Abstract
Climate change is accelerating the spread of plants and their associated species to new ranges. The differences in range shift capacity of the various types of species may disrupt long-term co-evolved relationships especially those belowground, however, this may be less so for seed-borne endophytic microbes. We collected seeds and soil of the range-expanding Centaurea stoebe and the congeneric Centaurea jacea from three populations growing in Slovenia (native range of both Centaurea species) and the Netherlands (expanded range of C. stoebe, native range of C. jacea). We isolated and identified endophytic fungi directly from seeds, as well as from roots of the plants grown in Slovenian, Dutch or sterilized soil to compare fungal endophyte composition. Furthermore, we investigated whether C. stoebe hosts a reduced community composition of endophytes in the expanded range due to release from plant-species specific fungi while endophyte communities in C. jacea in both ranges are similar. We cultivated 46 unique and phylogenetically diverse endophytes. A majority of the seed endophytes resembled potential pathogens, while most root endophytes were not likely to be pathogenic. Only one endophyte was found in both roots and seeds, but was isolated from different plant species. Unexpectedly, seed endophyte diversity of southern C. stoebe populations was lower than of populations from the north, while the seed endophyte community composition of northern C. stoebe populations was significantly different southern C. stoebe as well as northern and southern C. jacea populations. Root endophyte diversity was considerably lower in C. stoebe than in C. jacea independent of plant and soil origin, but this difference disappeared when plants were grown in sterile soils. We conclude that the community composition of fungal endophytes not only differs between related plant species but also between populations of plants that expand their range compared to their native habitat. Our results suggest that fungal endophytes of two Centaurea species are not able to systemically infect plants. We highlight that endophytes remain poorly studied and further work should investigate the functional importance of endophytes.
Keywords: endophytes, fungi, range expanding plant species, cultivation, phylogeny, soil, seeds, soil sterilization
Introduction
Ongoing anthropogenic global climate warming has enabled many plant species to expand their natural range (Walther et al., 2002; Parmesan, 2006) leading to an increase of non-native plant species in more northern, previously unsuitable, latitudes (Tamis et al., 2005). However, plants are more or less tightly linked to numerous associated organisms, which differ in their capacity to shift their range (Berg et al., 2010). Interactions between migrating plants and associated organisms have been studied extensively on introduced exotic plant species that have moved across continents. Some of these plant species become invasive, which is often attributed to a relaxation of plant interactions with specialized natural enemies, such as fungal pathogens (Keane and Crawley, 2002; Mitchell and Power, 2003). In the past decade a number of studies have suggested that such enemy release may also occur during intracontinental range shifts (Van Grunsven et al., 2007; Engelkes et al., 2008; Morriën et al., 2010), which has been demonstrated in several cases by comparing plant responses to soil from the original and new ranges (Van Grunsven et al., 2010; Dostálek et al., 2016). However, the plant holobiome (Mitter et al., 2016) consists of a much wider range of functionally diverse organisms and the holobiome concept most adequately matches to those organisms that are in intimate symbiotic relations with plants, such as endophytes. Little is known about how plant endophyte communities may respond to climate warming-induced range shifts within continents.
Endophytes are defined as organisms that asymptotically inhabit other organisms, without causing readily visible disease symptoms as pathogens do, or promoting plant performance as arbuscular mycorrhizal fungi (AMF) do (Stone and Bacon, 2000). Among the main groups of endophytes are fungi, hereafter simplified as ‘endophytes.’ Many, if not all plant species usually host several endophyte species simultaneously (Vandenkoornhuyse et al., 2002). In contrast to AMF, the phylogenetic diversity of endophytes is, enormous and spans the entire fungal kingdom, although most endophytes belong to the phylum Ascomycota (Jumpponen and Trappe, 1998; Arnold et al., 2000; Wehner et al., 2014; Glynou et al., 2016). Many fungal endophytes are highly plant host-specific, which explains their immense diversity (U’Ren et al., 2012; Wehner et al., 2014). Consequently, endophyte communities are suggested to resemble the phylogeny of their host plant (Johnston-Monje and Raizada, 2011) more than fungi that colonize the plant surface only.
In spite of the definition suggesting that endophytes are commensals rather than enemies or symbionts, endophytes can affect plant performance, especially under stress (Newsham, 2011). For instance, endophytes increase plant performance under thermal stress (Redman et al., 2002), induce plant resistance resulting in a reduction of root feeding nematodes (Martínez-Medina et al., 2017) and herbivores (Cosme et al., 2016). Under ambient conditions, endophytes might have both positive (Newsham, 2011) or negative effects on plant performance (Mayerhofer et al., 2012; Kia et al., 2017). Effects of endophytes are likely more important under stress. In addition to their role in plant performance, many endophytes can survive and grow as saprophytes in soils (Peay et al., 2016) and include species that are primary decomposers of infected plant material (Song et al., 2017). Therefore, the exact functions of most endophyte species remains largely unknown (Newsham, 2011), whereas their role in climate warming-induced plant range shifts has been completely unstudied.
Endophyte communities differ not only between plant species, but also between plant tissues. For instance, roots host communities of endophytes that largely differ from those in stems, leafs and shoots (Fisher and Petrini, 1992; Rodriguez et al., 2009), with only few endophytes being capable of systematically infecting the host plant (Johnston-Monje and Raizada, 2011). Further, plants interact with different communities of fungi during their life time and are most strongly affected in early (seed and seedling) growth stages (Gure et al., 2005; Müller et al., 2016). Fungi reduce survival of competing plants at early plant stages, while promoting adult plants resulting in higher plant diversity (Bennett and Cahill, 2016). Most plant-endophyte studies have been performed in agricultural settings, on grasses and on trees (Schardl et al., 2004; Rodriguez et al., 2009), while the effects of fungal endophytes in wild plants have rarely been studied (Rodriguez et al., 2009; Herrera Paredes and Lebeis, 2016) as most studies have. The tight connection of endophytes and their hosts has resulted in specific coevolutionary adaptations. Many endophytes can disperse ‘vertically,’ i.e., via seeds produced by their hosts, and mutualistic seedborne endophytes can promote germination (Ernst et al., 2003; Schardl et al., 2004). However, many pathogenic organisms that use seeds as vectors for dispersal may suppress seed germination (Elmer, 2001; Schardl et al., 2004). As belowground organisms have a limited dispersal capacity (Berg et al., 2010), this might be an important strategy to spread along with their plant host, which, in case of pathogens, can reduce the success of range expanding plant species.
To study endophytes in relation to climate warming-induced range expansion of a wild plant species we isolated endophytes from the seeds of the range expanding plant species Centaurea stoebe and the congeneric native Centaurea jacea. Both plants are native in Slovenia, whereas C. jacea is also native in the Netherlands, which is the expanded range of C. stoebe. We grew the seeds in sterilized soil, as well as in sterilized soil inoculated with soils collected from Slovenia and the Netherlands, and isolated fungal endophytes from the roots of the adult plants. We tested the hypotheses that (1) seed endophytic community will resemble the core root endophytic community as seed endophytes could quickly infect germinating roots; (2) endophyte community isolated from the roots or seeds of C. jacea will differ from endophyte community from C. stoebe as plant pathogens generally are species specific; and (3) southern and northern populations of C. jacea will host equal number and similar communities of endophyte taxa while northern populations of the range expander C. stoebe will host less and distinct endophyte taxa than southern populations due to the (partial) release from plant-species specific fungi during range shift.
Materials and Methods
Plant Species
Climate change such as global climate warming threatens many plant species (Thomas et al., 2004) but also enables plants to expand their naturalized habitat to previously unsuitable climate zones (Hooftman et al., 2006; Van der Putten, 2012). Among them is C. stoebe L, a perennial in the family Asteraceae that co-occurs with the common C. jacea L. in riverine habitats in the Netherlands. C. stoebe originates from Central and South-Eastern Europe (original range) and has expanded its range to higher latitudes in the past century. It has been established in the Netherlands (new range) since 1950 (Sparrius, 2014) but it is still considered a rare species in the novel range, as it occurs only in a few locations.
Seed Collection and Germination
In total more than 1000 seeds of more than 50 C. jacea and C. stoebe plants were collected from three randomly chosen populations in the Netherlands and Slovenia (Supplementary Table S1). From each population, 100 seeds were surface-sterilized in 5% household bleach (Dunne bleek, OKE, the Netherlands) for 3 min followed by washing with sterile demineralized H2O. Seeds were germinated on glass beads with sterile demineralized H2O in a growth cabinet at a 20/10°C; 16/8 h light/dark day/night regime under 60% humidity.
Soil Collection
Soil was collected from three different locations within a riverine area in Slovenia (35 kg; N45° 58′- 46° 09′; E014° 32′- E014° 45′) and the Netherlands (500 kg; N51° 51′; E5° 53′; Supplementary Table S2). The higher amount of soil needed from the Netherlands was because we only inoculated 10% alive soil (either from Slovenia or the Netherlands to 90% sterile soils (see below for further details). The soil was collected from 5 to 20 cm layer below the soil surface and transported cool (4°C) to the laboratory. In the laboratory, soil was sieved through a 5 mm × 5 mm mesh to remove larger stones, insect larvae, and earthworms. Then, the individual soil samples were formed by pooling three soil subsamples from the same riverine area. All pooled soils were sterilized by autoclaving (high pressure saturated steam at 121°C for 20–40 min) three times with 24 h interval. The live soils were kept in the dark climate room (4°C) before the use in the experiment.
Greenhouse Experiment
To create soil treatments 36 1-L pots were filled with a mix of live and sterilized Dutch soil (1:9; NL soil treatment); 36 pots were filled with a mix of live Slovenian and sterilized Dutch soil (1:9; SLO soil treatment); 36 pots were filled with a mix of sterilized Slovenian and Dutch soils (1:19; STERILE soil treatment). All pots received the same amount of soil (950 g) calculated based on dry weight of different soils. Germinated plant seedlings of similar size were individually planted in each pot. The pots were randomly placed on movable carts in a greenhouse at 21/15°C; 16/8 h light/dark; 60% humidity. Soil moisture was kept constant throughout the experiment by adjusting the pot weight to 1 kg with sterile demineralized H2O every second day. The carts were rotated weekly to avoid effects of variable conditions in the greenhouse. Rarely germinating weeds (in the live soil treatments) were instantly removed. No nutrients were added to the pots.
Eleven weeks after planting seedlings, shoots of all plants were clipped. Roots were carefully removed from the soil and thoroughly rinsed. Then, five randomly collected root fragments of approximately 5 cm length each were cut into pieces of 0.5 cm and stored in 2 mL centrifuge tubes filled with sterilized demineralized H2O.
Isolation of Fungal Endophytes from Seeds
To cultivate fungi, three seeds of all populations from both plant species (Supplementary Table S1) were placed on 1.6% H2O-agar pH 6.7 containing 50 μg/ml streptomycin in 10 cm Petri dishes. The seeds were first surface sterilized using two sterilization protocols; (1) rinsing with sterile demineralized H2O for 5 min; (2) thoroughly sterilizing by soaking seeds in 5% household bleach solution for 5 min followed by incubation in 70% ethanol for 3 min and washing with sterile demineralized H2O. For each plant species, five replicates per population with were initiated. The resulting cultures of endophytes were transferred to 0.5x potato-dextrose agar pH 6.7 (PDA; Oxoid) (Larone, 1987).
We also used another procedure aiming at isolating oomycetes. For that, five rinsed and thoroughly surface sterilized seeds (as explained before) of all populations from both plant species (Supplementary Table S1) were placed in 6 cm Petri dishes filled with a mix of sterile pond water filtered through cheesecloth and sterile demineralized H2O (1:1) containing grass leaves (Agrostis capillaris, 2–3 cm) to bait zoospore forming oomycetes (Pettitt et al., 2002). After incubation overnight at room temperature, grass leafs were transferred on 1.6% H2O-agar pH 6.7 containing 50 μg/ml streptomycin in a 6 cm diameter Petri dish. Growing cultures were transferred to a 6 cm diameter Petri dish containing 0.5x potato-dextrose agar pH 6.7 (PDA; Oxoid). To reduce the number of potentially duplicated isolates for both fungi and oomycetes, only one culture isolated from the same plant replicate was kept in case they were morphologically indistinguishable (Bosshard, 2011).
Isolation of Fungal Endophytes from Roots
To surface sterilize the roots, the collected root pieces were thoroughly washed in sterile demineralized H2O, transferred to new sterilized centrifuge tubes filled with 70% ethanol and incubated for 7 min. Root pieces were washed again in sterile demineralized H2O, and then placed in sterile demineralized H2O in new centrifuge tubes, followed by surface drying on sterile tissue paper under sterile conditions in a flow cabinet. Three root pieces were placed on 1.6% H2O-agar pH 6.7 containing 50 μg/ml streptomycin in 10 cm diameter Petri dishes. Five replicates were initiated and stored at room temperature.
The remaining root pieces were divided into three and placed into a 6 cm diameter Petri dishes filled with a mix of sterile pond water filtered through cheesecloth and sterile demineralized H2O (1:1) together with three grass leafs (Agrostis capillaris, 2–3 cm). The Petri dishes were placed at room temperature, incubated overnight. Grass leaves were transferred onto a 10 cm Petri dish containing 1.6% H2O-agar pH 6.7 containing 50 μg/ml streptomycin. All H2O-agar containing Petri dishes were checked for fungal and oomycete growth and newly formed colonies were transferred to Petri dishes containing 0.5x potato-dextrose agar pH 6.7 (PDA; Oxoid) (Larone, 1987). To reduce the number of potentially duplicated isolates, only one culture isolated from the same plant replicate was kept in case they were morphologically indistinguishable (Bosshard, 2011).
Molecular Work
DNA was extracted from all cultures using the Zymo Research Fungal/Bacterial miniprep kit according to the manufacturer’s instructions. The ITS region of all cultures was PCR amplified in 25 μl volume containing 3.125 μL 2 mM dNTPs, 1 μL 25 mM MgCl2, 2.5 μL 10x buffer with MgCl2, 0.125 μL 5 U [μL]-1 FastStart Taq DNA polymerase (Roche Diagnostics GmbH, Mannheim, Germany), 15.25 μL ddH2O, 1 μL 10 μM of both primers ITS1 and ITS4 (White et al., 1990) and 1 μL template DNA. PCR conditions were composed of an initial denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 56°C for 30 s and elongation at 72°C for 60 s with a final elongation at 72°C for 10 min. Amplified products were send for sequencing (LGC Genomics, Berlin, Germany).
Sequence Analyses
Obtained chromatograms from all cultures were manually curated in Chromas Lite v 2.11. Curated sequences were aligned using MAFFT (Katoh and Standley, 2013) and visualized in Seaview v4.6.1 (Gouy et al., 2010).
The resulting 46 consensus sequences were subjected to BLASTn searched against the NCBI nucleotide database2. For all consensus sequences, two best matches of known fungal species (three in case different species showed identical matches to best Blast matches of cultivated taxa, or unknown sequences), were aligned using MAFFT and visualized in Seaview. This resulted in an alignment containing 130 sequences.
Maximum likelihood (ML) and Bayesian analyses were performed to assess the phylogenetic relatedness of all cultivated fungi. Maximum likelihood analyses were run in RAxML v8. (Stamatakis, 2014) using the GTR+gamma model with eight rate categories. Rapid hill-climbing tree search algorithm with 1000 bootstrap replicates were used to build and assess the most stable shape of phylogenetic relationships. The phylogenetic tree was visualized in FigTree (Rambaut, 2007) and labeling of the final branches optimized in Gimp.
Data Depository
All sequence data has been submitted to GenBank under the accession numbers MF687671–MF687716.
Data Analyses
For seed endophyte data the total number of unique cultures was calculated per Petri dish (n = 24, one Petri dish per one treatment combination) and this data were used for the analyses. For root endophyte data the total number of unique cultures (cultures with distinct sequences) was calculated per pot (n = 108, three pots per one treatment combination). Therefore, to avoid pseudoreplication we first averaged the data per treatment combination and used this data for further analyses. To test the effect of plant species and seed origin on the number of seed endophyte cultures that were identified as being taxonomically different, we used general linear model with plant species (C. jacea and C. stoebe), seed origin (North – collected in The Netherlands and South – collected in Slovenia) and sterilization treatment (non-sterilized and sterilized) as fixed factors. Differences in the number of root cultures between plant species, seed and soil origins were analyzed using general linear model with plant species (C. jacea and C. stoebe), seed origin (North and South) and type of soil inoculum (Dutch soil -NL, Slovenian soil -SLO and Sterile soil) as fixed factors. In both analyses, populations were treated as true replicates.
To test whether the seed and root endophyte community composition was affected by plant species, seed origin and type of soil inoculum we used detrended correspondence analyses (DCA) and canonical correspondence analyses (CCA). For this we used presence-absence data of endophyte cultures. Detrending by segments was used in the DCA. Populations were treated as true replicates. For seed endophyte community analyses, all endophytes were included in the analyses. For root endophyte community analyses, the endophytes with more than two occurrences were included in the analyses because there were a large number of endophytes with one occurrence only.
Multivariate analyses were performed using CANOCO, version 5.03 (Šmilauer and Lepš, 2014) and all other analyses were executed using R, version 3.2.3 (R Core Team, 2015). To fulfill the assumptions of normality and homogeneity of variances total number of root endophyte cultures was log-transformed.
Results
Taxonomic Diversity of Endophytes
After removing morphologically and phylogenetically identical species from the respective plant populations grown in southern and northern soils, we obtained 91 distinct seed and 72 unique root endophytes. Many of those shared identical sequences resulting in 46 unique sequences (Supplementary Table S3).
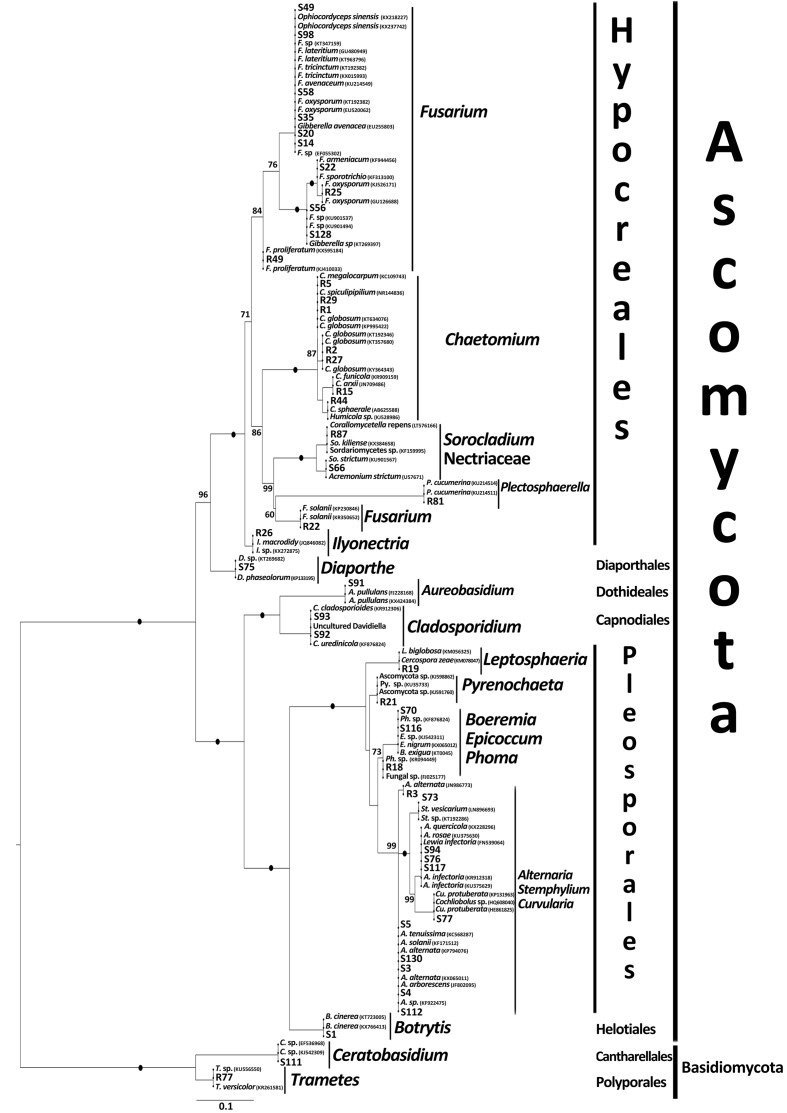
All cultures except two basidiomycetes resembled fungi of the Ascomycota (Figure 1 and Supplementary Table S3). The 46 unique sequences were phylogenetically diverse and placed in 19 genera, 15 families, 10 orders, and 4 fungal classes (Figure 1 and Supplementary Table S3).
FIGURE 1.
Maximum likelihood phylogenetic tree of all cultivated fungal endophytes (bold) and their best blast matches showing their phylogenetic affinities. For additional details see Supplementary Table S3.
Comparison between the Species Identities and Community Composition of Seed and Root Endophytes
The surface sterilization we used was highly efficient as on average less than one different culture (0.67) was obtained from inside roots, while sterilized seeds harbored more cultures (3.79) per population. The seed endophyte community yielded 28 unique cultures, whereas the root endophyte community yielded 17 unique cultures. There was no overlap in endophyte composition of roots and seeds, besides one culture with perfect match to Fusarium oxysporum f. sp. cumini that was collected from southern C. stoebe seeds, as well as from southern C. jacea roots (Supplementary Table S3).
Plant Species, Seed Origin, and Sterilization Effects on the Diversity and Community Composition of Seed Endophytes
Potential pathogens dominated the community composition of seed endophytes, with especially Fusarium and Alternaria species representing more than 50% of the different isolates (Supplementary Table S3).
Seed endophyte taxa richness was significantly affected by the interaction between plant species and seed origin (F1,16 = 5.23, P = 0.036; Figure 2). In particular, the number of cultures isolated from the C. stoebe seeds collected in the southern range was lower than in the northern range whereas the number of cultures isolated from the C. jacea seeds did not differ between origins (Figure 2).
FIGURE 2.
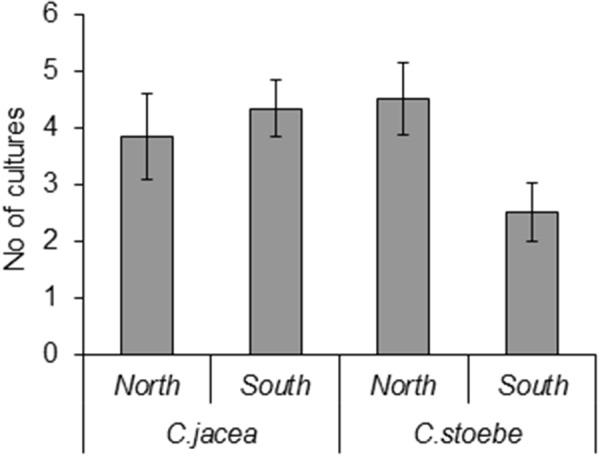
Numbers of different fungal endophyte taxa isolated from southern and northern seeds of Centaurea jacea, which is native in both ranges, and the range expander Centaurea stoebe, which is native in the south.
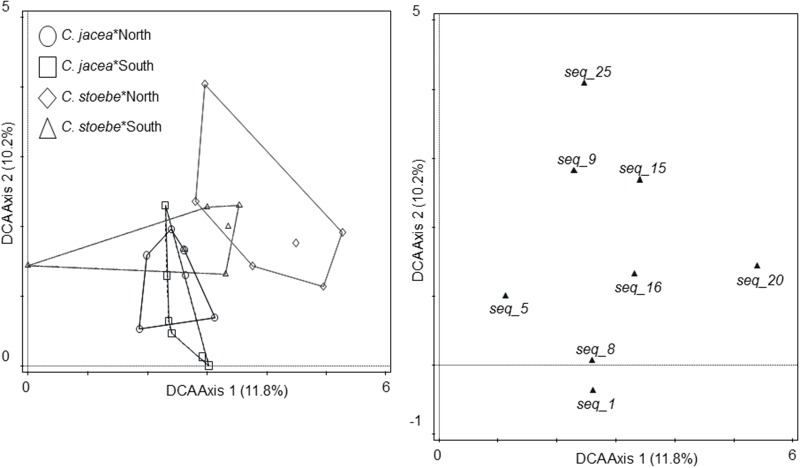
The community composition of seed endophytes was affected by the two-way interactions between plant species and seed origin (F = 1.5; P = 0.001; 18.3% explained variation, adjusted explained variation 6.0%; Figure 3). Seed sterilization did not affect the community composition of endophytes (F = 0.6; P = 0.997).
FIGURE 3.
Ordination diagrams of detrended correspondence analysis (DCA) of endophyte community isolated from seeds of C. jacea collected in The Netherlands (“North,” circles) and in Slovenia (“South,” squares), as well as seeds of C. stoebe collected in The Netherlands (“North,” diamonds) and in Slovenia (“South,” open triangles, left panel). All endophyte cultures with more than 10% fit are shown (right panel). Percentages of total explained variation by DCA axes are given in parentheses.
Plant Species, Seed Origin, and Soil Origin Effects on the Diversity and Community Composition of Root Endophytes
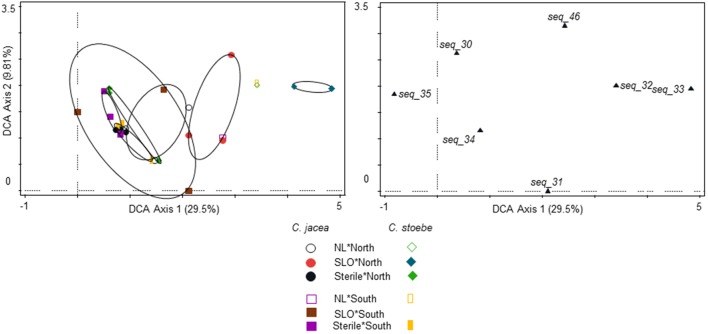
The most common (present in more than 10% of the samples) root endophytes were Chaetomium spp. (Supplementary Table S3). Range-expanding C. stoebe hosted a lower diversity of root endophytes than C. jacea plants in NL and Slo soils whereas in sterilized soil there was no difference in the diversity of root endophytes between the two plants species (F2,24 = 7.71, P = 0.0026; Figure 4). Root endophyte community composition was affected by a combination of plant species, soil and seed origin when singleton endophytes were not included in the analyses (by three-way interaction; F = 2.4; P = 0.005, 61.1% explained variation; 35.2% adjusted explained variation; Figure 5).
FIGURE 4.
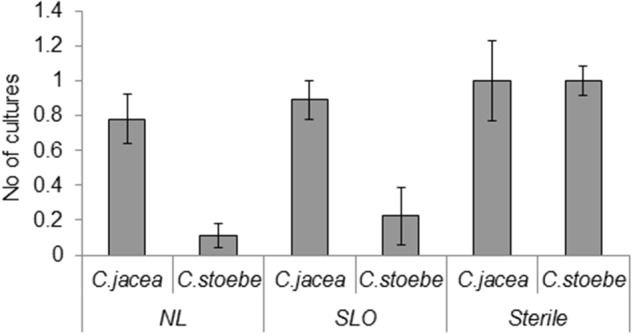
Numbers of different fungal endophyte taxa isolated from roots from the common C. jacea and the range expander C. stoebe grown in northern (NL), southern (SLO), and sterile soils (Sterile) expander C. stoebe.
FIGURE 5.
Ordination diagrams of DCA of endophyte community isolated from roots of northern C. jacea plants grown in northern (open black circles), southern (filled red circles), and sterilized soils (filled black circles); from roots of southern C. jacea plants grown in northern (open pink squares), southern (brown filled squares), and sterile soils (pink filled squares); from roots of northern C. stoebe plants grown in northern (open green diamonds), southern (filled blue diamonds), and sterile soils (filled green diamonds); from roots of southern C. stoebe plants grown in northern (open yellow rectangles) and sterile soils (filled yellow rectangles). All endophyte cultures with are shown (right panel). Percentages of total explained variation by DCA axes are given in parentheses. The overlapping symbols have been shifted by 0.1 unit to improve the visibility.
Discussion
We here show that there is no overlap of root-inhabiting and seed-inhabiting fungal endophytes in neither the range expanding plant species C. stoebe nor its common congener C. jacea. This strongly suggests that seeds might not serve as vehicles for (pathogenic) root endophytes to spread to new ranges.
High Diversity of Mainly Ascomycete Fungal Endophytes in Seeds and Roots
Almost all endophytes cultivated from both seeds and roots belong to Ascomycota, which is in line with many previous studies on endophytes in different parts of the plant (Jumpponen and Trappe, 1998; Arnold et al., 2000; Rodriguez et al., 2009). Interestingly, most seed endophytes, in contrast to root endophytes, most closely resembled potential pathogens. The overall function of endophytes is being debated (Newsham, 2011; Mayerhofer et al., 2012) and remains largely unknown (Vandenkoornhuyse et al., 2002). We assigned potential functions based on sequence identity without experimentally testing the vast amount of cultures. This approach is well-accepted to get an overall understanding of likely functions (Nguyen et al., 2016). Therefore, our results suggest that endophytes differ in their functioning between infected plant tissues with most endophytes inhabiting seeds likely being negative while root endophytes being neutral or positive for plant performance.
No Overlap between Seed and Root Endophytes
In line with potential functional differences between seed and root endophytes, seed and roots hosted a fundamentally different community of endophytes. Only one sequence resembling F. oxysporum f. sp. cumini was shared between an endophyte isolated from a seed and another one isolated from the root of southern plants. The presence of this fungi is not surprising as F. oxysporum f. sp. cumini generally has the ability to infect and damage host roots and shoots (Özer and Bayraktar, 2015). However, differentiating F. oxysporum subspecies and especially races is difficult and relies on other markers than the one we used here (ITS) as a general fungal barcode (Gordon and Martyn, 1997; Lievens et al., 2008). Considering that even races within F. oxysporum f. sp. cumini are differentially impacting plant performance (Özer and Bayraktar, 2015) and the fact that the sequence-identical cultures we obtained were originating from cultures isolated from two different plant species, ecological function of both cultures could be dissimilar. This further shows that root and seed endophyte communities are fundamentally different, suggesting that most endophytes have a restricted localization in the plant tissues in at least in the Centaurea species studied here (Rodriguez et al., 2009). The restriction of belowground endophytic fungi to root tissue also reduces the possibility that these belowground taxa spread (Berg et al., 2010). This would give the plant a competitive advantage in case they escape their more specialized pathogens – some of which are considered as being endophytic (Saikkonen et al., 1998).
Seed Endophytes Differ in C. stoebe Plants in the Expanded Range
Species specific seed endophytes seem to be generally plant pathogenic, which could control plant growth and in turn population dynamics, independent whether the plant is native or not (Blaney and Kotanen, 2001). Interestingly, we found the lowest diversity of (potentially pathogenic) fungi in the native range of C. stoebe compared to all other plant seed populations including its expanded range, while endophyte diversity in seeds of C. stoebe in the expanded range was highest. Fungi that are more generalist and less pathogenic might therefore infect seeds in the expanded range that have less negative impact on plant growth, which is in line with fungi having more negative effect on plant seeds in the native compared to the invaded range (Halbritter et al., 2012). These pathogenic endophytes could therefore be among the “enemies” that drive range expansion according to the enemy release hypothesis, which is among the key hypotheses to explain the success of plants coming to a new range (Keane and Crawley, 2002; Reinhart et al., 2003). These results indicate that C. stoebe cannot only expand, but can only perform well in new ranges indirectly by “escaping” their associated endophytes. While endophytes are directly impacted by climate change (Giauque and Hawkes, 2013) and soil biota are known to affect the performance of range expanding plants (Van Nuland et al., 2017), we lack an integrated understanding on the importance of endophytes in climate induced range expansions. Functional studies on a range of endophytes and their hosts in both ranges are needed to confirm that this hypothesis equally holds for seed endophytes.
Root Endophytes
In contrast to seed endophytes, root endophytes were mostly assigned as being non-pathogenic. Still, and in line with the seed endophytes, the diversity of root endophytes was much lower in C. stoebe than in C. jacea when plants were grown in Dutch or Slovenian soils. This suggests that compared with C. jacea, the range expanding plant C. stoebe is more antagonistic toward soil organisms as previously shown for fungi and root feeding nematodes (Wilschut et al., 2016), but nematodes might not always be reduced (Morriën et al., 2012; Viketoft and van der Putten, 2015). C. stoebe is known to produce a wide range of secondary metabolites that are more negative toward soil organisms in a new range, making it a noxious invader, especially in the United States, where it threatens natural systems (Ortega and Pearson, 2005; Ridenour et al., 2008). Production of secondary metabolites might be the key underlying factor that acts against plant-specialized soil pathogenic organisms and act as a novel weapon to change the microbial community structure to their own favor (Callaway et al., 2008; Verhoeven et al., 2009). This would also lead to a reduced infection potential of fungal endophytes as observed here.
In line with the reduction of seed endophytes in southern seeds of C. stoebe, no endophytes were cultured when those seeds where grown in southern soils. This suggests that southern seeds of C. stoebe seem particularly strong in defending themselves especially in their native soil habitat. This allows C. stoebe to defend against specialized native pathogens and, when expanding, benefit from more general, less harmful interactions, which is supported by a profound increase in infection in sterilized soils. Northern plants which escaped specialized pathogen pressure decades ago host higher endophyte diversities suggesting a trade-off toward losing costly-to-produce chemical defenses in favor of growth, which also allows them to benefit from more generalist mutualistic interactions including AMF (Bunn et al., 2015) and potentially endophytes as suggested here. Furthermore, range expanding plants could benefit in plant communities by accumulating pathogens that are deleterious for native plants (Mangla and Callaway, 2008). This suggests that range expanding plants benefit initially mostly from a release of specialist pathogens, while later from other mechanisms including increase of mutualists.
The profound reduction of endophytes in C. stoebe compared with C. jacea that is lost in sterile soil suggests that the native rhizosphere microbiome represents an intimate component of the plants ability to cope with incoming organisms. This is in line with more reduced mechanistic studies that show that invasive microbes are less likely to establish when a more diverse naïve microbiome is present (Mallon et al., 2015; Wei et al., 2015). This result further suggests that experiments done in sterile soils under non-sterile conditions can miss patterns that are actually present under more natural conditions. We therefore propose to avoid using experimental setups with entirely sterilized soils and always inoculate at least a fraction of natural soil to reduce the random effects of invasive microorganisms.
Conclusion
We conclude that there is no overlap in the taxonomic composition of endophytes in seeds and roots in the range expanding (C. stoebe) and its common congener (C. jacea), suggesting that root-inhabiting organisms, including plant pathogens, cannot spread along with the plant to infect roots in the new range. We further show that the range expanding plant hosts a reduced diversity of endophytes in roots, but that this difference is not present when soils are sterilized. This suggests that range expanding plants only in combination with a diverse microbiome obtained from soils have an increased defense against specialized soilborne organisms including pathogens, which provides them with a competitive advantage to establish in plant communities. Overall the distribution of endophytes in different plant species, especially their presence in different plant parts remains little studied. Moreover, there are no studies focusing on endophytes in range expanding plant species. As these can adapt distinct ecological roles their functional importance to affect range expanding plant species remains to be elucidated.
Author Contributions
SG, OK, and WP designed the study; BV, MC, FH, and SG conducted the experimental work, SG and OK analyzed data; SG, MC, OK, and WP drafted the manuscript supplemented with comments from FH.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Kim Magnee for collecting seeds from one population of C. stoebe and performing part of the experiment on aboveground interactions; Carolin Weser for technical assistance.
Funding. This work was conducted with support from the ERC advanced grant (ERC-Adv 260-55290) awarded to WP. This is NIOO-KNAW publication 6353.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01645/full#supplementary-material
References
- Arnold A. E., Maynard Z., Gilbert G. S., Coley P. D., Kursar T. A. (2000). Are tropical fungal endophytes hyperdiverse? Ecol. Lett. 3 267–274. 10.1016/j.ympev.2012.06.019 [DOI] [Google Scholar]
- Bennett J. A., Cahill J. F. (2016). Fungal effects on plant-plant interactions contribute to grassland plant abundances: evidence from the field. J. Ecol. 104 755–764. 10.1111/1365-2745.12558 [DOI] [Google Scholar]
- Berg M. P., Kiers E. T., Driessen G., Van Der Heijden M., Kooi B. W., Kuenen F., et al. (2010). Adapt or disperse: understanding species persistence in a changing world. Glob. Change Biol. 16 587–598. 10.1111/j.1365-2486.2009.02014.x [DOI] [Google Scholar]
- Blaney C. S., Kotanen P. M. (2001). Effects of fungal pathogens on seeds native of plants exotic: a test using congeneric pairs. J. Appl. Ecol. 38 1104–1113. 10.1046/j.1365-2664.2001.00663.x [DOI] [Google Scholar]
- Bosshard P. P. (2011). Incubation of fungal cultures: How long is long enough? Mycoses 54 e539–e545. 10.1111/j.1439-0507.2010.01977.x [DOI] [PubMed] [Google Scholar]
- Bunn R. A., Ramsey P. W., Lekberg Y., Van Der Heijden M. (2015). Do native and invasive plants differ in their interactions with arbuscular mycorrhizal fungi? A meta-analysis. J. Ecol. 103 1547–1556. 10.1111/1365-2745.12456 [DOI] [Google Scholar]
- Callaway R. M., Cipollini D., Barto K., Thelen G. C., Hallett S. G., Prati D., et al. (2008). Novel weapons: invasive plant suppresses fungal mutualists in America but not in its native Europe. Ecology 89 1043–1055. 10.1890/07-0370.1 [DOI] [PubMed] [Google Scholar]
- Cosme M., Lu J., Erb M., Stout M. J., Franken P., Wurst S. (2016). A fungal endophyte helps plants to tolerate root herbivory through changes in gibberellin and jasmonate signaling. New Phytol. 211 1065–1076. 10.1111/nph.13957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostálek T., Münzbergová Z., Kladivová A., Macel M. (2016). Plant–soil feedback in native vs. invasive populations of a range expanding plant. Plant Soil 399 209–220. 10.1007/s11104-015-2688-x [DOI] [Google Scholar]
- Elmer W. H. (2001). Seeds as vehicles for pathogen importation. Biol. Invasions 3 263–271. 10.1023/A:1015217308477 [DOI] [Google Scholar]
- Engelkes T., Morriën E., Verhoeven K. J. F., Bezemer T. M., Biere A., Harvey J. A., et al. (2008). Successful range-expanding plants experience less above-ground and below-ground enemy impact. Nature 456 946–948. 10.1038/nature07474 [DOI] [PubMed] [Google Scholar]
- Ernst M., Mendgen K. W., Wirsel S. G. R. (2003). Endophytic fungal mutualists: seed-borne Stagonospora spp. enhance reed biomass production in axenic microcosms. Mol. Plant Microbe Interact. 16 580–587. 10.1094/MPMI.2003.16.7.580 [DOI] [PubMed] [Google Scholar]
- Fisher P., Petrini O. (1992). Fungal saprobes and pathogens as endophytes of rice (Oryza sativa L.). New Phytol. 120 137–143. 10.1111/j.1469-8137.1992.tb01066.x [DOI] [Google Scholar]
- Giauque H., Hawkes C. V. (2013). Climate affects symbiotic fungal endophyte diversity and performance. Am. J. Bot. 100 1435–1444. 10.3732/ajb.1200568 [DOI] [PubMed] [Google Scholar]
- Glynou K., Ali T., Buch A. K., Haghi Kia S., Ploch S., Xia X., et al. (2016). The local environment determines the assembly of root endophytic fungi at a continental scale. Environ. Microbiol. 18 2418–2434. 10.1111/1462-2920.13112 [DOI] [PubMed] [Google Scholar]
- Gordon T., Martyn R. (1997). The evolutionary biology of Fusarium oxysporum. Annu. Rev. Phytopathol. 35 111–128. 10.1146/annurev.phyto.35.1.111 [DOI] [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O. (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27 221–224. 10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- Gure A., Wahlström K., Stenlid J. (2005). Pathogenicity of seed-associated fungi to Podocarpus falcatus in vitro. For. Pathol. 35 23–35. 10.1111/j.1439-0329.2004.00387.x [DOI] [Google Scholar]
- Halbritter A. H., Carroll G. C., Güsewell S., Roy B. A. (2012). Testing assumptions of the enemy release hypothesis: generalist versus specialist enemies of the grass Brachypodium sylvaticum. Mycologia 104 34–44. 10.3852/11-071 [DOI] [PubMed] [Google Scholar]
- Herrera Paredes S., Lebeis S. L. (2016). Giving back to the community: microbial mechanisms of plant–soil interactions. Funct. Ecol. 30 1043–1052. 10.1111/1365-2435.12684 [DOI] [Google Scholar]
- Hooftman D. A. P., Oostermeijer J. G. B., Den Nijs J. C. M. (2006). Invasive behaviour of Lactuca serriola (Asteraceae) in the Netherlands: spatial distribution and ecological amplitude. Basic Appl. Ecol. 7 507–519. 10.1016/j.baae.2005.12.006 [DOI] [Google Scholar]
- Johnston-Monje D., Raizada M. N. (2011). Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE 6:e20396 10.1371/journal.pone.0020396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumpponen A. R. I., Trappe J. M. (1998). Dark septate endophytes: a review of facultative biotrophic root-colonizing fungi. New Phytol. 140 295–310. 10.1046/j.1469-8137.1998.00265.x [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane R. M., Crawley M. J. (2002). Exotic plant invasions and the enemy release hypothesis. Trends Ecol. Evol. 17 164–170. 10.1016/S0169-5347(02)02499-0 [DOI] [Google Scholar]
- Kia S. H., Glynou K., Nau T., Thines M., Piepenbring M., Macia-Vicente J. G. (2017). Influence of phylogenetic conservatism and trait convergence on the interactions between fungal root endophytes and plants. ISME J. 11 777–790. 10.1038/ismej.2016.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larone D. H. (1987). Medically Important Fungi: A Guide to Identification. New York, NY: Elsevier. [Google Scholar]
- Lievens B., Rep M., Thomma B. P. H. J. (2008). Recent developments in the molecular discrimination of formae speciales of Fusarium oxysporum. Pest Manag. Sci. 64 781–788. 10.1002/ps.1564 [DOI] [PubMed] [Google Scholar]
- Mallon C. A., Van Elsas J. D., Salles J. F. (2015). Microbial invasions: the process, patterns, and mechanisms. Trends Microbiol. 23 719–729. 10.1016/j.tim.2015.07.013 [DOI] [PubMed] [Google Scholar]
- Mangla S., Callaway R. M. (2008). Exotic invasive plant accumulates native soil pathogens which inhibit native plants. J. Ecol. 96 58–67. 10.1093/aob/mcs061 [DOI] [Google Scholar]
- Martínez-Medina A., Fernandez I., Lok G. B., Pozo M. J., Pieterse C. M. J., Van Wees S. C. M. (2017). Shifting from priming of salicylic acid- to jasmonic acid-regulated defences by Trichoderma protects tomato against the root knot nematode Meloidogyne incognita. New Phytol. 213 1363–1377. 10.1111/nph.14251 [DOI] [PubMed] [Google Scholar]
- Mayerhofer M. S., Kernaghan G., Harper K. A. (2012). The effects of fungal root endophytes on plant growth: a meta-analysis. Mycorrhiza 23 119–128. 10.1007/s00572-012-0456-9 [DOI] [PubMed] [Google Scholar]
- Mitchell C. E., Power A. G. (2003). Release of invasive plants from fungal and viral pathogens. Nature 421 625–627. 10.1038/nature01317 [DOI] [PubMed] [Google Scholar]
- Mitter B., Pfaffenbichler N., Sessitsch A. (2016). Plant–microbe partnerships in 2020. Microb. Biotechnol. 9 635–640. 10.1111/1751-7915.12382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morriën E., Duyts H., Van Der Putten W. H. (2012). Effects of native and exotic range-expanding plant species on taxonomic and functional composition of nematodes in the soil food web. Oikos 121 181–190. 10.1111/j.1600-0706.2011.19773.x [DOI] [Google Scholar]
- Morriën E., Engelkes T., Macel M., Meisner A., Van Der Putten W. H. (2010). Climate change and invasion by intracontinental range-expanding exotic plants: the role of biotic interactions. Ann. Bot 105 843–848. 10.1093/aob/mcq064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller G., Horstmeyer L., Rönneburg T., Van Kleunen M., Dawson W. (2016). Alien and native plant establishment in grassland communities is more strongly affected by disturbance than above- and below-ground enemies. J. Ecol. 104 1233–1242. 10.1111/1365-2745.12601 [DOI] [Google Scholar]
- Newsham K. K. (2011). A meta-analysis of plant responses to dark septate root endophytes. New Phytol. 190 783–793. 10.1111/j.1469-8137.2010.03611.x [DOI] [PubMed] [Google Scholar]
- Nguyen N. H., Song Z., Bates S. T., Branco S., Tedersoo L., Menke J., et al. (2016). FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20 241–248. 10.1016/j.funeco.2015.06.006 [DOI] [Google Scholar]
- Ortega Y. K., Pearson D. E. (2005). Weak vs. strong invaders of natural plant communities: assessing invasibility and impact. Ecol. Appl. 15 651–661. 10.1890/04-0119 [DOI] [Google Scholar]
- Özer G., Bayraktar H. (2015). Intraspecific variation within Fusarium oxysporum f. sp. cumini from Cuminum cyminum in Turkey. Int. J. Agric. Biol. 17 375–380. [Google Scholar]
- Parmesan C. (2006). Ecological and evolutionary responses to recent climate change. Annu. Rev. Ecol. Evol. Syst. 37 637–669. 10.1146/annurev.ecolsys.37.091305.110100 [DOI] [Google Scholar]
- Peay K. G., Kennedy P. G., Talbot J. M. (2016). Dimensions of biodiversity in the Earth mycobiome. Nat. Rev. Microbiol. 14 434–447. 10.1038/nrmicro.2016.59 [DOI] [PubMed] [Google Scholar]
- Pettitt T. R., Wakeham A. J., Wainwright M. F., White J. G. (2002). Comparison of serological, culture, and bait methods for detection of Pythium and Phytophthora zoospores in water. Plant Pathol. 51 720–727. 10.1046/j.1365-3059.2002.00759.x [DOI] [Google Scholar]
- R Core Team (2015). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; Available at: http://www.R-project.org/ [Google Scholar]
- Rambaut A. (2007). FigTree, A Graphical Viewer of Phylogenetic Trees. Available at: http://tree.bio.ed.ac.uk/software/figtree [Google Scholar]
- Redman R. S., Sheehan K. B., Stout R. G., Rodriguez R. J., Henson J. M. (2002). Thermotolerance generated by plant/fungal symbiosis. Science 298 1581 10.1126/science.1072191 [DOI] [PubMed] [Google Scholar]
- Reinhart K. O., Packer A., Van Der Putten W. H., Clay K. (2003). Plant–soil biota interactions and spatial distribution of black cherry in its native and invasive ranges. Ecol. Lett. 6 1046–1050. 10.1046/j.1461-0248.2003.00539.x [DOI] [Google Scholar]
- Ridenour W. M., Vivanco J. M., Feng Y., Horiuchi J.-I., Callaway R. M. (2008). No evidence for trade-offs: Centaurea plants from America are better competitors and defenders. Ecol. Monogr. 78 369–386. 10.1890/06-1926.1 [DOI] [Google Scholar]
- Rodriguez R. J., White J. F., Jr., Arnold A. E., Redman R. S. (2009). Fungal endophytes: diversity and functional roles. New Phytol. 182 314–330. 10.1111/j.1469-8137.2009.02773.x [DOI] [PubMed] [Google Scholar]
- Saikkonen K., Faeth S. H., Helander M., Sullivan T. J. (1998). Fungal endophytes: a continuum of interactions with host plants. Annu. Rev. Ecol. Evol. Syst. 29 319–343. 10.1146/annurev.ecolsys.29.1.319 [DOI] [Google Scholar]
- Schardl C. L., Leuchtmann A., Spiering M. J. (2004). Symbioses of grasses with seedborne fungal endophytes. Annu. Rev. Plant Biol. 55 315–340. 10.1146/annurev.arplant.55.031903.141735 [DOI] [PubMed] [Google Scholar]
- Šmilauer P., Lepš J. (2014). Multivariate Analysis of Ecological Data Using CANOCO 5, 2nd Edn New York, NY: Cambridge University Press. [Google Scholar]
- Song Z., Kennedy P. G., Liew F. J., Schilling J. S. (2017). Fungal endophytes as priority colonizers initiating wood decomposition. Funct. Ecol. 31 407–418. 10.1111/1365-2435.12735 [DOI] [Google Scholar]
- Sparrius L. (2014). FLORIVON. v11.5. Dutch Foundation for Botanical Research (FLORON). Available at: http://www.verspreidingsatlas.nl:8085/ipt/resource?r=florivon&v=11.5 [Google Scholar]
- Stamatakis A. (2014). RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J. K., Bacon C. W. (2000). “An overview of endophytic microbes: endophytism defined,” in Microbial Endophytes, eds Bacon C. W., White J. F., Jr. (New York, NY: Marcel Dekker; ), 29–33. [Google Scholar]
- Tamis W. M., Zelfde M. T., Meijden R., Haes H. U. (2005). Changes in vascular plant biodiversity in the Netherlands in the 20th century explained by their climatic and other environmental characteristics. Clim. Change 72 37–56. 10.1007/s10584-005-5287-7 [DOI] [Google Scholar]
- Thomas C. D., Cameron A., Green R. E., Bakkenes M., Beaumont L. J., Collingham Y. C., et al. (2004). Extinction risk from climate change. Nature 427 145–148. 10.1038/nature02121 [DOI] [PubMed] [Google Scholar]
- U’Ren J. M., Lutzoni F., Miadlikowska J., Laetsch A. D., Arnold A. E. (2012). Host and geographic structure of endophytic and endolichenic fungi at a continental scale. Am. J. Bot. 99 898–914. 10.3732/ajb.1100459 [DOI] [PubMed] [Google Scholar]
- Van der Putten W. H. (2012). Climate change, aboveground-belowground interactions, and species’ range shifts. Annu. Rev. Ecol. Evol. Syst. 43 365–383. 10.1146/annurev-ecolsys-110411-160423 [DOI] [Google Scholar]
- Van Grunsven R. H. A., Van Der Putten W. H., Bezemer T. M., Tamis W. L. M., Berendse F., Veenendaal E. M. (2007). Reduced plant–soil feedback of plant species expanding their range as compared to natives. J. Ecol. 95 1050–1057. 10.1007/s00442-009-1526-3 [DOI] [Google Scholar]
- Van Grunsven R. H. A., Van Der Putten W. H., Martijn Bezemer T., Berendse F., Veenendaal E. M. (2010). Plant–soil interactions in the expansion and native range of a poleward shifting plant species. Glob. Change Biol. 16 380–385. 10.1111/j.1365-2486.2009.01996.x [DOI] [Google Scholar]
- Van Nuland M. E., Bailey J. K., Schweitzer J. A. (2017). Divergent plant–soil feedbacks could alter future elevation ranges and ecosystem dynamics. Nat. Ecol. Evol. 1:0150 10.1038/s41559-017-0150 [DOI] [PubMed] [Google Scholar]
- Vandenkoornhuyse P., Baldauf S. L., Leyval C., Straczek J., Young J. P. W. (2002). Extensive fungal diversity in plant roots. Science 295 2051–2051. 10.1038/s41559-017-0150 [DOI] [PubMed] [Google Scholar]
- Verhoeven K. J. F., Biere A., Harvey J. A., Van Der Putten W. H. (2009). Plant invaders and their novel natural enemies: Who is naïve? Ecol. Lett. 12 107–117. 10.1111/j.1461-0248.2008.01248.x [DOI] [PubMed] [Google Scholar]
- Viketoft M., van der Putten W. H. (2015). Top-down control of root-feeding nematodes in range-expanding and congeneric native plant species. Basic Appl. Ecol. 16 260–268. 10.1016/j.baae.2014.12.006 [DOI] [Google Scholar]
- Walther G. R., Post E., Convey P., Menzel A., Parmesan C., Beebee T. J. C., et al. (2002). Ecological responses to recent climate change. Nature 416 389–395. 10.1038/416389a [DOI] [PubMed] [Google Scholar]
- Wehner J., Powell J. R., Muller L. A. H., Caruso T., Veresoglou S. D., Hempel S., et al. (2014). Determinants of root-associated fungal communities within Asteraceae in a semi-arid grassland. J. Ecol. 102 425–436. 10.1111/1365-2745.12197 [DOI] [Google Scholar]
- Wei Z., Yang T., Friman V.-P., Xu Y., Shen Q., Jousset A. (2015). Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nat. Commun. 6:8413 10.1038/ncomms9413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T. J., Bruns T., Lee S., Taylor J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. (New York, NY: Academic Press; ),k315–322. [Google Scholar]
- Wilschut R. A., Geisen S., Ten Hooven F. C., Van Der Putten W. H. (2016). Interspecific differences in nematode control between range-expanding plant species and their congeneric natives. Soil Biol. Biochem. 100 233–241. 10.1016/j.soilbio.2016.06.025 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.