX may affect Myc activity. To examine whether activation of Myc could sensitize cells to CPX treatment, we transduced NIH3T3 cells, a murine fibroblast cell line, with GFP or N-Myc (Figure 4F). After transduction with N-Myc, NIH3T3 cells did not proliferate significantly faster than the control (proliferation index 3.0 vs 2.8) (Figure 4F), although the cell morphology changed. Interestingly, cells transduced with N-Myc were much more sensitive to CPX treatment (Figure 4G), indicating that Myc reprogrammed cells were less tolerant to the inhibition of KDMs.
Figure 4. CPX targets the Myc pathway.
(A) After 48 hours of 2.5µM CPX treatment of BE(2)-C, NB-1691 and SK-N-AS cells, GSEA analysis revealed that genes downregulated by CPX were enriched for Myc targets.
(B–C) GSEA motif analysis showed that genes inhibited by CPX have Myc/Max binding sites.
(D) RT-PCR showed that 2.5µM CPX treatment of BE(2)-C and SK-N-AS cells for 48 hours inhibited expression of miR-19a. This experiment was carried out in triplicate.
(E) ChIP-PCR showed that 2.5µM CPX treatment of BE(2)-C cells for 48 hours increased histone methylation of H3K9 but reduced acetylation. This experiment was carried out in triplicate.
(F) NIH3T3 cells were transduced with retroviral MSCV-GFP and MSCV-N-Myc-GFP, with comparable proliferation indices.
(G) Cells from F were stained with crystal violet after 2.5 or 5 µM of ciclopirox treatment for 4 days (left). Colony number was quantified by averaging counts from 5 different views under microscope. All data are shown as mean ± S.D.; **P<0.01.
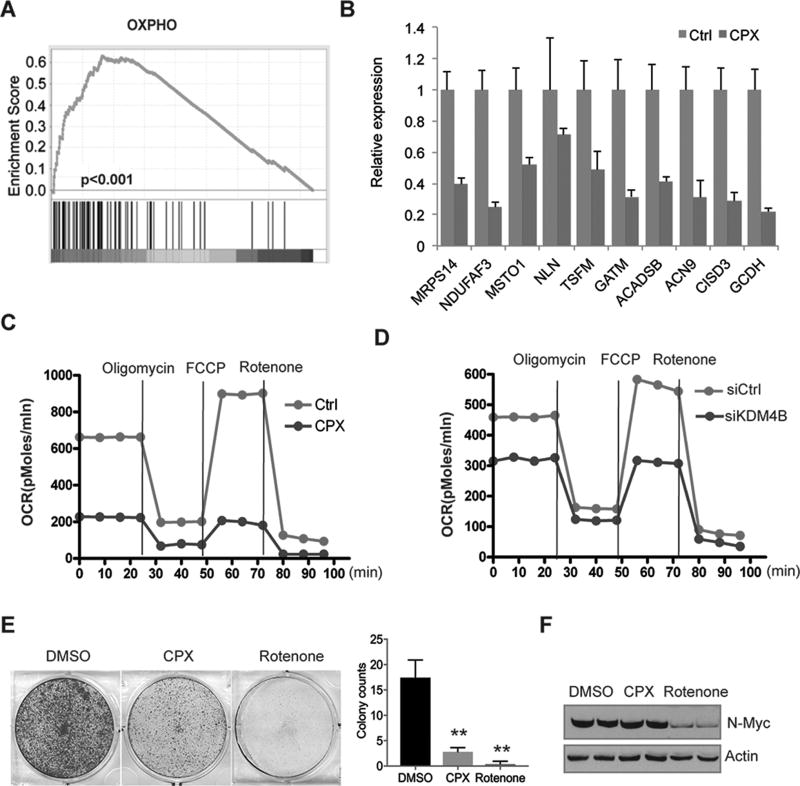
CPX suppresses oxidative phosphorylation
Myc is involved in mitochondria biogenesis and energy metabolism (26–29). Consistent with the function of Myc in promoting OXPHOS under some settings, GSEA revealed that CPX suppressed genes involved in mitochondrial oxidative phosphorylation or mitochondrial respiration (Figure 5A, Supplemental Figure 5, A–C). Next we picked the top ranked genes from GSEA analysis and performed RT-PCR to validate these findings (Figure 5B and Supplemental Figure 5D). We then measured the mitochondrial oxygen consumption rate in real-time with oligomycin (an inhibitor of ATP synthase), FCCP (an uncoupler of oxidative phosphorylation) and rotenone (an inhibitor of the electron transport of mitochondrial complex I). The results showed that CPX decreased mitochondrial oxygen consumption (Figure 5C and Supplemental Figure 5E), indicating that CPX inhibits the mitochondria respiratory chain reaction. We then tested whether KDM4B and N-Myc were involved in regulating mitochondrial function in neuroblastoma cells. Indeed, KDM4B and N-Myc depletion using siRNAs resulted in downregulation of the oxygen consumption rate (Figure 5D; Supplemental Figure 5, F–G), suggesting that CPX targets cancer cell metabolism at least partially through KDM4B.
Figure 5. CPX inhibits oxidative phosphorylation.
(A) GSEA analysis revealed that 2.5µM CPX treatment of BE(2)-C cells for 48 hours suppresses genes involved in oxidative phosphorylation.
(B) RT-PCR assessment of genes involved in oxidative phosphorylation after 2.5µM CPX treatment of BE(2)-C cells for 48 hours. This experiment was carried out in triplicate.
(C) Oxygen consumption analysis showing a decline effected by 2.5µM CPX treatment of BE(2)-C cells for 48 hours.
(D) Oxygen consumption analysis showing a decline by 48-hour depletion of KDM4B with siRNA in BE(2)-C cells.
(E) BE(2)-C cells were stained with crystal violet after 0.5 µM of rotenone or 2.5 of ciclopirox treatment for 3 days (left). Colony number was quantified by averaging counts from 5 different views under microscope. All data are shown as mean ± S.D.; **P<0.01.
(F) Western blotting assessment of N-Myc expression after a 48h treatment of BE(2)-C cells with 0.5 µM of rotenone or 2.5 of ciclopirox.
The inhibition of oxidative phosphorylation by CPX raised the possibility that the down-regulation of Myc target genes in response to CPX may be a consequence of the anti-proliferative activity due to the inhibition of energetics. We therefore compared the cellular effects of CPX and rotenone, the inhibitor to the complex I of electron transport chain (ETC). Unlike CPX, we found that rotenone rapidly induced cell death within 24 hours and killed nearly all cells after 72h treatment (Figure 5E). We further examined the N-Myc expression and found that rotenone but not CPX induced a dramatic reduction of N-Myc protein (Figure 5F), probably due to a secondary effect. These data suggest that CPX inhibits the Myc pathway through a different mechanism from rotenone which directly inhibits ETC and consequently causes Myc reduction.
CPX suppresses tumor growth and dissemination
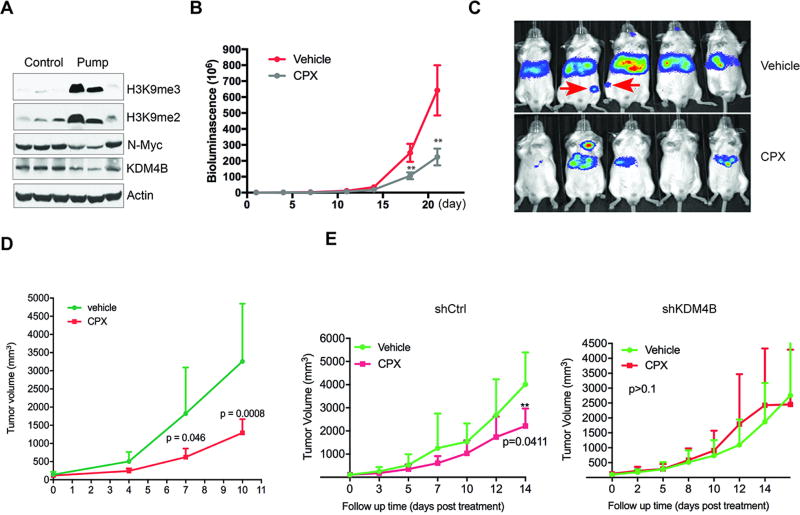
We have established that CPX targets KDMs, which are essential to neuroblastoma cell survival. We therefore tested the in vivo anti-neuroblastoma activity of CPX in a disseminated disease model established previously in our lab (17). The burden of disease after tail vein injection of NB-1691 cells engineered to stably express luciferase was assessed by bioluminescence imaging (30). Because of the short half-life of CPX in mice (31), we first chose to deliver the drug via a subcutaneously implanted, continuous release pump. The control mice received an alzet pump with vehicle. The length of time during which drug was released from the pumps was approximately four weeks; the calculated plasma concentration of CPX was approximately 2.5µM. Pharmacodynamic studies in which tumor histone methylation status was assessed by western blot analysis, showed successful target inhibition and epigenetic modification with increased H3K9me3/me2 (Figure 6A). Importantly, continuous delivery of CPX via the subcutaneous pump significantly reduced the tumor burden, as assessed by the intensities of bioluminescence (Figure 6, B and C). In addition, two mice in the control group showed distant metastasis in the femur (Figure 6C), while the CPX treatment group did not, suggesting that CPX inhibits or prevents disease dissemination.
Figure 6. CPX suppresses tumor growth.
(A) Pharmacodynamic analysis of neuroblastoma xenografts by western blot showed that continuous delivery of CPX via a subcutaneous pump resulted in an increase of H3K9me3/me2 marks in vivo.
(B) CPX inhibited NB-1691 neuroblastoma xenograft growth in a disseminated disease model (n=10/group). Curves show the average bioluminescence signals over time. (p<0.01, Student’s t test)
(C) Bioluminescence images from the control (right, top row) and pump treatment (right, bottom row) animals at 4 weeks in (B). The red arrow indicates bone or bone marrow metastasis.
(D) Growth of NB-1691 subcutaneous xenografts (n=5/group) treated with 20mg/kg of CPX twice daily via oral gavage (**p<0.01, Student’s t test).
(E) BE2-(C) xenografts from shRNA control group (n=5) or shKDM4B group (n=5) were treated with 20mg/kg CPX or excipient twice daily via oral gavage. P value was computed by Student’s t test.
To further assess the anti-tumor activity of CPX, we established NB-1691 subcutaneous xenografts. After tumors reached 100 mm3 in size, CPX was given orally twice daily via gavage. CPX treatment significantly reduced tumor growth (Figure 6D). Finally, since CPX may impact other targets in addition to KDM4B, we sought to determine the importance of KDM4B inhibition by CPX in impairing tumor growth by testing the anti-tumor effectiveness of CPX against tumors in which KDM4B had already been genetically ablated. Knockdown of KDM4B in BE2(C) cells significantly slowed tumor growth and extended survival (Supplemental Figure 6A). However, when the control and knockdown groups were treated with CPX via oral gavage twice daily once tumor size was about 100mm3, tumor growth in the control group was significantly delayed by CPX treatment and survival extended (Figure 6E, left; Supplemental Figure 6B), whereas there was no difference in the knockdown group (Figure 6E, right), suggesting that CPX impacts tumor growth largely through inhibition of KDM4B in BE2(C) cells.
Discussion
The Myc proto-oncogenic transcriptional factor plays an important role in tumorigenesis and tumor maintenance in a variety of cancers including neuroblastoma. Myc activity is determined by local chromatin histone methylation status (32) and thus we predicted that perturbations of histone methylation could interfere with the transcriptional function of Myc. In addition, Myc itself is able to regulate chromatin modification such as H3K9me3 and other histone marks (33–35), indicating that Myc may recruit histone methylation modifiers to establish the epigenetic status for its function. One recent study showed that c-Myc interacts with histone demethylases KDM4B and KDM4C to regulate mouse embryonic cell stemness (36). The histone demethylase KDM1A has been shown to reprogram the transcriptome of neuroblastoma cells and regulates neuroblastoma xenograft growth (37), cooperating with N-Myc to repress tumor suppressor genes in neuroblastoma (38). We have recently identified that KDM4B physically interacts with N-Myc and maintains the low levels of repressive H3K9me3/me2 marks at Myc target gene loci in neuroblastoma, thereby facilitating N-Myc activity (14). Interestingly, one study has shown that loss-of-function of histone demethylase KDM5D is synthetically lethal to human mammary epithelial cells transduced with Myc(39). These data indicate that Myc-driven cancer may be addicted to KDM for survival. In this study, we have identified additional histone demethylases C14orf169, JMJD6, KDM2B, KDM4D, KDM4E, KDM5C, KDM6B and PHF2 that regulate neuroblastoma cell viability, are correlated with MYCN expression and are associated with poor prognosis. Thus, different histone demethylases with diverse functions are implicated in regulation of neuroblastoma cell survival. Although the molecular mechanisms of these histone demethylases in neuroblastoma need to be further studied, these data indicate that targeting these histone demethylases with pharmacological inhibitors may achieve therapeutic efficacy. For example, a recent study indicates that a non-specific KDM6B inhibitor, GSKJ4, suppresses Notch-driven T-ALL (40). A pan-KDM inhibitor, JIB-04, also shows antitumor activity (23).
In this study, we tried to identify novel inhibitors against histone demethylases, including KDM4B, to target the Myc pathway using a unique strategy. “Inverse gene expression profiling” resulting from pharmacological treatment suggest that cancer driver genes can be suppressed by the identified drugs, thereby providing a rationale for transcriptome-based drug screening (18). In addition, drug sensitivity screening has shown that gene transcriptional features show a correlation with drug sensitivity that is equal to or stronger than those observed with gene mutation (41). We exploited this strategy to identify small molecules whose pharmacological transcriptomes inversely matched advanced neuroblastoma and KDM4B signatures. Strikingly, we found that ciclopirox, originally designed as an antifungal drug, was a novel histone demethylase inhibitor that bound KDM4B and inhibited its activity. The biological profile of ciclopirox is very similar to JIB-04, a compound recently identified as a general KDM inhibitor (42). Importantly, CPX suppresses Myc function. A number of Myc-responsive genes were downregulated by ciclopirox that caused an increase in global histone methylation of H3K9me3/me2, characteristic of transcriptional repressive marks. In addition, NIH3T3 cells transduced with N-Myc were more sensitive to CPX than the GFP transduced control, indicating that KDM plays an important role in Myc reprogrammed cells. Intriguingly, CPX also significantly compromised oxidative phosphorylation by inhibiting mitochondrial gene expression. Depletion of KDM4B or N-Myc resulted in similar effects, consistent with the role of Myc in mitochondria biosynthesis (26), suggesting that CPX is able to epigenetically reprogram mitochondrial metabolism at least partially through KDM4B. Consistent with its in vitro activity, CPX inhibited neuroblastoma growth and metastasis in a disseminated disease model. Interestingly, recent drug screening also identified CPX as an effective agent for solid tumors, acute myeloid leukemia and myeloma (20, 43–45). In neuroblastoma, the dependency on multiple histone demethylases of cancer cells indicates that the pan-KDM inhibition may contribute to its overall anti-tumor effect.
The structure modeling and in vitro biochemical studies indicate that CPX inhibits JmjC histone demethylases through chelating the iron in the catalytic domain. Neuroblastoma cells have been shown to be very sensitive to the depletion of iron as shown with the treatment with iron chelator deferoxamine although the mechanism was not clear (46–48). Our studies may explain, at least in part, why depletion of iron leads to anti-neuroblastoma activity.
KDMs may directly target non-histone proteins such as p53(49). To test whether N-Myc is subject to lysine methylation which can be affected by CPX, we purified N-Myc protein for mass spectrometry analysis after cells treated with CPX. We were unable to detect any lysine methylation from N-Myc, but interestingly, we discovered two arginines (R138 and R160) were methylated. The biological functions of M-Myc arginine methylation in neuroblastoma need to be further studied. Nevertheless, CPX had no effect on these two sites.
In summary, we have demonstrated that KDMs play an important role in regulation of neuroblastoma cell survival. Using an informatics approach, we have identified a novel pan-histone demethylase inhibitor, ciclopirox, that targets the Myc pathway and cancer cell metabolism, hence achieving therapeutic efficacy. Unfortunately, the pharmacokinetics of CPX is unfavorable as it has a very short half-life in vivo (50). In the future, ciclopirox could be used as a scaffold to develop more potent KDM inhibitors for cancer therapeutics.
Supplementary Material
Acknowledgments
We are grateful to the staff of the Cell and Tissue Imaging Center and Hartwell Center for Bioinformatics and Biotechnology at St Jude Children’s Research Hospital for technical assistance. We are grateful to the Vector Core Lab at St Jude Children’s Research Hospital for making lentivirus vectors and the Animal Imaging Center for assistance with animal experiments.
Financial Support: This work was supported by the Assisi Foundation of Memphis, the US Public Health Service Childhood Solid Tumor Program Project Grant No. CA23099, the Cancer Center Support Grant No. 21766 from the National Cancer Institute, and by the American Lebanese Syrian Associated Charities (ALSAC) to Andrew M Davidoff.
References
- 1.Gustafson WC, Weiss WA. Myc proteins as therapeutic targets. Oncogene. 2010;29(9):1249–59. doi: 10.1038/onc.2009.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. Embo J. 1997;16(11):2985–95. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu S, Lee JS, Guo F, Shin J, Perez-Atayde AR, Kutok JL, et al. Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer Cell. 2012;21(3):362–73. doi: 10.1016/j.ccr.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45(3):279–84. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hojfeldt JW, Agger K, Helin K. Histone lysine demethylases as targets for anticancer therapy. Nat Rev Drug Discov. 2013;12(12):917–30. doi: 10.1038/nrd4154. [DOI] [PubMed] [Google Scholar]
- 6.Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–79. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 7.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48(4):491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–9. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amente S, Bertoni A, Morano A, Lania L, Avvedimento EV, Majello B. LSD1-mediated demethylation of histone H3 lysine 4 triggers Myc-induced transcription. Oncogene. 2010;29(25):3691–702. doi: 10.1038/onc.2010.120. [DOI] [PubMed] [Google Scholar]
- 10.Sankar S, Bell R, Stephens B, Zhuo R, Sharma S, Bearss DJ, et al. Mechanism and relevance of EWS/FLI-mediated transcriptional repression in Ewing sarcoma. Oncogene. 2013;32(42):5089–100. doi: 10.1038/onc.2012.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong PP, Miranda F, Chan KV, Berlato C, Hurst HC, Scibetta AG. Histone demethylase KDM5B collaborates with TFAP2C and Myc to repress the cell cycle inhibitor p21(cip) (CDKN1A) Mol Cell Biol. 2012;32(9):1633–44. doi: 10.1128/MCB.06373-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawazu M, Saso K, Tong KI, McQuire T, Goto K, Son DO, et al. Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS One. 2011;6(3):e17830. doi: 10.1371/journal.pone.0017830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rui L, Emre NC, Kruhlak MJ, Chung HJ, Steidl C, Slack G, et al. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell. 2010;18(6):590–605. doi: 10.1016/j.ccr.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J, AlTahan AM, Hu D, Wang Y, Cheng PH, Morton CL, et al. The role of histone demethylase KDM4B in Myc signaling in neuroblastoma. J Natl Cancer Inst. 2015;107(6):djv080. doi: 10.1093/jnci/djv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor MD, Poppleton H, Fuller C, Su X, Liu Y, Jensen P, et al. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell. 2005;8(4):323–35. doi: 10.1016/j.ccr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Ferrick DA, Neilson A, Beeson C. Advances in measuring cellular bioenergetics using extracellular flux. Drug Discov Today. 2008;13(5–6):268–74. doi: 10.1016/j.drudis.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 17.Morton CL, Papa RA, Lock RB, Houghton PJ. Preclinical chemotherapeutic tumor models of common childhood cancers: solid tumors, acute lymphoblastic leukemia, and disseminated neuroblastoma. Curr Protoc Pharmacol. 2007 doi: 10.1002/0471141755.ph1408s39. Chapter 14:Unit14 8. [DOI] [PubMed] [Google Scholar]
- 18.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–35. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 19.Minden MD, Hogge DE, Weir SJ, Kasper J, Webster DA, Patton L, et al. Oral ciclopirox olamine displays biological activity in a phase I study in patients with advanced hematologic malignancies. Am J Hematol. 2013 doi: 10.1002/ajh.23640. [DOI] [PubMed] [Google Scholar]
- 20.Eberhard Y, McDermott SP, Wang X, Gronda M, Venugopal A, Wood TE, et al. Chelation of intracellular iron with the antifungal agent ciclopirox olamine induces cell death in leukemia and myeloma cells. Blood. 2009;114(14):3064–73. doi: 10.1182/blood-2009-03-209965. [DOI] [PubMed] [Google Scholar]
- 21.Sinha KM, Yasuda H, Coombes MM, Dent SY, de Crombrugghe B. Regulation of the osteoblast-specific transcription factor Osterix by NO66, a Jumonji family histone demethylase. Embo J. 2010;29(1):68–79. doi: 10.1038/emboj.2009.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minden MD, Hogge DE, Weir SJ, Kasper J, Webster DA, Patton L, et al. Oral ciclopirox olamine displays biological activity in a phase I study in patients with advanced hematologic malignancies. Am J Hematol. 2014;89(4):363–8. doi: 10.1002/ajh.23640. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, Chang J, Varghese D, Dellinger M, Kumar S, Best AM, et al. A small molecule modulates Jumonji histone demethylase activity and selectively inhibits cancer growth. Nat Commun. 2013;4:2035. doi: 10.1038/ncomms3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23(24):2839–49. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23(24):2806–11. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res. 2009;15(21):6479–83. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O'Donnell KA, et al. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25(14):6225–34. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12(2):108–13. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrish F, Neretti N, Sedivy JM, Hockenbery DM. The oncogene c-Myc coordinates regulation of metabolic networks to enable rapid cell cycle entry. Cell Cycle. 2008;7(8):1054–66. doi: 10.4161/cc.7.8.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dickson PV, Hamner B, Ng CY, Hall MM, Zhou J, Hargrove PW, et al. In vivo bioluminescence imaging for early detection and monitoring of disease progression in a murine model of neuroblastoma. J Pediatr Surg. 2007;42(7):1172–9. doi: 10.1016/j.jpedsurg.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 31.Weir SJ, Patton L, Castle K, Rajewski L, Kasper J, Schimmer AD. The repositioning of the anti-fungal agent ciclopirox olamine as a novel therapeutic agent for the treatment of haematologic malignancy. Journal of clinical pharmacy and therapeutics. 2011;36(2):128–34. doi: 10.1111/j.1365-2710.2010.01172.x. [DOI] [PubMed] [Google Scholar]
- 32.Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall' Olio V, et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8(7):764–70. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- 33.Martinato F, Cesaroni M, Amati B, Guccione E. Analysis of Myc-induced histone modifications on target chromatin. PLoS One. 2008;3(11):e3650. doi: 10.1371/journal.pone.0003650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu CH, van Riggelen J, Yetil A, Fan AC, Bachireddy P, Felsher DW. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc Natl Acad Sci U S A. 2007;104(32):13028–33. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. Embo J. 2006;25(12):2723–34. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Das PP, Shao Z, Beyaz S, Apostolou E, Pinello L, De Los Angeles A, et al. Distinct and combinatorial functions of Jmjd2b/Kdm4b and Jmjd2c/Kdm4c in mouse embryonic stem cell identity. Mol Cell. 2014;53(1):32–48. doi: 10.1016/j.molcel.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schulte JH, Lim S, Schramm A, Friedrichs N, Koster J, Versteeg R, et al. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res. 2009;69(5):2065–71. doi: 10.1158/0008-5472.CAN-08-1735. [DOI] [PubMed] [Google Scholar]
- 38.Amente S, Milazzo G, Sorrentino MC, Ambrosio S, Di Palo G, Lania L, et al. Lysine-specific demethylase (LSD1/KDM1A) and MYCN cooperatively repress tumor suppressor genes in neuroblastoma. Oncotarget. 2015;6(16):14572–83. doi: 10.18632/oncotarget.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 2012;335(6066):348–53. doi: 10.1126/science.1212728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ntziachristos P, Tsirigos A, Welstead GG, Trimarchi T, Bakogianni S, Xu L, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature. 2014;514(7523):513–7. doi: 10.1038/nature13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483(7391):570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L, Chang J, Varghese D, Dellinger M, Kumar S, Best AM, et al. A small molecule modulates Jumonji histone demethylase activity and selectively inhibits cancer growth. Nat Commun. 2013;4:2035. doi: 10.1038/ncomms3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim Y, Schmidt M, Endo T, Lu D, Carson D, Schmidt-Wolf IG. Targeting the Wnt/beta-catenin pathway with the antifungal agent ciclopirox olamine in a murine myeloma model. In Vivo. 2011;25(6):887–93. [PubMed] [Google Scholar]
- 44.Song S, Christova T, Perusini S, Alizadeh S, Bao RY, Miller BW, et al. Wnt inhibitor screen reveals iron dependence of {beta}-catenin signaling in cancers. Cancer Res. 2011 doi: 10.1158/0008-5472.CAN-11-2745. [DOI] [PubMed] [Google Scholar]
- 45.Zhou H, Shen T, Luo Y, Liu L, Chen W, Xu B, et al. The antitumor activity of the fungicide ciclopirox. Int J Cancer. 2010;127(10):2467–77. doi: 10.1002/ijc.25255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Becton DL, Bryles P. Deferoxamine inhibition of human neuroblastoma viability and proliferation. Cancer Res. 1988;48(24 Pt 1):7189–92. [PubMed] [Google Scholar]
- 47.Blatt J, Stitely S. Antineuroblastoma activity of desferoxamine in human cell lines. Cancer Res. 1987;47(7):1749–50. [PubMed] [Google Scholar]
- 48.Richardson DR, Ponka P. The iron metabolism of the human neuroblastoma cell: lack of relationship between the efficacy of iron chelation and the inhibition of DNA synthesis. J Lab Clin Med. 1994;124(5):660–71. [PubMed] [Google Scholar]
- 49.Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449(7158):105–8. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 50.Minden MD, Hogge DE, Weir SJ, Kasper J, Webster DA, Patton L, et al. Oral ciclopirox olamine displays biological activity in a phase I study in patients with advanced hematologic malignancies. Am J Hematol. 2014;89(4):363–8. doi: 10.1002/ajh.23640. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.