Abstract
Background: Disrupted-in-schizophrenia 1 (DISC1) regulates neurogenesis and is a genetic risk factor for major psychiatric disorders. However, how DISC1 dysfunction affects neurogenesis and cell cycle progression at the molecular level is still unknown. Here, we investigated the role of DISC1 in regulating proliferation, migration, cell cycle progression and apoptosis in mouse neural stem/progenitor cells (MNSPCs) in vitro.
Methods: MNSPCs were isolated and cultured from mouse fetal hippocampi. Retroviral vectors or siRNAs were used to manipulate DISC1 expression in MNSPCs. Proliferation, migration, cell cycle progression and apoptosis of altered MNSPCs were analyzed in cell proliferation assays (MTS), transwell system and flow cytometry. A neurogenesis specific polymerase chain reaction (PCR) array was used to identify genes downstream of DISC1, and functional analysis was performed through transfection of expression plasmids and siRNAs.
Results: Loss of DISC1 reduced proliferation and migration of MNSPCs, while an increase in DISC1 led to increased proliferation and migration. Meanwhile, an increase in the proportion of cells in G0/G1 phase was concomitant with reduced levels of DISC1, but significant changes were not observed in the number MNSPCs undergoing apoptosis. Paired box gene 5 (Pax5), sex determining region Y-box 2 (Sox2), delta-like1 (Dll1) and Neurogenin2 (Neurog2) emerged as candidate molecules downstream of DISC1, and rescue experiments demonstrated that increased or decreased expression of either molecule regulated proliferation and migration in DISC1-altered MNSPCs.
Conclusion: These results suggest that Pax5, Sox2, Dll1 and Neurog2 mediate DISC1 activity in MNSPC proliferation and migration.
Keywords: DISC1, proliferation, migration, Pax5, Sox2, Dll1, Neurog2
Introduction
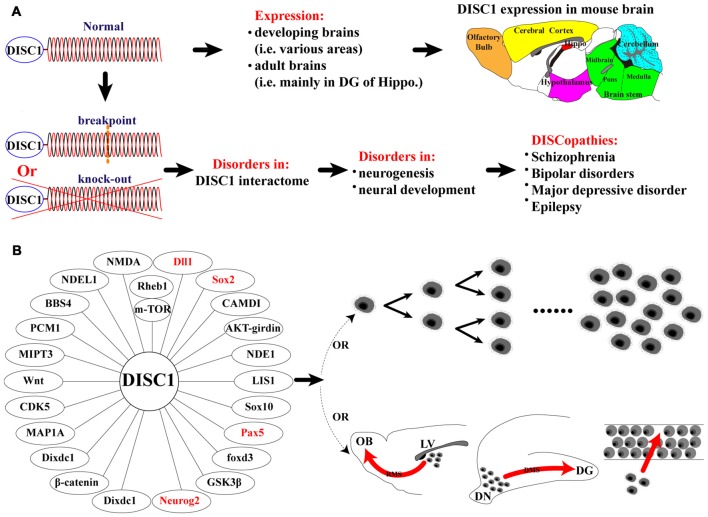
Until recently, many psychiatric disorders were undefined at the molecular level. These included schizophrenia (SZ; Le Strat et al., 2009), bipolar disorder (Blackwood et al., 2001b), major depressive disorder (Brandon et al., 2009) and epilepsy (Fournier et al., 2009, 2010). The discovery of disrupted-in-schizophrenia 1 (DISC1) as a genetic risk factor for these disorders marked a breakthrough in understanding psychiatric illness. Genetic analysis of a Scottish family revealed that family members affected by SZ inherited a balanced translocation between chromosomes 1 and 11 (t(1:11) (q42.1:q14.3); Millar et al., 2000), which encodes a protein now known as DISC1 (Blackwood et al., 2001a; Millar et al., 2004). DISC1 dysfunction is confirmed as a mechanism underlying these psychiatric disorders, now broadly termed “DISCopathies” (Figure 1A; Korth, 2009).
Figure 1.
Summary of findings of the present study and functions of disrupted-in-schizophrenia 1 (DISC1) on proliferation and migration. (A) Schematic representation of normal DISC1 protein and the translocation in the affected Scottish family, and physiological and pathological conditions for normal and disrupted DISC1; (B) Schematic representation of proteins that are known to interact with DISC1 regulating the proliferation and migration of mouse neural stem/progenitor cells (MNSPCs), neurons and other cell types.
DISC1 regulates almost all aspects of neurogenesis and neural development at various stages of brain development (Kang et al., 2011; Wu et al., 2013; Yerabham et al., 2013; Muraki and Tanigaki, 2015; Lee et al., 2015), including proliferation, migration, differentiation, neurite growth, morphogenesis and synaptogenesis. The molecular interaction network involving DISC1 has been referred to as the “DISC1 interactome” and plays a major role in neurogenesis and neural development (Figure 1A; Camargo et al., 2007; Korth, 2009). Members of this network include DISC1 binding partners such as Nuclear distribution protein nudE-like 1 (NDEL1), Fasciculation and elongation protein zeta-1 (FEZ1), lissencephaly (LIS1), Glycogen synthase kinase 3 beta (GSK3β), Girdin, Dixdc1 (Dixin), Growth factor receptor-bound protein 2 (Grb2), Ras homolog enriched in the brain 1 (Rheb1) and intracellular/extracellular signaling pathways, such as the AKT-mTOR, ERK, GSK3β, Wnt, NMDA-R, Reelin and GABA pathways (Fournier et al., 2010; Kang et al., 2011, 2015; Kim et al., 2012; Wu et al., 2013).
Despite our understanding of the DISC1 interactome (Camargo et al., 2007), few studies have focused on the role of DISC1 in the proliferation, migration, cell cycle progression and apoptosis of mouse neural stem/progenitor cells (MNSPCs). In this investigation, embryonic mouse hippocampal NSPCs were used as an in vitro cellular model to overexpress or silence DISC1 with retroviral expression vectors or siRNAs, and the biology of these altered MNSPCs was examined in a series of functional assays. To further understand the mechanisms of DISC1 regulated neurogenesis, a neurogenesis quantitative polymerase chain reaction (PCR) array was used to identify potential genes or signaling pathways associated with altered expression of DISC1. Paired box gene 5 (Pax5), sex determining region Y-box 2 (Sox2), Delta-like1 (Dll1) and Neurogenin2 (Neurog2) were among the candidate genes identified and were further examined in functional assays to determine whether they mediated DISC1 activity.
Materials and Methods
Ethics Statement
Animal protocols were approved by the ethics committee associated with Central South University (Changsha, Hunan, China). All experiments were performed in accordance with official recommendations by the Chinese animal community, and efforts were made to minimize the number of animals used and animal suffering.
Mice
C57/BL6 pregnant female mice (n = 4; P 14.5; Slac Laboratory Animal Co. Ltd, Shanghai, China) were used for the isolation and culture of MNSPCs.
Cell Culture
MNSPCs were isolated from the hippocampus of C57/BL6 fetal mouse brains as detailed in previous experiments (Yoneyama et al., 2011; Louis et al., 2013). Pregnant female mice (P 14.5) were anesthetized by i.p. injection with 10% chloral hydrate (0.1–0.2 mL/10 g), then the fetal mice were isolated from the uterus. The hippocampi of fetal mice were isolated using a dissecting microscope (Olympus). The hippocampi were washed in sterile cold phosphate buffered saline (PBS) in a 10 cm petri dish and cut into pieces using tissue scissors. The tissues were blown gently into smaller pieces using a 1 mL pipette, transferred into a 15 mL centrifuge tube, then D-HBSS was added, mixed into 10 mL suspension, and the tissues were incubated in room temperature (RT; 20–25°C) for 20 min with 1 mL ACCUTASE (A6964, Sigma). The tissue chunks were then spun down in a low-speed centrifuge at 700 g for 1 min at RT, and the supernatant was removed and transferred to a new 15 mL centrifuge tube. We determined the number of cells using a blood cell counting plate, seeded the cells at a density of 1 × 105/mL in a petri dish, and cultured the cells in NeuroCult™ Prol iferation Kit (Stem Cell; Vancouver, BC, Canada) to keep them in an undifferentiated, proliferative state.
siRNA Interference
Five siRNAs for each gene were designed against the functional sequences of DISC1, Pax5, Sox2, Dll1 and Neurog2 and sense and antisense sequences were chemically synthesized (Sigma). MNSPCs were seeded at a density of 2 × 105/mL in 24-well plates, and after 24 h were transfected with siRNAs at a concentration of 100 nM in Lipofectamine 2000 Reagent (Invitrogen, New York, NY, USA). The medium was replaced 48 h later, and the efficiency of gene knockdown was evaluated using real-time PCR (RT-PCR; Supplementary Figure S1).
Retroviral Infection
Packaging of Retroviral Vectors and Determination of Titers
A full-length mouse DISC1 sequence was ligated into the retroviral vector pCAG-EGFP (a generous gift from Dr. Chunmei Zhao, Laboratory of Genetics, The Salk Institute for Biological Studies, La Jolla, CA, USA; also available for purchase from Addgene; Cambridge, MA, USA), linearized with Pmel and Sfil restriction sites to generate the DISC1 expression construct CAG-DISC1-EGFP. The DISC1 retroviral construct and pCAG-EGFP were packaged into 293T cells with packaging plasmids pCMV-vsv-g and pCMV-gp (Addgene) and titered (as high as 108 c.f.u./mL) as previously described (Tashiro et al., 2006; Zhao et al., 2006; Jessberger et al., 2007).
Multiplicity of Infection (MOI)
Viral titers were serially diluted (104, 105 and 106) and applied to MNSPCs to determine the multiplicity of infection (MOI) prior to experimentation. MOI was assessed under fluorescence microscopy with expression of EGFP encoded by the retroviral vector. More than 60% of the MNSPCs were EGFP-positive after incubation with virus for 4 days at 104 c.f.u./mL and an MOI of 100.
Infection of Cells
MNSPCs (2 × 105/mL) were plated, and the medium was replaced 24 h later with 0.5 mL of 104 c.f.u./mL diluted, filtered virus containing polybrene at a concentration of 6 μg/mL (Sigma; St. Louis, MO, USA). The medium was changed after 2 days, and EGFP-positive colonies were collected for experiments.
Transfection of Cells by Electroporation
Expression plasmids for Pax5 (pPax5), Sox2 (pSox2), Dll1 (pDll1) and Neurog2 (pNeurog2) were constructed. One hundred microliter of cell suspension (107 cells/mL) was pipetted into a 4-mm electroporation cuvette and plasmids were added to a final concentration of 100–200 nM (unless otherwise specified) immediately before electroporation. Electroporation was performed with a Fischer electroporator (Fischer; Heidelberg, Germany) with a rectangle pulse of 330 V for 10 min. Cells were incubated for 15 min at RT, diluted 20-fold with culture medium, and incubated at 37°C in 5% CO2.
Real-Time PCR (RT-PCR)
RT-PCR was used to assess interference efficiency 48 h after cells were transfected with retrovirus vector, plasmids, or siRNAs. Total RNA was isolated with Trizol Reagent per the manufacturer’s protocols (Life Technologies; Grand Island, NY, USA). The samples were incubated with 1 μL DNase I (Life Technologies) for 30 min at 37°C to remove contaminating genomic DNA. cDNA was synthesized from RNA (200 ng) in a 20 μL reverse transcription reaction (Promega; Madison, WI, USA). RT-PCR was performed with SYBR Green qPCRSuperMix (Life Technologies) with the recommended program of one cycle at 95°C for 15 min, followed by 40 cycles at 95°C for 30 s and 60°C for 30 s on the ABI PRISM® 7500 Sequence Detection System. Primers used for RT-PCR of DISC1, Pax5, Sox2, Dll1 and Neurog2 are listed in Table 1.
Table 1.
Pairs of primer sequences for Disrupted-in-schizophrenia 1 (DISCI1), Paired box gene 5 (Pax5), sex determining region Y-box 2 (Sox2), deltalike1 (Dll1) and Neurogenin2 (Neurog2).
| Name | Fwd | Rev |
|---|---|---|
| DISC1 | 5′CTCGGAGCCATGTACAGTCA 3′ | 5′ACCAGCTGTCGGATAGGAA 3′ |
| Pax5 | 5′CAACAGGATCATTCGGACAA 3′ | 5′AGGATGCCACTGATGGAGTA 3′ |
| Sox2 | 5′CAAGATGCACAACTCGGAGAT 3′ | 5′TCATGAGCGTCTTGGTTTTC 3′ |
| Dll1 | 5′CTGTGGACTATAACCTCGTT 3′ | 5′ATCTTACACCTCAGTCGCTA 3′ |
| Neurog2 | 5′ATGGTCAAAGAGGACTATGG 3′ | 5′ATTCCCTCTGAGAGATTCAC 3′ |
| GAPDH | 5′GGCCTCCAAGGAGTAAGAAA 3′ | 5′GCCCCTCCTGTTATTATGG 3′ |
Relative expression levels were calculated as ratios normalized with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression. The RT-PCR data were analyzed using the ∆Ct method (Wang et al., 2010), and results were determined as the mean ± SD of three independent experiments.
Western Blotting
Equal protein quantities of lysates prepared from MNSPCs were subjected to SDS-PAGE and transferred to PVDF membranes. Membranes were blocked for 2 h in blocking buffer (5% non-fat dry milk in PBS with 0.1% Tween 20) at RT, incubated overnight at 4°C with primary antibodies (rabbit anti-DISC1 (1:1000; Life Technologies), rabbit anti-Pax5 (1:1000, Abcam; Cambridge, MA, USA), rabbit anti-Sox2 (1:1000, Cell Signaling Technology; Danvers, MA, USA), rabbit anti-Dll1 (1:500, Abcam), and rabbit anti-Neurog2 (1:10,000, Abcam)), followed by incubation with a conjugated secondary antibody (goat anti-rabbit IgG-HRP (1:10,000; Thermo Scientific; Pittsburgh, PA, USA)). Membranes were rinsed three times for 10 min after each incubation with Tris-buffered saline (TBS) containing 0.1% Tween-20. Protein bands were visualized using ECL reagent (Thermo Scientific), and densitometric analysis of protein bands was performed with Image-Pro Plus 6.0 software. All western blotting data are representative of at least three independent experiments.
Cell Proliferation Assay (MTS Assay)
MTS assay was performed to detect proliferation of MNSPCs. Approximately 10,000 cells in 100 μL of culture medium were seeded into three 96-well plates. Proliferation was determined at 0 h, 24 h, 48 h and 72 h following a 4 h incubation with 10 μL of MTS (3-(4,5-dimethylthiazol- 2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetra- zolium) solution (2 mg/mL, Promega, G3582). Colorimetric evaluation was performed at 490 nm in a mELISA microplate reader (Thermo Fisher Scientific, Waltham, MA, USA).
Flow Cytometric Detection of Apoptosis
Trypsinized cells (without EDTA) were rinsed twice in PBS and re-suspended in 1x binding buffer at a concentration of 1 × 106 cells/mL. Cells (100 μL of solution containing ~1 × 105 cells) were transferred to a 5 mL culture tube, and Annexin V-FITC antibody (5 μL) and propidium iodide (5 μL; BD Biosciences; San Jose, CA, USA) were added and incubated for 15 min at RT in the dark. Binding buffer (1×; 400 μL) was added, and the samples were analyzed by flow cytometry within 1 h on the FACSCalibur (Becton Dicknson; Franklin Lakes, NJ, USA).
Cell Cycle Analysis
Cells were harvested, rinsed twice with PBS, and re-suspended in the residual PBS. Fixation was performed with cold 75% ethanol (10 mL) added dropwise while vortexing, and cells were incubated at −20°C for a minimum of 2 h. Cells were pelleted by centrifugation, rinsed twice with PBS, and adjusted to a final concentration of 1 × 107 cells/mL. Aliquots (100 μL; 1 × 106 cells) of the cell suspension were then transferred into 12 × 75 mm tubes and incubated with PI-PBS (500 μL; 50 μg/mL PI, 100 μg/mL RNase A, 0.2% Triton X-100) after gentle vortexing for 30 min at 4°C. Samples were analyzed by flow cytometry on the FACSCalibur (Becton Dicknson, Franklin Lakes, NJ, USA), and data were analyzed with ModFit LT software (Becton Dickinson, Franklin Lakes, NJ, USA).
Transwell Migration Assay
Trypsinized cells suspended in serum-free DMEM medium were seeded onto untreated BD Falcon Cell Culture Inserts (8.0 μm Polyethylene terephthalate (PET) membranes; 0.8 × 106 pores 1 cm2) at a density of approximately 2.5 × 104 cells/cm2. The inserts were placed in the wells of BD Falcon multi-well plates containing 600 μL 10% FBS DMEM and incubated at 37°C in 5% CO2. After 24 h and 48 h, the cells were wiped off of the inserts with a cotton swab. The remaining cells migrating to the other side of the membrane were fixed with 4% paraformaldehyde (PFA) for 15 min and stained with Crystal Violet for 10 min, and then the average number of those cells was calculated to evaluate their migrated ability. Images were captured with the Olympus microscope.
Mouse Neurogenesis PCR Array
The Mouse Neurogenesis RT2Profiler™ PCR Array (PAHM 404Z, Qiagen, Shanghai, China) was used to profile 84 genes related to the process of neurogenesis (Supplementary Table S1). RNA was isolated with the RNasy Mini Kit and RNase-Free DNase Set (Qiagen; Valencia, CA, USA). The RT2 First Strand Kit (Qiagen) was used to synthesize cDNA, and RT-PCR was performed using SYBR Green Master mixes with the arrays on the Applied Bio-systems 7500HT system per the manufacturer’s instructions. Expression of each gene was normalized against the following endogenous control genes: β-actin (Actb), β2 microglobulin (B2m), GAPDH, β-Glucuronidase (Gusb), and heat shock protein 90 α, cytosolic, class B (Hsp90ab1). The Ct values of all the wells were exported to a blank Excel® spread sheet and analyzed with the SABiosciences (Qiagen) PCR Array Data Analysis Web-based software (RT2 Profiler PCR Array Data Analysis version 3.5).
Statistical Analysis
All statistical analyses were performed with Statistical Package for the Social Sciences (SPSS Inc. 2007, version 16.0; Chicago, IL, USA). Results are expressed as the mean ± SD. Data were analyzed with Student’s t-test, and p-value < 0.05 were considered statistically significant.
Results
Silence or Overexpression of DISC1 and Pax5, Sox2, Dll1 and Neurog2
To investigate the effect of DISC1, Pax5, Sox2, Dll1 and Neurog2 on various biological parameters in vitro, siRNAs were transfected into MNSPCs to knock down expression of the corresponding genes. Using RT-PCR, decreases in endogenous mRNA levels of DISC1, Pax5, Sox2, Dll1 and Neurog2 were observed in MNSPCs. DISC1siRNA (100 nM), Pax5siRNA (50 nM) and Sox2siRNA (100 nM), Dll1siRNA (50 nM), and Neurog2siRNA (50 nM) were effective at inhibiting the expression of DISC1, Pax5, Sox2, Dll1 and Neurog2. mRNA levels decreased by 90%, 72.5%, 72.7%, 80.7% and 80.9% respectively (Supplementary Figure S1).
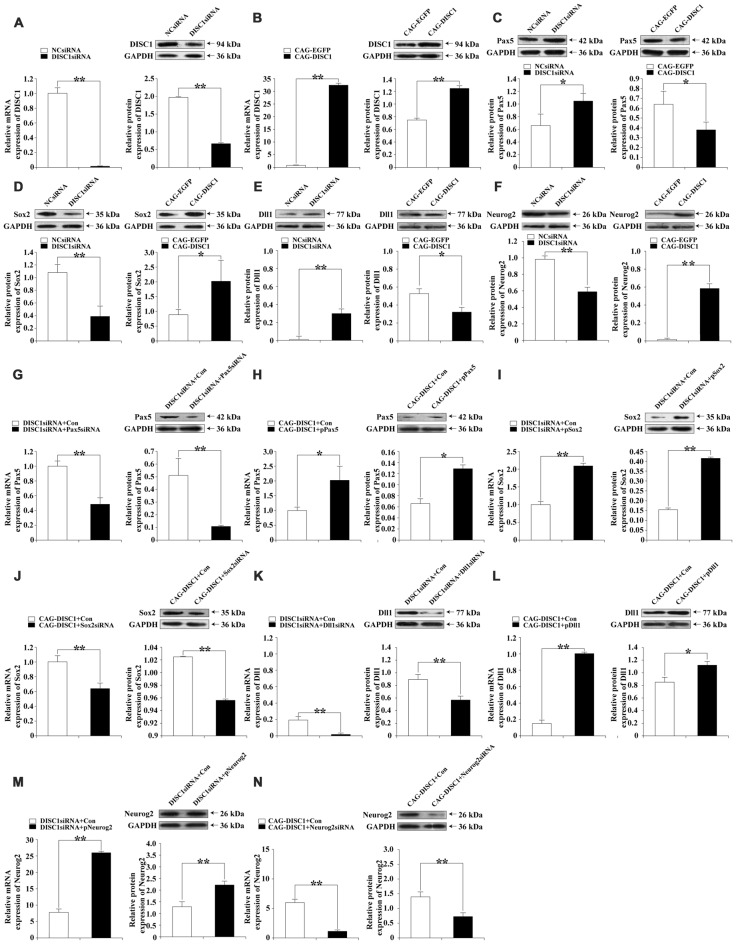
To determine whether the siRNAs (negative control siRNA (NCsiRNA) and DISC1siRNA) and expression constructs (CAG-EGFP and CAG-DISC1) affected protein levels, DISC1 protein was examined by Western blotting. RT-PCR and Western blotting were performed to detect the expression of DISC1 48 h after transfection with siRNAs or the expression constructs. siRNA led to decreases in DISC1 mRNA (Figure 2A, Left) and protein (Figure 2A, Right) levels by 53.2-fold and 2.9-fold, respectively, compared to controls. In contrast, transfection of the retroviral vector CAG-DISC1 increased mRNA (Figure 2B, Left) and protein (Figure 2B, Right) expression by 32.5- and 1.7-fold respectively. Changes in protein levels demonstrated that protein function may be altered as a result of transfection with siRNA or the expression construct.
Figure 2.
mRNA and protein expression levels of DISC1, Paired box gene 5 (Pax5), sex determining region Y-box 2 (Sox2), delta-like1 (Dll1) and Neurogenin2 (Neurog2) in knockdown and exogenous expression experiments. Real-time polymerase chain reaction (RT-PCR) and Western blotting were performed on cells where gene expression was modulated with siRNA, plasmid, or retroviral vector. RT-PCR and Western blots are paired according to the molecule analyzed for each gene interference experiment, except in (C–F) where only Western blots are shown. Bar graphs are used to display quantification by RT-PCR and Western blotting. The relative intensities of DISC1, Pax5, Sox2, Dll1 and Neurog2 by Western blot were normalized to the internal reference protein glyceraldehyde-3-phosphate dehydrogenase (GAPDH; KangChen Bio-tech, Shanghai, China). (A) DISC1, DISC1siRNA; (B) DISC1, CAG-DISC1; (C) Pax5, DISC1siRNA (Left); Pax5, CAG-DISC1 (Right); (D) Sox2, DISC1siRNA (Left); Sox2, CAG-DISC1 (Right); (E) Dll1, DISC1siRNA (Left); Dll1, CAG-DISC1 (Right); (F) Neurog2, DISC1siRNA (Left); Neurog2, CAG-DISC1 (Right); (G) Pax5, DISC1siRNA + Pax5siRNA; (H) Pax5, CAG-DISC1 + pPax5; (I) Sox2, DISC1siRNA + pSox2; (J) Sox2, CAG-DISC1 + Sox2siRNA; (K) Dll1, DISC1siRNA + Dll1siRNA; (L) Dll1, CAG-DISC1 + pDll1; (M) Neurog2, DISC1siRNA + pNeurog2; (N) Neurog2, CAG-DISC1 + Neurog2siRNA *p < 0.05; **p < 0.01 (controls-NCsiRNA, CAG-EGFP, DISC1siRNA + Con, CAG-DISC1 + Con). All results are from three independent experiments.
DISC1 Promotes Proliferation of MNSPCs
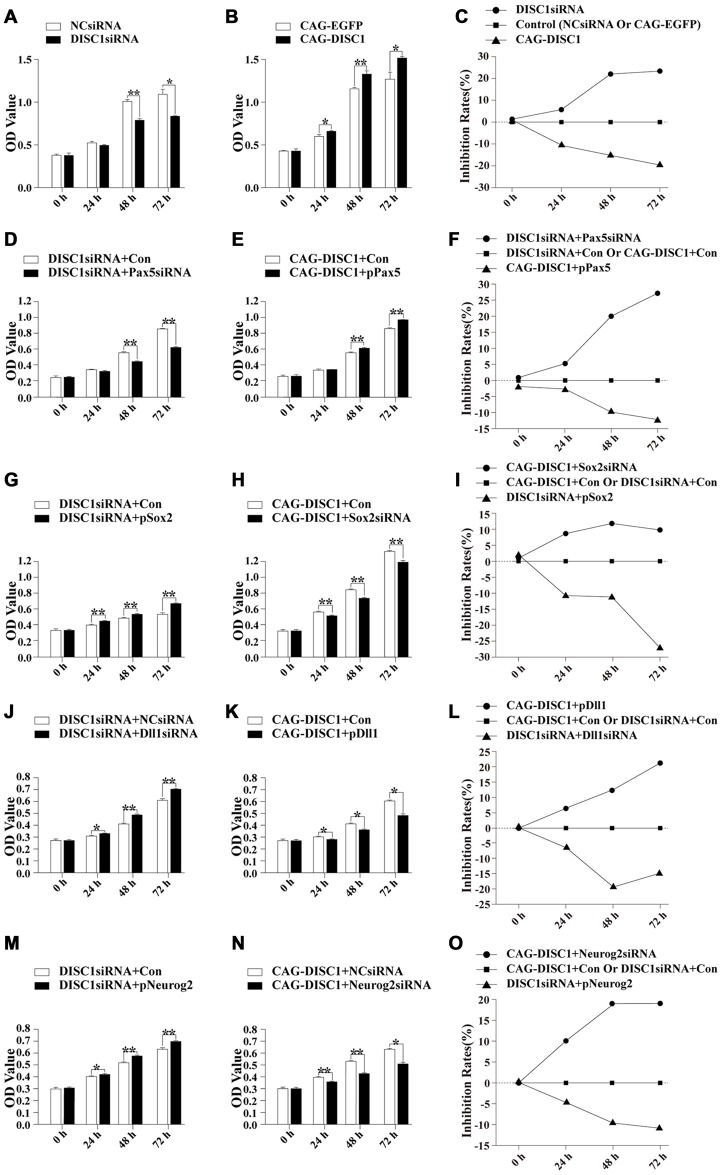
To examine the role of DISC1 on MNSPC proliferation, MTS assays were performed in cells transfected with siRNAs or expression construct. Proliferation of MNSPCs significantly diminished after siRNA knock-down of DISC1, whereas proliferation increased with increased DISC1 expression. We observed that siRNA inhibited proliferation of MNSPCs over time by 5.7%, 22% and 23.4% after 24 h, 48 h and 72 h respectively. In contrast, increased expression of DISC1 promoted cell growth by 10.6%, 15.2% and 19.6% after 24 h, 48 h, and 72 h, respectively (Figures 3A–C). These results indicate that the DISC1 gene had a role in promoting growth of MNSPCs in a time-dependent manner.
Figure 3.
DISC1 promotes proliferation of MNSPCs. Bar graphs representing OD values derived from MTS assays at 0, 24, 48, and 72 h and line charts are used to present rates of inhibition in different gene interference experiments in MNSPCs. Interference is performed with siRNA and/or expression vectors indicated. (A) OD, DISC1siRNA; (B) OD, CAG-DISC1; (C) inhibition rates by line chart, DISC1siRNA and CAG-DISC1; (D) OD, DISC1siRNA + Pax5siRNA; (E) OD, CAG-DISC1 + pPax5; (F) inhibition rates presented in line charts, Pax5siRNA or pPax5; (G) OD, DISC1siRNA + pSox2; (H) OD, CAG-DISC1 + Sox2siRNA; (I) inhibition rates presented in line charts, Sox2siRNA or pSox2; (J) OD, DISC1siRNA + Dll1siRNA; (K) OD, CAG-DISC1 + pDll1; (L) inhibition rates presented in line charts, Dll1siRNA or pDll1; (M) OD, DISC1siRNA + pNeurog2; (N) OD, CAG-DISC1 + Neurog2siRNA; (O) inhibition rates presented in line charts, Neurog2siRNA or pNeurog2. *p < 0.05, **p < 0.01. The rate of proliferation is expressed as a relative percentage of inhibition, which was calculated as follows: cell growth/proliferation inhibition rates (%) = [(A control − A sample)/A control] × 100% (controls-NCsiRNA, CAG-EGFP, DISC1siRNA + Con, CAG-DISC1 + Con). All results are from three independent experiments.
DISC1 Promotes MNSPC Migration
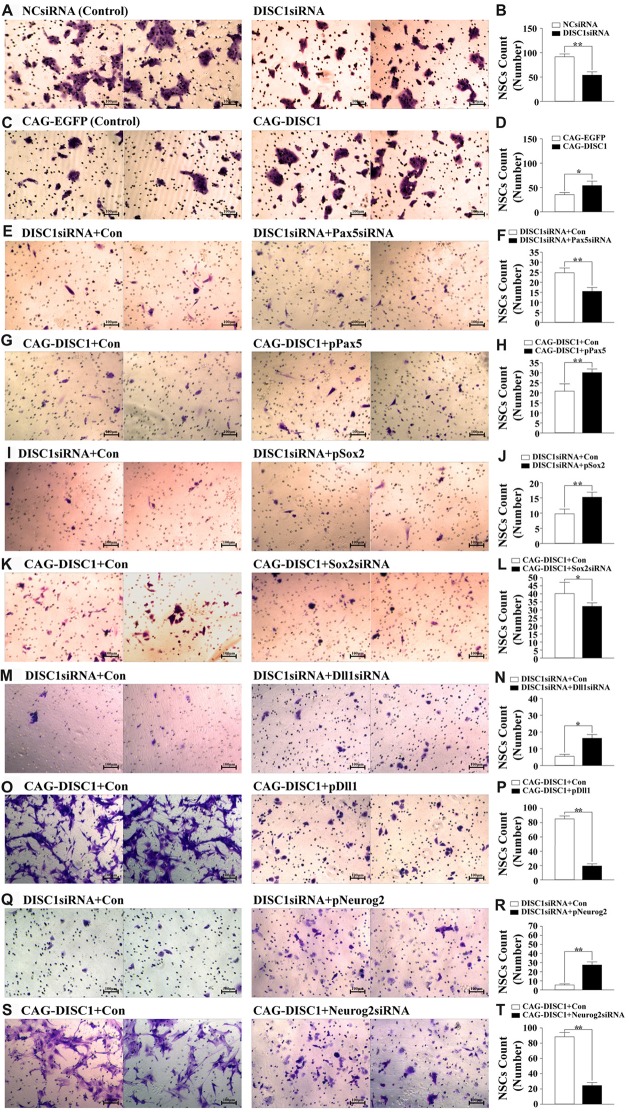
To determine how DISC1 influences migration, the ability of transfected MNSPCs to cross the Transwell membranes was examined. The average number of MNSPCs that migrated across the membrane after siRNA administration was fewer relative to controls (53.5 vs. 90.7 cells; p < 0.05; Figures 4A,B). DISC1 overexpression, however, led to increased MSPNC migration relative to the control group (53.7 vs. 34.8 cells; p < 0.01; Figures 4C,D). These results show that DISC1 mediates MNSPC.
Figure 4.
DISC1 promotes the migration of MNSPCs. Migration of control or altered MNSPCs was examined in transwell assays, and migrated cells were stained with crystal violet. Migrated cell counts are represented as bar graphs. Gene interference molecule (siRNA or expression construct) are as indicated. (A) Crystal violet stain, negative control siRNA (NCsiRNA) (control); DISC1siRNA; (B) bar graph, NCsiRNA (control) vs. DISC1siRNA; (C) crystal violet, CAG-EGFP (control); CAG-DISC1; (D) bar graph, CAG-EGFP (control) vs. CAG-DISC1; (E) crystal violet, DISC1siRNA (control); DISC1siRNA + Pax5siRNA; (F) bar graph, DISC1siRNA (control) vs. DISC1siRNA + Pax5siRNA; (G) crystal violet, CAG-DISC1 (control); CAG-DISC1 + pPax5; (H) bar graph, CAG-DISC1 (control) vs. CAG-DISC1 + pPax5; (I) crystal violet, DISC1siRNA (control); DISC1siRNA + pSox2; (J) bar graph, DISC1siRNA (control) vs. DISC1siRNA + pSox2; (K) crystal violet, CAG-DISC1 (control); CAG-DISC1 + Sox2siRNA; (L) bar graph, CAG-DISC1 (control) vs. CAG-DISC1 + Sox2siRNA; (M) crystal violet, DISC1siRNA (control); DISC1siRNA + Dll1siRNA; (N) bar graph, DISC1siRNA (control) vs. DISC1siRNA + Dll1siRNA; (O) crystal violet, CAG-DISC1 (control); CAG-DISC1 + pDll1; (P) bar graph, CAG-DISC1 (control) vs. CAG-DISC1 + pDll1; (Q) crystal violet, DISC1siRNA (control); DISC1siRNA + pNeurog2; (R) bar graph, DISC1siRNA (control) vs. DISC1siRNA + pNeurog2; (S) crystal violet, CAG-DISC1 (control); CAG-DISC1 + Neurog2siRNA; (T) bar graph, CAG-DISC1 (control) vs. CAG-DISC1 + Neurog2siRNA. *p < 0.05, **p < 0.01 (controls-NCsiRNA, CAG-EGFP, DISC1siRNA + Con, CAG-DISC1 + Con). All results are from three independent experiments.
DISC1 Influences Apoptosis and Cell Cycle Progression of MNSPCs
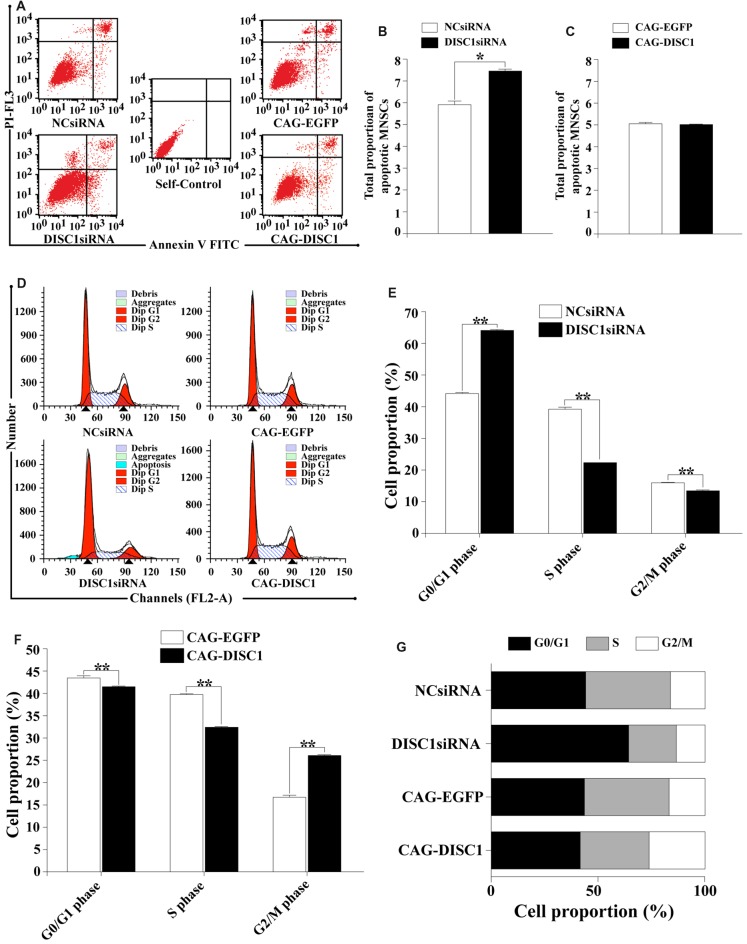
Apoptosis is an important cell function inhibiting cell growth. Flow cytometry was used to determine how DISC1 expression influences apoptosis in MNSPCs. Based on the total number of apoptotic events calculated in the four MNSPC groups, only DISC1siRNA altered apoptosis in MNSPCs. The percentage of MNSPCs undergoing apoptosis was greater with transfection of DISC1siRNA (7.5%) than NCsiRNA, CAG-EGFP and CAG-DISC1 (5%–6%; Figures 5A–C). These results suggest that loss of DISC1 only slightly increases apoptosis of MNSPCs.
Figure 5.
Interference of DISC1 expression alters apoptosis and cell cycle progression of MNSPCs. MNSPCs were analyzed by flow cytometry for apoptosis or cell cycle progression at the time points indicated following introduction of siRNA or expression constructs. (A) Measurement of apoptotic MNSPCs by flow cytometry after introduction of DISC1siRNA and CAG-DISC1. Quadrants for flow cytometry analysis are defined as the lower left (LL), lower right (LR), upper left (UL), and upper right (UR). Cells in LL (annexin V−-PI+) represent living cells; LR (annexin V+-PI−), early apoptotic cells or cytoclasis; UR (annexin V+-PI+), late apoptotic cells; UL (annexin V−-PI−), debris/cells with membrane only. UL + UR quadrants represent total MNSPCs undergoing apoptosis; (B,C) total proportion of MNSPCs undergoing apoptosis represented in bar graphs. *p < 0.05; (D) cell cycle analysis of MNSPCs transfected by NCsiRNA (control), DISC1siRNA, CAG-EGFP (control) and CAG-DISC1, respectively. Typical cell cycle profiles are presented where the first red peak corresponds to G1 phase, and the second peak, the G2 phase. The hatched blue region represents MNSPCs in S phase; (E,F) proportion of MNSPCs in G1/G0, S and G2/M presented in bar graphs. Constructs used are indicated; controls are compared to (E) DISC1siRNA and (F) CAG-DISC1. **p < 0.01; (G) cell cycle distribution of MNSPCs with DISC1siRNA or CAG-DISC1. Percentage of cells in G1 (black), S (gray), and G2/M (white) are represented in each bar for MNSPCs treated with the constructs indicated (controls-NCsiRNA, CAG-EGFP). All results are from three independent experiments.
Interestingly, both decreased and increased DISC1 levels altered cell cycle progression. Decreased expression of DISC1 inhibited cell growth as the average percentage of cells in G0/G1 phase increased from 44.2% to 64.2% (p < 0.01) and those in the S phase decreased from 39.7% to 22.3% at 48 h (p < 0.01). Cells in the G2/M phase had a decrease from 16.2% to 13.5% (p < 0.01; Figures 5D,E,G). In contrast, increased expression of DISC1 reduced the average percentage of cells in the G0/G1 phase (43.5% to 41.5%; p < 0.01) as well as in the S phase (39.6% to 32.3%; p < 0.01) relative to controls. The proportion of cells in the G2/M fraction, however, increased from 16.9% to 26.2% (p < 0.01; Figures 5D,F,G). These results show that alterations in DISC1 levels affect nearly all phases of the cell cycle in MNSPCs, with DISC1 reduction resulting in some form of G1 arrest.
DISC1 Levels Affect Neurogenic Specific Gene Expression
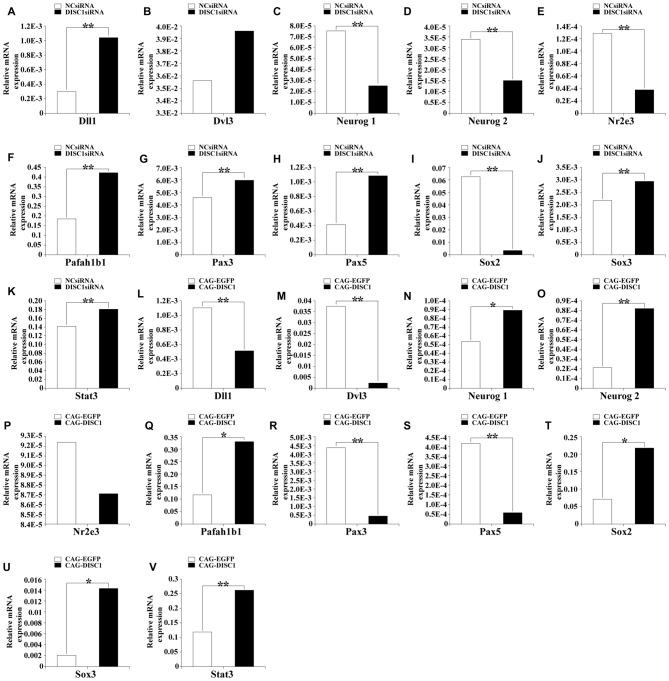
To identify candidate genes downstream of DISC1, expression levels of 84 neurogenesis-related genes in RNA isolated from transfected MNSPCs were determined using RT-PCR. In cells transfected with DISC1siRNA relative to controls, Neurog1, Neurog2, Nr2e3, and Sox2 were significantly decreased (3.0-, 2.3-, 3.3- and 16.6-fold, respectively; p < 0.01), whereas Dll1, Pafah1b1, and Pax5 were increased (3.4-, 2.3-, and 2.6-fold, respectively; p < 0.01; Figure 6). When DISC1 was overexpressed, mRNA levels of Dll1, Dvl3, Pax3, and Pax5 were down-regulated (2.1-, 14.3-, 9.1- and 6.9-fold, respectively; p < 0.01), whereas Neurog2, Pafah1b1, Sox2, Sox3, and Stat3 mRNA levels were upregulated (3.8-, 2.8-, 3.0-, 6.6- and 2.2-fold, respectively; Figure 6; p < 0.01). Taken together, these results show that Dll1, Neurog2, Pax5, and Sox2 are affected in opposite directions in response to increased or decreased DISC1 levels, while Pafah1b1 expression increases under both experimental conditions (Figures 6A–V).
Figure 6.
Pax5, Sox2, Dll1 and Neurog2 mRNA levels are regulated by DISC1, the levels were assessed by PCR array. Relative mRNA expression of several neurogenesis related genes after introduction of DISC1siRNA (A–K) or CAG-DISC1 (L–V) into MNSPCs (controls-NCsiRNA, CAG-EGFP). All results are from three independent experiments. *p < 0.05, **p < 0.01.
Pax5 or Sox2 or Dll1 or Neurog2 Protein Levels Are Regulated by DISC1
Based on the results of neurogenesis PCR array, expression of Pax5, Sox2, Dll1 and Neurog2 was further examined at the protein level. Western blotting was performed to determine how the protein levels of Pax5, Sox2, Dll1, or Neurog2 are affected by expression changes in DISC1 in MNSPCs. Transfection of MNSPCs with DISC1siRNA increased Pax5 (1.6-fold; p < 0.05; Figure 2C, Left) and Dll1 (9.7-fold; p < 0.01; Figure 2E, Left) protein levels after 48 h but decreased Sox2 (2.8-fold; p < 0.01; Figure 2D, Left) and Neurog2 (1.3-fold; p < 0.01; Figure 2F, Left) levels. In contrast, DISC1 overexpression resulted in a decrease in Pax5 (1.7-fold; p < 0.05; Figure 2C, Right) and Dll1 (1.6-fold; p < 0.05; Figure 2E, Right) levels after 48 h, but an increase in Sox2 (2.2-fold; p < 0.05; Figure 2D, Right) and Neurog2 (29.5-fold; p < 0.01; Figure 2F, Right) levels. Overall, Pax5, Sox2, Dll1, and Neurog2 levels parallel mRNA expression, suggesting that DISC1 regulates the expression of Pax5, Sox2, Dll1 and Neurog2 at both mRNA and protein levels.
Pax5, Sox2, Dll1 and Neurog2 siRNAs and Plasmids Alter mRNA and Protein Levels
RT-PCR and Western blotting were performed to determine mRNA and protein expression levels of Pax5, Sox2, Dll1, and Neurog2 48 h in MNSPCs where DISC1 levels were altered, after transfection with siRNA and expression plasmids. After transfection with their corresponding siRNA, Pax5 mRNA and protein levels decreased (2.0-fold and 4.1-fold, respectively; Figure 2G), as did Sox2 (1.6- and 1.1-fold, respectively; Figure 2J), Dll1 (15.8- and 1.6-fold, respectively; Figure 2K), and Neurog2 (5.9- and 1.9-fold, respectively; Figure 2N). Meanwhile, pPax5 expression plasmid increased Pax5 mRNA and protein levels (2.0- and 1.8-fold, respectively; Figure 2H), as did Sox2 (2.1- and 2.6-fold, respectively; Figure 2I), Dll1 (7.1- and 1.3-fold, respectively; Figure 2L), and Neurog2 (3.3- and 1.8-fold, respectively; Figure 2M). These results show that siRNAs down-regulate the expression of Pax5, Sox2, Dll1 and Neurog2, whereas pPax5, pSox2, pDll1 and pNeurog2 up-regulate the expression of Pax5, Sox2, Dll1 and Neurog2 in MNSPCs with increased or decreased DISC1.
Proliferation of MNSPCs in Pax5, Sox2, Dll1 or Neurog2 Rescue Experiment
The previous results show that DISC1siRNA: (1) decreases MNSPC proliferation; and (2) increases Pax5 and Dll1 while decreasing Sox2 and Neurog2 expression. Based on these observations, rescue experiments were designed to examine whether DISC1 regulates the MNSPC proliferation via Pax5, Sox2, Dll1, Neurog2, or a combination of the candidates. We expected that in DISC1siRNA MNSPCs, down-regulation of Pax5 by siRNA would enhance proliferation; surprisingly, proliferation was inhibited by 5.3%, 20.0%, and 27.1% at 24 h, 48 h and 72 h respectively. Conversely, we found that in CAG-DISC1 MNSPCs, up-regulation of Pax5 increased proliferation by 2.7%, 9.8%, and 12.3% after 24 h, 48 h and 72 h. These results suggest that Pax5 regulates the proliferation of MNSPCs independent of DISC1 (Figures 3D–F).
In the DISC1siRNA MNSPCs, up-regulation of Sox2 increased proliferation by 10.8%, 11.3%, and 26.8% after 24 h, 48 h and 72 h (Figures 3G,I), down-regulation of Dll1 increased proliferation by 6.5%, 19.5% and 14.8% (Figures 3J,L), up-regulation of Neurog2 increased proliferation by 5%, 9.6% and 11.1% (Figures 3M,O). In contrast, in CAG-DISC1 MNSPCs, down-regulation of Sox2 by siRNA inhibited proliferation by 8.6%, 11.8%, and 9.8% after 24 h, 48 h, and 72 h (Figures 3H,I), up-regulation of Dll1 inhibited proliferation by 6.7%, 12.2%, and 21.3% after 24 h, 48 h, and 72 h (Figures 3K,L), and down-regulation of Neurog2 inhibited proliferation by 10.0%, 18.9%, and 19.1% after 24 h, 48 h, and 72 h (Figures 3N,O). These results suggest that DISC1 and Sox2, Dll1 and Neurog2 co-regulate the MNSPC proliferation.
MNSPC Migration after Pax5, Sox2, Dll1 or Neurog2 Rescue Experiment
Rescue experiments were performed to investigate whether DISC1 regulates MNSPC migration via Pax5, Sox2, Dll1 or Neurog2. In DISC1siRNA MNSPCs, down-regulation of Pax5 by siRNA decreased the average number of migratory MNSPCs compared to the control (15.5 vs. 24.7; p < 0.01), contrary to our initial expectations. In CAG-DISC1 MNSPCs, up-regulation of Pax5 increased the average number of migratory MNSPCs compared to the control (30.2 vs. 21; p < 0.01). These results suggest that Pax5 regulates the migration of MNSPCs independent of DISC1 (Figures 4E–H).
In DISC1siRNA MNSPCs, up-regulation of Sox2 increased the average number of migratory MNSPCs (15.3 vs. 9.8; p < 0.01; Figures 4I,J), down-regulation of Dll1 increased the average number of migratory MNSPCs (15.8 vs. 4.8; p < 0.05; Figures 4M,N), and up-regulation of Neurog2 increased the average number of migratory MNSPCs compared to control (27.8 vs. 5.3; p < 0.01; Figures 4Q,R). In contrast, in CAG-DISC1 MNSPCs, down-regulation of Sox2 reduced the number of migratory MNSPCs (32 vs. 40; p < 0.05; Figures 4K,L), up-regulation of Dll1 decreased the average number of migratory MNSPCs (19 vs. 85.8; p < 0.01; Figures 4O,P), and down-regulation of Neurog2 reduced the average number of migratory MNSPCs compared to control (25 vs. 88.5; p < 0.01; Figures 4S,T). These results suggest that DISC1 regulates the migration of MNSPCs with Sox2, Dll1 or Neurog2.
Discussion
We used an in vitro model to characterize the function of DISC1 during neural development. We show that DISC1 promotes the proliferation and migration of MNSPCs, and loss of this protein profoundly alters cell cycle progression does not significantly alter apoptosis. Furthermore, experiments with knockdown or overexpression of Pax5, Sox2, Dll1 and Neurog2 suggest that DISC1 regulates the MNSPC proliferation and migration through one or more of these transcription factors.
Our observation that DISC1 positively regulates the proliferation and migration of MNSPCs corroborates previous studies performed in vivo on the embryonic and developing brain (Brandon and Sawa, 2011; Ishizuka et al., 2011). Neuronal proliferation and migration are essential to the process of neurogenesis and neural development (Tomita et al., 2011). DISC1 mediates the proliferation of neuronal progenitors in the developing cortex through the Wnt/β-catenin pathway (Mao et al., 2009), thus the phosphorylation of DISC1 may be a key molecular event from maintaining the proliferation of mitotic progenitor cells to activating the migration of post-mitotic neurons in the developing cortex (Ishizuka et al., 2011). However, the function of DISC1 on neuronal migration is complicated. DISC1 deficiency generally results in migration defects. For example, DISC1 knockdown results in migration delay in the developing neocortex (Duan et al., 2007; Kubo et al., 2010), but, DISC1 has opposite effects on neuronal migration in the developing vs. adult hippocampal neurons. While DISC1 deficiency delays migration in the developing hippocampal neurons, loss of DISC1 induces inappropriate migration in newly generated neurons in the adult hippocampus (Duan et al., 2007; Enomoto et al., 2009; Kim et al., 2009). DISC1 function also affects migration in a temporal manner. DISC1 knockdown does not alter migration of pyramidal cells in the developing CA1 at postnatal day 2 (P2; Meyer and Morris, 2009), whereas migration appears to be especially sensitive to DISC1 suppression from P2 to P3 (Meyer and Morris, 2009; Tomita et al., 2011). Moreover, DISC1 is known to regulate migration in a cell type specific manner. Loss of function of DISC1 hinders granule cell migration in the embryonic hippocampus, but fails to influence neuronal migration of pyramidal neurons in CA1 (Meyer and Morris, 2009), and DISC1 seems to have no impact on postnatal neuroblast migration within the rostral migratory stream (RMS; Wang et al., 2011). Further complicating the issue is an underlying difference in mouse strains due to a spontaneous 25-base pair (bp) deletion of DISC1. Swiss Webster, ICR, and 129S6/SvEv mice, for example, lose production of some specific isoforms of DISC1, which leads to a reduced or no apparent effect on neuronal migration, compared to C57BL/6 mice without the deletion (Ishizuka et al., 2006; Koike et al., 2006; Meyer and Morris, 2009). Taken together, the influence of DISC1 on neuronal migration seems to be affected by temporal and spatial differences, cell type specificity, and animal model. Although DISC1 is known to interact with several protein partners and extracellular/intracellular signaling pathways to co-regulate mainly neuronal proliferation and migration (Figure 1B; Kamiya et al., 2005; Duan et al., 2007; Namba et al., 2011), the exact molecular mechanisms remain to be elucidated.
In the present study, dysregulation of DISC1 strongly affects cell cycle progression of MNSPCs but only moderately affects apoptosis (Figure 5). Expression of mutated DISC1 has been previously associated with abnormalities in the expression of cell cycle genes and oligodendroglial progenitor markers (Katsel et al., 2011). DISC1 suppression reduces MNSPC proliferation and leads to premature exit from the cell cycle and differentiation (Mao et al., 2009). In addition, a partner of DISC1, the FEZ1, has been associated with the cell cycle and apoptosis in neuronal development (Assmann et al., 2006). A role for DISC1 in the cell cycle and apoptosis is observed in diseases of the central nervous system (CNS). For example, mitochondria involved in apoptosis are the predominant site of DISC1 expression, suggesting that the effect of DISC1 on apoptosis is related to neuronal disturbances in the pathogenesis of SZ (James et al., 2004). Furthermore, DISC1 is shown to be a target of miR-181b, a miRNA which may have a role in the pathogenesis of SZ (Carroll et al., 2013). Therefore, understanding the role of DISC1 in the cell cycle and apoptosis in different cell types requires further investigation.
Expression array analysis reveals Sox2, Pax5, Dll1 and Neurog2 as potential downstream or partner mediators of DISC1 (Figures 6A,D,H,I,L,O,S,T). Overexpression of the gene leads to increased Sox2 and Neurog2 but decreased Pax5 and Dll1 levels while suppression of DISC1 induces the opposite expression profile (Figures 2C–F). Therefore, we propose that DISC1 regulates proliferation and migration through one or more of these transcription factors. The results of Sox2, Dll1 and Neurog2 interference in functional assays parallel DISC1 expression, hence Sox2, Dll1 and Neurog2 appear to be necessary for proliferation and migration induced by DISC1 (Figures 3G–O, 4I–T). However, alterations in Pax5 expression yielded unexpected results in the functional assays, namely that increased Pax5 also promoted proliferation and migration rather than inhibiting these functions in response to DISC1 activity (Figures 3D–F, 4E–H). These results suggest that: (1) DISC1 regulates the proliferation and migration of MNSPCs through Sox2, Dll1 and Neurog2 and that exogenous intervention of Sox2, Dll1 and Neurog2 may help maintain the ability of MNSPCs to proliferate and migrate; and (2) Pax5 regulates the proliferation and migration of MNSPCs independent of DISC1, and endogenous changes in Pax5 levels may be a protective factor against DISC1 dysfunction, and thus Pax5 may act as a partner or co-regulator of DISC1.
Sox2 is a transcription factor that is expressed in MNSPCs and mature neurons in the mouse CNS and is necessary to maintain self-renewal or pluripotency of undifferentiated embryonic stem cells (Fong et al., 2008). Activated Sox2 promotes cell proliferation and differentiation in diverse cell types (Ferri et al., 2004; Que et al., 2009; Tompkins et al., 2011), and changes in levels of Sox2 transcription leads to abnormalities in the growth and development of stem cells (Bani-Yaghoub et al., 2006; Suh et al., 2007). In this study, Sox2 was also found to promote proliferation (Figures 3G–I) and migration (Figures 4I–L) of MNSPCs. Taken together, the data suggest that DISC1 regulates the proliferation and migration of MNSPCs directly or indirectly through Sox2.
Pax5 was originally identified as the B-cell-specific activator protein (BSAP; Adams et al., 1992), a transcription factor expressed at early but not late stages of B lymphocyte differentiation (Urbánek et al., 1994; Eberhard and Busslinger, 1999). Although expression of this gene was originally associated with proliferation of immune cell types, the phenotype of knock-out mice reveals a role for Pax5 in neural development. Transient expression of Pax5 occurs during embryogenesis in the mesencephalon and spinal cord in a spatial and temporal manner (Adams et al., 1992). Functionally, Pax5 over-expression reduces migration and motility of invasive MDA-MB231 cells, whereas the depletion of endogenous Pax5 enhances the migration of MCF-7 cells (Vidal et al., 2010). Moreover, Pax5 modulates the transcription of genes involved in the migration and adhesion of B cells, and promotes the adhesion of intercellular junctions in tumor cells (Schebesta et al., 2007). Our study found that Pax5 regulates the proliferation (Figures 3D–F) and migration (Figures 4E–H) of MNSPCs with DISC1. These findings suggest that Pax5 may stimulate proliferation and migration in a variety of cell types.
Notch ligand Dll1 may also be associated with DISC1-regulated neurogenesis. In the developing brain, steady Dll1 expression inhibits the proliferation of neural progenitors and accelerates neurogenesis (Shimojo et al., 2016), whereas oscillatory expression of Dll1 during embryonic development of the mammalian telencephalon is critical in neural fate decision (Barton and Fendrik, 2013) and cell differentiation in neural progenitor cells (Formosa-Jordan et al., 2012, Barton and Fendrik, 2013) through Notch signaling. In this study, Dll1 was found to regulate proliferation (Figures 3J–L) and migration (Figures 4M–P) of MNSPCs. Taken together, the data suggest that DISC1 regulates the proliferation and migration of MNSPCs directly or indirectly through Dll1.
Neurog2 is a member of the neurogenin subfamily of basic helix-loop-helix (bHLH) transcription factor genes that plays an important role in neurogenesis (Heng et al., 2008). Neurog2 is involved in the regulation of proliferation (Vied et al., 2014), migration, and apical progression (Kawaue et al., 2014) of the ventricular zone during neocortical development, as well as the control of successive steps of neurogenesis in the embryonic cerebral cortex (Azzarelli et al., 2014; Li et al., 2014). In this study, we found that Neurog2 also regulates proliferation (Figures 3M–O) and migration (Figures 4Q–T) of MNSPCs with DISC1, but how they work together requires further investigation.
The results from the expression array further suggest Pax3, Sox3, Dvl3 and Neurog1 as potential partners/mediators of DISC1 in regulating the proliferation or migration of MNSPCs. These observations provide new molecular clues for future studies of the “DISC1 interactome”. Pax3 mediates the migration of myoblasts in the developing limb bud through a signaling pathway which includes the activation of the c-Met (MET or Methylnitronitrosoguanidine HOS Transforming gene) receptor (Bottaro et al., 1991; Galland et al., 1992; Bober et al., 1994; Goulding et al., 1994; Bladt et al., 1995; Epstein, 1996). In addition, Pax3 is required for fine-tuning migration behavior of cardiac neural crest cells, but the protein is not essential for regulating their migration (Epstein et al., 2000). Pax3 was also found to be necessary for migration rather than differentiation in limb muscle precursors of mice (Daston et al., 1996). Although the exact role of Pax3 in MNSPC migration is unclear, Pax3 is considered as a candidate gene associated with DISC1 function based on our study.
Sox3 belongs to the high mobility group (HMG) family of transcription factors and is expressed in neural progenitor cells throughout the developing CNS. The protein is essential for the proliferation of neuroepithelial precursors (Rizzoti et al., 2004) and existing progenitors (Rogers et al., 2009). Furthermore, Xnr5, an early zygotic gene regulated by β-catenin/VegT, is a direct target of Sox3, suggesting that Sox3 mediates cell proliferation through Wnt signaling pathways (Zhang et al., 2003). Taken together, DISC1 appears to shape an intricate network associated with Sox2, Sox3 and Wnt signaling pathways which regulate cell proliferation.
Dvl3 is a Wnt signaling related gene whose mRNA levels changed in response to DISC1 in our study. A previous study reports that disheveled (Dvl; three isoforms) not only promotes β-catenin signaling in the Wnt pathway by stimulating GSK-3β phosphorylation but also promotes AKT signaling pathways by facilitating AKT phosphorylation. DISC1 participates in the WNT signaling pathway and in WNT-mediated cell proliferation, either in primary neural stem/progenitor in vitro or in embryonic brains (Mao et al., 2009). In addition to Dvl3, Pafah1b1 (also named lis1) has been shown to participate in neurodevelopmental processes such as neuronal precursor proliferation and differentiation, neuronal migration and neurite outgrowth, possibly in together with DISC1 through direct or indirect interactions. However, the exact mechanism remains to be defined (Lipska et al., 2006; Bradshaw et al., 2008, 2011; Rastogi et al., 2009; Bradshaw and Porteous, 2012). Additionally, Neurog1 is shown to influence the differentiation and migration of neural crest stem/progenitor cells and fate specification of embryonic stem cells (Velkey and O’Shea, 2013). Whether Neurog1 regulates neurogenesis via DISC1 requires further investigation.
Conclusion
Our findings reveal an important role for DISC1 in neurogenesis. First, DISC1 is required for the proliferation and migration of MNSPCs: DISC1 deficiency results in proliferation and migration defects, while DISC1 overexpression promotes both activities. Second, DISC1 regulates the proliferation and migration of MNSPCs in vitro with Pax5, Sox2, Dll1, and Neurog2, which may act as downstream mediators of DISC1 activity. Third, Pax3, Sox3, Dvl3, Neurog1 are potential partners of DISC1 in the regulation of neurogenesis.
DISC1 appears to be a crucial regulator not only of neural development but also of specific neurogenesis-related genes/molecules and signaling pathways. Further investigation of the molecular components of the DISC1 regulatory complex, as well as the nature of more detailed molecular interactions of DISC1 with Pax5, Sox2, Dll1 and Neurog2 in regulating migration, proliferation and the processes of neurogenesis such as differentiation is needed.
Author Contributions
QW: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript, provision of study material. WT: conception and design, collection and assembly of data, data analysis and interpretation, final approval of manuscript. ZL: conception and design, data analysis and interpretation, final approval of manuscript. YL, YS and ZY: conception and design, data analysis and interpretation. BX and LF: conception and design, financial support, final approval of manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81601134, 81100967, 81071048, 81371435, 81000553), and the Doctoral Scientific Research Foundation of First Affiliated Hospital of Kunming Medical University (2015BS007). We thank Chunmei Zhao for giving us suggestion for reconstructing the retroviral vector CAG-EGFP, and her gift of part of CAG-EGFP plasmid. We also thank Charlie W. Zhao for editing the final version of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fncel.2017.00261/full#supplementary-material
mRNA levels of Disrupted-in-schizophrenia 1 (DISC1), Paired box gene 5 (Pax5), and sex determining region Y-box 2 (Sox2) knocked down with siRNA. (A–E) siRNAs designed against each gene were transfected into cells and assessed for efficiency of knockdown by Real-time-polymerase chain reaction (RT-PCR); (F) Sense and anti-sense for each gene. All results are from three independent experiments. *p < 0.05, **p < 0.01.
References
- Adams B., Dürfler P., Aguzzi A., Kozmik Z., Urbánek P., Maurer-Fogy I., et al. (1992). Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes Dev. 6, 1589–1607. 10.1101/gad.6.9.1589 [DOI] [PubMed] [Google Scholar]
- Assmann E. M., Alborghetti M. R., Camargo M. E., Kobarg J. (2006). FEZ1 dimerization and interaction with transcription regulatory proteins involves its coiled-coil region. J. Biol. Chem. 281, 9869–9881. 10.1074/jbc.M513280200 [DOI] [PubMed] [Google Scholar]
- Azzarelli R., Pacary E., Garg R., Garcez P., van den Berg D., Riou P., et al. (2014). An antagonistic interaction between PlexinB2 and Rnd3 controls RhoA activity and cortical neuron migration. Nat. Commun. 5:3405. 10.1038/ncomms4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bani-Yaghoub M., Tremblay R. G., Lei J. X., Zhang D., Zurakowski B., Sandhu J. K., et al. (2006). Role of Sox2 in the development of the mouse neocortex. Dev. Biol. 295, 52–66. 10.1016/j.ydbio.2006.03.007 [DOI] [PubMed] [Google Scholar]
- Barton A., Fendrik A. J. (2013). Sustained vs. oscillating expressions of Ngn2, Dll1 and Hes1: a model of neural differentiation of embryonic telencephalon. J. Theor. Biol. 328, 1–8. 10.1016/j.jtbi.2013.03.004 [DOI] [PubMed] [Google Scholar]
- Blackwood D. H., Fordyce A., Walker M. T., St Clair D. M., Porteous D. J., Muir W. J. (2001a). Schizophrenia and affective disorders—cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am. J. Hum. Genet. 69, 428–433. 10.1086/321969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood D. H., Visscher P. M., Muir W. J. (2001b). Genetic studies of bipolar affective disorder in large families. Br. J. Psychiatry Suppl. 41, s134–s136. 10.1192/bjp.178.41.s134 [DOI] [PubMed] [Google Scholar]
- Bladt F., Riethmacher D., Isenmann S., Aguzzi A., Birchmeier C. (1995). Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature 376, 768–771. 10.1038/376768a0 [DOI] [PubMed] [Google Scholar]
- Bober E., Franz T., Arnold H. H., Gruss P., Tremblay P. (1994). Pax-3 is required for the development of limb muscles: a possible role for the migration of dermomyotomal muscle progenitor cells. Development 120, 603–612. [DOI] [PubMed] [Google Scholar]
- Bottaro D. P., Rubin J. S., Faletto D. L., Chan A. M., Kmiecik T. E., Vande Woude G. F., et al. (1991). Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science 251, 802–804. 10.1126/science.1846706 [DOI] [PubMed] [Google Scholar]
- Bradshaw N. J., Ogawa F., Antolin-Fontes B., Chubb J. E., Carlyle B. C., Christie S., et al. (2008). DISC1, PDE4B, and NDE1 at the centrosome and synapse. Biochem. Biophys. Res. Commun. 377, 1091–1096. 10.1016/j.bbrc.2008.10.120 [DOI] [PubMed] [Google Scholar]
- Bradshaw N. J., Porteous D. J. (2012). DISC1-binding proteins in neural development, signalling and schizophrenia. Neuropharmacology 62, 1230–1241. 10.1016/j.neuropharm.2010.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw N. J., Soares D. C., Carlyle B. C., Ogawa F., Davidson-Smith H., Christie S., et al. (2011). PKA phosphorylation of NDE1 is DISC1/PDE4 dependent and modulates its interaction with LIS1 and NDEL1. J. Neurosci. 31, 9043–9054. 10.1523/JNEUROSCI.5410-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon N. J., Millar J. K., Korth C., Sive H., Singh K. K., Sawa A. (2009). Understanding the role of DISC1 in psychiatric disease and during normal development. J. Neurosci. 29, 12768–12775. 10.1523/JNEUROSCI.3355-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon N. J., Sawa A. (2011). Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nat. Rev. Neurosci. 12, 707–722. 10.1038/nrn3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo L. M., Collura V., Rain J. C., Mizuguchi K., Hermjakob H., Kerrien S., et al. (2007). Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol. Psychiatry 12, 74–86. 10.1038/sj.mp.4001880 [DOI] [PubMed] [Google Scholar]
- Carroll A. P., Tooney P. A., Cairns M. J. (2013). Design and interpretation of microRNA-reporter gene activity. Anal. Biochem. 437, 164–171. 10.1016/j.ab.2013.02.022 [DOI] [PubMed] [Google Scholar]
- Daston G., Lamar E., Olivier M., Goulding M. (1996). Pax-3 is necessary for migration but not differentiation of limb muscle precursors in the mouse. Development 122, 1017–1027. [DOI] [PubMed] [Google Scholar]
- Duan X., Chang J. H., Ge S., Faulkner R. L., Kim J. Y., Kitabatake Y., et al. (2007). Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell 130, 1146–1158. 10.1016/j.cell.2007.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard D., Busslinger M. (1999). The partial homeodomain of the transcription factor Pax-5 (BSAP) is an interaction motif for the retinoblastoma and TATA-binding proteins. Cancer Res. 59, 1716s–1724s; discussion 1724s–1725s. [PubMed] [Google Scholar]
- Enomoto A., Asai N., Namba T., Wang Y., Kato T., Tanaka M., et al. (2009). Roles of disrupted-in-schizophrenia 1-interacting protein girdin in postnatal development of the dentate gyrus. Neuron 63, 774–787. 10.1016/j.neuron.2009.08.015 [DOI] [PubMed] [Google Scholar]
- Epstein J. A. (1996). Pax3, neural crest and cardiovascular development. Trends Cardiovasc. Med. 6, 255–260. 10.1016/s1050-1738(96)00110-7 [DOI] [PubMed] [Google Scholar]
- Epstein J. A., Li J., Lang D., Chen F., Brown C. B., Jin F., et al. (2000). Migration of cardiac neural crest cells in Splotch embryos. Development 127, 1869–1878. [DOI] [PubMed] [Google Scholar]
- Ferri A. L., Cavallaro M., Braida D., Di Cristofano A., Canta A., Vezzani A., et al. (2004). Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development 131, 3805–3819. 10.1242/dev.01204 [DOI] [PubMed] [Google Scholar]
- Fong H., Hohenstein K. A., Donovan P. J. (2008). Regulation of self-renewal and pluripotency by Sox2 in human embryonic stem cells. Stem Cells 26, 1931–1938. 10.1634/stemcells.2007-1002 [DOI] [PubMed] [Google Scholar]
- Formosa-Jordan P., Ibañes M., Ares S., Frade J. M. (2012). Regulation of neuronal differentiation at the neurogenic wavefront. Development 139, 2321–2329. 10.1242/dev.076406 [DOI] [PubMed] [Google Scholar]
- Fournier N. M., Andersen D. R., Botterill J. J., Sterner E. Y., Lussier A. L., Caruncho H. J., et al. (2010). The effect of amygdala kindling on hippocampal neurogenesis coincides with decreased reelin and DISC1 expression in the adult dentate gyrus. Hippocampus 20, 659–671. 10.1002/hipo.20653 [DOI] [PubMed] [Google Scholar]
- Fournier N. M., Caruncho H. J., Kalynchuk L. E. (2009). Decreased levels of disrupted-in-schizophrenia 1 (DISC1) are associated with expansion of the dentate granule cell layer in normal and kindled rats. Neurosci. Lett. 455, 134–139. 10.1016/j.neulet.2009.03.051 [DOI] [PubMed] [Google Scholar]
- Galland F., Stefanova M., Lafage M., Birnbaum D. (1992). Localization of the 5′ end of the MCF2 oncogene to human chromosome 15q15→q23. Cytogenet. Cell Genet. 60, 114–116. 10.1159/000133316 [DOI] [PubMed] [Google Scholar]
- Goulding M., Lumsden A., Paquette A. J. (1994). Regulation of Pax-3 expression in the dermomyotome and its role in muscle development. Development 120, 957–971. [DOI] [PubMed] [Google Scholar]
- Heng J. I., Nguyen L., Castro D. S., Zimmer C., Wildner H., Armant O., et al. (2008). Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature 455, 114–118. 10.1038/nature07198 [DOI] [PubMed] [Google Scholar]
- Ishizuka K., Kamiya A., Oh E. C., Kanki H., Seshadri S., Robinson J. F., et al. (2011). DISC1-dependent switch from progenitor proliferation to migration in the developing cortex. Nature 473, 92–96. 10.1038/nature09859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka K., Paek M., Kamiya A., Sawa A. (2006). A review of Disrupted-In-Schizophrenia-1 (DISC1): neurodevelopment, cognition, and mental conditions. Biol. Psychiatry 59, 1189–1197. 10.1016/j.biopsych.2006.03.065 [DOI] [PubMed] [Google Scholar]
- James R., Adams R. R., Christie S., Buchanan S. R., Porteous D. J., Millar J. K. (2004). Disrupted in Schizophrenia 1 (DISC1) is a multicompartmentalized protein that predominantly localizes to mitochondria. Mol. Cell. Neurosci. 26, 112–122. 10.1016/j.mcn.2004.01.013 [DOI] [PubMed] [Google Scholar]
- Jessberger S., Zhao C., Toni N., Clemenson G. D., Jr., Li Y., Gage F. H. (2007). Seizure-associated, aberrant neurogenesis in adult rats characterized with retrovirus-mediated cell labeling. J. Neurosci. 27, 9400–9407. 10.1523/JNEUROSCI.2002-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya A., Kubo K., Tomoda T., Takaki M., Youn R., Ozeki Y., et al. (2005). A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat. Cell Biol. 7, 1167–1178. 10.1038/ncb1328 [DOI] [PubMed] [Google Scholar]
- Kang E., Burdick K. E., Kim J. Y., Duan X., Guo J. U., Sailor K. A., et al. (2011). Interaction between FEZ1 and DISC1 in regulation of neuronal development and risk for schizophrenia. Neuron 72, 559–571. 10.1016/j.neuron.2011.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang E., Kim J. Y., Liu C. Y., Xiao B., Chen P. Y., Christian K. M., et al. (2015). Rheb1 mediates DISC1-dependent regulation of new neuron development in the adult hippocampus. Neurogenesis (Austin) 2:e1081715. 10.1080/23262133.2015.1081715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsel P., Tan W., Abazyan B., Davis K. L., Ross C., Pletnikov M. V., et al. (2011). Expression of mutant human DISC1 in mice supports abnormalities in differentiation of oligodendrocytes. Schizophr. Res. 130, 238–249. 10.1016/j.schres.2011.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaue T., Sagou K., Kiyonari H., Ota K., Okamoto M., Shinoda T., et al. (2014). Neurogenin2–d4Venus and Gadd45g-d4Venus transgenic mice: visualizing mitotic and migratory behaviors of cells committed to the neuronal lineage in the developing mammalian brain. Dev. Growth Differ. 56, 293–304. 10.1111/dgd.12131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraki K., Tanigaki K. (2015). Neuronal migration abnormalities and its possible implications for schizophrenia. Front. Neurosci. 9:74. 10.3389/fnins.2015.00074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. Y., Duan X., Liu C. Y., Jang M. H., Guo J. U., Pow-anpongkul N., et al. (2009). DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron 63, 761–773. 10.1016/j.neuron.2009.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. Y., Liu C. Y., Zhang F., Duan X., Wen Z., Song J., et al. (2012). Interplay between DISC1 and GABA signaling regulates neurogenesis in mice and risk for schizophrenia. Cell 148, 1051–1064. 10.1016/j.cell.2011.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike H., Arguello P. A., Kvajo M., Karayiorgou M., Gogos J. A. (2006). Disc1 is mutated in the 129S6/SvEv strain and modulates working memory in mice. Proc. Natl. Acad. Sci. U S A 103, 3693–3697. 10.1073/pnas.0511189103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korth C. (2009). DISCopathies: brain disorders related to DISC1 dysfunction. Rev. Neurosci. 20, 321–330. 10.1515/revneuro.2009.20.5-6.321 [DOI] [PubMed] [Google Scholar]
- Kubo K., Tomita K., Uto A., Kuroda K., Seshadri S., Cohen J., et al. (2010). Migration defects by DISC1 knockdown in C57BL/6, 129X1/SvJ, and ICR strains via in utero gene transfer and virus-mediated RNAi. Biochem. Biophys. Res. Commun. 400, 631–637. 10.1016/j.bbrc.2010.08.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Strat Y., Ramoz N., Gorwood P. (2009). The role of genes involved in neuroplasticity and neurogenesis in the observation of a gene-environment interaction (GxE) in schizophrenia. Curr. Mol. Med. 9, 506–518. 10.2174/156652409788167104 [DOI] [PubMed] [Google Scholar]
- Lee H., Kang E., GoodSmith D., Yoon D. Y., Song H., Knierim J. J., et al. (2015). DISC1-mediated dysregulation of adult hippocampal neurogenesis in rats. Front. Syst. Neurosci. 9:93. 10.3389/fnsys.2015.00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Mattar P., Dixit R., Lawn S. O., Wilkinson G., Kinch C., et al. (2014). RAS/ERK signaling controls proneural genetic programs in cortical development and gliomagenesis. J. Neurosci. 34, 2169–2190. 10.1523/JNEUROSCI.4077-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipska B. K., Peters T., Hyde T. M., Halim N., Horowitz C., Mitkus S., et al. (2006). Expression of DISC1 binding partners is reduced in schizophrenia and associated with DISC1 SNPs. Hum. Mol. Genet. 15, 1245–1258. 10.1093/hmg/ddl040 [DOI] [PubMed] [Google Scholar]
- Louis S. A., Mak C. K., Reynolds B. A. (2013). Methods to culture, differentiate, and characterize neural stem cells from the adult and embryonic mouse central nervous system. Methods Mol. Biol. 946, 479–506. 10.1007/978-1-62703-128-8_30 [DOI] [PubMed] [Google Scholar]
- Mao Y., Ge X., Frank C. L., Madison J. M., Koehler A. N., Doud M. K., et al. (2009). Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3β/β-catenin signaling. Cell 136, 1017–1031. 10.1016/j.cell.2008.12.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. D., Morris J. A. (2009). Disc1 regulates granule cell migration in the developing hippocampus. Hum. Mol. Genet. 18, 3286–3297. 10.1093/hmg/ddp266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar J. K., James R., Brandon N. J., Thomson P. A. (2004). DISC1 and DISC2: discovering and dissecting molecular mechanisms underlying psychiatric illness. Ann. Med. 36, 367–378. 10.1080/07853890410033603 [DOI] [PubMed] [Google Scholar]
- Millar J. K., Wilson-Annan J. C., Anderson S., Christie S., Taylor M. S., Semple C. A., et al. (2000). Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet. 9, 1415–1423. 10.1093/hmg/9.9.1415 [DOI] [PubMed] [Google Scholar]
- Namba T., Ming G. L., Song H., Waga C., Enomoto A., Kaibuchi K., et al. (2011). NMDA receptor regulates migration of newly generated neurons in the adult hippocampus via Disrupted-In-Schizophrenia 1 (DISC1). J. Neurochem. 118, 34–44. 10.1111/j.1471-4159.2011.07282.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que J., Luo X., Schwartz R. J., Hogan B. L. (2009). Multiple roles for Sox2 in the developing and adult mouse trachea. Development 136, 1899–1907. 10.1242/dev.034629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi A., Zai C., Likhodi O., Kennedy J. L., Wong A. H. (2009). Genetic association and post-mortem brain mRNA analysis of DISC1 and related genes in schizophrenia. Schizophr. Res. 114, 39–49. 10.1016/j.schres.2009.06.019 [DOI] [PubMed] [Google Scholar]
- Rizzoti K., Brunelli S., Carmignac D., Thomas P. Q., Robinson I. C., Lovell-Badge R. (2004). SOX3 is required during the formation of the hypothalamo-pituitary axis. Nat. Genet. 36, 247–255. 10.1038/ng1309 [DOI] [PubMed] [Google Scholar]
- Rogers C. D., Harafuji N., Archer T., Cunningham D. D., Casey E. S. (2009). Xenopus Sox3 activates sox2 and geminin and indirectly represses Xvent2 expression to induce neural progenitor formation at the expense of non-neural ectodermal derivatives. Mech. Dev. 126, 42–55. 10.1016/j.mod.2008.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schebesta A., McManus S., Salvagiotto G., Delogu A., Busslinger G. A., Busslinger M. (2007). Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity 27, 49–63. 10.1016/j.immuni.2007.05.019 [DOI] [PubMed] [Google Scholar]
- Shimojo H., Isomura A., Ohtsuka T., Kori H., Miyachi H., Kageyama R. (2016). Oscillatory control of Delta-like1 in cell interactions regulates dynamic gene expression and tissue morphogenesis. Genes Dev. 30, 102–116. 10.1101/gad.270785.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H., Consiglio A., Ray J., Sawai T., D’Amour K. A., Gage F. H. (2007). In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell 1, 515–528. 10.1016/j.stem.2007.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro A., Zhao C., Gage F. H. (2006). Retrovirus-mediated single-cell gene knockout technique in adult newborn neurons in vivo. Nat. Protoc. 1, 3049–3055. 10.1038/nprot.2006.473 [DOI] [PubMed] [Google Scholar]
- Tomita K., Kubo K., Ishii K., Nakajima K. (2011). Disrupted-in-Schizophrenia-1 (Disc1) is necessary for migration of the pyramidal neurons during mouse hippocampal development. Hum. Mol. Genet. 20, 2834–2845. 10.1093/hmg/ddr194 [DOI] [PubMed] [Google Scholar]
- Tompkins D. H., Besnard V., Lange A. W., Keiser A. R., Wert S. E., Bruno M. D., et al. (2011). Sox2 activates cell proliferation and differentiation in the respiratory epithelium. Am. J. Respir. Cell Mol. Biol. 45, 101–110. 10.1165/rcmb.2010-0149oc [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbánek P., Wang Z. Q., Fetka I., Wagner E. F., Busslinger M. (1994). Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell 79, 901–912. 10.1016/0092-8674(94)90079-5 [DOI] [PubMed] [Google Scholar]
- Velkey J. M., O’Shea K. S. (2013). Expression of Neurogenin 1 in mouse embryonic stem cells directs the differentiation of neuronal precursors and identifies unique patterns of down-stream gene expression. Dev. Dyn. 242, 230–253. 10.1002/dvdy.23920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal L. J., Perry J. K., Vouyovitch C. M., Pandey V., Brunet-Dunand S. E., Mertani H. C., et al. (2010). PAX5α enhances the epithelial behavior of human mammary carcinoma cells. Mol. Cancer Res. 8, 444–456. 10.1158/1541-7786.MCR-09-0368 [DOI] [PubMed] [Google Scholar]
- Vied C. M., Freudenberg F., Wang Y., Raposo A. A., Feng D., Nowakowski R. S. (2014). A multi-resource data integration approach: identification of candidate genes regulating cell proliferation during neocortical development. Front. Neurosci. 8:257. 10.3389/fnins.2014.00257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Liu X., Wu H., Ni P., Gu Z., Qiao Y., et al. (2010). CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 38, 5366–5383. 10.1093/nar/gkq285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Kaneko N., Asai N., Enomoto A., Isotani-Sakakibara M., Kato T., et al. (2011). Girdin is an intrinsic regulator of neuroblast chain migration in the rostral migratory stream of the postnatal brain. J. Neurosci. 31, 8109–8122. 10.1523/jneurosci.1130-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Li Y., Xiao B. (2013). DISC1-related signaling pathways in adult neurogenesis of the hippocampus. Gene 518, 223–230. 10.1016/j.gene.2013.01.015 [DOI] [PubMed] [Google Scholar]
- Yerabham A. S., Weiergraber O. H., Bradshaw N. J., Korth C. (2013). Revisiting disrupted-in-schizophrenia 1 as a scaffold protein. Biol. Chem. 394, 1425–1437. 10.1515/hsz-2013-0178 [DOI] [PubMed] [Google Scholar]
- Yoneyama M., Kawada K., Shiba T., Ogita K. (2011). Endogenous nitric oxide generation linked to ryanodine receptors activates cyclic GMP/protein kinase G pathway for cell proliferation of neural stem/progenitor cells derived from embryonic hippocampus. J. Pharmacol. Sci. 115, 182–195. 10.1254/jphs.10290fp [DOI] [PubMed] [Google Scholar]
- Zhang C., Basta T., Jensen E. D., Klymkowsky M. W. (2003). The β-catenin/VegT-regulated early zygotic gene Xnr5 is a direct target of SOX3 regulation. Development 130, 5609–5624. 10.1242/dev.00798 [DOI] [PubMed] [Google Scholar]
- Zhao C., Teng E. M., Summers R. G., Jr., Ming G. L., Gage F. H. (2006). Distinct morphological stages of dentate granule neuron maturation in the adult mouse hippocampus. J. Neurosci. 26, 3–11. 10.1523/JNEUROSCI.3648-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
mRNA levels of Disrupted-in-schizophrenia 1 (DISC1), Paired box gene 5 (Pax5), and sex determining region Y-box 2 (Sox2) knocked down with siRNA. (A–E) siRNAs designed against each gene were transfected into cells and assessed for efficiency of knockdown by Real-time-polymerase chain reaction (RT-PCR); (F) Sense and anti-sense for each gene. All results are from three independent experiments. *p < 0.05, **p < 0.01.