Abstract
SSCP and heteroduplex analysis (HA) continue to be the most popular methods of mutation detection due to their simplicity, high sensitivity and low cost. The advantages of these methods are most clearly visible when large genes, such as BRCA1 and BRCA2, are scanned for scattered unknown mutations and/or when a large number of DNA samples is screened for specific mutations. Here we describe a novel combined SSCP/duplex analysis adapted to the modern capillary electrophoresis (CE) system, which takes advantage of multicolor labeling of DNA fragments and laser-induced fluorescence detection. In developing this method, we first established the optimum conditions for homoduplex and heteroduplex analysis by CE. These were determined based on comprehensive analysis of representative Tamra-500 markers and BRCA1 fragments at different concentrations of sieving polymer and temperatures in the presence or absence of glycerol. The intrinsic features of DNA duplex structures are discussed in detail to explain differences in the migration rates between various types of duplexes. When combined SSCP/duplex analysis was carried out in single conditions, those found to be optimal for analysis of duplexes, all 31 BRCA1 and BRCA2 mutations, polymorphisms and variants tested were detected. It is worth noting that the panel of analyzed sequence variants was enriched in base substitutions, which are usually more difficult to detect. The sensitivity of mutation detection in the SSCP portion alone was 90%, and that in the duplex portion was 81% in the single conditions of electrophoresis. As is also shown here, the proposed combined SSCP/duplex analysis by CE has the potential of being applied to the analysis of pooled genomic DNA samples, and to multiplex analysis of amplicons from different gene fragments. These modifications may further reduce the costs of analysis, making the method attractive for large scale application in SNP scanning and screening.
INTRODUCTION
More than 1000 genes implicated in inherited human diseases have been identified thus far, and over 20 000 different mutations in these genes have been reported to the Human Gene Mutation Database (1). In just two genes, though perhaps the most extensively studied, BRCA1 and BRCA2, the total number of different alterations exceeds 1700 and continues to increase steadily (2,3). These numbers, although impressive, show that the process of identifying disease genes and characterizing their mutations is still in its infancy, and that demands for efficient and cost-effective mutation detection methods coming from both research and clinical laboratories will increase.
Among the various techniques developed for mutation detection, single-strand conformation polymorphism (SSCP) (4,5) and heteroduplex analysis (HA) (6–8) are widely used. SSCP detects base changes in single-stranded DNA, whereas HA does the same in double-stranded DNA subjected to electrophoresis in non-denaturing conditions. In SSCP analysis, the PCR product is denatured, and separated strands adopt folded structures determined by their nucleotide sequences. A single base alteration is detected by SSCP when the folding of the single strand changes sufficiently to alter its electrophoretic mobility. The most frequently claimed deficiency of the SSCP method is its variable mutation detection rate. The sensitivity of this method usually reported is about 80% when single conditions of electrophoresis are used (9–12), and reaches 95% and more when several different conditions are applied (10,11,13) for analysis of PCR products of an optimum length of 150–300 bp (9,14). In HA, the PCR-amplified DNA fragments are denatured and re-annealed to give a mixture of four duplexes, two homoduplexes and two heteroduplexes in the heterozygote samples. Heteroduplexes have an aberrant, distorted structure with bubbles or bulges at the sites of mismatched bases, and generally move more slowly in gel than homoduplexes (15). The mutation detection rate of the HA method is also about 80% (15,16), and PCR products analyzed by this method are usually of a length similar to that used in SSCP (17–19), or longer (20). Among the drawbacks of the HA method is its lower sensitivity in detecting base substitutions compared to detection of insertions and deletions (21–23).
The original protocols of SSCP and HA have undergone many modifications over the past decade (reviewed in 18,24,25). One of the most important improvements was their adaptation to automated DNA sequencing machines, which not only afforded greater convenience and safety, but also greater sensitivity in detecting mutations. SSCP analysis was performed with PCR products labeled with fluorescent primers (26–28) or with fluorescent deoxynucleotides either during PCR (29) or after PCR (30,31). SSCP systems based on either slab-gel electrophoresis (26–29,32) or capillary electrophoresis (CE) (33–37) have been developed. The latter offer important advantages compared to slab-gel systems, including higher analysis speed and lower reagent consumption. Automation of SSCP analysis by CE makes the method attractive for clinical genetic laboratories, and with the advent of multi-capillary systems, the instruments no longer have a lower throughput than slab gels. Enhancements in HA include improvement of its sensitivity by running gels in mildly denaturing conditions (15), multiplex analysis (17) and adaptation of the method to a fluorescent platform (23,38–41).
In this paper, we show the successful adaptation of CE for a combined SSCP/HA. To achieve this goal, we first optimized conditions for the HA alone by CE in a standard ABI 310 genetic analyzer. When the combined SSCP/HA was run in these optimum conditions, all sequence variants tested were detected. Our results show that in the absence of a single perfect method to screen or scan for mutations, a combination of two methods, such as SSCP and HA in the modern CE system with laser-induced multicolor fluorescence detection may be very efficient in mutation detection.
MATERIALS AND METHODS
DNA samples
Genomic DNA used in this study was extracted from whole blood samples of Polish breast and/or ovarian cancer patients, women with family histories of these cancers, and healthy controls with characterized BRCA1 and BRCA2 mutations, polymorphisms and variants (42–46).
Five artificial BRCA1 mutants were generated by PCR using a template of the wild-type fragment 11.22, which was reamplified with primers 11.22RmutA and 11.22RmutG in combination with primer 11.22F. The mutagenic primers contained at position six from their 3′-end nucleotides A and G, respectively, non-complementary to the template, which changed position 186 in fragment 11.22 in their complementary T and C. The obtained products were diluted 20 000 times and served as templates for amplification with primers 11.22F and 11.22R. In order to obtain all possible mismatched variants, the PCR products with artificially introduced mutations 3667T and 3667G were mixed at a 1:1 ratio with the PCR products of naturally occurring variants 3667A and 3667G.
Polymerase chain reaction
For HA only one PCR primer of the pair was labeled with a fluorescent dye, while for combined SSCP/HA both primers contained fluorescent labels, as shown in Table 1. All unlabeled and labeled primers were synthesized at MWG Biotech, Ebersberg, Germany. Amplifications were conducted on a standard thermocycler, Perkin Elmer 9600 or 9700, in a volume of 5 µl, using a protocol for fast and economical PCR (47). Briefly, 1 s denaturation at 94°C, 1 s extension at 72°C and 1 s annealing were used in all amplifications. In four cases, where amplified fragments were longer than 500 bp, the extension time was increased to 30 s. The applied annealing temperatures are specified in Table 1.
Table 1. Primers used for HA and combined SSCP/HA.
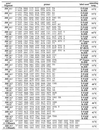 |
*Primers used for HA in unlabeled form.
Sample preparation for capillary electrophoresis
To determine migration rates of the 100 and 500 bp DNA fragments, a commercially available DNA length standard Tamra 500 (PE Applied Biosystems) was diluted 50 times in deionized water, and 5 µl aliquots were subjected to analysis. The BRCA1 fragments used for HA were prepared as follows. The PCR products were diluted with deionized water 40–160-fold depending on efficiency of amplification, and 4.5 µl aliquots of this solution were mixed in the ABI 310 sample tube with 0.5 µl of 2× diluted ROX 500 or the Tamra 500 marker. For combined SSCP/HA, the single-strand conformers and duplexes were prepared separately and combined before analysis. The 3 µl aliquots of the 40–160 times diluted PCR product were denatured at 95°C for 30 s in a 0.2 ml ABI 310 sample tube, and placed on ice to generate single-strand conformers. Then a 1.5 µl sample of diluted PCR product (duplex DNA) and 0.5 µl of 2× diluted DNA length standard were added. DNA samples used for multiplex analysis were prepared by mixing samples prepared for analysis of the individual gene fragments. Genomic DNA pools were prepared by combining a heterozygous sample with the equal volumes of one or more homozygous samples. Before pooling, the concentrations of all genomic DNA samples were adjusted by UV measurements and agarose gel electrophoresis tests to achieve the same value of 50 ng/µl.
Conditions of capillary electrophoresis
CE was performed on a ABI 310 genetic analyzer (PE Applied Biosystems) equipped with an argon laser, which emits the greatest intensity of light at 488 and 514.5 nm. The instrument has a capacity of 48 samples, which are injected automatically during the run. The samples were electro-injected at 15 kV for 10 s to a 42 cm long diameter (∅) 50 µm capillary (PE Applied Biosystems) filled with GeneScan polymer diluted to an appropriate concentration with 1× TBE buffer either containing 10% glycerol or not. Electrophoresis was at 13 kV at temperatures ranging from 30 to 65°C. Virtual filter A, allowing signal detection at wavelengths 531, 560, 580 and 610 nm, was used in all separations. The results were collected using the Data Collection program and analyzed using GeneScan software. To obtain reproducible results all electrophoregrams were calibrated by fixing the positions of peaks produced by the DNA length standard. Positions of the analyzed peaks were determined by the Local Southern method (GeneScan software, PE Applied Biosystems).
RESULTS
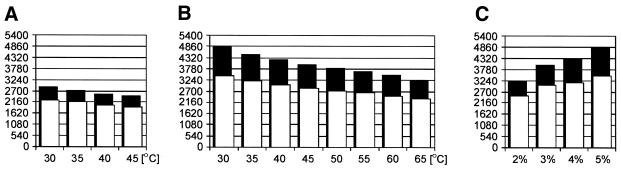
The initial selection of appropriate conditions for duplex analysis by CE was done using a Tamra 500 marker containing, among others, the 100 and 500 bp DNA fragments. These two fragments represented the lower and upper size limit of DNA fragments analyzed by the combined SSCP/heteroduplex method. The influence of a GeneScan polymer concentration, the role of the presence of glycerol in the polymer and buffer, and the effect of temperature on separation between these two DNA size markers were investigated. It is apparent from the results shown in Figure 1 that the rate with which the fragments migrate decreases gradually with either increases in the GeneScan polymer concentration or with decreases in the temperature at which the separation occurs. Moreover, the migration rate of the analyzed fragments is nearly two times lower in a polymer containing 10% glycerol. It turned out from the results shown in Figure 1 that a 5% concentration of a GeneScan polymer, an electrophoresis temperature of 30°C and the presence of 10% glycerol in the separating medium result in the largest difference in the migration rates between the markers. In these conditions, the 100 bp marker requires 12.5 min and the 500 bp marker 18 min to pass through a signal detection window located 31 cm from the sample loading site when electrophoresis is performed at 13 kV. This difference in the migration rates seemed sufficient to allow satisfactory separation of duplexes of different BRCA1 and BRCA2 fragments at a reasonably high velocity.
Figure 1.
Migration rates of 100 bp (white bars) and 500 bp (black bars) fragments of Tamra 500 DNA size standard, as expressed in the number of scans, in different conditions of electrophoresis. (A) Temperature dependence (30–45°C) at a constant 5% GeneScan polymer concentration; (B) temperature dependence (30–65°C) at a constant 5% GeneScan polymer concentration containing 10% glycerol; (C) influence of different concentrations of GeneScan polymer (2–5%) containing 10% glycerol at a constant temperature (30°C).
Analysis of duplexes of BRCA1 fragments
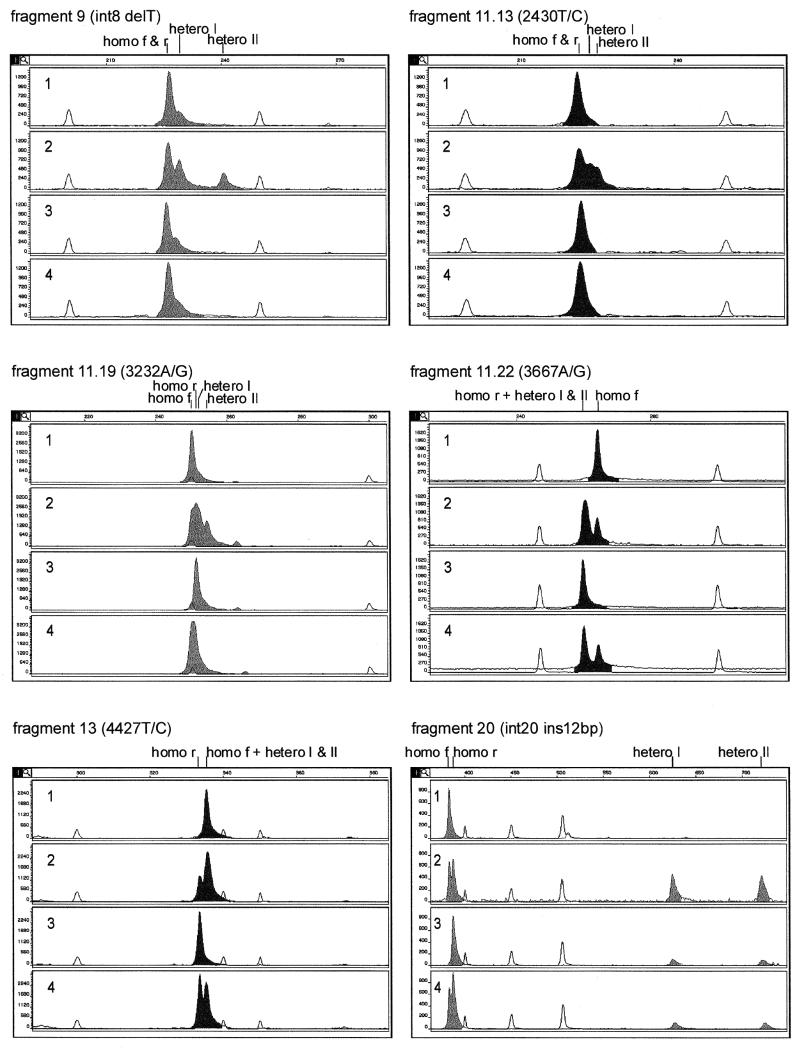
Eight different polymorphic fragments of the BRCA1 gene were analyzed by CE in a duplex form. Six of these fragments contained single base substitutions, one harbored single base deletion and in one the 12 bp insertion was present. Four different DNA samples were analyzed for each of the eight fragments in the following order: (i) homozygous for the more frequent polymorphic variant, (ii) heterozygous and (iii) homozygous for the less frequent polymorphic variant and (iv) a mixture of two homozygous samples. Two homoduplexes and two heteroduplexes were identified as follows. By comparing peak positions in the electrophoregrams of samples (i) and (iii) with those in (iv), the peaks corresponding to the two homoduplexes were assigned. The two heteroduplexes were distinguished from the homoduplexes by comparing electrophoregrams of samples (ii) and (iv). The migration rates for all types of duplexes were expressed in scans relative to the mobility of the internal standard present in each analyzed sample. The results obtained for six BRCA1 fragments analyzed in a single condition are shown in Figure 2. All but one BRCA1 fragment analyzed gave more than one duplex peak in these conditions, which indicated the presence of a sequence variant. For example, fragments 20, 9 and 11.22 gave, respectively, 4, 3 and 2 well-resolved peaks. Only fragment 17 showed a single duplex peak at 30°C, but a good separation of duplexes occurred after increasing the temperature to 65°C (electrophoregram not shown, data presented in Fig. 3).
Figure 2.
DA of six sequence variants of the BRCA1 gene. The characteristics of samples analyzed in panels 1–4 are described in Results. Positions of homoduplexes of the more frequent polymorphic variant (f), less frequent polymorphic variant (r), as well as the positions of two heteroduplexes I and II are indicated above each set of electrophoregrams. Duplexes labeled with 6-carboxy-2′,7′-dimethoxy-4′,5′-dichlorofluorescein (JOE) and FAM are shown as dark and light peaks, respectively. DNA size standards are shown as white peaks.
Figure 3.
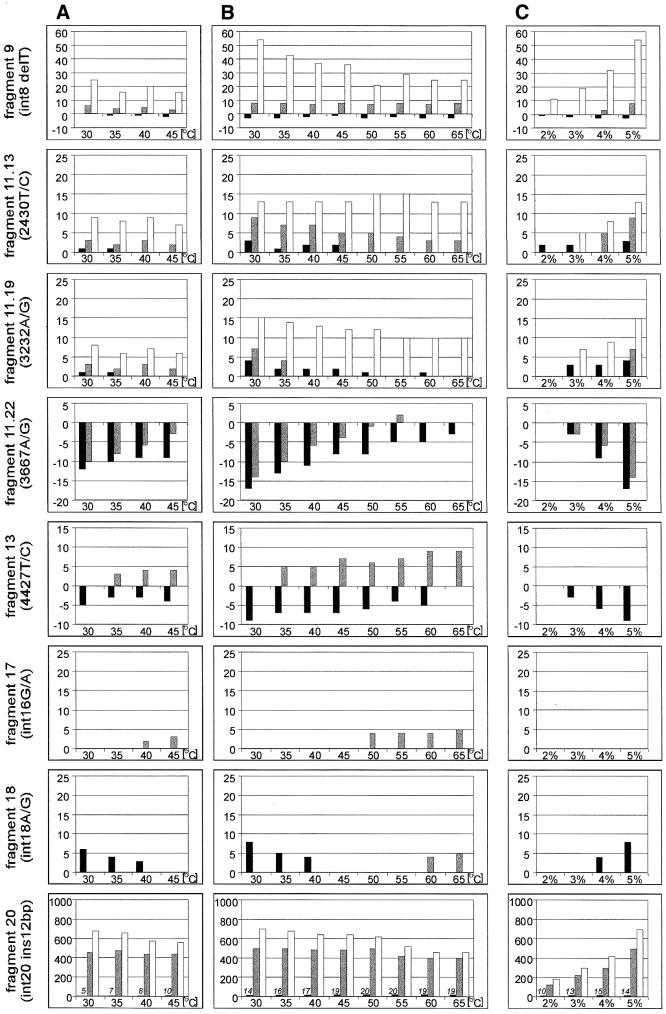
Separations of duplexes of eight fragments of the BRCA1 gene in different conditions of electrophoresis. The graphs show the differences in migration rates between the homoduplex of the less frequent polymorphic variant (black bars), as well as the faster and slower migrating heteroduplexes (gray and white bars, respectively) relative to the homoduplex of the more frequent polymorphic variant. Bars above and below the x-axis indicate, respectively, duplexes migrating slower and faster than the homoduplex of the more frequent polymorphic variant. (A), (B) and (C) are denoted as in the legend to Figure 1. In the case of BRCA1 fragment 20 with 12 bp insertion, in which migration of heteroduplexes is much slower than that of homoduplexes, the number of scans representing the distance between two homoduplexes is indicated above the corresponding bars.
As for the Tamra 500 marker, the influence of such variables as the GeneScan polymer concentration, temperature and glycerol was analyzed for each of the eight fragments of the BRCA1 gene. To compare the migration rates of all duplexes that could be distinguished from each other, all peak positions were related to the one corresponding to the homozygote of the more frequent polymorphic variant (Fig. 3). The heights of the bars positively correlate with the distance between the reference homoduplex and other duplexes, and bars with negative signs; those below the x-axis represent duplexes migrating faster than the reference homoduplex.
It appears from the data shown in Figure 3 that the temperature of electrophoresis influences the separation of different duplexes to a variable extent. In all analyzed BRCA1 fragments, the separation between the two homoduplexes is highest at 30°C, the lowest temperature applied. On the contrary, at 65°C the separation is strongly reduced (fragment 11.22) or disappears completely (fragments 11.19, 13 and 18). Homoduplexes of fragments differing in length (fragments 9 and 20) are an exception. At higher temperatures the differences in their migration rates reach constant values, reflecting the difference in their lengths. In the case of BRCA1 fragment 20, the distance between the two homoduplexes differing by 12 bp in length is small, only 14 scans, which corresponds to 4 bp at 30°C, but reaches 19 scans, equal to 9 bp, at 65°C.
As far as heteroduplexes are concerned, the influence of temperature on the rate of their electrophoretic migration is more complex. In four fragments (11.22, 13, 17 and 18), an increase in temperature decreases the mobility of the heteroduplexes (Fig. 3). Among these fragments, the electrophoretic mobility of the heteroduplexes derived from fragment 11.22 is unusual. At the lowest temperature studied, both heteroduplexes and homoduplex of the less frequent polymorphic variant migrate faster than the homoduplex of the more frequent polymorphic variant. As temperature increases, the heteroduplexes become less mobile than their accompanying homoduplex, gradually separate from it and migrate closer to the other homoduplex. In fragments 9, 11.13, 11.19 and 20, two separated heteroduplexes are observed. While the migration rate of one of these heteroduplexes relative to the homoduplexes remains constant with increases in temperature, the other heteroduplex increases its mobility until it becomes similar to that of both homoduplexes.
In summary, the best separation of duplexes of all analyzed BRCA1 fragments is achieved in the highest concentration of GeneScan polymer used. The presence of 10% glycerol does not significantly influence the patterns of separation between homoduplexes and heteroduplexes, or within them either. The order in which various duplexes migrate remains unchanged, but the separation between them is approximately two times larger and migration rates are nearly two times lower in the presence of glycerol. The influence of temperature on the separation of duplexes in the presence or absence of glycerol is similar.
Combined SSCP/heteroduplex analysis
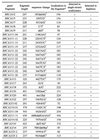
To evaluate the sensitivity of the combined SSCP/heteroduplex method in mutation detection, 31 different mutations, polymorphisms and variants present in 24 different BRCA1 and BRCA2 fragments were analyzed. They included 22 base substitutions and nine insertions/deletions. Detailed characteristics of all these sequence variants and the gene fragments harboring them is shown in Table 2. All naturally occurring variants of each fragment that were available in our DNA bank were used for analysis, i.e. mutant fragments along with wild-types, and both homozygotes and heterozygote for each common polymorphism.
Table 2. Characteristics of analyzed gene fragments and sequence variants and summary of mutation detection results.
 |
One technical problem that had to be solved at the very beginning was the way in which the DNA sample was prepared for SSCP/HA. Several options were investigated and in the one chosen the crude PCR product, without any purification with an expensive cartridge, was diluted with water and an aliquot was subjected to heating and cooling to generate single-strand conformers. At a very low salt concentration, these conformers remained stable for a long period of time and did not reform duplexes. The ssDNA fraction was mixed with the diluted and untreated PCR product (duplexes) and with an appropriate marker, which served as an internal DNA size standard in all electrophoresis runs.
Some representative results of combined SSCP/HA by CE are shown in Figure 4. They were obtained in conditions shown earlier to be optimal for HA. The combined SSCP/HA of a single sample from a series required from 18 to 30 min, depending on the length and sequence of the analyzed fragment. This included the time required to refill and wash the capillary between runs (∼1 min for each step). Homoduplexes usually migrated faster than the heteroduplexes and these were ∼3–10 min faster than the single-strand conformers. For example, in the case of the shortest fragment 11.04 (201 bp), duplexes were detected after 12.5 min, and single-strand conformers after 16 min of electrophoresis. In the case of fragment 20 (401 bp), the homo- and heteroduplexes were detected between 16.5–19 min, whereas the single-strand conformers were detected after 23 min. Duplexes of the longest BRCA2 fragment 11.11 (650 bp) were detected after 20 min and single-strand conformers after 30 min.
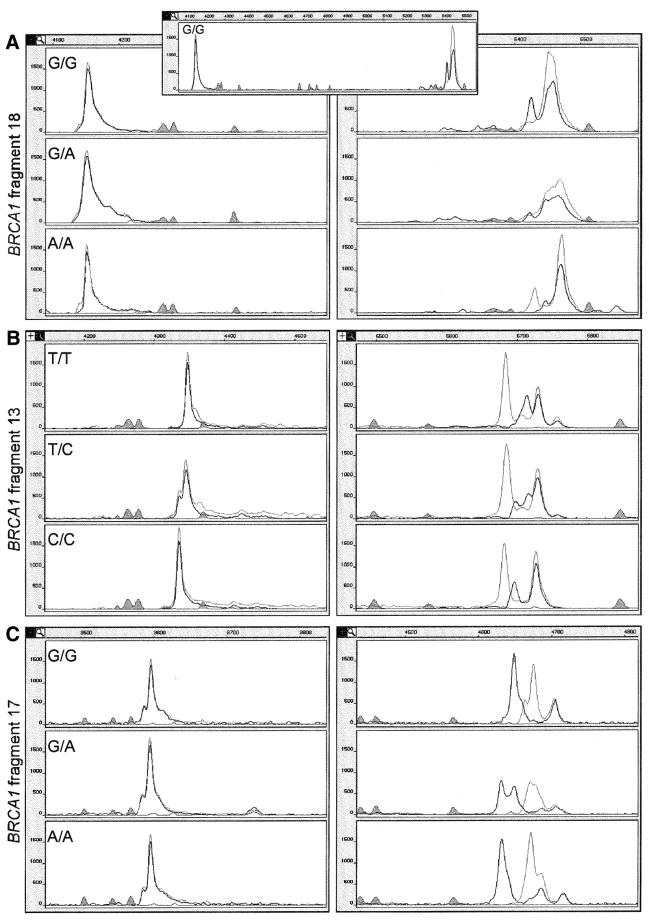
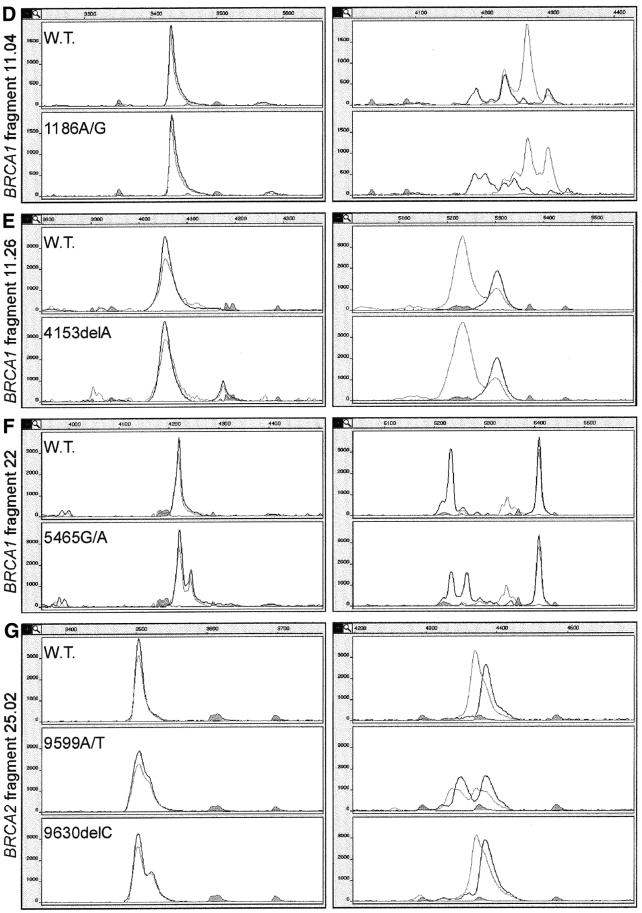
Figure 4.
(Opposite and above) Examples of combined SSCP/DA of seven different fragments of the BRCA1 and BRCA2 genes. Left and right panels show in magnification the duplex and SSCP portions of electrophoregrams, respectively. In addition, in the inset to (A), the real distance between duplex and SSCP portions is shown in a continuous fragment of electrophoregram. The peaks drawn with black and gray lines represent strands labeled with FAM or Tamra and with JOE or 6-carboxy-X-rhodamine (ROX), respectively. The shaded peaks represent fragments of the DNA size standard. Electrophoregrams of BRCA1 fragments spanning common polymorphism sites are shown (A–C). The electrophoregram representing homozygote for the more frequent polymorphic variant is shown in the upper panel, that representing the homozygote for the less frequent polymorphic variant in the lower panel, and that of the heterozygote is displayed in the central panel. Electrophoregrams of gene fragments spanning mutation or rare sequence variant sites are shown (D–G). In each case, the electrophoregrams representing the wild-type sequence or the common sequence variant (upper panel) are shown along with electrophoregrams representing the heterozygous sample (lower panel). Genotypes of the analyzed mutations, polymorphisms and variants are specified in each electrophoregram.
Out of the 31 investigated sequence variants, three were not detected in the single-strand conformer portion and six escaped detection in the duplex portion. When both single-strand conformers and duplexes were taken into account, all mutations, polymorphisms and variants were detected in single experimental conditions (Table 2). The SSCP or duplex patterns were considered changed only when a major shift took place in the position of a peak, or when the number of observed peaks was increased or decreased. Two of the three mutations detected only in the duplex form (BRCA1 4153delA and BRCA2 3134delC) are shown in Figure 4E and G. One extra duplex peak appears in both fragments, whereas the single-strand conformer pattern remains unchanged upon mutation. Two out of the six variants detected only in the single-strand conformer portion are shown in Figure 4C and D. The single strands of fragment 11.04 variant form multiple conformers, and a few of them are absent in the pattern from the corresponding wild-type sequence. Four out of the 22 sequence variants which were detected in both single-stranded and double-stranded DNA portions (IVS18A/G, 4427T/C, 5465G/A in BRCA1 and 9599A/T in BRCA2) are presented in Figure 4A, B, F and G, respectively. The examples of electrophoretic separations displayed in this figure show some of the variety of ways in which base substitutions influence the SSCP pattern. In BRCA1 fragments 11.04 (Fig. 4D) and 11.19 (data not shown) as well as in BRCA2 fragment 25.2 A/T (Fig. 4G), all peaks derived from both single strands change their positions as a result of a mutation. In BRCA1 fragments 13 and 22 (Fig. 4B and F), only one of the single-strand conformers changes its structure and mobility, whereas in fragment 17 (Fig. 4C) only the ratio between the two conformers changes, and their mobility is not altered. Interestingly, several electrophoregrams show peaks labeled with two colors. This effect, detected due to different labeling of the two PCR primers, occurs among others in BRCA1 fragments 11.04, 13, 17 and 22 (Fig. 4D, B, C and F). This is most likely caused by specific hybridization of primers to some of the single-strand conformers.
Multiplexing, pooling and screening for selected BRCA1 SNPs
Although the 24 different fragments of the BRCA1 and BRCA2 genes analyzed in this study were not specifically designed for multiplex SSCP/HA, such analysis can be conducted by CE.
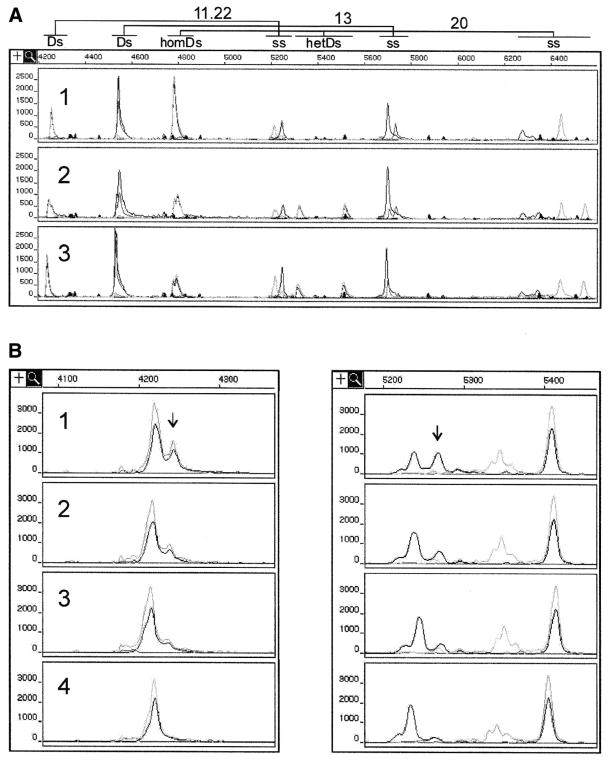
Screening for three BRCA1 polymorphisms was performed in a large sample of Polish women. Two of the eight linked intragenic polymorphisms (45,48–50) were analyzed in fragments 11.22 and 13, along with the IVS20+48ins12 variant occurring in fragment 20. Three different genotypes observed are shown in Figure 5A. The 29% frequency of the less common BRCA1 haplotype obtained from analysis of 480 control DNA samples is within the range found in other populations (3,49). On the other hand, the frequency of the IVS20+48ins12 is significantly higher among Polish women (1.9%) (43,46) than in other populations, which implies that this variant may have originated in Poland.
Figure 5.
Modifications increasing throughout of combined SSCP/DA. (A) Multiplex analysis of three BRCA1 fragments: 11.22, 13 and 20. The electrophoregrams represent three possible combinations of genotypes in BRCA1 fragments 11.22, 13 and 20: 1, aa, aa, aa; 2, ab, ab, ab; and 3, bb, bb, ab, respectively. The symbols aa, ab and bb represent the more frequent homozygote, heterozygote and less frequent homozygote, respectively. (B) DNA pooling prior to PCR. 1, SSCP/duplex pattern of the BRCA1 5465 G/A heterozygous mutant; 2 and 3, the corresponding patterns of the mutant sample diluted, respectively, two and three times with DNA samples of the wild-type sequence 5465G; 4, SSCP/duplex pattern of DNA fragment with the pure wild-type sequence (5465G). Note that the peak shown by arrow distinguishing the mutant sequence pattern from that of the wild-type sequence contributes to 25% of the total duplex signal and to ∼50% of the signal produced by one of the conformers of the FAM-labeled strand (black line).
Moreover, to further accelerate the process of screening many samples for specific mutations or scanning them for unknown mutations, DNA pooling can be considered (42). In this case, multiple samples of genomic DNA from different individuals can be combined for analysis before the PCR step, as long as the peak or peaks specific for mutant sequences can be detected. Figure 5B shows the example of the BRCA1 fragment 22, in which the applied pooling factor 3 still enables easy detection of mutation.
DISCUSSION
The rationale behind this study was the straightforward reasoning that the sensitivity of mutation detection will increase when the detection power of the SSCP technique is combined with that of the heteroduplex method. Although first applications of this attractive idea have been described for traditional slab-gel electrophoresis (e.g. 12,42,51–53), here we show the first successful adaptation of combined SSCP/HA to a modern CE system.
Homoduplexes and heteroduplexes of BRCA1 fragments
The presence of a base substitution, insertion or deletion mutation in a heterozygous sample gives rise to four different duplexes, two homoduplexes and two heteroduplexes. The mutation is detected when at least two different duplexes can be distinguished. It is common that one or both heteroduplexes are separated from the homoduplexes, as in BRCA1 fragments 9, 11.13, 11.19 and 20 (Fig. 2). It may happen, however, that both heteroduplexes comigrate with one homoduplex, and only the separated second homoduplex forms the basis for mutation detection, as in the case of BRCA1 fragments 13 and 11.22 (Fig. 2). This is the reason why the commonly used term HA should be replaced by a more correct one, duplex analysis (DA), which also includes mutation detection by differentiation of homoduplexes. Analysis of the latter is also known as a double strand conformation analysis (DSCA) (54–57). Of the eight BRCA1 variant fragments analyzed in this study, seven show different migrations of homoduplexes in optimal conditions of analysis: 5% GeneScan polymer with 10% glycerol, at a temperature of 30°C. Further decrease in electrophoresis temperature down from 30°C would most likely improve the sensitivity of mutation detection, but this would require implementation of an extra cooling system (33,36) with which the standard ABI 310 analyzer is not equipped. On the other hand, different migration rates of heteroduplexes are observed in all analyzed fragments, but in three of them (fragments 13, 17 and 18) the difference becomes apparent only at higher temperatures. Six out of the eight analyzed variants contain base substitution mutations, which are usually more difficult to detect (23). In spite of this, all mutations were found when electrophoresis was performed at both the lowest and the highest temperatures of 30 and 65°C, respectively. Seven variants were detected in single conditions at 30°C. This is a satisfactory result similar to those reported by other authors who performed duplex analysis using the traditional slab-gel system (15,16). The high mutation detection rate obtained shows that duplex analysis by CE may be considered an independent, efficient method of mutation detection. Among the advantages of this method is single primer labeling, which allows the use of other colors to label several different gene fragments for multiplex analysis. Also, the short time of analysis, usually <20 min, makes this method attractive for clinical laboratories. The high efficiency of the method in detecting insertion and deletion mutations is also an important factor, as the majority of mutations in BRCA1 and BRCA2 genes are of this type (3).
Structural basis for duplex separations
In trying to understand the variety of ways in which base mutations influence the electrophoretic migration of duplexes, questions still remain. Why do some mutations change the migration of homoduplexes and others alter the mobility of heteroduplexes? Why in some cases do both heteroduplexes migrate with the same rate and in others with different rates? Why do the majority of heteroduplexes migrate slower than homoduplexes, whereas the heteroduplex of fragment 11.22 migrates in between the faster and slower migrating homoduplex? And finally, why does the distance between homoduplexes usually decreases with an increase in temperature, while between homoduplexes and heteroduplexes it usually increases? Six base substitution mutations analyzed in detail (Fig. 3) form two types of mismatches in heteroduplexes, the T:G and C:A. They influence the migration of heteroduplexes to a different extent, which is consistent with earlier results (15). Although our data do not allow us to distinguish between individual heteroduplexes, we observe slower migration of both heteroduplexes relative to homoduplexes in BRCA1 fragments 11.13 and 11.19 (Figs 2 and 3). This pattern of migration changes at higher temperatures, i.e. in conditions resembling these in which conformation sensitive gel electrophoresis (15) is performed. At elevated temperatures one of the heteroduplexes migrates with a rate similar to that of homoduplexes, whereas migration of the other heteroduplex remains slow. Lowering the melting temperature of a heteroduplex in the vicinity of mismatched bases is perhaps the most important factor to be taken into account when interpreting different migration rates at different temperatures. Heteroduplexes of BRCA1 fragment 13 show a mobility similar to that of homoduplexes at 30°C, but as temperature increases, their migration rate becomes lower. Contrary to that, heteroduplexes of fragment 11.19 migrate slower than the corresponding homoduplexes already at 30°C. The reason why two homoduplexes of the same length migrate with different rates is most likely their different curvature resulting from a single base pair change (58,59). In the case of six substitution mutations studied in more detail (Fig. 3), the separation of homoduplexes is always highest at 30°C and a 5% GeneScan polymer concentration, decreases at a moderately elevated temperature and disappears at 60–65°C. Also, in the case of fragments with a deletion or insertion mutation, their migration rate corresponds well to their length only at higher temperatures.
Combined SSCP/duplex analysis by CE
The promising results of duplex analysis described above and recent results of other authors (40,41) followed several years of successful applications of CE to SSCP (33–35,37). These facts taken together imply that combined SSCP/DA can also be adapted to the CE platform. The first goal was then to find the optimal way to prepare a DNA sample for analysis that would contain both the single-strand conformers and duplexes in the required proportions. The second goal was to find suitable conditions in which mutations could be detected with a high sensitivity in both DNA fractions. In known CE–SSCP protocols, the PCR product was denatured either in a formamide solution (33,36,60), in NaOH solution (34,37) or in water (35). In our experiments with DA, the PCR product was diluted with water before electro-injection. It turned out that a PCR product at the same dilution forms stable single-strand conformers after denaturation and cooling. Thus, the sample containing single-strand conformers and that containing duplexes could be simply mixed for analysis at the required ratio depending on the structures of the duplex and SSCP peaks. The stability of the single-strand conformers in a very low salt water solution is an important issue for combined SSCP/DA by CE in which multiple samples are analyzed one by one, and analysis of a single sample takes ∼25 min. In the case of the 48 samples analyzed in a ABI 310 apparatus (full sample tray), the last one is injected to the capillary 20 h after beginning the analysis. During the time delay between sample preparation and its analysis, the contribution of single-strand conformers in the sample remains unchanged. Concerning the conditions of electrophoresis, the CE parameters that we found optimal for DA were highly similar to those reported by other authors as optimal for SSCP (33–35). Therefore, in combined SSCP/DA, the conditions worked out for DA were used.
Among the different effects of base changes on the SSCP pattern, the one observed for BRCA1 fragment 17 is noteworthy (Fig. 4C). The 5-carboxyflurescein (FAM)-labeled strand (gray line) forms one major and one minor conformer in each of the homozygous samples. The major conformer of the G/G homozygote shows a migration rate identical to that of the minor conformer of the A/A homozygote and vice versa. In the electrophoregram of the heterozygous sample, two peaks of similar intensity are observed. This example illustrates the fact that the base mutation may not only result in the appearance of a new conformer but also, as in homozygotes of fragment 17, may shift the equilibrium between the existing conformers. Another interesting observation is the presence of peaks marked with two different colors in several SSCP patterns. The frequency at which this effect occurs and the precise peak overlapping suggest that hybridization of PCR primers to single strands may be the explanation (61,62). Less complex SSCP patterns are usually obtained when primers are removed from PCR products during sample desalting on a suitable cartridge. However, more complex SSCP patterns, which may include a contribution from the primer hybridization to their complexity, may facilitate mutation detection by the SSCP method.
The combined SSCP/DA by CE proposed in this study detected all 31 BRCA1 and BRCA2 sequence variants tested in single conditions (Table 2), indicating that these methods based on different principles complement each other perfectly. The mutation detection rate for SSCP analysis alone is 90%, and the sensitivity of DA is 81%. Among the three mutations undetected by SSCP, two are base substitutions and one is a single base deletion. On the other hand, the 2 bp deletion and five base substitutions (two C/T and two A/G transitions, and one G/C transversion) are undetected by DA. Two of the undetected mutations are located a short distance (<50 bp) from the duplex end (Table 2). Among all 22 base substitutions analyzed in this study there are 15 transitions, four transversions A/C, two A/T and one C/G, which form the following pairs of mismatches A:C and G:T, A:G and C:T, A:A and T:T, C:C and G:G.
Considering the advantages and drawbacks of the various mutation detection methods, the cost of analysis is one of the most important criterions. In the case of the combined SSCP/DA method proposed here, the cost is the same as for each of the methods used independently, which means it is at least one order of magnitude lower than the cost of direct DNA sequencing. As also shown in this study, the cost of analysis may be further reduced and its speed increased by performing multiplex analysis, or by DNA sample pooling before PCR, whenever using these modifications is appropriate. The multiplex analysis shown in Figure 5A was successfully used in this study to determine the frequency of BRCA1 intragenic haplotypes in the Polish population and to confirm high frequency of the IVS20+48ins12 BRCA1 variant observed earlier among Polish women (43,45). We have shown earlier (42) the advantages of DNA sample pooling in the analysis of radiolabeled BRCA1 fragments by combined SSCP/DA. It was concluded that a pooling factor of 4–5 can be safely applied, although some mutations could be detected in mutant samples as many as eight times diluted with the wild-type sequence. The result of our present analysis, which makes use of CE, suggests a slightly lower pooling factor of 3 (Fig. 5B). This corroborates well with recently published results (63).
Another possibility for cutting the costs of mutation detection by combined SSCP/duplex method is the analysis of longer PCR products obtained either from genomic DNA or cDNA. Four sequence variants analyzed in this study occur in fragments of length 320–401 bp and an additional four occur in fragments 503–650 bp. These fragments are two to three times longer than those considered optimal for SSCP and DA. Importantly, mutations were detected in both single-strand and duplex portions of all long fragments (Table 2). The question of what is the maximum length of DNA fragments in which mutations can be detected by the F-SSCP/duplex/CE method needs to be answered in another study designed specifically to address this point.
Finally, the combined SSCP/DA by CE, which was developed using a single capillary apparatus ABI 310, can be readily implemented to a higher throughput platform i.e. the 16 and 96 capillary systems ABI 3100 and ABI 3700, respectively. They would allow analysis of up to 1000 and 6000 single samples per day. Each of the modifications described here, multiplex analysis and DNA pooling, may further increase throughput of analysis by a factor of 3 making the method highly efficient in screening and scanning for SNPs and mutations.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by the State Committee for Scientific Research, Grants No 6P04B 00212 and 6P04B 03118 and Foundation for Polish Science, Grants No 117/96 and 8/2000.
References
- 1.Krawczak M., Ball,E.V., Fenton,I., Stenson,P.D., Abeysinghe,S., Thomas,N. and Cooper,D.N. (2000) Human gene mutation database-a biomedical information and research resource. Hum. Mutat., 15, 45–51. [DOI] [PubMed] [Google Scholar]
- 2.Shen D. and Vadgama,J.V. (1999) BRCA1 and BRCA2 gene mutation analysis: visit to the Breast Cancer Information Core (BIC). Oncol. Res., 11, 63–69. [PubMed] [Google Scholar]
- 3.Szabo C., Masiello,A., Ryan,J.F. and Brody,L.C. (2000) The breast cancer information core: database design, structure and scope. Hum. Mutat., 16, 123–131. [DOI] [PubMed] [Google Scholar]
- 4.Orita M., Suzuki,Y., Sekiya,T. and Hayashi,K. (1989) Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction. Genomics, 5, 874–879. [DOI] [PubMed] [Google Scholar]
- 5.Orita M., Iwahana,H., Kanazawa,H., Hayashi,K. and Sekiya,T. (1989) Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc. Natl Acad. Sci. USA, 86, 2766–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagamine C.M., Chan,K. and Lau,Y.F. (1989) A PCR artifact: generation of heteroduplexes. Am. J. Hum. Genet., 45, 337–339. [PMC free article] [PubMed] [Google Scholar]
- 7.Keen J., Lester,D., Inglehearn,C., Curtis,A. and Bhattacharya,S. (1991) Rapid detection of single base mismatches as heteroduplexes on Hydrolink gels. Trends Genet., 7, 5. [DOI] [PubMed] [Google Scholar]
- 8.Keen T.J., Inglehearn,C.F., Lester,D.H., Bashir,R., Jay,M., Bird,A.C., Jay,B. and Bhattacharya,S.S. (1991) Autosomal dominant retinitis pigmentosa: four new mutations in rhodopsin, one of them in the retinal attachment site. Genomics, 11, 199–205. [DOI] [PubMed] [Google Scholar]
- 9.Murakami Y., Katahira,M., Makino,R., Hayashi,K., Hirohashi,S. and Sekiya,T. (1991) Inactivation of the retinoblastoma gene in a human lung carcinoma cell line detected by single-strand conformation polymorphism analysis of the polymerase chain reaction product of cDNA. Oncogene, 6, 37–42. [PubMed] [Google Scholar]
- 10.Michaud J., Brody,L.C., Steel,G., Fontaine,G., Martin,L.S., Valle,D. and Mitchell,G. (1992) Strand-separating conformational polymorphism analysis: efficacy of detection of point mutations in the human ornithine delta-aminotransferase gene. Genomics, 13, 389–394. [DOI] [PubMed] [Google Scholar]
- 11.Vidal-Puig A. and Moller,D.E. (1994) Comparative sensitivity of alternative single-strand conformation polymorphism (SSCP) methods. Biotechniques, 17, 490–492, 494, 496. [PubMed] [Google Scholar]
- 12.Ravnik-Glavac M., Glavac,D. and Dean,M. (1994) Sensitivity of single-strand conformation polymorphism and heteroduplex method for mutation detection in the cystic fibrosis gene. Hum. Mol. Genet., 3, 801–807. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi K. (1991) PCR-SSCP: a simple and sensitive method for detection of mutations in the genomic DNA. PCR Methods Appl., 1, 34–38. [DOI] [PubMed] [Google Scholar]
- 14.Sheffield V.C., Beck,J.S., Kwitek,A.E., Sandstrom,D.W. and Stone,E.M. (1993) The sensitivity of single-strand conformation polymorphism analysis for the detection of single base substitutions. Genomics, 16, 325–332. [DOI] [PubMed] [Google Scholar]
- 15.Ganguly A., Rock,M.J. and Prockop,D.J. (1993) Conformation-sensitive gel electrophoresis for rapid detection of single-base differences in double-stranded PCR products and DNA fragments: evidence for solvent-induced bends in DNA heteroduplexes. Proc. Natl Acad. Sci USA, 90, 10325–10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perry D.J. and Carrell,R.W. (1992) Hydrolink gels: a rapid and simple approach to the detection of DNA mutations in thromboembolic disease. J. Clin. Pathol., 45, 158–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gayther S.A., Harrington,P., Russell,P., Kharkevich,G., Garkavtseva,R.F. and Ponder,B.A. (1996) Rapid detection of regionally clustered germ-line BRCA1 mutations by multiplex heteroduplex analysis UKCCCR. Familial Ovarian Cancer Study Group. Am. J. Hum. Genet., 58, 451–456. [PMC free article] [PubMed] [Google Scholar]
- 18.Cotton R.G.H. (1997) Heteroduplex analysis (HA). In Cotton,R.G.H. (ed.), Mutation Detection. Oxford University Press, Oxford, New York, Tokyo, pp. 58–67.
- 19.Maynard J.H. and Upadhyaya,M. (1998) High-throughput screening for the detection of unknown mutations: improved productivity using heteroduplex analysis. Biotechniques, 25, 648–651. [DOI] [PubMed] [Google Scholar]
- 20.Boyd M., Lanyon,W.G. and Connor,J.M. (1993) Screening for molecular pathologies in Lesch-Nyhan syndrome. Hum. Mutat., 2, 127–130. [DOI] [PubMed] [Google Scholar]
- 21.Lohmann D.R., Brandt,B., Hopping,W., Passarge,E. and Horsthemke,B. (1996) The spectrum of RB1 germ-line mutations in hereditary retinoblastoma. Am. J. Hum. Genet., 58, 940–949. [PMC free article] [PubMed] [Google Scholar]
- 22.Abernathy C.R., Rasmussen,S.A., Stalker,H.J., Zori,R., Driscoll,D.J., Williams,C.A., Kousseff,B.G. and Wallace,M.R. (1997) NF1 mutation analysis using a combined heteroduplex/SSCP approach. Hum. Mutat., 9, 548–554. [DOI] [PubMed] [Google Scholar]
- 23.Wikman F.P., Katballe,N., Christensen,M., Laurberg,S. and Orntoft,T.F. (2000) Efficient mutation detection in mismatch repair genes using a combination of single-strand conformational polymorphism and heteroduplex analysis at a controlled temperature. Genet. Test., 4, 15–21. [DOI] [PubMed] [Google Scholar]
- 24.Cotton R.G.H. (1997) Single strand conformation analysis (SSCA). In Cotton,R.G.H. (ed.), Mutation Detection. Oxford University Press, Oxford, New York, Tokyo, pp. 45–58.
- 25.Hayashi K. (1999) Recent enhancements in SSCP. Genet. Anal., 14, 193–196. [DOI] [PubMed] [Google Scholar]
- 26.Makino R., Yazyu,H., Kishimoto,Y., Sekiya,T. and Hayashi,K. (1992) F-SSCP: fluorescence-based polymerase chain reaction-single-strand conformation polymorphism (PCR-SSCP) analysis. PCR Methods Appl., 2, 10–13. [DOI] [PubMed] [Google Scholar]
- 27.Ellison J., Dean,M. and Goldman,D. (1993) Efficacy of fluorescence-based PCR-SSCP for detection of point mutations. Biotechniques, 15, 684–691. [PubMed] [Google Scholar]
- 28.Iwahana H., Yoshimoto,K., Mizusawa,N., Kudo,E. and Itakura,M. (1994) Multiple fluorescence-based PCR-SSCP analysis. Biotechniques, 16, 296–297, 300–305. [PubMed] [Google Scholar]
- 29.Iwahana H., Fujimura,M., Takahashi,Y., Iwabuchi,T., Yoshimoto,K. and Itakura,M. (1996) Multiple fluorescence-based PCR-SSCP analysis using internal fluorescent labeling of PCR products. Biotechniques, 21, 510–514, 516–519. [DOI] [PubMed] [Google Scholar]
- 30.Iwahana H., Adzuma,K., Takahashi,Y., Katashima,R., Yoshimoto,K. and Itakura,M. (1995) Multiple fluorescence-based PCR-SSCP analysis with postlabeling. PCR Methods Appl., 4, 275–282. [DOI] [PubMed] [Google Scholar]
- 31.Inazuka M., Tahira,T. and Hayashi,K. (1996) One-tube post-PCR fluorescent labeling of DNA fragments. Genome Res., 6, 551–557. [DOI] [PubMed] [Google Scholar]
- 32.Dobson-Stone C., Cox,R.D., Lonie,L., Southam,L., Fraser,M., Wise,C., Bernier,F., Hodgson,S., Porter,D.E., Simpson,A.H. and Monaco,A.P. (2000) Comparison of fluorescent single-strand conformation polymorphism analysis and denaturing high-performance liquid chromatography for detection of EXT1 and EXT2 mutations in hereditary multiple exostoses. Eur. J. Hum. Genet., 8, 24–32. [DOI] [PubMed] [Google Scholar]
- 33.Inazuka M., Wenz,H.M., Sakabe,M., Tahira,T. and Hayashi,K. (1997) A streamlined mutation detection system: multicolor post-PCR fluorescence labeling and single-strand conformational polymorphism analysis by capillary electrophoresis. Genome Res., 7, 1094–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atha D.H., Wenz,H.M., Morehead,H., Tian,J. and O’Connell,C.D. (1998) Detection of p53 point mutations by single strand conformation polymorphism: analysis by capillary electrophoresis. Electrophoresis, 19, 172–179. [DOI] [PubMed] [Google Scholar]
- 35.Ren J. and Ueland,P.M. (1999) Temperature and pH effects on single-strand conformation polymorphism analysis by capillary electrophoresis. Hum. Mutat., 13, 458–463. [DOI] [PubMed] [Google Scholar]
- 36.Larsen L.A., Christiansen,M., Vuust,J. and Andersen,P.S. (1999) High-throughput single-strand conformation polymorphism analysis by automated capillary electrophoresis: robust multiplex analysis and pattern-based identification of allelic variants. Hum. Mutat., 13, 318–327. [DOI] [PubMed] [Google Scholar]
- 37.Tian H., Jaquins-Gerstl,A., Munro,N., Trucco,M., Brody,L.C. and Landers,J.P. (2000) Single-strand conformation polymorphism analysis by capillary and microchip electrophoresis: a fast, simple method for detection of common mutations in BRCA1 and BRCA2. Genomics, 63, 25–34. [DOI] [PubMed] [Google Scholar]
- 38.Ganguly T., Dhulipala,R., Godmilow,L. and Ganguly,A. (1998) High throughput fluorescence-based conformation-sensitive gel electrophoresis (F-CSGE) identifies six unique BRCA2 mutations and an overall low incidence of BRCA2 mutations in high-risk BRCA1-negative breast cancer families. Hum. Genet., 102, 549–556. [DOI] [PubMed] [Google Scholar]
- 39.Blesa J.R. and Hernandez-Yago,J. (2000) Adaptation of conformation-sensitive gel electrophoresis to an ALFexpress DNA sequencer to screen BRCA1 mutations. Biotechniques, 28, 1019–1025. [DOI] [PubMed] [Google Scholar]
- 40.Tian H., Brody,L.C. and Landers,J.P. (2000) Rapid detection of deletion, insertion and substitution mutations via heteroduplex analysis using capillary- and microchip-based electrophoresis. Genome Res., 10, 1403–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian H., Brody,L.C., Mao,D. and Landers,J.P. (2000) Effective capillary electrophoresis-based heteroduplex analysis through optimization of surface coating and polymer networks. Anal. Chem., 72, 5483–5492. [DOI] [PubMed] [Google Scholar]
- 42.Kozlowski P., Sobczak,K., Napierala,M., Wozniak,M., Czarny,J. and Krzyzosiak,W.J. (1996) PCR-SSCP-HDX analysis of pooled DNA for more rapid detection of germline mutations in large genes. The BRCA1 example. Nucleic Acids Res., 24, 1177–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sobczak K., Kozlowski,P., Napierala,M., Czarny,J., Wozniak,M., Kapuscinska,M., Losko,M., Koziczak,M., Jasinska,A., Powierska,J., Braczkowski,R., Breborowicz,J., Godlewski,D., Mackiewicz,A. and Krzyzosiak,W. (1997) Novel BRCA1 mutations and more frequent intron-20 alteration found among 236 women from Western Poland. Oncogene, 15, 1773–1779. [DOI] [PubMed] [Google Scholar]
- 44.Grzybowska E., Zientek,H., Jasinska,A., Rusin,M., Kozlowski,P., Sobczak,K., Sikorska,A., Kwiatkowska,E., Gorniak,L., Kalinowska,E., Utracka-Hutka,B., Wloch,J., Chmielik,E. and Krzyzosiak,W.J. (2000) High frequency of recurrent mutations in BRCA1 and BRCA2 genes in Polish families with breast and ovarian cancer. Hum. Mutat., 16, 482–490. [DOI] [PubMed] [Google Scholar]
- 45.Kozlowski P., Sobczak,K., Jasinska,A. and Krzyzosiak,W.J. (2000) Allelic imbalance of BRCA1 transcript in the IVS20 12-bp insertion carrier. Hum. Mutat., 16, 371. [DOI] [PubMed] [Google Scholar]
- 46.Jasinska A. and Krzyzosiak,W.J. (2001) Prevalence of BRCA1 founder mutations in western Poland. Hum. Mutat., 17, 75. [DOI] [PubMed] [Google Scholar]
- 47.Sobczak K., Kozlowski,P. and Krzyzosiak,W.J. (1995) Faster and cheaper PCR on a standard thermocycler. Acta Biochim. Pol., 42, 363–366. [PubMed] [Google Scholar]
- 48.Neuhausen S.L., Mazoyer,S., Friedman,L., Stratton,M., Offit,K., Caligo,A., Tomlinson,G., Cannon-Albright,L., Bishop,T., Kelsell,D., Solomon,E., Weber,B., Couch,F., Struewing,J., Tonin,P., Durocher,F., Narod,S., Skolnick,M.H., Lenoir,G., Serova,O., Ponder,B., Stoppa-Lyonnet,D., Easton,D., King,M.C. and Goldgar,D.E. (1996) Haplotype and phenotype analysis of six recurrent BRCA1 mutations in 61 families: results of an international study. Am. J. Hum. Genet., 58, 271–280. [PMC free article] [PubMed] [Google Scholar]
- 49.Shattuck-Eidens D., Oliphant,A., McClure,M., McBride,C., Gupte,J., Rubano,T., Pruss,D., Tavtigian,S.V., Teng,D.H., Adey,N., Staebell,M., Gumpper,K., Lundstrom,R., Hulick,M., Kelly,M., Holmen,J., Lingenfelter,B., Manley,S., Fujimura,F., Luce,M., Ward,B., Cannon-Albright,L., Steele,L., Offit,K. and Thomas,A. (1997) BRCA1 sequence analysis in women at high risk for susceptibility mutations. Risk factor analysis and implications for genetic testing. JAMA, 278, 1242–1250. [PubMed] [Google Scholar]
- 50.Scholl T., Pyne,M.T., Ward,B. and Pruss,D. (1999) Biochemical and genetic characterisation shows that the BRCA1 IVS20 insertion is a polymorphism. J. Med. Genet., 36, 571–572. [PMC free article] [PubMed] [Google Scholar]
- 51.Gerrard B. and Dean,M. (1998) Single-stranded conformation polymorphisms and heteroduplex analysis. In Cotton,R.G.H., Edkins,E. and Forrest,S. (eds), Mutation Detection. A Practical Approach. IRL Press at Oxford University Press, Oxford, New York, Tokyo, pp. 25–34.
- 52.Larsen L.A., Andersen,P.S., Kanters,J.K., Jacobsen,J.R., Vuust,J. and Christiansen,M. (1999) A single strand conformation polymorphism/heteroduplex (SSCP/HD) method for detection of mutations in 15 exons of the KVLQT1 gene, associated with long QT syndrome. Clin. Chim. Acta, 280, 113–125. [DOI] [PubMed] [Google Scholar]
- 53.Liechti-Gallati S., Schneider,V., Neeser,D. and Kraemer,R. (1999) Two buffer PAGE system-based SSCP/HD analysis: a general protocol for rapid and sensitive mutation screening in cystic fibrosis and any other human genetic disease. Eur. J. Hum. Genet., 7, 590–598. [DOI] [PubMed] [Google Scholar]
- 54.Pieneman W.C., Reitsma,P.H. and Briet,E. (1993) Double strand conformation polymorphism (DSCP) detects two point mutations at codon 280 (AAC→ATC) and at codon 431 (TAC→AAC) of the blood coagulation factor VIII gene. Thromb. Haemost., 69, 473–475. [PubMed] [Google Scholar]
- 55.Kirkpatrick B.W., Huff,B.M. and Casas-Carrillo,E. (1993) Double-strand DNA conformation polymorphisms as a source of highly polymorphic genetic markers. Anim. Genet., 24, 155–161. [DOI] [PubMed] [Google Scholar]
- 56.Saad F.A., Halliger,B., Muller,C.R., Roberts,R.G. and Danieli,G.A. (1994) Single base substitutions are detected by double strand conformation analysis. Nucleic Acids Res., 22, 4352–4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saad F.A., Vita,G., Toffolatti,L. and Danieli,G.A. (1994) A possible missense mutation detected in the dystrophin gene by Double-Strand Conformation Analysis (DSCA). Neuromusc. Disord., 4, 335–341. [DOI] [PubMed] [Google Scholar]
- 58.Ulanovsky L.E. and Trifonov,E.N. (1987) Estimation of wedge components in curved DNA. Nature, 326, 720–722. [DOI] [PubMed] [Google Scholar]
- 59.Bolshoy A., McNamara,P., Harrington,R.E. and Trifonov,E.N. (1991) Curved DNA without A-A: experimental estimation of all 16 DNA wedge angles. Proc. Natl Acad. Sci. USA, 88, 2312–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghozzi R., Morand,P., Ferroni,A., Beretti,J.L., Bingen,E., Segonds,C., Husson,M.O., Izard,D., Berche,P. and Gaillard,J.L. (1999) Capillary electrophoresis-single-strand conformation polymorphism analysis for rapid identification of Pseudomonas aeruginosa and other gram-negative nonfermenting bacilli recovered from patients with cystic fibrosis. J. Clin. Microbiol., 37, 3374–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cai Q.Q. and Touitou,I. (1993) Excess PCR primers may dramatically affect SSCP efficiency. Nucleic Acids Res., 21, 3909–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hennessy L.K., Teare,J. and Ko,C. (1998) PCR conditions and DNA denaturants affect reproducibility of single-strand conformation polymorphism patterns for BRCA1 mutations. Clin. Chem., 44, 879–882. [PubMed] [Google Scholar]
- 63.Amos C.I., Frazier,M.L. and Wang,W. (2000) DNA pooling in mutation detection with reference to sequence analysis. Am. J. Hum. Genet., 66, 1689–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]