Abstract
Cell-free circulating linear DNA is being explored for non-invasive diagnosis and management of tumors and fetuses, the so-called liquid biopsy. Previously, we observed the presence of small extrachromosomal circular DNA (eccDNA), called microDNA, in the nuclei of mammalian tissues and cell lines. Now, we demonstrate that cell-free microDNA derived from uniquely mapping regions of the genome is detectable in plasma and serum from both mice and humans and that they are significantly longer (30–60% >250 bases) than cell-free circulating linear DNA (~150 bases). Tumor-derived human microDNA is detected in the mouse circulation in a mouse xenograft model of human ovarian cancer. Comparing the microDNA from paired tumor and normal lung tissue specimens reveals that the tumors contain longer microDNA. Consistent with human cancers releasing microDNA into the circulation, serum and plasma samples (12 lung and 11 ovarian cancer) collected prior to surgery are enriched for longer cell free microDNA compared to samples from the same patient obtained several weeks after surgical resection of the tumor. Thus, circular DNA in the circulation is a previously unexplored pool of nucleic acids that could complement microRNAs (miRs) and linear DNA for diagnosis and for intercellular communication.
Keywords: Circular DNA, microDNA, Cell free DNA, Cancer, Biomarker
Introduction
Circulating nucleic acids are being investigated as minimally-invasive disease biomarkers for a patient-friendly method for the diagnosis and prognosis of diseases like cancer (1). The detection of linear fetal DNA in cell-free DNA (cfDNA) from the blood of a pregnant woman for identifying genetic abnormalities in the fetus has also been a success (1–3). Currently, the potential of linear cfDNA as a cancer biomarker is being explored (4). The technological advancement in high-throughput sequencing has provided an opportunity to study cfDNA in an unbiased genome-wide manner. cfDNA, present in normal individuals, are believed to be released into the blood after cell death (5). In cancer patients, cancer-specific cfDNA can be categorized and quantitated based on cancer-specific somatic mutations in oncogenes (5,6). In addition, methylation patterns in the cfDNA have been used to identify the epigenetic state, and thus the tissue of origin, of cfDNA (7,8). The cfDNA identified are usually of a size that is protected by a nucleosome (<166 bases) and therefore mostly likely the product of cell death followed by degradation of the chromatinized DNA to nucleosomes (9,10). Though majority of linear cfDNA are of mononucleosome size (11) a small proportion of fragments detected in the plasma, especially from patients with tumors, are of di- and tri-nucleosomal lengths (4).
Circulating nucleic acids also include RNA, particularly microRNAs, and in this case not only have they been explored for diagnosis (12), reviewed in (13)) but also implicated as potential messengers for intercellular communication (reviewed in (14))
The re-sequencing and comparison of DNA isolated from normal and tumor tissue by high-throughput sequencing has identified many tumor-specific somatic mutations in genomic DNA (15–17). Tumor cells die at a higher rate compared to normal cells due to uncontrolled cell growth and therefore fragments from the tumor genome are expected to be present in cfDNA (18,19). There are numerous reports using high-throughput sequencing to identify tumor-specific cfDNA present in the serum or plasma (reviewed in (4)). Furthermore, the concentration of cell-free circulating tumor DNA was positively correlated with tumor burden, suggesting measurement of the quantity of tumor-specific cfDNA could monitor disease progression (19–21).
However, these observations have been restricted to linear DNA. We recently reported the presence of naturally occurring small extrachromosomal circular DNA, microDNA, in the nuclei of cells (22,23). In this study, we examine the presence of circular microDNAs in cell-free circulating DNA. MicroDNA are present in both serum and plasma and their properties are very similar to the microDNA identified in various human and mouse tissues and cell lines. Most interesting, xenografts of human tumors release microDNA into the mouse circulation. Further, the longer microDNA in tumor versus paired normal lung tissue allowed us to ask whether human cancers in situ release microDNA into the circulation. Indeed, the microDNA in the serum/plasma of patients carrying tumors were often longer (in two third of patients) than that from the same patients following tumor resection, suggesting that microDNA are released from tumors to the circulation. Circular DNA is expected to be more stable compared to linear DNA molecules and this is the first evidence of such DNA being released from normal tissues and cancers into the circulation.
Materials and Methods
MicroDNA isolation and purification
Total circulating DNA was isolated from 1 mL of mouse serum (normal or SKOV3 xenograft) or 1 mL of human lung cancer patient serum or plasma using the Plasma/Serum Circulating DNA Purification Midi Kit (Norgen Biotek Corporation, Product # 51200). The DNA was eluted in 100 μL of elution buffer, after which the resulting DNA was digested with proteinase K (Ambion, AM2546), followed by Exonuclease VII (1U/μg extrachromosomal DNA, Epicentre, EN510100) to remove linear single-stranded DNA, another proteinase K digestion, ATP-dependent DNase (4U/μg extrachromosomal DNA, Epicentre, E3101K) to remove linear double-stranded DNA, and a final proteinase K digestion.
The starting volume of plasma or serum to isolate circular DNA from humans was 2 and 1 ml for ovarian and lung cancer sample respectively. The yields of DNA after DNase treatment were in the range of 27–231 ng, but (a) the Nanodrop method used to estimate these amounts is highly inaccurate in this range and (b) we cannot rule out contamination of the circulating circular DNA with linear DNA not digested completely by exonucleases. Thus, these measures of yield are highly inaccurate.
To ensure that we can isolate circulating microDNA from humans before proceeding to valuable patient-derived samples, 2 mL of plasma or serum from a healthy human volunteer was processed to yield DNase resistant microDNA. We performed Sanger sequencing on 12 clones from the rolling circle amplification products and confirmed that we can detect the tandem repeated sequences diagnostic of rolling circle amplicons derived from microDNA (Table S1).
Sample Information
Details of the patients whose serum or plasma was used in this study are given in Table S2 (lung cancer) and Table S3 (ovarian cancer). The detailed protocol for serum or plasma isolation is given in the supplementary method. This study was performed under approval of the IRB of the University of Virginia: protocol #11613 for ovarian cancer samples and #15204 for lung cancer samples
MicroDNA library preparation and sequencing
eccDNA was amplified using the REPLI-g Mini Kit (Qiagen, 150023), utilizing Multiple Displacement Amplification with random primers. EccDNA (≥ 20 ng) in 5 μL Buffer EB was denatured by incubation for 3 min with 5 μL Buffer D1, Denaturation was stopped by addition of 10 μL Buffer N1, after which 29 μL Repli-g Mini reaction buffer and 1 μL Repli-g Mini DNA polymerase were added and the reaction mixture incubated at 30°C for 16 hrs. After inactivating the polymerase at 65°C for 3 min, the buffer was exchanged using a Microcon YM-100 (Millipore, 42413) or DNA Fast Flow (Millipore, MRCF0R100) columns. 5 μg amplified DNA was used for paired-end library construction using the NEBNext DNA Library prep Master Mix set for Illumina (New England Biolabs, E6040S). Paired-end high-throughput sequencing (50 cycles) was performed according to the manufacturer’s protocol (Illumina) either on the Illumina Genome Analyzer IIX at the University of Virginia DNA Sciences Core (Charlottesville, VA, USA), the HiSeq2000 at BGI (Hong Kong), or the HiSeq2500 at HudsonAlpha Institute for Biotechnology (Huntsville, AL, USA).
MicroDNA identification in mouse and human
Pair-end high throughput sequencing reads were mapped using Novoalign software. We use mm10 for mouse and hg38 genome build for human. The algorithm used for the identification of microDNAs from paired-end sequencing data is the same as described in Shibata et al (22). The detailed instructions, source code and scripts for microDNA identification can be found at GitHub (https://github.com/pk7zuva/Circle_finder).
Results
MicroDNA are present in mouse serum
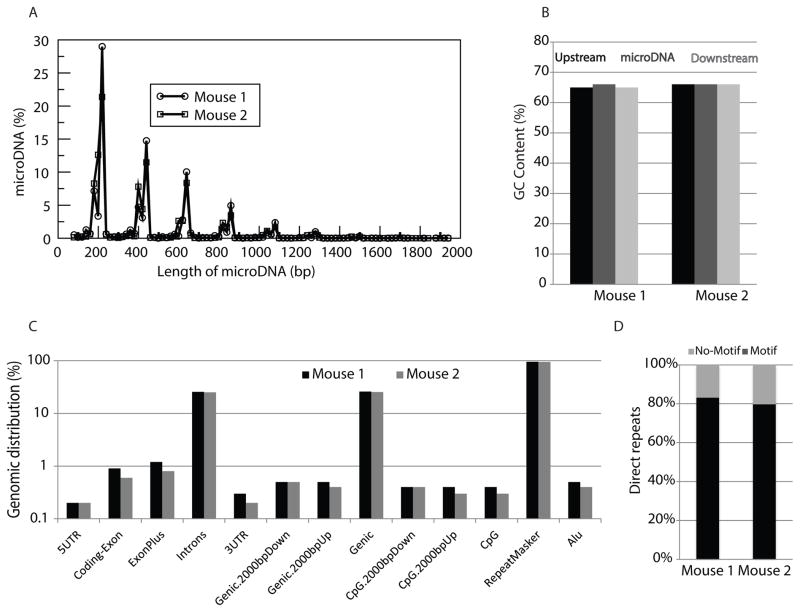
To determine if microDNA are present outside of the cell and in the circulation, circular DNA was isolated from mouse serum as previously described with slight modifications. Briefly, circular DNA were isolated from the sera of two normal adult C57BL/6 mice, treated with proteinase and exonucleases to remove linear DNA, and subjected to rolling circle amplification to increase the yield. Paired-end high-throughput sequencing of the amplified product and analysis to identify the junctions created by the circularization of genomic segments was performed as previously described (22). MicroDNA originating from tens of thousands of unique regions within the mouse genome were identified in the sera of the two mice (Table 1), demonstrating for the first time that microDNA are present within an organism outside of the cell and in circulation. The majority of cfmicroDNA in the mouse serum share characteristics with microDNA previously identified within the cell, including a length of less than 600 bases (Fig. 1A) and a GC content (60%) significantly higher than the genomic average (46%) (Fig. 1B). Furthermore, cfmicroDNA are derived from locations throughout the mouse genome, including both genic and intergenic regions (Fig. 1C). The sequences directly flanking the parental genomic locus of 80% of the circulating microDNA had 2 to 15-bp direct repeats (Fig. 1D). Since the properties of cfmicroDNA are very similar to microDNA previously observed in mouse tissue (22,23), this implies that cfmicroDNA are produced inside intact tissue cells.
Table 1.
Summary of mouse serum circulating microDNA sequencing and mapping to the mouse genome.
| Mouse 1 | Mouse 2 | |
|---|---|---|
| Paired End Reads | 23,380,919 | 42,324,947 |
| Pairs Aligned | 1,617,326 | 9,364,265 |
| Total Reads | 46,761,838 | 84,649,894 |
| Uniquely Aligned Reads | 6,953,203 | 34,969,005 |
| Multi-Mapped Reads | 719,571 | 4,819,280 |
| Unique microDNA | 22,666 | 62,245 |
Figure 1. Properties of circulating microDNA from mouse serum are similar to those observed in mouse tissues.
(A) Length distribution of microDNAs identified in the serum of two mice. (B) Median percent GC content of microDNAs and the genomic sequences of equal length upstream or downstream of the microDNA source loci are enriched relative to the average GC content of the mouse genome (dashed line). (C) Distribution of microDNA in the indicated genomic regions. (D) Percentage of microDNA with (black) or without (gray) 2- to 15-bp direct repeats flanking the microDNA locus at the genomic source.
Linear circulating cfDNA in patients with tumors is primarily 136–166 bases in length, suggesting they are derived from the digestion of chromatinized genomic DNA by nucleases to mononucleosomes, most likely during apoptosis (4,5,11), although in normal humans about 30% of the circulating linear cfDNA can be >200 bases (11). In contrast most of the circulating cfmicroDNA (>70%) are longer than 200 bases, suggesting that the circles are resistant to digestion by these nucleases. On the other hand, the striking 200-base periodicity of cfmicroDNA and cellular microDNA length (22,23)), suggests that wrapping of DNA around one or more nucleosomes contributes in some fashion to the formation of microDNA.
Human-derived microDNA are present in the sera of mice carrying human ovarian cancer xenografts
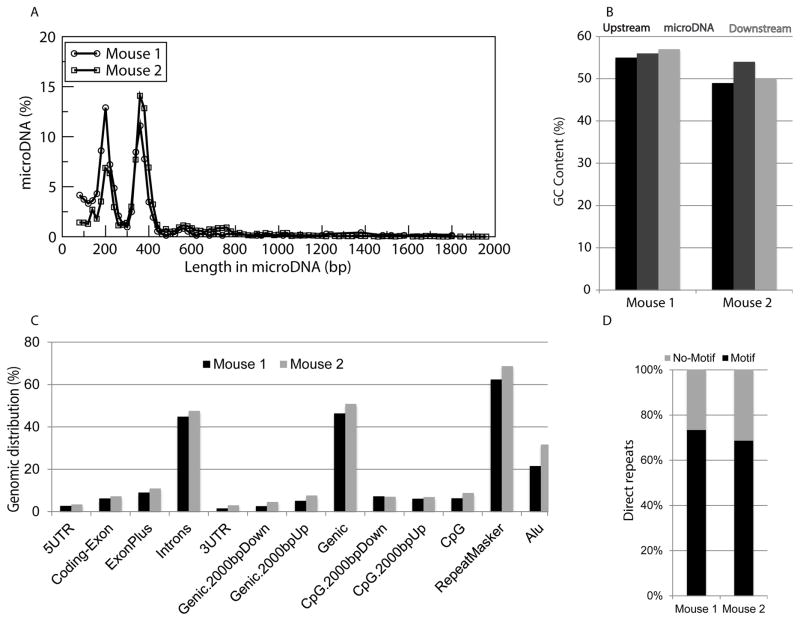
To determine if tumor cells release microDNA into the circulation, circular DNA was isolated from the sera of two SCID mice with human SKOV3 ovarian cancer tumors 19 days after establishment of the xenograft. A majority of the circulating cfmicroDNA mapped to the mouse genome (mm10) (Table 2). As seen in the normal mice, mouse-derived cfmicroDNA from the serum of SKOV3 xenograft mice display characteristics (length distribution, high GC content, genomic distribution, and direct repeats flanking the microDNA producing loci) consistent with our current knowledge of microDNA (Fig. S1) (22,23). In addition, a significant number of the paired-end reads mapped to the human genome (hg38), corresponding to thousands of human-derived microDNA, which could only have originated from the human xenograft ovarian tumor (Table 2). The human-derived cfmicroDNAs also share the same general characteristics as microDNA previously observed in SKOV3 cell lines in culture (23), including length distribution, GC content, genomic distribution, and a high frequency of direct repeats flanking the microDNA-producing genomic locus (Fig. 2).
Table 2.
Summary of SKOV3 xenograft mouse serum circulating microDNA sequencing and mapping to the mouse and human genomes.
| Mouse Genome | Human Genome | |||
|---|---|---|---|---|
| Mouse 1 | Mouse 2 | Mouse 1 | Mouse 2 | |
| Paired End Reads | 51,163,118 | 62,660,388 | 51,163,118 | 62,660,388 |
| Pairs Aligned to Species Genome | 7,219,103 | 10,237,364 | 316,626 | 6,513,724 |
| Total Reads | 102,326,236 | 125,320,776 | 102,326,236 | 125,320,776 |
| Uniquely Aligned Reads to Species Genome | 32,172,669 | 42,514,110 | 1,315,087 | 25,709,112 |
| Multi-Mapped Reads to Species Genome | 4,011,645 | 3,568,502 | 73,195 | 1,591,304 |
| Unique microDNA from Species | 120,608 | 57,417 | 721 | 7,214 |
Figure 2. Properties of human-derived microDNA in serum of mice with human ovarian cancer xenografts.
(A) Length distribution of human microDNAs identified in the serum of two mice carrying xenografts of the human ovarian cancer SKOV3 cell line. (B) Median percent GC content of human-derived microDNAs and the genomic sequences of equal length upstream or downstream of the microDNA source locus are enriched relative to the average GC content of the human genome (dashed line). (C) Distribution of human-derived microDNA in the indicated genomic regions. (D) Percentage of microDNA with (black) or without (gray) 2- to 15-bp direct repeats flanking the microDNA sequence at the genomic source loci.
Since there are many genomic regions in mouse and human that are identical in sequence (24), the mapping of microDNA to the human genome was cross-checked against an in-house database of identified mouse microDNA. Less than 10 out of the 7,935 human-derived cfmicroDNA isolated from the SKOV3 xenograft mouse serum were ambiguous in origin, eight of which mapped to ribosomal DNA that are known to be very similar in sequence between mouse and human. The detection of human-derived microDNA in the serum of mice harboring tumors of human origin is the first evidence suggesting that microDNA are released by tumor cells, most likely after tumor cell death, and are present in the circulation.
MicroDNA from human lung cancers are longer than from the paired normal tissue
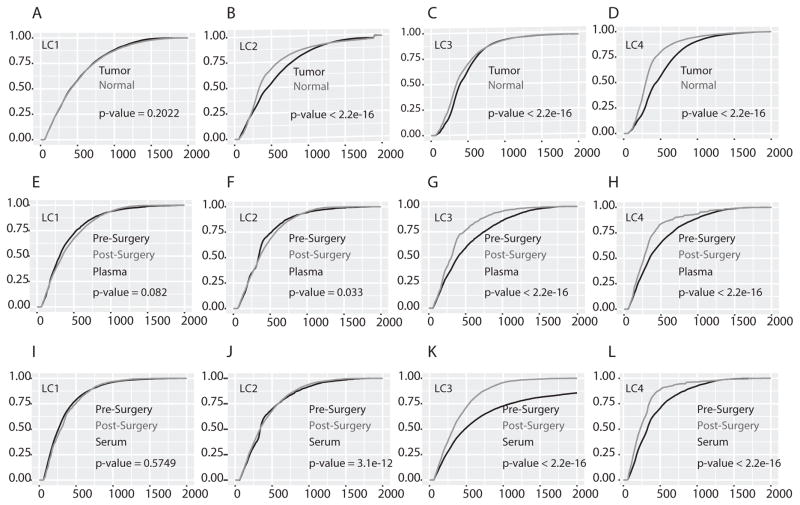
Circulating cfDNAs are derived from the dead cells of various tissues throughout the body and a variety of techniques have been used to determine the origin of cfDNA fragments. Tissue-specific cfDNAs have been identified through analysis of methylation pattern in the cfDNA fragments (7,8,25). Tumor-derived cfDNA fragments have been identified by detecting the presence of tumor-specific mutations in a given cfDNA fragment (1,8,19). Additionally, cfDNA length has been used since the length of cfDNA fragments can differ between normal and tumor tissue (4,5,9,20). In order to determine if microDNA properties could be altered in normal versus tumor tissues, microDNA were analyzed in lung cancer tissue samples and adjoining normal lung tissue from four lung cancer patients (Table S2 & S4). Tens of thousands of microDNA were identified in each of the paired tumor and normal lung tissues (Table S4). The majority of the microDNA properties (including GC content, genomic distribution, and direct repeats flanking the microDNA-producing locus) were similar regardless of their origin in normal or tumor tissue (Fig. S2A–C). However, the lengths of microDNA identified in tumor samples were longer compared to that from matched normal tissue in 3 out of 4 patients (Fig. 3A–D; patients LC2, LC3 and LC4: the CDF plot for tumor microDNA is shifted to the right of the CDF plot for normal tissue microDNA). This is the first report of a difference in length distribution of any circular DNA between normal and tumor tissue and is consistent with our previous observation the microDNA from human cancer cell lines had a significant population of microDNAs longer than 380 bases, in contrast to the microDNA from immortalized mouse cell lines and from mouse tissues (22).
Figure 3. Cumulative distribution function (CDF) plot of microDNA lengths in lung tumor and matched normal tissue.
(A–D) Plot showing the CDF of microDNA lengths in tumor (black line) and matched normal tissue (gray line) in each patient. The cumulative frequency was calculated by using ecdf function in R. The further to the right a plot is, the longer the microDNA in that sample. (E–H) Plot showing the CDF of microDNA in pre surgery (black line) and post surgery (gray line) plasma sample in each patient. (I–L) Plot showing the CDF of microDNA length in pre surgery (black line) and post surgery (gray line) serum sample in each patient. In all plots the statistical significance of the length difference in tumor vs. normal or pre surgery vs. post surgery sample was calculated by a one-sided k-s test (tumor microDNA plot is below normal microDNA plot; pre-surgery microDNA plot is below post-surgery microDNA plot) and the p-value indicated.
The longer circulating microDNA in patients with a tumor are decreased following resection of the tumor
If microDNA from lung tumors is present in circulating cfmicroDNA, we predicted that for the three lung cancer patients where tumor microDNA was longer than normal tissue microDNA (Fig. 3B–D), the cfmicroDNA isolated from plasma or serum obtained prior to surgical resection of the tumor would be longer compared to cfmicroDNA isolated from serum or plasma obtained at several weeks post-surgery. Indeed, the pre-surgery circulating cfmicroDNA was longer compared to post-surgery cfmicroDNA in the sera of all three lung cancer patients (LC2, LC3 and LC4) and the plasma of two patients (LC3 and LC4) (Fig. 3F–L). Consistent with our hypothesis, we did not find any difference in length of the circulating cfmicroDNA isolated from the pre- and post-surgery plasma or serum in patient LC1, who displayed no significant difference in microDNA length distribution between normal and tumor lung tissues (Fig. 3E and I). The statistically significant difference in the length of cfmicroDNA isolated from pre- and post-surgery plasma or serum persisted even when we pooled the results from patients LC2–LC4 (Fig. S2D–E).
Since circulating microDNA were identified in the plasma, this confirms that microDNAs detected in circulation are not the product of clotting-induced lysis of circulating cells, but were released as cell-free DNA from cells in normal tissues and tumors.
Comparison of circulating cfmicroDNA isolated before surgery and after surgery in lung and ovarian cancer patients
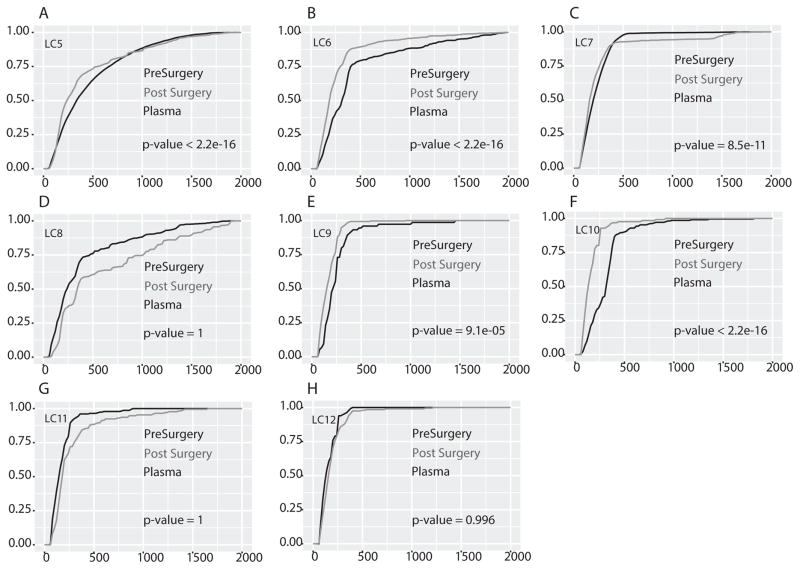
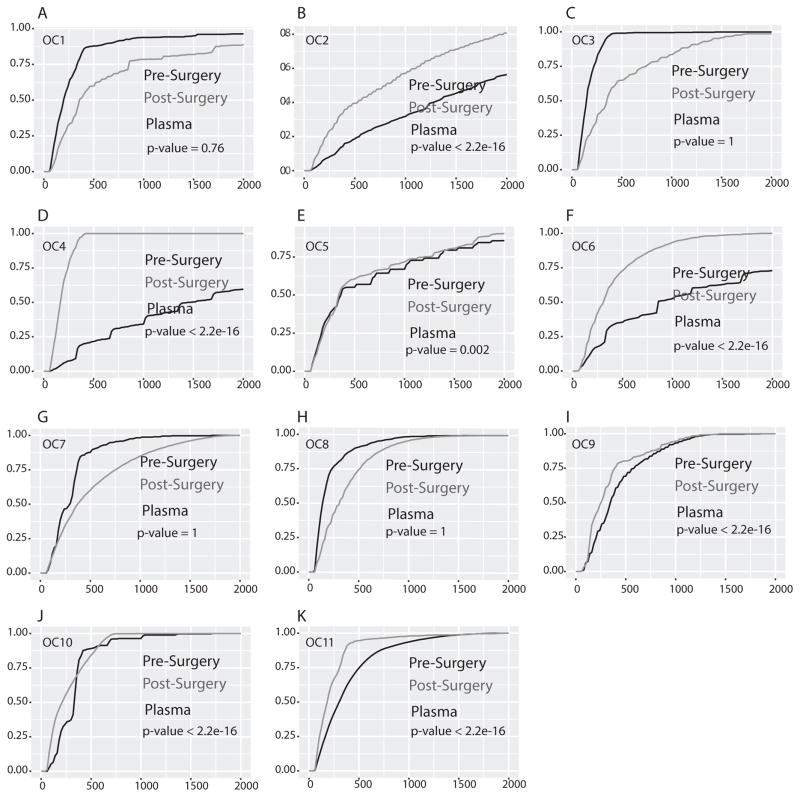
To confirm the possibility that circulating microDNA in patients with tumors could be longer than in the same patients after resection of the tumors, we expanded our study to a larger cohort: pre- and post-surgery (several weeks after surgery) plasma from eight additional lung cancer patients (LC5–LC12) (Table S2) and eleven ovarian cancer patients (OC1–OC11) (Table S3). Circulating cfmicroDNA was isolated from the serum and plasma and sequenced (Table S2 & S3). Of the eight lung cancer patients, five (63%) had significantly longer circulating microDNA pre-surgery compared to post-surgery (Fig. 4A–C, E–F). In three patients microDNAs are longer in post–surgery samples and thus showed an opposite trend with significant p-value (Fig. 4D, G & H). Similarly, seven (64%) out of eleven ovarian cancer patients had significantly longer circulating microDNA in pre-surgery plasma compared to post-surgery plasma (Fig. 5B, D–F, I–K). In four patients microDNAs are significantly longer in post–surgery samples (Fig. 5A, C, G & H). Taken together, out of twenty-three patients (12 with lung cancer and 11 with ovarian cancer) fifteen (65%, 8 with lung cancer and 7 with ovarian cancer) had significantly longer circulating cfmicroDNA before surgery than after surgery. These results support the possibility that cancer-derived microDNA are released into the circulation even when the tumor is growing in situ. Note that our IRB protocol does not allow us to follow up the patients to see if the one-third of patients who had longer microDNA post surgery showed signs of tumor recurrence.
Figure 4. Cumulative distribution function (CDF) plot of length of microDNA isolated from plasma of lung cancer patients pre surgery and post surgery.
(A–H) Plot showing the CDF of lengths of microDNA in pre surgery (black line) and post surgery (gray line) plasma sample in each patient. The rest as in Fig. 3. In five out of eight samples microDNA length in pre surgery plasma sample were significantly longer.
Figure 5. Cumulative length distribution (CDF) plot of microDNA isolated from plasma of ovarian cancer patients pre surgery and post surgery.
(A–K) Plot showing the CDF of lengths of microDNA in pre surgery (black line) and post surgery (gray line) plasma sample in each patient. Rest as in Fig. 3. In seven out of eleven samples microDNA length in pre surgery plasma sample were significantly long.
Discussion
This study was designed to determine if microDNA released from cells can be detected in serum or plasma, and if so, whether tumor-derived microDNA can be detected in circulation. We show for the first time that microDNA are present in circulation and detectable in both serum and plasma from mice and humans. These circulating cfmicroDNA were found to share the features (length, GC content, genomic distribution, direct repeats, etc.) of microDNA previously characterized in mouse tissues, human cancer cell lines, and chicken lymphoma cells (22,23). Human-derived cfmicroDNA were detectable in the sera of mice harbouring human ovarian xenograft tumors, suggesting that microDNA from xenografted tumor cells are released into circulation. Incidentally, this is the first report comparing microDNA in paired human tumor and normal tissue, allowing for the direct analysis of changes to microDNA generation following tumorigenesis. The similarities of the microDNA identified in human tumor and normal tissue to that in our previous reports confirm that the microDNA that we previously identified in human cancer cell lines are not a product of cells growing under artificial conditions in vitro.
Although we did not observe any significant difference in the microDNA from paired tumor and normal lung tissue with regards to GC content, genomic source, or the presence of direct repeats flanking the microDNA genomic locus, there was a surprising trend in 3 out of 4 lung cancer patients where microDNA from the tumor was significantly longer than that in the normal tissue. Additionally, when comparing the length of circulating microDNA in lung and ovarian cancer patients before surgery and several weeks after surgical resection of the tumor, we found that nearly two-thirds of the patients displayed a reduction in the length of their circulating microDNA after tumor resection. This shortening of circulating microDNA post-resection suggests that human tumors growing in situ release tumor-specific longer microDNA into the circulation. We note, however, that one-third of the patients showed the longer circulating microDNA after surgery. Since this was a preliminary study, we did not have IRB approval to follow up on these patients and so do not know if these patients showed signs of recurrence after surgery. Either way it is important to do a similar analysis on a large sample size with follow up of the patients before reaching a final conclusion as to whether the decrease in length of circulating microDNA post-surgery can be used as a measure of successful tumor eradication.
cfDNAs have a short half life (15 minutes to several hours) because they are cleared from the blood by the liver and kidney (26). Our results indicate that there is a significant release of microDNA into the circulation even in patients without active tumors. Circular DNA is resistant to digestion from RNAses and exonucleases, thus making it more stable and likely to survive much longer than RNA or linear DNA after release from cells into the blood (27). The source of the cfmicroDNAs in normal patients is unclear, and in the future we plan to perform methylation analysis on the cfmicroDNA (as done in (7,8)) to identify epigenetic marks that may indicate their tissue of origin. The method by which microDNA is released into the circulation is also unknown, but it is most likely following the death of normal or cancer cells. However, the release of small RNAs from live cells via exosomes (28,29) indicates that we should examine if nuclear microDNA are being similarly released from live cells. Another area of future inquiry is how microDNA are cleared from the circulation.
Circular DNA can be readily amplified by rolling-circle amplification with random primers. This ease of amplification allows us to avoid site-specific amplification with specific primers that is often used to detect linear cfDNA, and so the circular cfDNA can be analysed genome-wide without any pre-existing bias of their site of origin. The circular DNA identified in the circulation is also significantly longer than the linear DNA or microRNAs that have been studied till now. The analysis of cell-free circulating microDNA may offer an attractive biomarker for detection and monitoring of diseases like cancer throughout treatment and follow-up. For an individual patient a shortening of circulating microDNA after surgery may indicate a good outcome, and a subsequent lengthening of the circulating microDNA may indicate recurrence of the disease. However, the lengthening of microDNA in tumors was not observed in one patient and so these differences need to be explored in future studies. We also hypothesize that rapid death of tumor cells after chemo- or radiotherapy may be apparent as an increase in the length of the circulating microDNA population in the patient. Further research may indicate that certain tumors yield microDNA from specific genomic loci, and detecting such microDNA in circulation may also be useful for cancer diagnosis and prognosis.
An important question raised by our observation is whether the abundance of circulating microDNA is altered in patients with cancer. However, our present method of detecting microDNA involves rolling circle amplification with random primers and thus loses a significant amount of information related to the abundance of a specific microDNA. We hope to develop better methods for easily quantitating microDNA abundance to answer this question. Additional questions include whether cell free microDNA are present in exosomes or detected in other body fluids, and whether microDNA in circulation are taken up by cells and affect cellular phenotype.
In conclusion, this study demonstrates for the first time that microDNA, a novel class of small circular DNAs, are present and detectable in the circulation and that they are released both by normal and tumor tissue. Thus, microDNA should be added to the repertoire of circulating nucleic acids being studied as diagnostic biomarkers for cancers and fetal diseases. In addition, if the released microDNA are also taken up by cells and transcribed, then circulating microDNA may have a role in intercellular communication, much as suggested for circulating cell-free RNA.
Supplementary Material
Implications.
Extrachomosomal circular DNA (eccDNA) derived from chromosomal genomic sequence, first discovered in the nuclei of cells, are detected in the circulation, are longer than linear cell free DNA and are released from normal tissue and tumors into the circulation.
Acknowledgments
This work was supported by R01 CA60499 and CA166054 to A. Dutta, DRJ fund for lung cancer research, Ovarian Cancer Research Fund (290250 to L.W.Dillon.) and the Kaleidoscope of Hope Foundation (to L.W.Dillon.) philanthropic support from UVA Women’s Oncology and Joan Grossmann Fegely Foundation (to A. Jazaeri).
Financial support (source and number of grant for each author)
P. Kumar: W81XWH-13-1-0088 Prostate Cancer Research Program, U.S. Dept. of Defense
L.W. Dillon: Ovarian Cancer Research Fund (290250) and the Kaleidoscope of Hope Foundation Yoshiyuki Shibata: None
Amir Jazaeri: None
D. R. Jones: DRJ philanthropic funds at UVA
A. Dutta: R01 CA60499 and CA166054 from National Cancer Institute
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
References
- 1.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–37. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 2.Chandrasekharan S, Minear MA, Hung A, Allyse M. Noninvasive prenatal testing goes global. Sci Transl Med. 2014;6:231fs15. doi: 10.1126/scitranslmed.3008704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lo YM, Chiu RW. Genomic analysis of fetal nucleic acids in maternal blood. Annu Rev Genomics Hum Genet. 2012;13:285–306. doi: 10.1146/annurev-genom-090711-163806. [DOI] [PubMed] [Google Scholar]
- 4.Jiang P, Lo YM. The Long and Short of Circulating Cell-Free DNA and the Ins and Outs of Molecular Diagnostics. Trends Genet. 2016;32:360–71. doi: 10.1016/j.tig.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Giacona MB, Ruben GC, Iczkowski KA, Roos TB, Porter DM, Sorenson GD. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas. 1998;17:89–97. doi: 10.1097/00006676-199807000-00012. [DOI] [PubMed] [Google Scholar]
- 6.Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–54. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehmann-Werman R, Neiman D, Zemmour H, Moss J, Magenheim J, Vaknin-Dembinsky A, et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc Natl Acad Sci U S A. 2016;113:E1826–34. doi: 10.1073/pnas.1519286113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan KC, Jiang P, Chan CW, Sun K, Wong J, Hui EP, et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci U S A. 2013;110:18761–8. doi: 10.1073/pnas.1313995110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng YW, Chan KC, Sun H, Jiang P, Su X, Chen EZ, et al. Nonhematopoietically derived DNA is shorter than hematopoietically derived DNA in plasma: a transplantation model. Clin Chem. 2012;58:549–58. doi: 10.1373/clinchem.2011.169318. [DOI] [PubMed] [Google Scholar]
- 10.Nagata S. Apoptotic DNA fragmentation. Exp Cell Res. 2000;256:12–8. doi: 10.1006/excr.2000.4834. [DOI] [PubMed] [Google Scholar]
- 11.Underhill HR, Kitzman JO, Hellwig S, Welker NC, Daza R, Baker DN, et al. Fragment Length of Circulating Tumor DNA. PLoS Genet. 2016;12:e1006162. doi: 10.1371/journal.pgen.1006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13:358–69. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mittelbrunn M, Sanchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol. 2012;13:328–35. doi: 10.1038/nrm3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A. 2005;102:16368–73. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heitzer E, Auer M, Ulz P, Geigl JB, Speicher MR. Circulating tumor cells and DNA as liquid biopsies. Genome Med. 2013;5:73. doi: 10.1186/gm477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heitzer E, Ulz P, Belic J, Gutschi S, Quehenberger F, Fischereder K, et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 2013;5:30. doi: 10.1186/gm434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerr JF, Winterford CM, Harmon BV. Apoptosis. Its significance in cancer and cancer therapy. Cancer. 1994;73:2013–26. doi: 10.1002/1097-0142(19940415)73:8<2013::aid-cncr2820730802>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 19.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Umetani N, Giuliano AE, Hiramatsu SH, Amersi F, Nakagawa T, Martino S, et al. Prediction of breast tumor progression by integrity of free circulating DNA in serum. J Clin Oncol. 2006;24:4270–6. doi: 10.1200/JCO.2006.05.9493. [DOI] [PubMed] [Google Scholar]
- 21.Chan KC, Leung SF, Yeung SW, Chan AT, Lo YM. Persistent aberrations in circulating DNA integrity after radiotherapy are associated with poor prognosis in nasopharyngeal carcinoma patients. Clin Cancer Res. 2008;14:4141–5. doi: 10.1158/1078-0432.CCR-08-0182. [DOI] [PubMed] [Google Scholar]
- 22.Shibata Y, Kumar P, Layer R, Willcox S, Gagan JR, Griffith JD, et al. Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues. Science. 2012;336:82–6. doi: 10.1126/science.1213307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dillon LW, Kumar P, Shibata Y, Wang YH, Willcox S, Griffith JD, et al. Production of Extrachromosomal MicroDNAs Is Linked to Mismatch Repair Pathways and Transcriptional Activity. Cell Rep. 2015;11:1749–59. doi: 10.1016/j.celrep.2015.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki Y, Yamashita R, Shirota M, Sakakibara Y, Chiba J, Mizushima-Sugano J, et al. Sequence comparison of human and mouse genes reveals a homologous block structure in the promoter regions. Genome Res. 2004;14:1711–8. doi: 10.1101/gr.2435604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun K, Jiang P, Chan KC, Wong J, Cheng YK, Liang RH, et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci U S A. 2015;112:E5503–12. doi: 10.1073/pnas.1508736112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer--a survey. Biochim Biophys Acta. 2007;1775:181–232. doi: 10.1016/j.bbcan.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 27.Bendich A, Wilczok T, Borenfreund E. Circulating DNA as a Possible Factor in Oncogenesis. Science. 1965;148:374–6. doi: 10.1126/science.148.3668.374. [DOI] [PubMed] [Google Scholar]
- 28.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 29.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–79. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.