Abstract
Bruton tyrosine kinase (BTK) links the B-cell antigen receptor (BCR) and Toll-like receptors with Nuclear factor kappa B (NF-κB). The role of BTK in primary CNS lymphoma (PCNSL) is unknown. We performed a Phase 1 clinical trial with ibrutinib, the first-in-class BTK inhibitor, for patients with relapsed or refractory CNS Lymphoma. Clinical responses to ibrutinib occurred in 10/13 (77%) PCNSL patients, including five complete responses. The only PCNSL with complete ibrutinib resistance harbored a mutation within the coiled-coil domain of Caspase Recruitment Domain Family Member 11, a known ibrutinib resistance mechanism. Incomplete tumor responses were associated with mutations in the B-Cell Antigen Receptor-associated protein CD79B. CD79B-mutant PCNSLs showed enrichment of mammalian target of rapamycin (mTOR)-related gene sets and increased staining with Phosphatidylinositol 3-kinase (PI3K)/mTOR activation markers. Inhibition of the PI3K-isoforms p110α/p110δ or mTOR synergized with ibrutinib to induce cell death in CD79B-mutant PCNSL cells.
INTRODUCTION
Diffuse large-B-cell lymphoma (DLBCL) represents the most common type of malignant lymphoma, accounting for 30% – 40% of newly diagnosed adults cases. Central nervous system (CNS) involvement, or “secondary” CNS lymphoma (SCNSL), occurs in a subset of DLBCL patients. Primary CNS lymphoma (PCNSL) is an aggressive lymphoma manifesting exclusively in the CNS. Almost all PCNSLs are DLBCLs. Many patients with CNS lymphoma suffer from disease recurrence following initial tumor therapy. Other patients have refractory disease that fails to respond to first-line therapy. Patients with relapsed or refractory (r/r) CNS lymphoma respond poorly to currently available therapies (1–3).
Ibrutinib is a first-in-class, oral inhibitor of Bruton’s tyrosine kinase (BTK). BTK integrates B-cell antigen receptor (BCR) and Toll-like receptor (TLR)(4) signaling. Genes encoding members of these pathways frequently harbor mutations in DLBCL. These include the BCR-associated protein CD79B (5) and myeloid differentiation primary response 88 (MYD88)(6), a cytosolic adapter protein that links interleukin-1 and TLRs with the Nuclear factor kappa B (NF-κB)(7). Activating mutations in MYD88 and CD79B have also been reported in PCNSL (8–12). Ibrutinib induced death of DLBCL cells with deregulated BCR signaling (5) and showed promising activity in a Phase 1 trial of patients with a variety of B-cell malignancies (13). Subsequent clinical trials reported 70–90% response rates to single-agent ibrutinib in patients with Chronic Lymphocytic Leukemia (CLL) and Small Lymphocytic Lymphoma (14), Mantle-Cell Lymphoma (MCL)(15), and Waldenström Macroglobulinemia (WM)(16). Response rates were considerably lower (~ 25%) in patients with r/r systemic DLBCL (17). Burkitt’s lymphoma cells, which are derived from germinal centre B cells, do not require BTK for survival (4,18).
The goals of the current study were to evaluate the tolerability of ibrutinib in patients with recurrent or refractory (r/r) CNS lymphoma, assess drug concentrations in cerebrospinal fluid (CSF), determine overall response rates, and explore molecular determinants of treatment response.
RESULTS
Study Design and Patient Demographics
This open-label, non-randomized, single center, dose escalation study was designed to establish the MTD of single-agent ibrutinib in r/r PCNSL/SCNSL. The defined MTD was used in an expansion cohort to further assess toxicity and clinical activity (NCT02315326). We explored drug doses above the recommended Phase 2 dose of 560 mg daily because plasma levels of ibrutinib have been reported to increase proportionally from 420 to 840 mg per day and because higher doses of ibrutinib have been administered in prior studies without reaching a maximum tolerated dose (MTD).
The primary end-points were safety of ibrutinib in CNS lymphoma and overall response rate (ORR) defined as complete and partial responders. The secondary end points were progression-free survival (PFS) and pharmacokinetics. Ibrutinib was administered continuously until disease progression, intolerable toxicity or death. The starting dose was 560mg/day. Dose escalation among cohorts followed the "3+3" design and was allowed if, after 28 days of therapy, none of three or one of six patients had a DLT. Plasma and CSF samples were collected two hours after ibrutinib dosing on day 1 (cycle 1, day 1) and day 29 (cycle 2, day 1).
Twenty eligible patients (Table 1) with r/r CNS lymphoma were enrolled. Median age was 69 years (range, 21– 85). Twelve were women. The median ECOG score was 1 (range, 0–2). Thirteen had PCNSL and 7 had SCNSL; 14 patients had recurrent and 6 refractory disease. Seventeen had parenchymal brain lesions, three isolated CSF involvement, and four both. Median number of prior therapies was two (range, 1–8), including methotrexate (MTX) chemotherapy (100%), radiotherapy (15%), and hematopoietic cell transplantation (15%). Eight patients had failed prior MTX-based salvage therapy, currently the most effective therapy for recurrent CNS lymphoma (19). Three patients received 560mg ibrutinib and 13 patients received 840mg (Supplementary Tables S1/S2).
Table 1.
Baseline Characteristics of Patients (n=20)
| Characteristics | All (n=20) | PCNSL (n=13) | SCNSL (n=7) |
|---|---|---|---|
| Age - years | No. | No. | No. |
| Median | 69 | 69 | 61 |
| Range | 21–85 | 60–80 | 21–85 |
| Gender | no (%) | no (%) | no (%) |
| Male | 8 (40) | 5 (38) | 3 (43) |
| Female | 12 (60) | 8 (62) | 4 (57) |
| ECOG | No. | No. | No. |
| Median | 1 | 1 | 1 |
| Range | 0–2 | 0–2 | 0–2 |
| CNS Lymphoma | No. (%) | No. (%) | No. (%) |
| Primary (PCNSL) | 13 (65) | 13 (100) | n/a |
| Secondary (SCNSL) | 7 (35) | n/a | 7 (100) |
| Disease Status | No. (%) | No. (%) | No. (%) |
| Recurrent | 14 (70) | 10 (77) | 4 (57) |
| Refractory | 6 (30) | 3 (23) | 3 (43) |
| CNS Involvement | No. (%) | No. (%) | No. (%) |
| Brain | 13 (65) | 10 (77) | 3 (43) |
| Cerebrospinal fluid (CSF) | 3 (15) | 2 (15) | 1 (14) |
| Brain and CSF | 4 (20) | 1 (8) | 3 (43) |
| Prior Treatment | No. (%) | No. (%) | No. (%) |
| Chemotherapy | 20 (100) | 13 (100) | 7 (100) |
| Radiation | 3 (15) | 2 (15) | 1 (14) |
| Stem cell transplant | 3 (15) | 0 (0) | 3 (43) |
| Number of Prior Regimens | No. | No. | No. |
| Median | 2 | 2 | 2 |
| Range | 1–8 | 1–8 | 1–3 |
ECOG PS: Eastern Cooperative Oncology Group Performance Status, CNS: Central Nervous System, PCNSL: Primary Central Nervous System Lymphoma, SCNSL: Secondary Central Nervous System Lymphoma
Ibrutinib Tolerability
Ibrutinib was tolerated with manageable adverse events (Table 2). No dose-limiting toxicity occurred during the dose-escalation portion of the study. The most common adverse events were hyperglycemia, thrombocytopenia, anemia, and hypertriglyceridemia. In the dose escalation cohort (n=10), three patients received 560mg and experienced one grade 4 neutropenia (4 months after drug initiation), one grade 3 thrombocytopenia, and one grade 3 ALT elevation. Seven patients received 840mg and developed one grade 4 lymphopenia and seven grade 3 toxicities, all except one (grade 3) occurring in the same patient (#5). In the expansion cohort (n=10), ten patients received 840mg. Four grade 4 toxicities (neutropenia (2), sepsis, lymphopenia) were observed in four patients and resolved after drug was held. Fifteen grade 3 toxicities were observed in six patients. Treatment was discontinued due to a fungal infection (grade 3) in one patient with MTX-refractory disease (patient #11) and chronic corticosteroids treatment for 17 weeks prior to enrollment. The patient developed pulmonary aspergillosis six weeks after initiating ibrutinib. New ring-enhancing lesions were detected on routine response evaluation, but CSF fungal and bacterial cultures were negative. Ibrutinib was held during antifungal therapy and the patient succumbed to disease progression 77 days after the last dose of ibrutinib. This was the only patient where treatment was discontinued due to toxicity. The ibrutinib dose was reduced in one patient (patient #13) with colitis from 840 to 560mg.
Table 2.
Treatment related adverse events
| Adverse Event* | Grade 1–2 | Grade 3–4 | Total |
|---|---|---|---|
| Number of patients (percent) | |||
| Hyperglycemia | 16 (80) | 3 (15) | 19 (95) |
|
| |||
| Thrombocytopenia | 12 (60) | 2 (10) | 14 (70) |
|
| |||
| Anemia | 13 (65) | 1 (5) | 14 (70) |
|
| |||
| Hypertriglyceridemia | 13 (65) | 1 (5) | 14 (70) |
|
| |||
| Cholesterol high | 12 (60) | 0 | 12 (60) |
|
| |||
| Alanine aminotransferase increased | 8 (40) | 2 (10) | 10 (50) |
|
| |||
| Hypoalbuminemia | 10 (50) | 0 | 10 (50) |
|
| |||
| Hyponatremia | 7 (35) | 1 (5) | 8 (40) |
|
| |||
| Aspartate aminotransferase increased | 8 (40) | 0 | 8 (40) |
|
| |||
| White blood cell decreased | 4 (20) | 2 (10) | 6 (30) |
|
| |||
| Hypocalcemia | 5 (25) | 1 (5) | 6 (30) |
|
| |||
| Activated partial thromboplastin time prolonged | 4 (20) | 2 (10) | 6 (30) |
|
| |||
| Alkaline phosphatase increased | 6 (30) | 0 | 6 (30) |
|
| |||
| Creatinine increased | 6 (30) | 0 | 6 (30) |
|
| |||
| Neutropenia | 2 (10) | 3 (15) | 5 (25) |
|
| |||
| Blood bilirubin increased | 4 (20) | 1 (5) | 5 (25) |
|
| |||
| Diarrhea | 5 (25) | 0 | 5 (25) |
|
| |||
| Lymphopenia | 1 (5) | 4 (20) | 5 (25) |
|
| |||
| Edema limbs | 4 (20) | 0 | 4 (20) |
|
| |||
| Mucositis | 4 (20) | 0 | 4 (20) |
|
| |||
| Cramping | 4 (20) | 0 | 4 (20) |
|
| |||
| Headache | 3 (15) | 0 | 3 (15) |
|
| |||
| Urinary tract infection | 2 (10) | 1(5) | 3 (15) |
|
| |||
| Anorexia | 2 (10) | 0 | 2 (10) |
|
| |||
| Epistaxis | 2 (10) | 0 | 2 (10) |
|
| |||
| Hyperkalemia | 2 (10) | 0 | 2 (10) |
|
| |||
| Hypernatremia | 2 (10) | 0 | 2 (10) |
|
| |||
| Sinusitis | 2 (10) | 0 | 2 (10) |
|
| |||
| Lung infection | 0 | 2 (10) | 2 (10) |
|
| |||
| Rash | 2 (10) | 0 | 2 (10) |
|
| |||
| Colitis | 0 | 1 (5) | 1 (5) |
|
| |||
| Encephalitis | 0 (0) | 1 (5) | 1 (5) |
|
| |||
| Febrile neutropenia | 0 | 1 (5) | 1 (5) |
|
| |||
| Sepsis | 0 | 1 (5) | 1 (5) |
National Cancer Institute Common Terminology Criteria for Adverse Events, Version 4.0
Ibrutinib concentration in CSF
We measured CSF ibrutinib concentrations two hours post-dose in 18/20 patients and 14/20 patients at day 1 and day 29, respectively. The two hour time point was chosen based on the reported median time of ibrutinib peak plasma concentrations (14). CSF was collected from an Ommaya reservoir for patient #1, #3, and #18. CSF was collected through lumbar puncture in all other patients. Mean CSF ibrutinib concentration was 0.77ng/ml (1.7nM) and 1.95ng/ml (4.4nM) in patients receiving 560mg and 840mg, respectively with a trend to higher CSF concentrations after one month of therapy (1.65ng/ml and 3.18ng/ml for patients receiving 560mg and 840mg respectively)(Supplementary Fig. S1).
Frequent Tumor Responses to Ibrutinib
Twelve of 13 PCNSL patients were evaluated for response. One patient (#7) was not evaluable because drug was discontinued within 14 days of treatment due to personal choice. 10/13 (77%)(95% CI: 56%– 94%) patients showed a clinical response, including five patients with a complete response (CR) and five patients with a partial responses (PR). One additional patient (#19) experienced tumor regression that did not meet criteria for PR (Fig. 1A and B). All three patients with malignant cells in the CSF at study entry showed no malignant cells on follow-up evaluation. At a median follow-up of 479 days (range, 354–739), two patients are still receiving treatment (Fig. 1C). Both had a CR as their best response. The median progression-free survival (PFS) was 4.6 months (95% CI: 2.4–7.5 months). The median overall survival (OS) was 15 months (7/13 subjects still alive).
Figure 1. Clinical Response to Ibrutinib in recurrent/refractory CNS Lymphoma.
A, Representative tumor response to ibrutinib as determined by magnetic resonance imaging (MRI) T1+contrast sequences in a Primary CNS Lymphoma (PCNSL; patient #3). B, Best response to ibrutinib in PCNSL patients, assessed using International PCNSL Collaborative Group (IPCG) guidelines (47). Displayed is the change in target lesion diameter from baseline (%) by magnetic resonance imaging or clearance of cerebrospinal fluid; negative values indicate tumor shrinkage. Shown are subjects who underwent at least 15 days of drug treatment and one post-treatment evaluation. Dashed lines indicate 25% change. Black: progression of disease (PD); orange: stable disease (SD); blue: partial response (PR), and green: complete response (CR). C, Progression-free survival in PCNSL. ➙, patient still receiving ibrutinib; #, drug discontinued because of personal choice (#4, #7) or infection (#11); ◆, progression. CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease; n/a, not assessable. Two patients withdrew from treatment despite clinical and radiographic response (#4, #7). Treatment was discontinued due to a fungal infection (grade 3) in one patient (#11) with MTX-refractory disease. The ibrutinib dose was reduced in one patient with colitis (#13) from 840 to 560mg. D, Representative tumor response to ibrutinib in a Secondary CNS Lymphoma (SCNSL; patient #15). E, Best response to ibrutinib in SCNSL. Shown are subjects who underwent at least 15 days of drug treatment and one post-treatment evaluation. Dashed lines indicate 25% change. Black: progression of disease (PD); orange: stable disease (SD); blue: partial response (PR), and green: complete response (CR). F, Progression-free survival in SCNSL. ➙, patient still receiving ibrutinib; *, hematopoietic stem cell transplantation (in CR); ◆, progression. CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease; n/a, not assessable.
Five of seven (71%) SCNSL patients showed a clinical response, including 4 CRs (Fig. 1D and E). One patient is still receiving treatment and two patients were treated with high-dose chemotherapy and stem cell rescue after achieving a CR on single-agent ibrutinib. Four patients had CSF disease at enrollment. In 3/4 patients, malignant cells were no longer detectable on follow-up evaluation on ibrutinib. The median PFS in SCNSLs was 7.43 months with a median OS that has not been reached yet (5/7 subjects still alive) (Fig. 1F).
Of the ten patients requiring corticosteroids to control neurologic symptoms at enrollment, six (60%) were successfully tapered off corticosteroids after initiating ibrutinib. None of the complete responses to single-agent ibrutinib in PCNSL could be attributed to concurrent corticosteroid use (Supplementary Table S3).
Genomic Landscape of PCNSL
The antitumor activity of ibrutinib in r/r PCNSL was considerably greater than reported for patients with r/r DLBCL outside the CNS (overall response rate/ORR 25% and median OS 6.4 months)(17). To explore genetic differences between these two clinically distinct DLBCL entities, we examined the coding regions of 586 cancer-associated genes in 177 archival human PCNSL biopsies and compared our findings to the genomic landscape of DLBCL outside the CNS, as reported in prior studies (20–22).
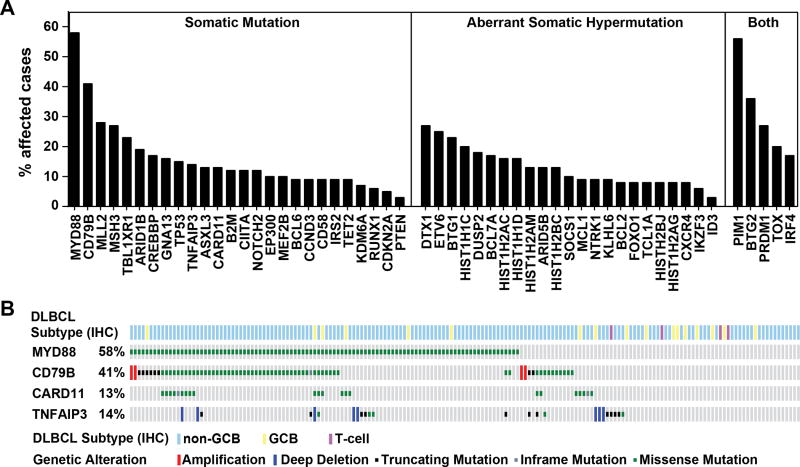
Twenty-six genes were recurrently mutated in PCNSL (Fig. 2A). Mutations in 23 additional genes could be attributed to aberrant somatic hypermutation (aSHM), the result of malfunctioning of the physiologic somatic hypermutation process (23). Five genes (PIM1, BTG2, PRDM1, TOX, and IRF4) scored as both recurrent mutations and targets of aSHM (Supplementary Table S4 and S5). Mutations in the Toll/IL-1 receptor (TIR) domain of the cytosolic adapter protein myeloid differentiation primary response 88 (MYD88) and the immunoreceptor tyrosine-based activation motif (ITAM) of the B-Cell Antigen Receptor-associated protein CD79B occurred in 58% and 41 % of PCNSLs, a frequency that is consistent with prior PCNSL studies and considerably higher than reported for DLBCL outside the CNS (Supplementary Table S6). In contrast, we observed similar mutation frequencies for all other examined genes, including genes involved in immune escape (B2M, CD58), DNA and histone modifications (MLL2, CREBBP), and B cell differentiation (PRDM1)(Supplementary Fig. S2).
Figure 2. Genomic Landscape of PCNSL.
A, Shown are mutation frequencies in PCNSLs (n=177), grouped into genes with recurrent somatic mutations, genes affected by aberrant somatic hypermutation, and genes meeting both criteria (see Supplementary Table S5). B, Relationship between PCNSL disease subtypes and mutations in BCR pathway members in PCNSL. The disease subtype is shown in the top row and was determined by IHC (GCB: germinal center B-cell like; non-GCB: non-germinal center B-cell like; T-cell). Missense mutations are displayed in green, in-frame mutations in grey, truncating mutations in black, amplifications in red and deletions in blue.
At the molecular level, DLBCL outside the CNS has been further classified into disease subgroups (24). Mutations in BCR pathway members have been shown to be enriched in the activated B-cell–like (ABC) DLBCL subgroup, but rarely found in the germinal center B-cell–like (GCB) subgroup (5,6,25–27). We examined in PCNSL the association between disease subgroup and mutations in the four BCR-NF-κB pathway members MYD88, CD79B, Caspase Recruitment Domain Family Member 11 (CARD11) and Tumor Necrosis Factor Alpha-Induced Protein 3 (TNFAIP3, also known as A20). Unlike the more balanced distribution of disease subgroups in DLBCL outside the CNS, the vast majority of PCNSLs (157/177) belonged to the non-germinal center (non-GCB) DLBCL subgroup (Supplementary Fig. S3) and we identified mutations in BCR-NF-κB pathway members in both GCB and non-GCB tumors (Fig. 2B).
Genomic Determinants of Ibrutinib Response in PCNSL
Our clinical trial findings and genomic analysis suggested that BTK dependence and BCR pathway mutations are distinguishing features of PCNSL compared to DLBCL outside the CNS. We therefore examined the relationship between BCR pathway mutations and clinical ibrutinib response in pretreatment tumor biopsies from our clinical trial patients. We observed a missense mutation within the coiled-coil domain of CARD11 (R179Q) in the only PCNSL patient with complete ibrutinib resistance (#5). Mutations in the coiled-coil domain of CARD11 have been shown to promote BTK-independent activation of NF-κB (25) and have been identified in patients with clinical ibrutinib resistance in DLBCL outside the CNS and in Mantle-Cell Lymphoma (17,28). Three other tumors with incomplete ibrutinib responsiveness showed a mutation in CARD11 (R337Q) or inactivating lesions in TNFAIP3 (deletion, frameshift mutation), a negative regulator of NF-κB (Table 3). Surprisingly, none of the PCNSLs with concurrent mutations in MYD88 and CD79B, a particularly common event in PCNSL (see Fig. 2B), showed a complete response to ibrutinib.
Table 3.
Molecular Determinants for Clinical Response to Ibrutinib
| ID | Disease | Best Response |
COO | BCR Pathway Mutations | ||||
|---|---|---|---|---|---|---|---|---|
| MYD88 | CD79B | CARD11 | TNFAIP3 | |||||
| #12 | PCNSL | CR | NGC | L265P | WT | WT | WT | |
| #4 | PCNSL | CR | NGC | L265P | WT | WT | WT | |
| #17 | PCNSL | CR | NGC | WT | WT | WT | WT | |
| #1 | PCNSL | CR | n/a | n/a | ||||
| #6 | PCNSL | CR | n/a | n/a | ||||
| #10 | PCNSL | PR | NGC | L265P | Y196H | WT | WT | |
| #11 | PCNSL | PR | GCB | L265P | Y196D | WT | L324Qfs*7 | |
| #20 | PCNSL | PR | NGC | L265P | Y196D | WT | DEL | |
| #16 | PCNSL | PR | NGC | V217F | A43V/M164I | R337Q | WT | |
| #3 | PCNSL | PR | NGC | M232T | Y196C | S622del | DEL | |
| #19 | PCNSL | SD | NGC | L265P | Y196S | WT | WT | |
| #5 | PCNSL | PD | NGC | WT | WT | R179Q | WT | |
| #7 | PCNSL | - | NGC | WT | WT | WT | WT | |
COO, cell-of-origin (as determined by immunohistochemistry); NGC, non-germinal center subtype; GCB, germinal center subtype; n/a, not assessed due to lack of tissue; WT, wild-type; DEL, deletion; CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease; - , not assessable.
PI3K-mTOR axis promotes survival in CD79B-mutant PCNSL
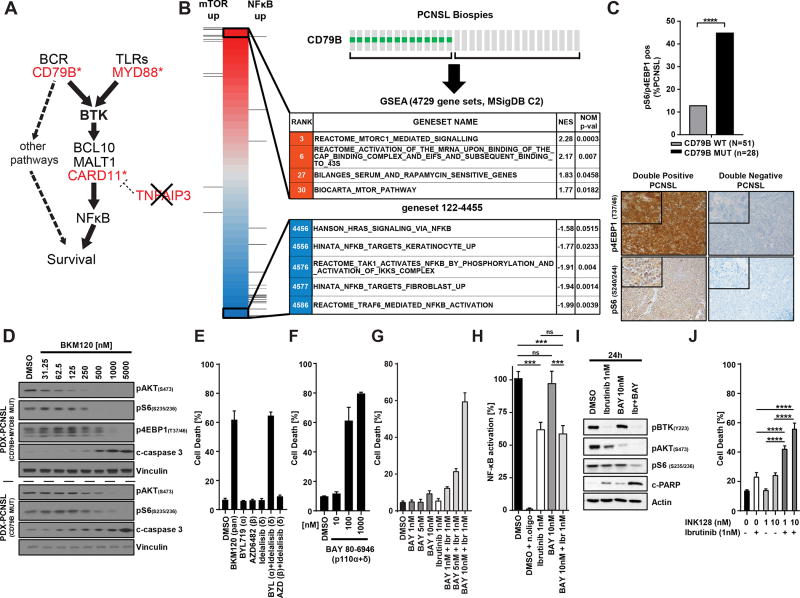
Mutations in the first tyrosine (Y196D) of the immunoreceptor tyrosine-based activation motif (ITAM) of CD79B have been shown to impair BCR downregulation (5). We hypothesized that these mutations might attenuate BTK dependence by diversifying BCR signal output and providing a BTK-independent survival signal (Fig. 3A). To identify such signals, we isolated RNA from PCNSL biopsies with known CD79B status and compared the transcriptomes of CD79B-mutant and CD79B-wildtype PCNSLs using gene-set enrichment analysis (Broad MSigDB C2 pathways containing 4929 gene sets). Multiple gene sets associated with activated mTOR signaling were among the most highly enriched pathways in CD79B-mutant PCNSLs whereas gene sets associated with activated NF-κB were negatively enriched (Fig. 3B and Supplementary Table S7). We also stained CD79B-mutant and CD79B-wildtype PCNSLs with antibodies against activated forms of the two PI3K/mTOR pathway components eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1, Threonine 37/46) and S6 Ribosomal Protein (S6RP, Serine 240). A significantly higher fraction of CD79B-mutant than CD79B-wildtype PCNSLs stained with both antibodies (Fig. 3C). We also derived xenograft models (PDX) from two CD79B-mutant PCNSL biopsies. In both orthotopic PCNSL models, we observed diffuse infiltration of the mouse brain by CD20-positive tumor cells which stained briskly with antibodies against phosphorylated forms of S6RP and 4E-BP1 (Supplementary Fig. S4). Treatment of slice cultures from these xenograft tumors with the PI3K inhibitor BKM120 (29) induced cell death at concentrations that inhibited phosphorylation of the serine/threonine kinase AKT (Serine 473) and the mTOR targets S6RP and 4E-BP1 (Fig. 3D). Taken together, these data indicate that the PI3K/mTOR-signaling axis is constitutively activated and promotes survival in CD79B-mutant PCNSL.
Figure 3. PI3K/mTOR pathway promotes survival in CD79B-mutant PCNSL.
A, Cartoon of the BCR/NFκB signaling axis. Genes harboring mutations in PCNSL are highlighted in red. B, Gene-set enrichment analysis (GSEA) of PCNSL biopsies shows enrichment of mTOR related gene sets in CD79B-mutated PCNSL. C, Activation of PI3K/mTOR in CD79B-mutant PCNSL. PCNSL tissue was stained with antibodies against 4EBP1 (T37/46) and S6 Ribosomal protein (S240/244). Tumors staining with both antibodies, labeled as “double positive”, were more common in CD79B-mutated PCNSL than in CD79B-wildtype PCNSL. (see bargraph on top ****: p<0.0001). The images below the bargraph show representative IHC images for a “double-positive” (left column) and “double-negative” (right column) PCNSL. D, BKM120, a pan-class I PI3K inhibitor, induces cell death in slice cultures from two CD79B-mutant PCNSL xenograft models. Shown are Western Blots of whole cell lysates after incubation for 24 hours with the indicated concentrations of BKM120. E, Combination of the PI3Kα-specific inhibitor BYL719 and the PI3Kδ-specific inhibitor Idelalisib, but neither inhibitor alone, induces cell death in a CD79B-mutant PCNSL cell line. F, The PI3Kα/δ specific inhibitor BAY 80-6946 induces cell death in PCNSL-MSK cells. G, Synergistic cell death induction by combination of ibrutnib and BAY 80-6946. H, Combination of ibrutnib and BAY 80-6946 is not associated with further NF-κB inhibition. n. oligo, oligonucleotides with the NF-κB binding domain are added and serve as a negative control for the assay. ***: p<0.001 I, Increased inhibition of the PI3K/mTOR pathway by combination of ibrutinib and BAY 80-6946; shown are Western Blots. J, Synergism between Ibrutinib and the dual mTOR inhibitor INK128 in PCNSL-MSK cells. ****: p<0.0001.
PI3K pathway activation in DLBCL occurs during both antigen-induced and antigen-independent (or “tonic”) BCR activation (30), but is particularly critical for survival during tonic BCR signaling (31). Class I PI3Ks are heterodimers that contain a p85 regulatory subunit and one of four catalytic subunits isoforms (class IA: p110α, p110β, p110δ; class IB: p110γ). While all catalytic isoforms mediate the same enzymatic reaction, they serve unique functions in specific cell types and signaling contexts (32,33). We examined the contribution of specific PI3K isoforms with PI3K isoform-selective inhibitors (34) in a CD79B-mutant PCNSL cell line derived from one of the two CD79B-mutant PCNSL PDX models (Supplementary Fig. S5). Unlike BKM120, which inhibits all class I PI3K isoforms, isoform-selective inhibitors of p110α (BYL719), p110β (AZD6482), or p110δ (idelalisib) did not induce cell death (Supplementary Fig. S6). In contrast, we observed robust cell death induction when we blocked the p110α and p110δ isoforms together, either using the combination of BYL719 and idelalisib (Fig. 3E) or the p110α/p110δ-inhibitor BAY80-6946 (Fig. 3F). These data indicate that p110α and p110δ PI3K isoforms provide redundant survival signals in CD79B-mutant PCNSL cells, reminiscent of the redundant pro-survival function of these two PI3K isoforms during tonic BCR signaling in normal B cells (35).
PI3K activates multiple downstream signaling pathways, including mTOR and BTK (36). We therefore wondered whether the survival signal mediated by PI3K p110α/p110δ was, at least in part, independent of BTK. Treatment of CD79B-mutant PCNSL cells with the BTK inhibitor ibrutinib resulted in dose-dependent reduction of BTK phosphorylation, NF-κB transcriptional activity, and tumor cell proliferation. However, we only observed cell death at ibrutinib concentrations around 10 nM (Supplementary Fig. S7), a concentration that was not achieved in cerebrospinal fluid for many PCNSL patients in our trial (see Supplementary Fig. S1). The combination of BAY80-6946 and ibrutinib, at concentrations at which neither drug induced cell death, resulted in synergistic cell death induction (Fig. 3G). Of note, the synergistic effect of the drug combination was not associated with further NF-κB inhibition (Fig. 3H), but more complete mTOR inhibition (Fig. 3I). We therefore also explored the combination of 1nM ibrutinib with the mTOR kinase inhibitor INK128 (37) and again observed synergistic cell death induction (Fig. 3J). Taken together, these results suggest that the survival signal provided by the PI3K/mTOR-axis is, at least in part, independent of BTK/NF-κB signaling.
DISCUSSION
Our study shows remarkable single-agent activity of ibrutinib in patients with recurrent or treatment-refractory PCNSL, including patients who had already failed prior MTX-based salvage therapy. Ibrutinib compared favorably to other therapies explored in this setting, including the mTOR inhibitor temsirolimus (PFS 2.1 months)(38), the combination of rituximab and temozolomide (PFS 1.6 months)(39), or topotecan (PFS 2 months)(40). While most PCNSL patients eventually developed resistance to ibrutinib, as seen with other kinase inhibitors in cancer (41), further evaluation of ibrutinib as part of combination therapies for PCNSL seems warranted. In contrast to a recent clinical trial that combined ibrutinib with temozolomide, etoposide, doxorubicin, dexamethasone, and rituximab for the treatment of PCNSL (42), we did not observe any treatment-related deaths and single-agent ibrutinib was tolerated with manageable toxicity in this heavily pretreated patient population.
Kinase inhibitors are typically less active against cancer in the CNS than outside the CNS. It is therefore remarkable that responses to ibrutinib were more frequent in r/r PCNSL (77% ORR) than the reported response rate in r/r DLBCL outside the CNS (25% ORR)(17). This observation points toward a divergent pathogenesis of these two related diseases, further supported by our genomic landscape analysis. The difference in intrinsic “BTK dependence” between PCNSL and DLBCL outside the CNS might be due to specific genetic alterations. MYD88 mutations, for example, have been linked to ibrutinib sensitivity in Waldenström Macroglobulinemia (16) and are significantly more common in PCNSL than in DLBCL outside the CNS. Alternatively, the brain microenvironment might augment BTK dependence of lymphoma cells through chronic antigen presentation and BCR activation (43,44), perhaps explaining the observed greater than expected response rate to ibrutinib in SCNSL patients. Further studies are needed to determine how genetic and tumor microenvironmental factors, alone or in combination, create intrinsic BTK dependence in different B cell malignancies (4,18). In the meantime, our data suggests that clinical trials with BTK inhibitors in recurrent/refractory CNS lymphoma should include patients whose tumors lack mutations in the B-cell receptor signaling axis.
Lastly, our study provides a first glimpse into genetic mechanisms of de-novo ibrutinib-resistance in PCNSL. The only PCNSL that was completely resistant to ibrutinib harbored a mutation in the coiled-coil domain of CARD11, a reported mechanism of ibrutinib resistance in other B cell malignancies (17,28). CD79B mutations, which are frequently associated with MYD88 mutations in PCNSL, appear to attenuate BTK “addiction” by providing a redundant survival signal, consistent with the recent observation of CD79B upregulation in ABC-DLBCL cell lines with reduced ibrutinib sensitivity (45). This resistance mechanism, while less dramatic than the effects of CARD11 mutations, is particularly relevant in the setting of incomplete BTK blockade. Our pharmacokinetic data suggests that clinically achievable ibrutinib concentrations in the CSF result in incomplete BTK inhibition. Direct measurement of BTK occupancy in CSF, if technically feasible, might provide further insight into this important question. Combination therapy of ibrutinib with inhibitors of p110α/p110δ PI3K or mTOR, which are in advanced stages of clinical testing, may offer a strategy to augment the depth and duration of ibrutinib responses in CD79B-mutant PCNSL and perhaps other DLBCL subgroups (46). Further studies are needed to evaluate if second generation BTK inhibitors with different pharmacokinetic properties and kinase selectivity might augment the remarkable clinical activity of ibrutinib in CNS lymphoma.
METHODS
Study oversight
Pharmacyclics LLC, an AbbVie company, provided the study drug. All authors collected the data and all the authors interpreted the results. This study was conducted with appropriate ethical approval. The protocol and its amendments were approved by the Institutional Review Board of Memorial Sloan Kettering Cancer Center. All participants provided written informed consent. This study was conducted in accordance with the Declaration of Helsinki and International Conference on Harmonization Guidelines for Good Clinical Practice. ClinicalTrials.gov: (NCT02315326) https://clinicaltrials.gov/ct2/show/NCT02315326
Eligibility
The trial population comprised patients with recurrent/refractory (r/r) PCNSL or SCNSL. All subjects had histopathologic confirmation of diagnosis. Patients met the following criteria: age ≥18; r/r disease on imaging or CSF; completion of at least one prior CNS-directed therapy; Eastern Cooperative Oncology Group (ECOG) performance status score of 0–2; adequate bone marrow and organ function; recovery to grade 1 toxicity from prior therapy. Patients with active non-CNS disease, prior ibrutinib therapy, or requiring >8mg of dexamethasone daily for neurologic disability were excluded.
Pharmacokinetic Evaluation
Plasma and CSF samples were collected two hours after ibrutinib dosing on day 1 (cycle 1, day 1) and day 29 (cycle 2, day 1). In patients with an Ommaya reservoir, plasma and CSF were collected 0, 1, 2, 3, 4, 6, and 24 hours after drug administration on day 1 (cycle 1, day 1) and 1, 2, 3, 4, and 6 hours after drug administration on day 29 (cycle 2, day 1).
Adverse Event Grading
Adverse events were graded using the NCI Common Terminology Criteria for Adverse Events (4.0). Dose limiting events (DLTs) were defined as any grade 4 hematologic toxicity, grade 3 febrile neutropenia and grade 3 thrombocytopenia associated with bleeding or any grade 3 non-hematologic toxicity that did not respond to supportive therapy and at least possibly related to treatment with ibrutinib.
Treatment Response Assessments
Evaluation of treatment response followed the International Primary CNS Lymphoma Collaborative Group guidelines (47).
Statistical Analysis
Descriptive statistics, including means, standard deviations, and medians for continuous variables and proportions for discrete variables, were used to summarize the findings in each of the defined cohorts. All analyses included patients who received the study drug. The ORR with a 95% confidence interval was calculated. The Kaplan–Meier method was used for time-to-event analysis. PFS was defined as time from ibrutinib start to progression, death or date of last follow-up scan with no documented progression. No imputation of missing values was performed. The primary objective of this phase I trial was to define the MTD of ibrutinib. Aim of the expansion portion was the assessment of ORR and progression-free survival at 24 weeks (PFS24w) rate of patients receiving ibrutinib. Patients from the expansion cohort as well as from the MTD were used to estimate the ORR and PFS rate. PFS24w was estimated along with a 95% confidence interval. PFS24w was defined as the percentage of patients that have not developed progression of disease or died without progression at 24 weeks from the start of treatment. All patients were followed for at least 24 weeks. Based on an accrual of 16 participants at the MTD the two-sided 95% binominal confidence intervals for a PFS24w of 60% (8/16 still on trial) were 24.1 and 75.4%. Additionally, if a complete response (CR) was seen in >1 patient, the investigational drug was deemed promising for additional clinical studies. Frequencies of single genes in the PCNSL and non-CNS DLBCL datasets were compared using the student T-test.
Immunohistochemistry (IHC)
Archival pretreatment tumor samples were obtained from patients participating in the clinical trial and from archival tissues. All samples were studied in accordance with a protocol approved by the Memorial Sloan Kettering Cancer Center Institutional Review Board. Formalin-fixed paraffin-embedded tissue from pre-treatment tumor biopsies from all study participants was used for IHC and sequencing. FFPE samples were sectioned at 5µm thickness. The first and last section was used for Hematoxylin and Eosin (H&E) staining to determine tumor content. All samples were reviewed by a neuropathologist (JTH) to confirm the histopathologic diagnosis of PCNSL.
We performed additional studies in archival tumor biopsies to evaluate the frequency of DLBCL subtypes and genetic alterations in a larger number of PCNSLs (n=177). These archival samples included tumors from MSK, Kaiser-Permanente Northern California, and the Radiation Therapy Oncology Group. Assignment to DLBCL subtype was based on established IHC criteria(48). To determine DLBCL subtype, FFPE tumor biopsies were stained with antibodies against CD10 (Vector Laboratories, Burlingame, CA), B-cell CLL/lymphoma 6 (BCL6)(GI191E/A8, Ventana/Roche), melanoma associated antigen (mutated) 1 (MUM1)(Dako/Agilent, Carpinteria, CA), phospho S6 Ribosomal protein (Ser 240/244)(Cell Signaling, Danverse, MA) and phospho 4EBP1 (Thr37/46)(Cell Signaling, Danverse, MA).
Genomic Analysis and in vitro experiments
Tumor biopsies were obtained before treatment from patients participating in the clinical trial and from archival tissues. All samples were studied in accordance with a protocol approved by the Memorial Sloan Kettering Cancer Center Institutional Review Board. Additional details on genomic analysis and in vitro experiments using PCNSL patient-derived xenograft models and cell line are available "online only".
Genomic analysis
All RNA and DNA sequencing of human tumor samples and Data Analysis was performed at Memorial Sloan Kettering Cancer Center. RNA was isolated from 31 primary CNS lymphoma samples and profiled on the Illumina Human HT12 V4 platform. To detect outliers, we used the detection p-value information of the raw data output and spearman correlation of each sample to a median pseudo-sample (49). Three samples with low detection and poor correlation to the median sample were excluded from further preprocessing and downstream analyses. The data were quantile normalized. A strong correlation was detected between the first principal component of the data and two annotation variables of presumed biological irrelevance: the time in paraffin and the source trial. This confounding signal was removed using the R language package “swamp” (https://cran.r-project.org/web/packages/swamp/index.html) prior to downstream analyses and visualization. We used the gene-set-enrichment analysis (GSEA) algorithm (50) and pathway annotation defined by the Molecular Signatures Database (MSigDB) C2 collection (version 2.5) of canonical signaling pathways, cellular processes, chemical and genetic perturbations, and human disease states(50) as previously described (51).
DNA isolation, sequencing library preparation and sequencing on a HiSeq 2500 was performed as described by Cheng DT et al. (52). Hybridization capture was preformed using the MSK-HemePACT targeted panel including the following 585 cancer genes (please see Supplementary Methods for complete gene list). All samples from study participants had a matching germline sample. The archival samples did not have a matching germline sample. Read alignment and processing were performed using standard parameters. The GATK tool was use for local realignment and quality score recalibration as of GATK best practices. Mutation analysis was performed using Mutect v 1.1.7(53) and GATK Haplotype Caller. Germline polymorphisms and sequencing artifacts were excluded by paired analysis if matched normal was available, and by filtering with a panel-of-normal samples. Known germline polymorphisms were excluded if present in the Exome Sequencing Project or at frequencies >1% in Exome Aggregation Consortium population databases. Initially, we eliminated known SNPs that are present in >1% of the population or a panel of normal controls and identified 5024 somatic mutations in 513 genes. We determined recurrently mutated genes using MutSig(54) and Oncodrive(55) algorithms. We used previously published criteria(56) to determine whether a mutation was the result of aberrant somatic hypermutation. DNA from a PCNSL cell line and patient primary tumor was used for array comparative genomic hybridization (array CGH) using a protocol provided by Agilent Technologies. aCGH data were analyzed and processed using Agilent Cytogenomics software version 1.5.
PCNSL Disease Models
Fresh samples were collected from patients undergoing diagnostic biopsy at MSKCC. The patient signed informed consent for use of tissue for research purposes under protocols approved by the Memorial Sloan Kettering (MSK) Cancer Center Institutional Review Board The samples were mechanically dissociated as well as enzymatically treated with Accumax (Innovative Cell Technologies). Cells were counted and checked for viability prior to injection into the striatum of SCID mice. At time of development of neurologic deficits, mice were sacrificed and brains were removed in a sterile fashion. 300µm thick tumor containing brain slices were generated by using a vibratome (Leica 1200 S). Tissue slices were cultured on organotypic inserts (Millipore). Organotypic inserts are Teflon membranes with 0.4-µm pores that allow preservation of 3D tissue structure in culture. Tissue culture was performed at 37 °C in a 5% CO2 humidified incubator using 1 ml of RPMI with 10% FBS (Omega Scientfic) and 100 U/mL penicillin/100 µg/mL streptomycin (Gemini Bio-Products). Tumor containing brain slices were treated with BKM120. DMSO was used in matched control samples. Tissue slices were harvested at 24h post-treatment and snap-frozen for western blot analysis. A PCNSL cell line (PCNSL-MSK) was derived from a patient derived xenograft model. PCNSL-MSK was cultured in RPMI, 5% fetal bovine serum and 1% penicillin/ streptomycin. NF-κB activity was measured in nuclear protein extracts by the TransAM NF-κB p65 protein assay (Active Motif, Carlsbad, CA, USA), an ELISA-based method to quantify NF-κB p65 subunit activation. The assay was performed according to the manufacturer's protocol and analyzed using a microplate absorbance reader. The BTK inhibitor ibrutinib and the PI3K pan and isoform inhibitors BKM120, BYL719, AZD6482, idelalisib, and BAY80-6946 were purchased from Selleck Chemicals. The cell lines Raji, Jurkat, SUDHL-4, SUDHL-5, SUDHL-6, SUDHL-10 were purchased from ATCC. The cell lines OCI-LY2, OCI-LY3, and OCI-LY13 were provided by Dr. Chaganti (MSKCC). All lymphoma cell lines were cultured in RPMI, 10% FBS. The cell line PCNSL#11 was generated from the orthotopic xenograft model PCNS#11 and maintained in RPMI and 10% FBS. Mouse B cells were isolated as previously described (57).
Immunoblotting
Snap-frozen tissues or cell culture cells were lysed and homogenized in 1% triton lysis buffer (#9803, Cell Signaling) containing fresh protease and phosphatase inhibitors by standard procedures. Protein concentrations were quantified with the BCA Protein Assay kit (Pierce Chemical Co.), and proteins were separated in a gradient (4–15%) SDSPAGE gel, transferred to nitrocellulose membranes, and hybridized with antibodies to the indicated antigens by standard procedures. Signals were detected by chemoluminescence using ECL detection reagents (Amersham Pharmacia Biotech). Primary antibodies to the following antigens were used: cleaved PARP (Asp214/D64E10 XP, #5625, Cell Signaling), cleaved caspase-3 (Asp175, #9661, Cell Signaling), Vinculin (clone hVIN-1, V9131, Sigma), BTK (#8546, Cell Signaling), phospho-BTK (#5083, Tyr223Y223, Cell Signaling), phospho-S6 Ribosomal protein (#2211, Ser 235/236, Cell Signaling), phospho-4EBP1 (#2855, Thr37/46, Cell Signaling) and phospho-AKT (#4060, Ser473, Cell Signaling).
Immunohistochemistry and computer-assisted image analysis of xenograft models
Paraffin-embedded sections of tumor xenografts were obtained at 5µm/slide. Antigen retrieval, immunohistochemical detection and counter staining were performed using the Ventana Discovery Ultra autostainer (Ventana) using primary antibodies against pS6 (S240/244), p4EBP1 (T37/46), and Ki67 (M7240, Dako). Digitized images were taken from representative areas within the xenografts.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
Ibrutinib has substantial activity in patients with relapsed or refractory B cell lymphoma of the CNS. Response rates in PCNSL were considerably higher than reported for DLBCL outside the CNS, suggesting a divergent molecular pathogenesis. Combined inhibition of BTK and PI3K/mTOR may augment the ibrutinib response in CD79B-mutant human PCNSLs.
Acknowledgments
This research was supported by grants from the National Institutes of Health (1R01NS080944–01 to I.K.M., P30-CA008748), the National Brain Tumor Society (I.K. Mellinghoff), the Geoffrey Beene Cancer Research Center (I.K. Mellinghoff), the Memorial Sloan Kettering Brain Tumor Center (C. Grommes), the Society of Memorial Sloan Kettering Cancer Center (C.G.), the American Brain Tumor Association Basic Research Fellowship Award (C.G.), the Lymphoma Research Foundation Career Development Award (C.G.), Susan and Peter Solomon Divisional Fund (C.G.), Cycle for Survival Equinox Innovation Award (C.G.), a MSK SPORE in Lymphoma (5 P50 CA 192937-02) Developmental Research Program (DRP) award (CG), a Mildred-Scheel Postdoctoral Research Fellowship of the Deutsche Krebshilfe e.V. (#111354) (A. P.), and the Robertson Foundation (N.S.). This research includes tissue contributed through a Radiation Therapy Oncology Group (RTOG) translational research program grant (TRP#176).
Footnotes
Conflict of interest: A. Noy reports receiving a commercial research grant and speakers bureau honoraria from Pharmacyclics. P.A. Hamlin reports receiving a commercial research grant from Janssen Pharmaceuticals. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Nogai H, Dorken B, Lenz G. Pathogenesis of non-Hodgkin's lymphoma. J Clin Oncol. 2011;29(14):1803–11. doi: 10.1200/JCO.2010.33.3252. [DOI] [PubMed] [Google Scholar]
- 2.Carnevale J, Rubenstein JL. The Challenge of Primary Central Nervous System Lymphoma. Hematol Oncol Clin North Am. 2016;30(6):1293–316. doi: 10.1016/j.hoc.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferreri AJ, Assanelli A, Crocchiolo R, Ciceri F. Central nervous system dissemination in immunocompetent patients with aggressive lymphomas: incidence, risk factors and therapeutic options. Hematol Oncol. 2009;27(2):61–70. doi: 10.1002/hon.881. [DOI] [PubMed] [Google Scholar]
- 4.Hendriks RW, Yuvaraj S, Kil LP. Targeting Bruton's tyrosine kinase in B cell malignancies. Nat Rev Cancer. 2014;14(4):219–32. doi: 10.1038/nrc3702. [DOI] [PubMed] [Google Scholar]
- 5.Davis RE, Ngo VN, Lenz G, Tolar P, Young RM, Romesser PB, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim KH, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470(7332):115–9. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gay NJ, Symmons MF, Gangloff M, Bryant CE. Assembly and localization of Toll-like receptor signalling complexes. Nat Rev Immunol. 2014;14(8):546–58. doi: 10.1038/nri3713. [DOI] [PubMed] [Google Scholar]
- 8.Montesinos-Rongen M, Godlewska E, Brunn A, Wiestler OD, Siebert R, Deckert M. Activating L265P mutations of the MYD88 gene are common in primary central nervous system lymphoma. Acta neuropathologica. 2011;122(6):791–2. doi: 10.1007/s00401-011-0891-2. [DOI] [PubMed] [Google Scholar]
- 9.Vater I, Montesinos-Rongen M, Schlesner M, Haake A, Purschke F, Sprute R, et al. The mutational pattern of primary lymphoma of the central nervous system determined by whole-exome sequencing. Leukemia. 2015;29(3):677–85. doi: 10.1038/leu.2014.264. [DOI] [PubMed] [Google Scholar]
- 10.Braggio E, McPhail ER, Macon W, Lopes MB, Schiff D, Law M, et al. Primary central nervous system lymphomas: a validation study of array-based comparative genomic hybridization in formalin-fixed paraffin-embedded tumor specimens. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17(13):4245–53. doi: 10.1158/1078-0432.CCR-11-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura T, Tateishi K, Niwa T, Matsushita Y, Tamura K, Kinoshita M, et al. Recurrent mutations of CD79B and MYD88 are the hallmark of primary central nervous system lymphomas. Neuropathology and applied neurobiology. 2016;42(3):279–90. doi: 10.1111/nan.12259. [DOI] [PubMed] [Google Scholar]
- 12.Montesinos-Rongen M, Schafer E, Siebert R, Deckert M. Genes regulating the B cell receptor pathway are recurrently mutated in primary central nervous system lymphoma. Acta neuropathologica. 2012;124(6):905–6. doi: 10.1007/s00401-012-1064-7. [DOI] [PubMed] [Google Scholar]
- 13.Advani RH, Buggy JJ, Sharman JP, Smith SM, Boyd TE, Grant B, et al. Bruton tyrosine kinase inhibitor ibrutinib (PCI-32765) has significant activity in patients with relapsed/refractory B-cell malignancies. J Clin Oncol. 2013;31(1):88–94. doi: 10.1200/JCO.2012.42.7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byrd JC, Furman RR, Coutre SE, Flinn IW, Burger JA, Blum KA, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. The New England journal of medicine. 2013;369(1):32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang ML, Rule S, Martin P, Goy A, Auer R, Kahl BS, et al. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. The New England journal of medicine. 2013;369(6):507–16. doi: 10.1056/NEJMoa1306220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Treon SP, Tripsas CK, Meid K, Warren D, Varma G, Green R, et al. Ibrutinib in previously treated Waldenstrom's macroglobulinemia. The New England journal of medicine. 2015;372(15):1430–40. doi: 10.1056/NEJMoa1501548. [DOI] [PubMed] [Google Scholar]
- 17.Wilson WH, Young RM, Schmitz R, Yang Y, Pittaluga S, Wright G, et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat Med. 2015;21(8):922–6. doi: 10.1038/nm.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young RM, Staudt LM. Targeting pathological B cell receptor signalling in lymphoid malignancies. Nature reviews Drug discovery. 2013;12(3):229–43. doi: 10.1038/nrd3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pentsova E, Deangelis LM, Omuro A. Methotrexate re-challenge for recurrent primary central nervous system lymphoma. Journal of neuro-oncology. 2014;117(1):161–5. doi: 10.1007/s11060-014-1370-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nature genetics. 2011;43(9):830–7. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morin RD, Mungall K, Pleasance E, Mungall AJ, Goya R, Huff RD, et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood. 2013;122(7):1256–65. doi: 10.1182/blood-2013-02-483727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(10):3879–84. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti RS, Kuppers R, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412(6844):341–6. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 24.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–11. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 25.Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319(5870):1676–9. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 26.Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459(7247):717–21. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kato M, Sanada M, Kato I, Sato Y, Takita J, Takeuchi K, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459(7247):712–6. doi: 10.1038/nature07969. [DOI] [PubMed] [Google Scholar]
- 28.Wu C, de Miranda NF, Chen L, Wasik AM, Mansouri L, Jurczak W, et al. Genetic heterogeneity in primary and relapsed mantle cell lymphomas: Impact of recurrent CARD11 mutations. Oncotarget. 2016;7(25):38180–90. doi: 10.18632/oncotarget.9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maira SM, Pecchi S, Huang A, Burger M, Knapp M, Sterker D, et al. Identification and characterization of NVP-BKM120, an orally available pan-class I PI3-kinase inhibitor. Mol Cancer Ther. 2012;11(2):317–28. doi: 10.1158/1535-7163.MCT-11-0474. [DOI] [PubMed] [Google Scholar]
- 30.Okkenhaug K, Vanhaesebroeck B. PI3K in lymphocyte development, differentiation and activation. Nature reviews Immunology. 2003;3(4):317–30. doi: 10.1038/nri1056. [DOI] [PubMed] [Google Scholar]
- 31.Srinivasan L, Sasaki Y, Calado DP, Zhang B, Paik JH, DePinho RA, et al. PI3 kinase signals BCR-dependent mature B cell survival. Cell. 2009;139(3):573–86. doi: 10.1016/j.cell.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol. 2010;11(5):329–41. doi: 10.1038/nrm2882. [DOI] [PubMed] [Google Scholar]
- 33.Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2015;15(1):7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yap TA, Bjerke L, Clarke PA, Workman P. Drugging PI3K in cancer: refining targets and therapeutic strategies. Curr Opin Pharmacol. 2015;23:98–107. doi: 10.1016/j.coph.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Limon JJ, Fruman DA. B cell receptor signaling: picky about PI3Ks. Science signaling. 2010;3(134):pe25. doi: 10.1126/scisignal.3134pe25. [DOI] [PubMed] [Google Scholar]
- 36.Lien EC, Dibble CC, Toker A. PI3K signaling in cancer: beyond AKT. Curr Opin Cell Biol. 2017;45:62–71. doi: 10.1016/j.ceb.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485(7396):55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korfel A, Schlegel U, Herrlinger U, Dreyling M, Schmidt C, von Baumgarten L, et al. Phase II Trial of Temsirolimus for Relapsed/Refractory Primary CNS Lymphoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2016;34(15):1757–63. doi: 10.1200/JCO.2015.64.9897. [DOI] [PubMed] [Google Scholar]
- 39.Nayak L, Abrey LE, Drappatz J, Gilbert MR, Reardon DA, Wen PY, et al. Multicenter phase II study of rituximab and temozolomide in recurrent primary central nervous system lymphoma. Leukemia & lymphoma. 2013;54(1):58–61. doi: 10.3109/10428194.2012.698736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischer L, Thiel E, Klasen HA, Birkmann J, Jahnke K, Martus P, et al. Prospective trial on topotecan salvage therapy in primary CNS lymphoma. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2006;17(7):1141–5. doi: 10.1093/annonc/mdl070. [DOI] [PubMed] [Google Scholar]
- 41.Chong CR, Janne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med. 2013;19(11):1389–400. doi: 10.1038/nm.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dunleavy K, Lai CE, Roschewski M, Brudno JN, Widemann B, Pittaluga S, et al. Phase I Study of Dose-Adjusted-Teddi-R with Ibrutinib in Untreated and Relapsed/Refractory Primary CNS Lymphoma. Blood. 2015;126(23) [Google Scholar]
- 43.Montesinos-Rongen M, Purschke FG, Brunn A, May C, Nordhoff E, Marcus K, et al. Primary Central Nervous System (CNS) Lymphoma B Cell Receptors Recognize CNS Proteins. J Immunol. 2015;195(3):1312–9. doi: 10.4049/jimmunol.1402341. [DOI] [PubMed] [Google Scholar]
- 44.Spies E, Fichtner M, Muller F, Krasemann S, Illerhaus G, Glatzel M, et al. Comment on "Primary Central Nervous System (CNS) Lymphoma B Cell Receptors Recognize CNS Proteins". J Immunol. 2015;195(10):4549–50. doi: 10.4049/jimmunol.1501964. [DOI] [PubMed] [Google Scholar]
- 45.Kim JH, Kim WS, Ryu K, Kim SJ, Park C. CD79B limits response of diffuse large B cell lymphoma to ibrutinib. Leukemia & lymphoma. 2016;57(6):1413–22. doi: 10.3109/10428194.2015.1113276. [DOI] [PubMed] [Google Scholar]
- 46.Paul J, Soujon M, Wengner AM, Zitzmann-Kolbe S, Sturz A, Haike K, et al. Simultaneous Inhibition of PI3Kdelta and PI3Kalpha Induces ABC-DLBCL Regression by Blocking BCR-Dependent and -Independent Activation of NF-kappaB and AKT. Cancer cell. 2017;31(1):64–78. doi: 10.1016/j.ccell.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 47.Abrey LE, Batchelor TT, Ferreri AJ, Gospodarowicz M, Pulczynski EJ, Zucca E, et al. Report of an international workshop to standardize baseline evaluation and response criteria for primary CNS lymphoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23(22):5034–43. doi: 10.1200/JCO.2005.13.524. [DOI] [PubMed] [Google Scholar]
- 48.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275–82. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 49.Hoshida Y, Villanueva A, Kobayashi M, Peix J, Chiang DY, Camargo A, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. The New England journal of medicine. 2008;359(19):1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palaskas N, Larson SM, Schultz N, Komisopoulou E, Wong J, Rohle D, et al. 18F-fluorodeoxy-glucose positron emission tomography marks MYC-overexpressing human basal-like breast cancers. Cancer Res. 2011;71(15):5164–74. doi: 10.1158/0008-5472.CAN-10-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. The Journal of molecular diagnostics : JMD. 2015;17(3):251–64. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature biotechnology. 2013;31(3):213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gonzalez-Perez A, Lopez-Bigas N. Functional impact bias reveals cancer drivers. Nucleic acids research. 2012;40(21):e169. doi: 10.1093/nar/gks743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khodabakhshi AH, Morin RD, Fejes AP, Mungall AJ, Mungall KL, Bolger-Munro M, et al. Recurrent targets of aberrant somatic hypermutation in lymphoma. Oncotarget. 2012;3(11):1308–19. doi: 10.18632/oncotarget.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harrow F, Ortiz BD. The TCRalpha locus control region specifies thymic, but not peripheral, patterns of TCRalpha gene expression. J Immunol. 2005;175(10):6659–67. doi: 10.4049/jimmunol.175.10.6659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.