Abstract
Background & Aims
The human hepatitis B virus (HBV) is a major cause of chronic hepatitis and hepatocellular carcinoma, but molecular mechanisms driving liver disease and carcinogenesis are largely unknown. We therefore studied cellular pathways altered by HBV infection.
Methods
We performed gene expression profiling of primary human hepatocytes infected with HBV and proved the results in HBV-replicating cell lines and human liver tissue using real-time polymerase chain reaction and Western blotting. Activation of signal transducer and activator of transcription (STAT3) was examined in HBV-replicating human hepatocytes, HBV-replicating mice, and liver tissue from HBV-infected individuals using Western blotting, STAT3-luciferase reporter assay, and immunohistochemistry. The consequences of STAT3 activation on HBV infection and cell survival were studied by chemical inhibition of STAT3 phosphorylation and small interfering RNA–mediated knockdown of STAT3.
Results
Gene expression profiling of HBV-infected primary human hepatocytes detected no interferon response, while genes encoding for acute phase and antiapoptotic proteins were up-regulated. This gene regulation was confirmed in liver tissue samples of patients with chronic HBV infection and in HBV-related hepatocellular carcinoma. Pathway analysis revealed activation of STAT3 to be the major regulator. Interleukin-6–dependent and –independent activation of STAT3 was detected in HBV-replicating hepatocytes in cell culture and in vivo. Prevention of STAT3 activation by inhibition of Janus tyrosine kinases as well as small interfering RNA–mediated knockdown of STAT3-induced apoptosis and reduced HBV replication and gene expression.
Conclusions
HBV activates STAT3 signaling in hepatocytes to foster its own replication but also to prevent apoptosis of infected cells. This very likely supports HBV-related carcinogenesis.
Keywords: Hepatitis B Virus Infection, STAT3 Signaling, Hepatocellular Carcinoma, Apoptosis
Abbreviations used in this paper: APR, acute phase response; cDNA, complementary DNA; cccDNA, covalently closed circular DNA; cRNA, complementary RNA; CRP, C-reactive protein; DMSO, dimethyl sulfoxide; FCS, fetal calf serum; HBeAg, hepatitis B early antigen; HBV, Hepatitis B virus; HBV pg RNA, hepatitis B pregenomic RNA; HBVtg, hepatitis B transgenic; HCC, hepatocellular carcinoma; HNF, hepatocyte nuclear factor; IFN, interferon; IL-6, interleukin 6; IRF3, interferon regulatory factor 3; mRNA, messenger RNA; NAC, N-acetyl-L-cysteine; p.i., postinfection; PCR, polymerase chain reaction; pgRNA, pregenomic RNA; PHH, primary human hepatocyte; pSTAT3, phosphorylated signal transducer and activator of transcription 3; ROS, reactive oxygen species; RT, reverse transcription; siRNA, small interfering RNA; STAT3, signal transducer and activator of transcription 3
Graphical abstract
Summary.
We show that hepatitis B virus infection activates STAT3 signaling that supports virus replication and prevents apoptosis of infected hepatocytes potentially supporting malignant transformation. Our findings provide new insides into hepatitis B virus–host interaction and open a new avenue to the development of drugs that control the infection and may help to prevent carcinoma development.
The hepatitis B virus (HBV) is a small, enveloped DNA virus characterized by a pronounced liver tropism and replication via reverse transcription (RT) of an RNA pregenome. Despite an effective prophylactic vaccine, HBV infection still is a major health problem, with more than 240 million chronically infected individuals, who are at high risk to develop liver cirrhosis, end-stage liver disease and hepatocellular carcinoma (HCC). The virus escapes efficient immune elimination by a very limited activation of innate and adaptive immune responses in the liver.1 With the introduction of antivirals, treatment options for chronic hepatitis B improved over the last years, but a curative treatment is still lacking.2 Although rates of HBV-related HCC are slowly decreasing,3 HCC still rates number 6 among the most frequent cancers and is the number 3 cause of cancer-related death with about half of all HCC related to HBV infection.4 Advanced liver disease with liver cirrhosis due to ongoing hepatocellular activation and inflammation are major risk factors.5, 6 Persistent viral replication, male sex and a positive family history increase the risk for HBV-related HCC.6
A strong correlation between HBV viral load and the risk of HCC development has been established in large clinical trials.5 In the absence of a dominant oncogene encoded by the HBV genome, the role of HBV in carcinogenesis is complex7 and still incompletely understood. Direct as well as indirect roles of HBV have been proposed.8 Integration of HBV DNA into the host genome occurs at early steps of clonal tumor expansion. This may activate cellular cancer-related genes and very likely induces the host chromosomal instability. Hereby, prolonged expression of the viral regulatory HBV X protein may contribute to de-regulating cellular transcription and influences protein degradation, cell proliferation and apoptotic signaling pathways (summarized in Neuveut et al,8 Ringelhan et al,9 and Tan10).
In a number of clinical settings and disease entities chronic liver inflammation seems to be sufficient to induce HCC development.11 Pioneering work by Nakamoto et al12 provided first experimental evidence that HBV-related HCC may develop in the absence of viral transactivation, insertional mutagenesis, and genotoxic chemicals12 solely triggered by the immune response to HBV and resulting chronic inflammation. Key signaling pathways contributing to HCC development, however, have only partially been identified.
Lymphotoxins (LTs) and their receptor are up-regulated in viral hepatitis and related HCC, and sustained triggering of the LT-beta receptor resulting in canonical and noncanonical NF-kb signaling leads to HCC development.13 We have previously demonstrated that nonparenchymal liver cells, particularly liver macrophages, recognize HBV particles resulting in an activation of NF-kb signaling and production of proinflammatory cytokines (eg, tumor necrosis factor-α, interleukin 6 [IL-6]).14 IL-6 induces signal transducer and activator of transcription (STAT3) signaling in hepatocytes, and NF-kB and STAT3 have been described to be key players in liver inflammation and cancer (reviewed in He and Karin15).
In this study, we aimed at identifying major signaling pathways activated by HBV infection in hepatocytes. As signaling cascades may be largely affected in immortalized cell lines as well as by overexpression of viral proteins, we studied the influence of HBV replication on cellular gene expression in infected primary human liver cell cultures prepared from different donors and in human liver tissues derived from patients with chronic hepatitis B and HBV-related HCC. Finally, we corroborated our findings in transgenic mice expressing the complete genome of HBV in hepatocytes16 and in mice challenged intravenously with adenoviral vectors encoding a replication competent HBV genome (Ad HBV).17, 18, 19
Methods
Chemicals
The pharmacological inhibitor AG-490 was purchased from Calbiochem (San Diego, CA). Dimethyl sulfoxide (DMSO) and N-acetyl-L-cysteine (NAC) were obtained from Sigma (St. Louis, MO).
Ethics Statement
The study followed the ethical guidelines of the Declaration of Helsinki and use of human material was approved by the local ethics committees of the University Hospital Cologne, the University of Zürich, and the Hospital rechts der Isar der Technischen Universität München. Liver cell cultures were prepared from surgical human liver biopsies after written informed consent of patients. Studies on mice were conducted in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (USA). Our animal protocol was approved by the Regierung Oberbayern (Munich, Germany).
Patients
Samples of HBV-positive tumor tissue or noncancerous HBV-infected liver tissue were recruited from the tissue bank of the Institute of Pathology, University of Cologne, and the Institute of Surgical Pathology, University Hospital Zürich, after obtaining written informed consent from all patients. All tissue samples stained positive for hepatitis B core antigen and hepatitis B surface antigen. Active HBV replication in all noncancerous HBV-infected liver tissue samples was confirmed by the detection of pregenomic RNA (pgRNA) using real-time RT polymerase chain reaction (PCR). All tumor samples were HBV-related HCC with Edmonson’s grade II or III of histological differentiation, whereas other causative agents for HCC were excluded. Noninfected (HBV negative, hepatitis C virus negative, human immunodeficiency virus negative) liver tissue was obtained from human liver grafts not suited for transplantation.
Infection of Primary Human Hepatocytes With HBV and Analysis of Infection
Primary human hepatocyte (PHH) cultures were prepared from surgical human liver biopsies after written informed consent of patients by a standard 2-step collagenase perfusion followed by a differential centrifugation as described previously.20 PHH cultures contained ≥85% hepatocytes and 3%–15% of liver sinusoidal endothelial cells or liver macrophages, Kupffer cells.14 HBV inoculum was concentrated from the medium of HepG2.2.15 cells21 using centrifugal filter devices (Centricon Plus-70, Biomax 100.000, Millipore Corp, Bedford, MA) and sedimented into a CsCl gradient to determine the amount of enveloped, DNA-containing viral particles as described before.22 Mock inoculum was concentrated from HepG2 cells accordingly. On day 3 after seedling, PHHs were infected with HBV at a multiplicity of infection of 250 enveloped DNA-containing viral particles per cell in the presence of 5% PEG 6000. At 24 hours after infection with HBV, the inoculum was removed, and cells were cultivated in PHH medium23 at 37°C in 5% CO2 for next 4 days.
Cell culture medium was collected from day 1 to day 4 postinfection (p.i.), and hepatitis B early antigen (HBeAg) secretion was determined using the HBeAg 2.0 Axsym immuno assay (Abbott Laboratories, Wiesbaden, Germany). In addition, IL-6 secretion was determined using IL-6 enzyme-linked immunosorbent assay (ELISA) (BD Biosciences, Heidelberg, Germany) and IFNα production was examined using interferon-α (IFNα) ELISA (PBL Biomedical Laboratories Piscataway, NJ).
Formation of HBV covalently closed circular DNA (cccDNA) was detected by real-time PCR on a LightCycler instrument (Roche Diagnostics, Mannheim, Germany) using specific primers ccc2760fw and HBVccc156rev selectively amplifying cccDNA over relaxed circular DNA as described previously.24 Total DNA extracted from mock- (control) or HBV-infected PHHs was subjected to real-time PCR analysis to specifically detect HBV cccDNA. DNA extracted from patient serum and from serum of an HBV-positive chimpanzee as well as total liver DNA extracted from HBV-positive patient served as negative and positive controls, respectively.
To estimate relative efficiency of HBV replication in PHHs, levels of HBV pgRNA were determined relative to those in PHH transduced with AdG-HBV1.3 vector24 using RT-PCR and pgRNA specific primers.23 After infection with AdG-HBV1.3, an adenoviral vector containing a linear 1.3-fold over length HBV genome and a green fluorescent protein expression cassette,24 at an multiplicity of infection of 1–2 infectious units per cell, virtually all hepatocytes were infected. The pgRNA levels were here defined as 100%.
Animals
HBV transgenic (HBVtg) C57BL/6J mice carrying a replication competent 1.3-fold HBV genome16 were examined in comparison to wild type C57BL/6J mice. In addition, C57BL/6J mice were infected with adenovirus type 5 vectors transferring an HBV genome (Ad HBV) or as control with an identical construct in which all open reading frames were knocked out by introducing stop codons (Ad HBV/ko).19 All animals received humane care equivalent to the “Guide for the Care and Use of Laboratory Animals” prepared by the U.S. Academy of Sciences and published by the National Institutes of Health.
Cell Culture
HepG2 cells (ATCC number: HB-8065), HBV-replicating HepG2.2.15,21 or HepG2-H1.3 cells25 were plated onto dishes coated with collagen type IV (Serva Electrophoresis, Heidelberg, Germany) and cultivated in Dulbecco’s modified Eagle medium containing 10% (w/v) fetal calf serum (FCS) as described.23 After achieving 80%–90% confluence, cell cultures were maintained in the Williams E / Dulbecco’s modified Eagle medium (1:1, v/v) containing 5% FCS. HepaRG cells were differentiated and infected with HBV as described previously.23 Cell viability upon treatment with AG-490 was analyzed by CellTiter-Blue Cell Viability Assay (Promega, Madison WI).
RNA Isolation, Microarray Procedure and Data Analysis
Total RNA from mock- or HBV-infected cells was isolated with Trizol reagent (Invitrogen/Thermo Fisher Scientific, Waltham, MA) and cleaned up using RNeasy mini kit including DNAse digestion (Qiagen, Hilden, Germany).
A total of 100 ng of total RNA was used to generate double-stranded complementary DNA (cDNA) with a T7(dT)24-oligonucleotide primer performed with the Two Cycle cDNA Synthesis Kit (Affymetrix, Santa Clara, CA) followed by an in vitro transcription reaction to amplify the cDNA using the MEGAScript T7 Kit (Ambion, Kaufungen, Germany). After purification with the Sample Cleanup Module, cRNA served as a template to prepare a second round of cDNA. After purification of the cDNA with the sample Cleanup Module, biotinylated cRNA was generated, using the GeneChip IVT Labeling Kit (all reagents from Affymetrix). Hybridization of complementary RNA (cRNA) to the high-density oligonucleotide microarrays (HG-U133A arrays; Affymetrix) was performed according to the manufacturer's GeneChip Expression Analysis Technical Manual.
For data collection, assessment and analysis we used Affymetrix Microarray Analysis Suite Version 5.0 program (MAS5.0). Transcripts were considered to be up-regulated, when the absolute call in HBV sample was not absent, and the difference call was increased or marginal increased and fold change in gene expression between HBV–infected and appropriate mock samples was ≥1.5. Transcripts were considered as down-regulated, when the absolute call of the mock control was not “absent,” the difference call was decreased or marginal decreased, and fold change in gene expression was ≥1.5.
Quantification of Gene Expression by Real-Time LightCycler PCR
Total RNA (650 ng) isolated from PHHs or hepatoma cells as described previously was reverse transcribed into cDNA using the Superscript III First-Strand Synthesis System (Invitrogen/Thermo Fisher Scientific). Real-time PCRs were performed with the LightCycler FastStart DNA MasterPLUS SYBR Green Kit using the LightCycler system and normalized to a dilution series of calibrator cDNA using the Relative Quantification Software (all Roche Diagnostics) as described.14, 25 Primers for target genes were chosen with the assistance of LightCycler primer design program (Roche Diagnostics) and listed in Table 1. Primers for the reference genes glyceraldehyde-3-phosphate dehydrogenase and 5-aminolevulinate synthase, for hepatocyte nuclear factor 4 alpha (HNF4α), hepatocyte nuclear factors 3 alpha and beta (HNF3α and HNF3β, respectively) were published earlier.14, 23 For expression analysis of selected genes in HBV-expressing liver tumor and nontumorous tissue and in the healthy liver, 1 μg of total RNA was reverse transcribed into cDNA and applied for real-time PCR and as described previously. cDNA levels were normalized for the expression of the ALAS1 gene.
Table 1.
Primer Sets Used for LightCycler Real-Time PCR
| Gene | LightCycler PCR Primers |
|---|---|
| cIAP2 | fw: 5′-TGAAGCTGTGTTATATGAGCA-3′ rev: 5′-ACGAACTGTACCCTTGAT-3′ |
| Mn-SOD | fw: 5′-AGCCCAGATAGCTCTTC-3′ rev: 5′-AGGTAGTAAGCGTGCTC-3′ |
| TNFAIP8 | fw: 5′-GCTTGCTTAGGGCTTC-3′ rev: 5′-GCTCATGTTCTTTAGCAGT-3′ |
| IER3 | fw: 5′-CGAGGCGCATAGAGAC-3′ rev: 5′-CTACTTTGCCGCAGTTC-3′ |
| IGFBP1 | fw: 5′-AGAGCACGGAGATAACT-3′ rev: 5′-TCCAAGGGTAGACGCA-3′ |
| CRP | fw: 5′-ACATTCACAGGGCTCT-3′ rev: 5′-ACAAGGTTCGTGTGGA-3′ |
| CCL2 | fw: 5′-AGATGCAATCAATGCCC-3′ rev: 5′-GTTGTGGAGTGAGTGTTC-3′ |
| IL-8 | fw: 5′-AAGAACTTAGATGTCAGTGC-3′ rev: 5′-ACTTCTCCACAACCCT-3′ |
| APOE | fw 1: 5′-CAAGGTGGAGCAAGCG-3′ rev 1: 5′-ATGGTCTCGTCCATCAG-3′ |
| CYP3A4 | fw: 5′-TCACCGTGACCCAAAG-3′ rev: 5′-TTTGAGGTCTCTGGTGT-3′ |
| 2‘5’OAS | fw: 5′-CAGTTAAATCGCCGGG-3′ rev: 5′-AGGTTATAGCCGCCAG-3′ |
| IP10 | fw: 5′-ACTGTACGCTGTACCT-3′ rev: 5′-TGGCCTTCGATTCTGGA-3′ |
| IL-6 | fw: 5′-AAACAACCTGAACCTTCC-3′ rev: 5′-CAGGGGTGGTTATTGC-3′ |
| STAT3 | fw: 5′-TTCGGAAAGTATTGTCGGC-3′ rev: 5′-GGGTTTGGCTGTGTGA-3′ |
NOTE. PCR, polymerase chain reaction.
Western Blot Analysis
Nuclear and cytoplasmic protein extracts from mock- or HBV-infected cells were isolated with NE-PER Nuclear and Cytoplasmic extraction Reagents (Pierce/Thermo Fisher Scientific). Purity of nuclear protein preparations was controlled by Western blotting using antialbumin antibodies. Total proteins were extracted with CHAPS buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 1% Chaps) containing phosphatase inhibitors sodium vadanate and sodium fluoride.
Proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and analyzed by Western blotting as described.23 The following primary antibodies were used: mouse monoclonal anti-cIAP2 and mouse monoclonal anti–poly(ADP-ribose) polymerase (BD Biosciences, San Diego, CA); mouse monoclonal anti-Phospho-STAT3-Tyr705, rabbit monoclonal anti-STAT3, rabbit monoclonal anti-Src and rabbit polyclonal anti-Phospho-src family (Tyr416) (Cell Signaling, Denvers, MA); rabbit polyclonal anti-Mn-SOD, rabbit polyclonal anti–interferon regulatory factor 3 (IRF3), rabbit polyclonal anti-lamin B1, and rabbit polyclonal antialbumin (Santa Cruz Biotechnology, Santa Cruz, CA); mouse monoclonal anti-β actin (Sigma); and rabbit polyclonal anti-HBV core.23, 26 The secondary antibodies were anti-mouse or anti-rabbit IgG horseradish peroxidase–coupled antibodies (Sigma). ImageJ software, version 1.51h (National Institutes of Health, Bethesda, MD) was applied for densitometric quantification of Western blot signals.
Immunohistochemistry
Fixed, pretreated 2-mm-thick, paraffin-embedded liver sections derived from human liver needle biopsies or from mouse liver tissue were decorated with a monoclonal antibody against phosphor-Stat3 (Tyr 705). The antibody was purchased from Cell Signaling (clone #D3A7, dilution 1:100). Pretreatment (H2(60)) and staining were performed on a Leica Bond-Max, detection with a Bond Polymer refine kit (Leica Biosystems, Heidelberg, Germany) using an anti-rabbit secondary antibody, coupled to horseradish peroxidase and 3,3’-diaminobenzidine as a substrate. Stainings were analyzed and pictures were taken with an Olympus BX53 and an Olympus DP72 camera (Olympus, Tokyo, Japan).
Detection of STAT3 Activation
To detect phosphor STAT3-Y705, total or nuclear protein extracts were analyzed by Western blotting using mouse monoclonal anti-Phospho-STAT3-Tyr705 antibodies (Cell Signaling) as described previously. To additionally measure STAT3 activity in cells, the Cignal STAT3 Reporter (luc) Assay (Qiagen) was applied. To this end, HepG2 or HepG2-H1.3 cells were transfected with either Cignal reporter or positive or negative control constructs using the fast-forward protocol provided by the manufacturer. The activity of STAT3-dependent Firefly luciferase and the activity of constitutively expressed Renilla luciferase used for internal normalization were measured by the Dual-Luciferase Reporter Assay System (Promega) according to manufacturer’s instructions.
Detection of Reactive Oxygen Species
HepG2, HepG2.2.15, or HepG2-H1.3 cells were plated on collagen-coated 96-dishes to reach confluence (day 1) and maintained in Williams E or Dulbecco’s modified Eagle medium (1:1, v/v) containing 1% FCS. HepaRG cells were plated on collagen-coated 96-dishes, differentiated and HBV- or mock-infected. Reactive oxygen species were measured using Cellular Reactive Oxygen Species Detection Assay Kit (Abcam, Cambridge, UK) according to manufacturer’s instructions. This assay measures hydroxyl, peroxyl, and other reactive oxygen species (ROS) activity within the cell. Specifically, HepG2, HepG2-2.15, HepG2-H1.3, and noninfected or HBV-infected HepaRG cells (1 × 104 cells per probe) were harvested by trypsinization and subsequently stained with 20 μM DCFDA in cell culture medium for 30 min followed by flow cytometry assay. To analyze the influence of ROS inhibition on STAT3 activity, HepG2 or HepG2-H1.3 cells were first transfected with either the cignal reporter- or positive- or negative control constructs (Qiagen) using the STAT3-luciferase reporter assay by the fast-forward protocol according to manufacturer’s instructions. 18 hours later, cell culture medium was changed and 5 mM ROS-inhibitor NAC was added to the culture medium of HepG2-H1.3 cells. Then, 48 hours thereafter, the activity of STAT3-dependent Firefly luciferase and the activity of constitutively expressed Renilla luciferase used for internal normalization were measured by the Dual-Luciferase Reporter Assay System (Promega) according to manufacturer’s instructions.
Small Interfering RNA Knockdown
HepG2-H1.3 cells were transfected with 5 nM of either STAT3-specific small interfering RNA (siRNA) (Hs_STAT3_7 FlexiTube siRNA, functionally verified siRNA directed against human STAT3; NM_003150, NM_139276, NM_213662) or nonsilencing siRNA using the HiPerFect transfection reagent (both from Qiagen) according to the fast-forward protocol provided by manufacturer. The knockdown of STAT3 was confirmed at the messenger RNA (mRNA) and protein level by real-time RT-PCR and Western blot analysis, respectively.
Caspase 3/7 Assay
The activity of 2 effector caspases, caspase-3 and caspase-7 (caspase 3/7), was measured by luminescent Caspase-Glo 3/7 assay (Promega). PHHs were seeded at a density of 4 × 105 cells per well in a 24-well culture plates. PHHs were treated with 100 μM of AG-490 or with 0.1% DMSO for 24 hours, and then lysed in 200 μL of cell culture lysis buffer (Promega). The luminescence was measured after addition of the caspase 3/7 substrate according to the manufacturer’s instructions.
Statistics
Statistical significance was determined by Student’s unpaired 2-tailed t test. Differences with P ≤ .05 were considered statistically significant.
Results
HBV Induces Up-Regulation of Genes Encoding Acute Phase Response Proteins and Proteins Promoting Cell Survival
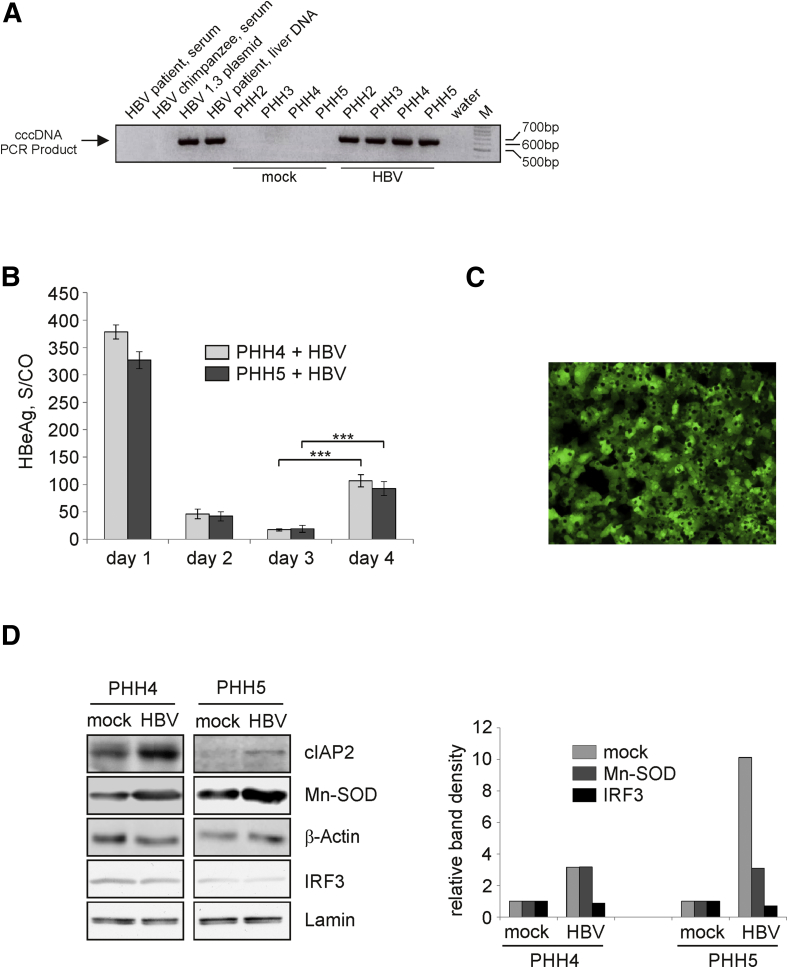
PHH cultures were prepared from 7 patients undergoing surgical liver resection and were either mock- or HBV-infected in parallel experiments. To monitor HBV infection, cell lysates and culture medium were collected daily from day 1 to day 4 p.i. On day 4 p.i., we detected the formation of HBV cccDNA by a selective, quantitative real-time PCR24 and the release of newly synthesized HBeAg into the cell culture medium of HBV-infected PHHs (Figure 1A and B, respectively) confirming successful HBV infection. To compare efficiency of HBV infection in different PHH cultures, total cellular RNA was extracted on day 4 p.i., and levels of HBV pgRNA were determined relative to those in PHHs transduced with AdG-HBV1.3 vector (Figure 1C) using quantitative real-time RT-PCR and pgRNA specific primers as described in the Methods. The efficiency of infection with HBV broadly varied between 5% and 45%. Three experiments, in which infection rates were ≥15%, named PHH1, PHH2, and PHH3 (Table 2), were selected for gene expression analysis.
Figure 1.
Up-regulation of apoptosis inhibitors induced by hepatitis B virus (HBV) replication in primary human hepatocyte (PHHs). (A) Formation of HBV covalently closed circular DNA (cccDNA) in HBV-infected PHHs was examined by real-time polymerase chain reaction (PCR) using total DNA extracted from mock- (control) or HBV-infected PHH cultures on day 4 postinjection (p.i.). DNA extracted from high-titer (>5 × 10e8/mL) serum of an HBV-positive patient and from serum of an HBV-positive chimpanzee containing HBV relaxed circular DNA were used as specificity (negative) controls. Total liver DNA extracted from HBV-positive patient and plasmid DNA encoding for HBV 1.3-overlength genome served as positive controls. PCR products were separated in a 2% agarose gel, water control, and DNA size marker (M) are indicated. (B) Hepatitis B early antigen (HBeAg) was measures in cell culture supernatants from day 1 to 4 p.i. of HBV-infected PHHs prepared from 2 different donors (PHH4 and PHH5) as signal-to-control ratio. Day 1 values reflect input virus. Values are shown as mean ± SD (n = 3; ***P < .001; Student's t test). (C) PHH cultures were infected with AdG-HBV1.3 at a multiplicity of infection of 1–2 infectious units per cell. Expression of green fluorescent protein was visualized by fluorescence microscopy at 24 hours p.i. (D) Expression of cIAP2, Mn-SOD, and β-actin (loading control) in mock- or HBV-infected PHH4 and PHH5 was analyzed by Western blot analysis of total protein extracts prepared on day 4 p.i. Levels of interferon regulatory factor 3 (IRF3) and lamin (loading control) in the nucleus of mock- or HBV-infected PHH4 and PHH5 were determined by Western blot analysis of nuclear proteins extracted on day 4 p.i. (left panel). Densitometric quantification (right panel) of cIAP2 or Mn-SOD relative to β-actin and interferon regulatory factor 3 (IRF3) relative to lamin was performed using ImageJ software. Protein levels are given relative to respective mock control (set to 1).
Table 2.
Gene Expression Profiling of HBV-Infected PHH Cultures
| Samplea | Transcripts up-regulatedb | Transcripts down-regulatedb | Relative efficiency of HBV replication (%)c | HBeAg (ng/mL)d |
|---|---|---|---|---|
| PHH1 | 192 | 416 | 15 | 11.1 |
| PHH2 | 894 | 1364 | 45 | 21.7 |
| PHH3 | 511 | 349 | 30 | 14.95 |
| Common regulated in all PHHs | 40 | 17 | — | — |
Primary human hepatocyte (PHH) prepared from different patients (1–3), RNA isolated on day 4 post–mock or hepatitis B virus (HBV) infection.
Up- or down-regulated transcripts in HBV-infected PHHs compared with mock-infected PHH, fold change ≥1.5.
Relative expression levels of HBV pregenomic RNA determined by real-time reverse transcription polymerase chain reaction on day 4 postinfection.
Levels of hepatitis B early antigen secreted into the culture medium on day 4 postinfection.
To detect changes in cellular gene expression and activation of appropriate cellular pathways at the beginning of productive HBV infection, we harvested HBV and mock infected PHHs on day 4 p.i., when HBV replication in infected cells had just started as indicated by the detection of newly synthesized HBeAg after formation of cccDNA (Figure 1A and B). We chose this time point to ensure HBV replication and to determine the influence of HBV replication on hepatocytes, but avoid influences or early pattern recognition of the HBV inoculum14 as well as dedifferentiation of the primary cells at later time points. cRNAs derived from mock- and HBV-infected samples were hybridized to HG-U133A Affymetrix oligonucleotide microarray containing 22,283 probe sets representing approximately 14,000 human genes.
The total number of transcripts regulated ≥1.5-fold upon HBV infection was relatively low, and correlated with the efficiency of infection. Comparative analysis of 3 HBV infection experiments detected 40 genes up-regulated and 17 genes down-regulated in all 3 experiments (Table 2). This minor overlap reflects the variability between primary hepatocytes prepared from different donors but also the high adaptation of HBV to the host cell environment. At the same time, it shows that HBV infection and HBV replication influence the host cell gene expression profile—even if only at a low level.
Based on results obtained from microarray analyses, we grouped genes with altered expression patterns according to their biological functions (Table 3). The largest group of genes, up-regulated upon HBV infection promoted cellular acute phase response (APR). Among them, C-reactive protein (CRP) was induced most strongly upon HBV infection. In contrast, several cellular genes implicated in lipid and xenobiotic metabolism as well as cytochrome P 450 (CYP450) family members involved in biotransformation and generation of ROS27 were down-regulated in HBV-infected hepatocytes.
Table 3.
Functional Grouping of Genes Identified by Affymetrix Analyses
| Functional relevance of regulated genes | Gene | Description | Public ID | Fold changea |
||
|---|---|---|---|---|---|---|
| PHH1b | PHH2b | PHH3b | ||||
| Positive APR | ||||||
| Major APR | CRPc | C-reactive protein, pentraxin-related | NM_000567 | 9.2 I | 45 I | 512 I |
| SAA1 | Serum amyloid 1 | NM_000331 | 2 I | 4.3 I | 2.5 I | |
| SAA2 | Serum amyloid 2 | NM_030754 | 2.3 I | 11.3 I | 3 I | |
| APCS | Amyloid P component, serum | NM_001639 | 2 I | 2.5 I | 1.6 I | |
| Coagulation proteins | FGA | Fibrinogen alpha chain | NM_000508 | 2.6 I | 7.5 I | 4.6 I |
| FGB | Fibrinogen beta chain | NM_005141 | 1.6 I | 2.1 I | 1.7 I | |
| FGG | Fibrinogen gamma chain | NM_000509 | 1.7 I | 2.3 I | 1.6 I | |
| FGL-1 | Fibrinogen-like protein | NM_004467 | 2.3 I | 2.5 I | 2.5 I | |
| Metal-binding proteins | HP | Haptoglobin | NM_005143 | 1.9 I | 1.7 I | 2 I |
| HPR | Haptoglobin-related protein | NM_020995 | 2.6 I | 1.7 I | 2.3 I | |
| Mn-SODc | Superoxide dismutase 2, mitochondrial | NM_000636 | 2 I | 3.7 I | 4.3 I | |
| Proteinase inhibitors | SERPINA 1 | Serpin peptidase inhibitor, clade A, member 1, anti-trypsin | NM_000295 | 1.8 I | 1.4 I | 1.9 I |
| SERPINA 3 | Serpin peptidase inhibitor, clade A, member 3, anti-trypsin | NM_001085 | 2.1 I | 2.2 I | 2 I | |
| Complement proteins | C4A | Complement component 4A | NM_007293 | 2.8 I | nc | 3 I |
| C9 | Complement component 9 | NM_001737 | 3.2 I | 3.7 I | 16 I | |
| Other proteins | LBP | Lipopolysaccharide-binding protein | NM_004139 | 4.3 I | 8 I | 9.2 I |
| ORM1 | Orosomucoid 1 | NM_000607 | 2.7 I | 2.3 I | 3.2 I | |
| ORM2 | Orosomucoid 2 | NM_000608 | 2.3 I | 1.8 I | 5.3 I | |
| Negative APR | ALB | Albumin | NM_000477 | nc | –2.5 D | –1.6 D |
| ITIH2 | Interalpha (globulin) inhibitor H2 | NM_002216 | nc | –2 D | –2.5 D | |
| HRG | Histidine-rich protein | NM_000412 | nc | –6.1 D | –1.5 D | |
| Chemokines | CXCL1 | Chemokine (C-X-C motif) ligand 1 | NM_001511 | 2.9 I | 15.3 I | 33 I |
| CXCL2 | Chemokine (C-X-C motif) ligand 2 | NM_002089 | 1.5 I | 82 I | 8 I | |
| CXCL5 | Chemokine (C-X-C motif) ligand 5 | NM_002994 | nc | 9.2 I | 7 I | |
| CXCL6 | Chemokine (C-X-C motif) ligand 6 | NM_002993 | 1.7 I | 13.3 I | 29 I | |
| CCL2c | Chemokine (C-C motif) ligand 2 | NM_002982 | 5.9 I | 11.9 I | 19.3 I | |
| CCL20 | Chemokine (C-C motif) ligand 20 | NM_004591 | 1.7 I | 26.7 I | 6.7 I | |
| Apoptosis | ||||||
| Negative regulators | cIAP2 | cIAP2 apoptosis inhibitor 2, Birc3 | NM_001165 | nc | 4.9 I | 4.6 I |
| IER3 | Immediate early response 3 | NM_003897 | nc | 4.4 I | 1.9 I | |
| TNFAIP8 | Tumor necrosis factor-α–induced protein 8, FLIP-like, SCC-S2 | NM_014350 | nc | 2.5 I | 2.3 I | |
| Mn-SOD | Superoxide dismutase 2, mitochondrial | NM_000636 | 2 I | 3.7 I | 4.3 I | |
| PBEF1 | Pre–B-cell colony enhancing factor 1 | NM_005746 | nc | 2.3 I | 3.3 I | |
| CRP | C-reactive protein, pentraxin-related | NM_000567 | 9.2 I | 45 I | 512 I | |
| IL-8 | Interleukin 8 | NM_000584 | nc | 21 I | 8 I | |
| CCL2 | Chemokine (C-C motif) ligand 2 | NM_002982 | 5.9 I | 11.9 I | 19.3 I | |
| IGFBP1 | Insulin-like growth factor binding protein 1 | NM_000596 | nc | 113.8 I | 4.3 I | |
| Positive regulators | APOEc | Apolipoprotein E | NM_000041 | nc | –10.5 D | –2.7 D |
| CASP4 | Caspase 4 | NM_001225 | nc | 3.6 I | 1.8 I | |
| Metabolism | ||||||
| Antioxidant proteins | Mn-SOD | Superoxide dismutase 2, mitochondrial | NM_000636 | 2 I | 3.7 I | 3.7 I |
| GPX2 | Glutathione peroxidase 2 | NM_002083 | 5.1 I | 3.1 I | 6.1 I | |
| CP | Ceruloplasmin (ferroxidase) | NM_000096 | nc | 4.8 I | 7 I | |
| Xenobiotic metabolism proteins | CYP3A4c | Cytochrome P450, Family 3, subfamily A, polypeptide 4 | NM_000776 | –2.8 D | –6.5 D | –7 D |
| UGT2B15 | UDP glucuronosyltransferase 2 family, polypeptide B15 | NM_001076 | –2.3 D | –16 D | –13 D | |
| UGT2B28 | UDP glucuronosyltransferase 2 family, polypeptide B28 | NM_053039 | nc | –6.4 D | –2.5 D | |
| Oxydoreductases and electron transport proteins | CYP3A4 | Cytochrome P450, Family 3, subfamily A, polypeptide 4 | NM_000776 | –2.8 D | –6.5 D | –7 D |
| CYP3A7 | Cytochrome P450, Family 3, subfamily A, polypeptide 7 | NM_000765 | –1.6 D | –2.5 D | –2 D | |
| CYP2C8 | Cytochrome P450, Family 2, subfamily C, polypeptide 8 | NM_030878 | –1.6 D | –8 D | –4.4 D | |
| CYP2A6 | Cytochrome P450, Family 2, subfamily A, polypeptide 6 | NM_000762 | nc | –4.9 D | –6.1 D | |
| CYP2C9 | Cytochrome P450, Family 2, subfamily C, polypeptide 9 | NM_000771 | –2 D | –3.5 D | –5.7 D | |
| CYP2C19 | Cytochrome P450, Family 2, subfamily C, polypeptide 19 | NM_000769 | –1.7 D | –5.3 D | –1.9 D | |
| GRHPR | Glyoxylate reductase, hydroxypyruvate reductase | NM_012203 | nc | –2.5 D | –2.8 D | |
| ACADL | Acyl-coenzyme A dehydrogenase, long chain | NM_001608 | –1.7 D | –2.6 D | –2 D | |
| BDH | 3-hydroxybutyrate dehydrogenase | NM_004051 | nc | –1.9 D | –5.5 D | |
| Lipid metabolism and transport proteins | APOE | Apolipoprotein E | NM_000041 | nc | –10.5 D | –2.7 D |
| ACADL | Acyl-coenzyme A dehydrogenase, long chain | NM_001608 | –1.7 D | –2.6 D | –2 D | |
| CYP3A4 | Cytochrome P450, Family 3, subfamily A, polypeptide 4 | NM_000776 | –2.8 D | –6.5 D | –7 D | |
| AKR1B10 | Aldo-keto reductase family 7, member B10 | NM_020299 | –3 D | –1.4 D | –4 D | |
APR, acute phase response; HBV, hepatitis B virus; PHH, primary human hepatocyte.
increased (I), decreased (D), or no change (nc) in gene expression in HBV-infected PHH compared to mock-infected cells.
PHH prepared from different patients (1-3).
Due to the dual functions, some of genes are listed twice.
A divergent regulation was identified for genes associated with apoptosis induction and cell survival. Importantly, in the 2 experiments with the highest efficiency of HBV infection (PHH2 and PHH3), 4 established apoptosis inhibitor genes: cIAP2 (apoptosis inhibitor 2),28, 29 IER3 (immediate early response protein 3),30 TNFAIP 8 (tumor necrosis factor-α–induced protein 8),31 and IGFBP1 (insulin-like growth factor binding protein 1)32 (Table 3) were up-regulated. In addition, genes encoding for PBEF1 (pre–B-cell colony enhancing factor 1),33 IL-8,34 Mn-SOD (manganese superoxide dismutase),35, 36 CCL2 (Chemokine [C-C motif] ligand 2 / monocyte chemoattractant protein-1),37 and CRP38 with reported antiapoptotic activities, were markedly up-regulated in all experiments (Table 3).
To confirm regulation of gene expression by real-time RT-PCR, PHH4 and PHH5 with HBV infection rates 36% and 29%, respectively, were additionally used. RNA expression levels for the 10 highest regulated genes from APR, antiapoptosis, and CYP450 groups were determined in PHH2, PHH3, PHH4, and PHH5 with and without HBV infection (Table 4). Real-time RT-PCR confirmed the regulation of all genes. Most importantly, the fold change of gene expression detected by real time RT-PCR was very comparable to that identified by microarray analysis (Tables 3 and 4).
Table 4.
Confirmation of Microarray Data by Real-Time Quantitative Reverse Transcription Polymerase Chain Reaction
| Gene | Fold changea in gene expression: PHHb/HBV vs PHHb/mock |
|||
|---|---|---|---|---|
| PHH2 | PHH3 | PHH4 | PHH5 | |
| cIAP2 | 5.1 I | 3.9 I | 5.1 I | 2.9 I |
| Mn-SOD | 3.9 I | 2 I | 4.1 I | 3.4 I |
| TNFAIP8 | 2 I | 2.3 I | 2.3 I | 3.1 I |
| IER3 | 5.7 I | 1.7 I | 1.5 I | 1.6 I |
| IGFBP1 | 116 I | 5.3 I | 23 I | 7.4 I |
| CRP | 18.5 I | 115 I | 269 I | 88 I |
| CCL2 | 4 I | 21 I | 6.7 I | 6.9 I |
| IL-8 | 24.7 I | 10.8 I | 8.5 I | 4.2 I |
| APOE | −19 D | −3.5 D | −1.4 D | −2.3 D |
| CYP3A4 | −4 D | −8.6 D | −4.3 D | −12.2 D |
| 2‘5’OAS | nc | −2 D | nc | −1.6 D |
| IP10 | nc | −1.7 D | −1.5 D | −2.5 D |
HBV, hepatitis B virus; PHH, primary human hepatocyte.
Increased (I) and decreased (D) compared with mock-infected PHH; mean values of at least 2 independent measurements per experiment are given. nc, no change in gene expression detected.
PHH prepared from different patient materials (PHH2–PHH5).
Next, we aimed at verifying alterations in HBV-infected PHH4 and PHH5 at the protein level. Western blot analyses for representative proteins cIAP2 and Mn-SOD confirmed their up-regulation in HBV-infected PHHs compared to respective mock controls (Figure 1D). As the microarray analysis did not reveal activation of any genes involved in a type I or II IFN response, we additionally examined expression levels of IFN type I-inducible 2’5’-oligoadenylatsynthetase and IFN-inducible protein 10 (IP10) genes. For both, real time RT-PCR analysis revealed rather down- than an up-regulation (Table 4), and we did neither detect IFNa by ELISA (detection limit 47 pg/mL) nor activation of IRF3 in HBV-infected PHH cultures (Figure 1D).
Thus, HBV up-regulated proteins involved in APR, oxygen detoxification, and apoptosis inhibition at the transcriptional and at the protein levels. Moreover, our data suggest that an IFN response is neither induced by HBV infection nor by HBV replication in PHHs.
Confirmation of HBV-Induced Changes in Cellular Gene Expression in HBV-Replicating Hepatoma Cells, Chronically HBV-Infected Human Liver Tissue, and HBV-Related HCC
To study whether HBV infection or HBV replication influenced gene expression, we compared gene expression profiles of HBV-replicating cell lines HepG2.2.15, established many years ago21 and HepG2-H1.3, a cell line established in our laboratory,23, 25 with parental HepG2 cells. Total RNA from these cell lines was examined for the expression of the cIAP2, TNFAIP8, IER3, IGFBP1, Mn-SOD, CRP, and CYP3A4 genes by real-time RT-PCR.
In contrast to PHH cultures, we found, that hepatoma cell lines did not express the CRP and CYP3A4 genes at a detectable level. The expression of the Mn-SOD gene was unaltered in HBV-replicating cells compared to HepG2 cells. But, in agreement with our data for HBV-infected PHH cultures, we found that cIAP2 was up-regulated in HepG2.2.15 cells, and TNFAIP8, IER3, and IGFBP1 genes were up-regulated in both HBV-replicating HepG2-H1.3 and HepG2.2.15 cells in comparison with parental HepG2 cells (Table 5).
Table 5.
Alteration of Gene Expression in HBV-Replicating Cell Lines
| Gene | Fold changea in gene expression |
|
|---|---|---|
| HepG2-H1.3 vs HepG2 | HepG2.2.15 vs HepG2 | |
| cIAP2 | nc | 2.7 ± 0.2 I |
| TNFAIP8 | 1.7 ± 0.4 I | 2.8 ± 0.5 I |
| IER3 | 2 ± 0.1 I | 3.5 ± 0.5 I |
| IGFBP1 | 3 ± 0.9 I | 7.1 ± 0.9 I |
| Mn-SOD | nc | nc |
| CRP | nd | nd |
| CYP3A4 | nd | nd |
| IL-6 | nd | nd |
NOTE. Mean ± SD values from at least 3 independent experiments are given.
HBV, hepatitis B virus.
I, increased; D, decreased compared with HepG2 cells; nc, no change in gene expression observed; nd, gene expression was not detectable by real-time quantitative reverse transcription polymerase chain reaction.
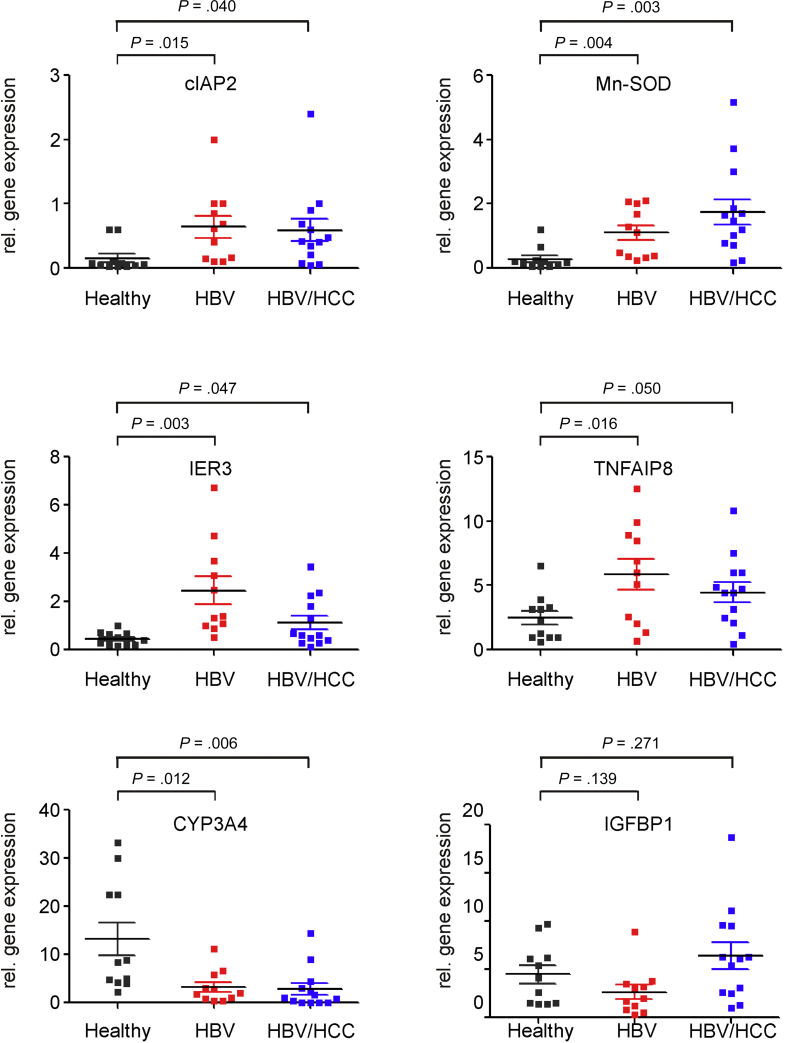
To determine whether these apoptosis- and carcinogenesis-related genes were similarly regulated by HBV in vivo and whether they may be implicated in carcinogenesis, we compared the mRNA expression levels of cIAP2, Mn-SOD, TNFAIP8, IER3, IGFBP1, and CYP3A4 in liver tissue samples from healthy HBV-naive subjects (n = 11) to those from chronically HBV-infected patients (n = 11) and patients who had developed HBV-related HCC (n = 13) (Figure 2).
Figure 2.
Changes in cellular gene expression in chronically hepatitis B virus (HBV)–infected human liver tissue and in HBV-related hepatocellular carcinoma (HCC). Liver tissue samples from healthy HBV-naive subjects (healthy, n = 11), from chronically HBV-infected patients (HBV, n = 11) and patients who had developed HBV-related HCC (HBV/HCC, n = 13) were analyzed for expression of the cIAP2, Mn-SOD, IER3, TNFAIP8, CYP3A4, and IGFBP1 genes by real-time reverse transcription polymerase chain reaction. Each data point represents the expression level determined in a single sample. Mean expression level ± SD in each group and statistical significance (Student's t test) are given.
Real time RT-PCR analysis did not reveal up-regulation of the IGFBP1 gene (Figure 2). However, in agreement with our in vitro findings, we observed an up-regulation of the cIAP2, Mn-SOD, TNFAIP8, and IER3 genes and down-regulation of the CYP3A4 gene in HBV-expressing tumor and, more importantly, in HBV-replicating nontumorous tissue in comparison to that from healthy donors (Figure 2). For all genes we observed a significant difference in the expression between HBV-positive nontumorous (HBV) and HCC tissue (HBV/HCC) and HBV-negative control liver tissue. These results confirmed the primary role of HBV in the induction of genes encoding for a set of apoptosis inhibitors, which may contribute to the prevention of apoptosis and to the transformation of HBV-infected cells.
Activation of STAT3 in HBV-Replicating Hepatocytes
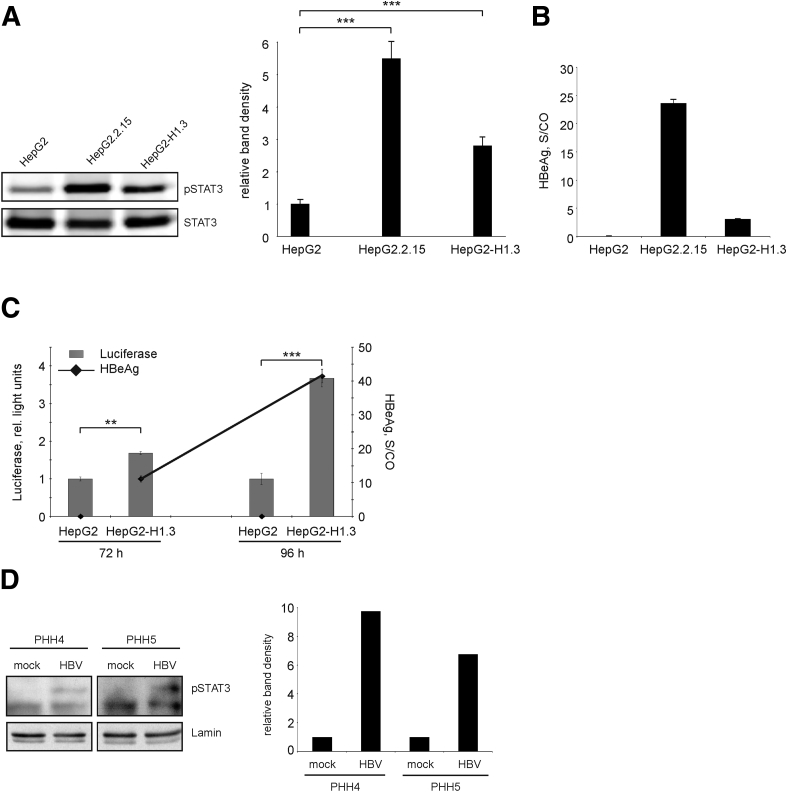
Next, we investigated the mechanisms underlying up-regulation of APR- and antiapoptotic- and down-regulation of pro-apoptotic genes. Since we observed expression changes of a number of STAT3-regulated genes, namely cIAP2, Mn-SOD, IER3, IGFBP1, and the numerous APR genes, we further examined the activation of STAT3 in HBV-replicating hepatoma cells and PHHs. STAT3 activation requires phosphorylation of STAT3 at tyrosine 705 (Y705) which results in STAT3 dimerization and translocation to the nucleus. We found that levels of phosphorylated STAT3 (pSTAT3) were significantly increased in HBV-replicating hepatoma cells in comparison to HBV-negative cells, as shown by specific detection of pSTAT3 by Western blotting (Figure 3A).
Figure 3.
Activation of signal transducer and activator of transcription 3 (STAT3) in hepatitis B virus (HBV) replicating cells. (A) Total cellular proteins were analyzed for the presence of tyrosine 705 phosphorylated STAT3 (pSTAT3) and total STAT3 by Western blotting. One representative Western blot of 3 is shown (left panel). Band density was quantified using ImageJ software and relative levels of pSTAT3 were calculated (right panel). Level of pSTAT3 in HepG2 cells was set to 1. Values are shown as mean ± SD (n = 3; ***P < .001; Student's t test). (B) Secretion of hepatitis B early antigen (HBeAg) into the medium of stably HBV-replicating HepG2.215 or HepG2-H1.3 cells was determined as signal-to-control (S/CO) ratio, mean ± SD from 3 independent experiments is given. HepG2 cells were used as negative control. (C) Activation of STAT3 in HepG2-H1.3 cells was analyzed by the STAT3-luciferase reporter assay. HepG2-H1.3 and HepG2 cells were transfected with STAT3 Cignal reporter, negative or positive control constructs. Cells were harvested 72 and 96 hours after transfection and analyzed by the Dual-Luciferase Cignal reporter assay. The activity of STAT3-dependent firefly luciferase is expressed in relative light units. The activity of constitutively expressed Renilla luciferase was used for internal normalization. Secretion of HBeAg into the culture medium of HepG2-H1.3 cells is indicated by dots. Values are shown as mean ± SD (n = 3; **P < .01; ***P < .001; Student's t test). (D) Nuclear proteins extracted from mock- or HBV-infected PHHs prepared from 2 different donors (PHH4 and PHH5) on day 4 postinfection were analyzed for the presence of pSTAT3 by Western blotting (left panel). Band densities for pSTAT3 and lamin (loading control for nuclear proteins) were quantified using ImageJ software (right panel), and levels of pSTAT3 were calculated relative to respective mock control (set as 1).
To confirm the activation of the STAT3 signaling pathway in HBV-replicating cells by an alternative method, we performed a STAT3-luciferase reporter assay. We observed a significantly higher activity of STAT3-dependent firefly luciferase in HepG2-H1.3 cells as compared to HepG2 cells (Figure 3C). As in hepatoma cells, STAT3 became activated in PHHs upon HBV infection (Figure 3D). Moreover, activation of STAT3-signaling increased over the time parallel to the increased HBV replication as detected by measuring HBeAg (Figure 3C). A positive correlation between levels of STAT3 activation and HBV replication was also observed when HepG2.2.15 and HepG2-H1.3 cells were compared (Figure 3A and B). Collectively, these data strongly suggest the involvement of HBV replication in STAT3 activation.
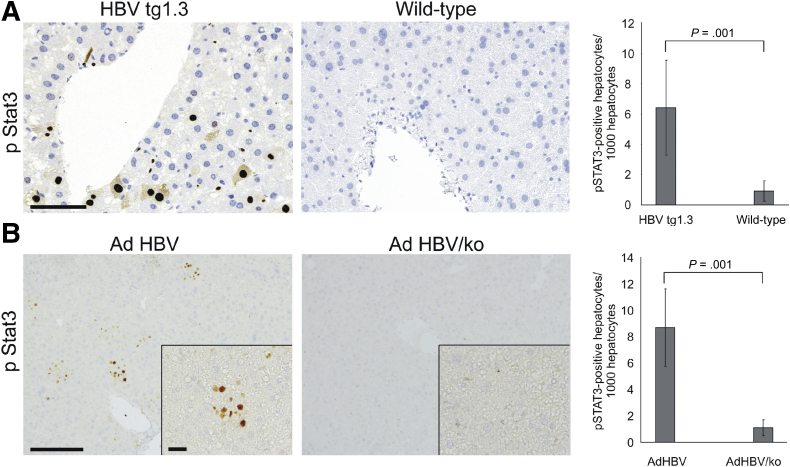
To test a causal relation between HBV replication and STAT3 phosphorylation, we studied STAT3 activation in liver tissue of HBVtg mice16 and mice treated intravenously with adenoviral vector encoding 1.3-fold overlength genome of HBV (Ad HBV)19 (Figure 4). For control, age matched wild-type mice (C57BL/6) or mice treated with control adenoviral vector (Ad HBV/ko), respectively, were used. HBVtg mice displayed a significant amount of STAT3 phosphorylation in hepatocytes around central veins (Figure 4A), where HBV is primarily replicating,16 while age-matched C57BL/6 mice hardly displayed or completely lacked pSTAT3 in hepatocytes (Figure 4A). Similarly, Ad HBV–treated mice displayed a significant amount of pSTAT3-positive hepatocytes when compared to control (Figure 4B) indicating that HBV replication activates STAT3 in vivo.
Figure 4.
Activation of signal transducer and activator of transcription 3 (STAT3) in mouse models of hepatitis B virus (HBV) infection. (A) Liver tissue samples from HBV transgenic (HBVtg1.3) (n = 5) or age- and sex-matched wild-type C57Bl6 (wild-type) (n = 4) mice were stained for pSTAT3. (B) C57Bl6 mice were transduced with adenoviral vectors either encoding a replication-competent 1.3-fold overlength genome of HBV (Ad HBV) (n = 3) or one in which all open reading frames were knocked-out (Ad HBV/ko) (n = 4). Liver tissue samples were isolated on day 7 after transduction and stained for pSTAT3. Number of pSTAT3-positive hepatocytes per 1000 hepatocytes was calculated from 6 independent tissue sample areas and is given in the right panels (A, B). For better comparison, ratio determined in wild-type or in Ad HBV k/o mice was set to 1. Statistical significance is shown (Student's t test).
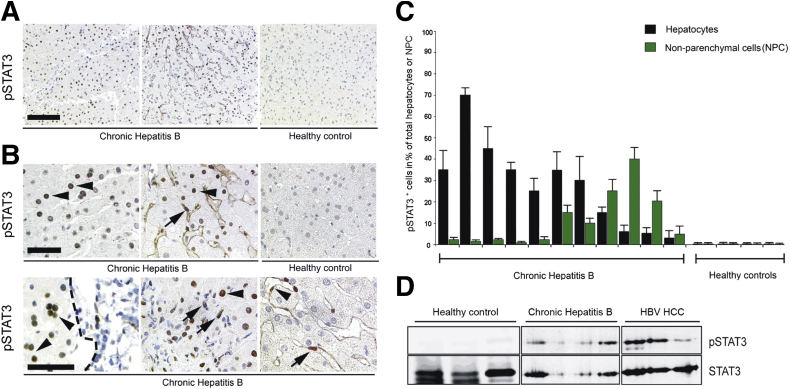
To confirm the relevance of this finding in the human situation, we assessed STAT3 phosphorylation and nuclear translocation in liver needle biopsies of patients with chronic hepatitis B. We investigated paraffin embedded liver sections of patients with ongoing hepatitis B (n = 11) and “healthy” control liver biopsies (n = 4) from liver transplant donors. All liver sections derived from patients with chronic hepatitis B stained positive for HBV surface antigen and/or HBV-core protein (data not shown). Immunohistochemical staining for pSTAT3 revealed that, indeed, in contrast to the controls, all liver sections from patients with chronic hepatitis B showed pSTAT3 phosphorylation in hepatocytes (Figure 5A). In 7 of 11 HBV-infected patients STAT3 phosphorylation and nuclear translocation were mainly detected in hepatocytes (with 5%–70% of all hepatocytes staining positive), while in 4 patients pSTAT3 was predominantly detected in nonparenchymal liver cells (pSTAT3 staining in 1%–15% of all nonparenchymal liver cells) (Figure 5C). pSTAT3-positive hepatocytes were either evenly distributed throughout large areas of the liver or occasionally distributed in a more focal manner, very often neighboring periportal inflammatory sites.
Figure 5.
Phosphorylation and nuclear translocation of signal transducer and activator of transcription 3 (STAT3) in human livers chronically infected with hepatitis B virus (HBV). (A, B) Immunohistochemical analysis for pSTAT3 and its subcellular localization in paraffin-embedded liver tissue sections of chronically HBV-infected patients and control individuals. (A) Scale bar: 150 mm. (B) Enlargement, scale bar: 50 mm. The upper row shows nuclear localization of pSTAT3 in hepatocytes, evenly distributed throughout large areas of the liver as it was detected in most sections. The lower row shows alternative, more focal distribution of pSTAT3-positive hepatocytes, very often nearby periportal inflammatory sites. A periportal inflammatory lesion is marked by a dashed line. Arrowheads depict pSTAT3-positive hepatocytes. In some cases, pSTAT3 and its nuclear localization were detected in hepatocytes (arrowhead) and at the same time in sinusoidal macrophages (arrows, middle panels). Occasionally, pSTAT3 was predominantly detected in endothelial cells (arrowhead) and NPCs, most likely Kupffer cells (lower row, right panel). Control livers lack STAT3 phosphorylation and nuclear translocation in hepatocytes or nonparenchymal liver cells (NPCs) in all liver sections investigated. (C) Quantification of pSTAT3-positive cells relative to total number of hepatocytes or NPCs is shown in percent. Black bars represent hepatocytes, green bars represent nonparenchymal liver cells. (D) Western blot analysis of total proteins prepared from healthy liver tissue, from chronically HBV-infected patients or from patients with HBV-related HCC for the presence of pSTAT3. Membranes were reprobed with anti-STAT3 antibodies to control STAT3 expression levels and protein loading.
To corroborate these data, we analyzed total protein extracts prepared from tumor- and nontumorous tissue of 3 chronically HBV-infected patients. In contrast to noninfected liver tissue, phosphorylation of STAT3 was detected in all analyzed HBV-replicating nontumorous and tumor samples (Figure 5D). Collectively, our data clearly demonstrate activation of STAT3 upon HBV infection not only in cell culture but also in vivo.
IL-6 Dependent and –Independent STAT3 Activation
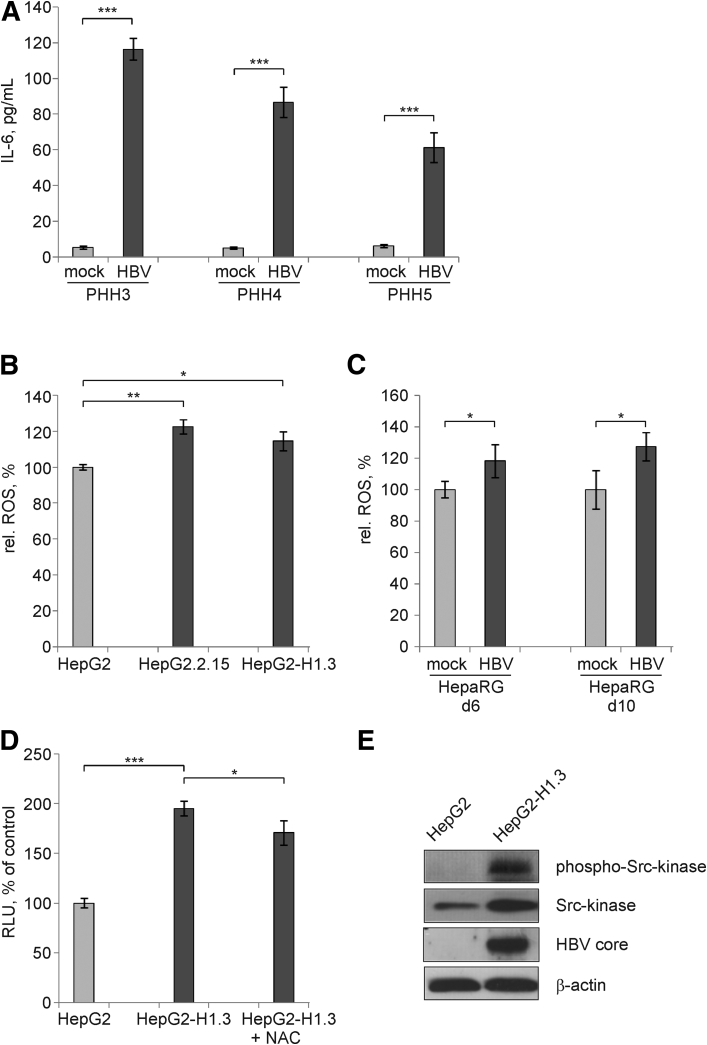
To understand how STAT3 may become activated by HBV infection, we first measured the concentration of IL-6—a well-known cytokine inducer of STAT3 signaling—in HBV-infected PHH cultures prepared from 3 different donors. In all PHH cultures, we observed a pronounced increase in IL-6 concentration after HBV infection (Figure 6A).
Figure 6.
Activation of signal transducer and activator of transcription 3 (STAT3) by interleukin 6 (IL-6)–dependent and –independent intracellular mechanisms. (A) Secretion of IL-6 into the medium of mock- or HBV-infected primary human hepatocyte (PHH) cultures prepared from 3 different patients was measured by enzyme-linked immunosorbent assay (ELISA) on day 4 p.i. Values are shown as mean ± SD (n = 3; ***P < .001; Student's t test). (B) Reactive oxygen species (ROS) were determined in HepG2, HepG2.2.15, and HepG2-H1.3 cells on day 7 after reaching confluency and (C) in mock- or HBV-infected HepaRG cells on day 6 and 10 p.i. by Cellular ROS Detection Assay Kit. Relative ROS levels in HepG2 cells or in mock-infected HepaRG cells were set to 100%. Values are shown as mean ± SD (n = 6; *P < .05; **P < .01; Student's t test). (D) Levels of STAT3 activity after treatment of HepG2-H1.3 cells with 5 mM of ROS-inhibitor N-acetyl-L-cysteine (NAC) were determined by the STAT3-luciferase reporter assay. Relative activity of firefly luciferase reflecting STAT3 activation in HepG2 cells was set to 100%. Values are shown as mean ± SD (n = 6; *P < .05; ***P < .001; Student's t test). (E) Phosphorylated (Tyr416) and nonphosphorylated Src-kinase were detected by Western blot analysis using total cellular proteins isolated from HepG2 or HepG2-H1.3 cells. To control HBV replication and protein loading, membranes were reprobed and stained with anti-HBV core or β actin antibodies, respectively. RLU, relative light units.
As the parental HepG2 cell line does not express IL-639, 40 and both HepG2 and HBV-replicating HepG2.2.15 and HepG2-H1.3 cells expressed the IL-6 gene only at very low levels below the real time RT-PCR quantification limits (Table 5), we looked for alternative pathways of STAT3 activation in the absence of IL-6. STAT3 signaling can also be induced by ROS. Indeed, we measured a significant increase of ROS in HBV-replicating HepG2.2.15 and HepG2-H1.3 cells in comparison to parental HepG2 cells (Figure 6B). In addition, ROS were released after infection of differentiated HepaRG cells with HBV (Figure 6C). Moreover, treatment of HepG2-H1.3 with the ROS-inhibitor NAC reduced STAT3 activity (Figure 6D) indicating that ROS induced by HBV contribute to activation of STAT3.
Activation of various tyrosine kinases can phosphorylate STAT3. JAK as well as Src kinase have been shown to activate STAT3.41 To this end, we detected activation of Src-kinase by Tyr416 phosphorylation in HBV-replicating HepG2-H1.3 cells as compared to parental HepG2 cells (Figure 6E). Thus, our data suggest that HBV replication activates STAT3 not only in a cytokine-dependent fashion but also in cytokine-independent by intracellular activation.
Activation of STAT3 Is Needed to Support HBV Replication and Prevent Apoptosis of HBV Infected Cells
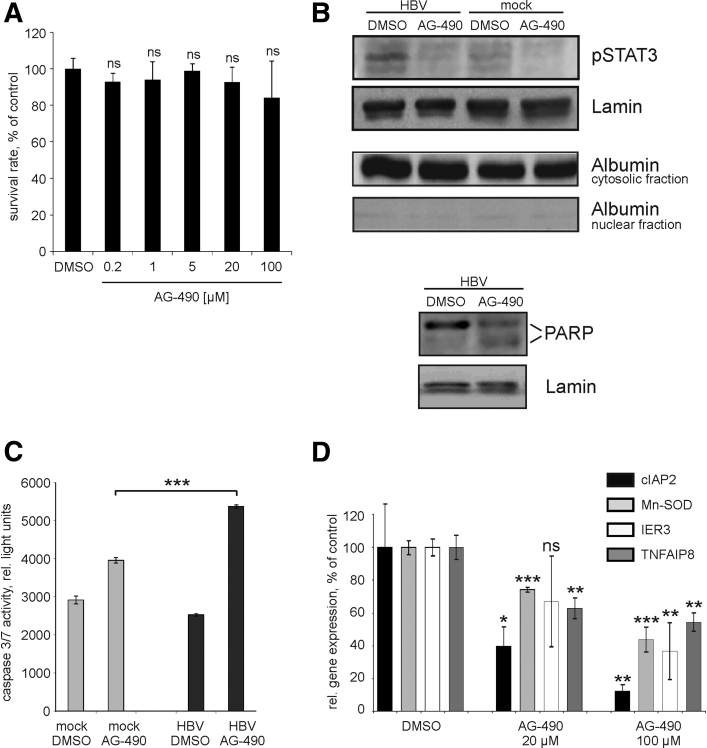
To examine whether STAT3 activation has any influence on HBV infection or on efficiency of HBV replication, we inhibited STAT3 phosphorylation with AG-490, a JAK-2 protein tyrosine kinase inhibitor that inhibits Y705 phosphorylation of STAT3.42 AG-490 dissolved in 0.1% of DMSO was added to HBV-infected HepaRG cells. HBV-infected cells treated with 0.1% of DMSO only were served as control. 24 hours after AG-490 administration we didn’t detect any significant effect of AG-490 on cell viability at concentrations up to 100 μM (Figure 7A). We therefore used 20 μM and 100 μM of AG-490 for further experiments. AG490 was added to mock- or HBV-infected PHHs on day 4 p.i. 24 hours after AG-490 treatment, cells were either harvested for RNA and protein isolation or lysed for measurement of caspase 3/7 activity. AG-490 inhibited phosphorylation of STAT3 in mock- and HBV-infected PHH cells as shown by Western blotting of nuclear protein preparations (Figure 7B). We here used expression of lamin as a loading control instead of total STAT3 because phosphorylated STAT3 drives the transcription of STAT343 and thus inhibition of STAT3 activation by AG-490 in turn leads to down-regulation of total STAT3. Purity of nuclear protein preparations was controlled by Western blotting using anti-albumin antibodies (Figure 7B). Induction of apoptosis in HBV-infected cells treated with AG-490 was indicated by cleavage of poly(ADP-ribose) polymerase, which is a well-known substrate of activated caspases44 (Figure 7B). While caspase activity also increased in AG-490 treated mock-infected cells, HBV-infected cells showed a markedly higher activation of effector caspases 3/7 in response to AG-490 (Figure 7C). Since antiapoptotic genes were up-regulated in HBV-infected hepatocytes and in HBV-expressing tumor and nontumorous tissue (Figure 1D, Tables 3 and 4 and Figure 2, respectively), we investigated their expression level in dependence of STAT3 phosphorylation. Quantitative real-time RT-PCR analysis of AG-490-treated HBV-infected PHHs revealed a significant down-regulation of cIAP2, Mn-SOD, IER3 and TNFAIP8 genes (Figure 7D) in comparison to DMSO-treated, HBV-infected cells. Together, these results strongly suggest that activation of STAT3 signaling is important for the prevention of apoptosis and the survival of HBV-infected cells.
Figure 7.
Inhibition of signal transducer and activator of transcription 3 (STAT3) by AG-490 and its consequences on apoptosis signaling in hepatocytes. (A) Hepatitis B virus (HBV)–infected differentiated HepaRG cells were either treated with 0.1% dimethyl sulfoxide (DMSO) or with different amounts of AG-490 (0.2 μM, 1 μM, 5 μM, 20 μM, and 100 μM); 24 hours after administration of AG-490, cell viability was analyzed by CellTiter-Blue Cell Viability Assay. Values are given as median ± SD (n = 3; statistical significance relative to DMSO-control; Student's t test). (B) Mock- or HBV-infected primary human hepatocytes (PHHs) were either incubated with 0.1% DMSO or with 100 μM of AG-490 for 24 hours. Nuclear or cytosolic proteins were prepared using the NE-PER Nuclear and Cytoplasmic extraction Reagents. Inhibition of STAT3 phosphorylation (upper panel) was analyzed by Western blotting using nuclear proteins and anti-pSTAT3 antibodies. Purity of nuclear protein preparations was controlled by Western blotting using antialbumin antibodies. Cleavage of poly(ADP-ribose) polymerase (PARP) (lower panel) was analyzed by Western blotting using nuclear proteins and anti–poly(ADP-ribose) polymerase (PARP) antibodies. Positions of 116 kDa uncleaved (upper band) and 85 kDa cleaved PARP (lower band) are indicated. Membranes were reprobed with anti-lamin antibodies to control equal protein loading. (C) Caspase 3/7 activity in mock- or HBV-infected PHHs treated with either DMSO or with 100 μM AG-490 was measured by luminescent caspase 3/7 assay. Mean ± SD is given (n = 3; ***P < .001; Student's t test). (D) Expression levels of cIAP2, Mn-SOD, IER3, and TNFAIP8 genes in HBV-infected PHHs treated with either DMSO or with AG-490 (20 or 100 μM) were determined by real-time reverse transcription polymerase chain reaction. Gene expression levels in HBV-infected cells treated with DMSO were set to 100%. Values are shown as mean ± SD, statistical significance was calculated relative to the respective DMSO-control sample (n = 3; *P < .05; **P < .01; ***P < .001; Student's t test). ns, not significant.
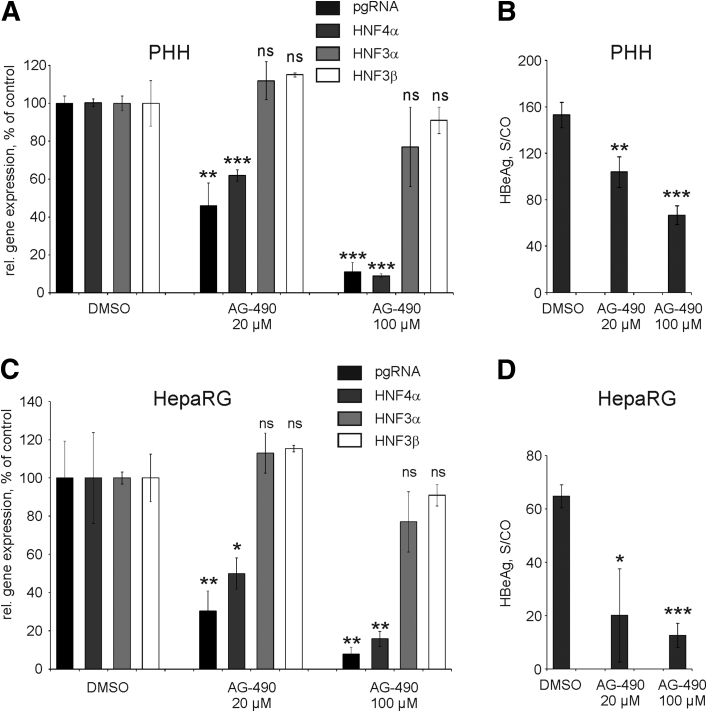
To investigate whether STAT3 activation has an effect on HBV replication, we determined the expression levels of HBV pgRNA in HBV-infected PHHs and in HBV-infected differentiated HepaRG cells treated either with DMSO or with AG-490 (Figure 8A and C, respectively). Inhibition of STAT3 in PHHs by 20 μM or 100 μM of AG-490 reduced relative levels of HBV pgRNA per surviving hepatocyte in a dose-dependent manner by 54% and 89%, respectively (Figure 8A). Accordingly, levels of secreted HBeAg dropped significantly after addition of AG-490 (Figure 8B). Similarly, we observed significantly decreased levels of HBV pgRNA and HBeAg in HBV-infected differentiated HepaRG cells after treatment with 20 μM or 100 μM of AG-490 (Figure 8C and D, respectively). To explain a strong decrease in the level of HBV pgRNA, we quantified HNF4α, a critical regulator of pgRNA transcription.23, 45 We found that HNF4α expression was strongly dependent on STAT3 activation (Figure 8A and C) while HNF3α and HNF3β, also playing a role in regulation of HBV gene expression46, 47, 48 remained unchanged in both cell types (Figure 8A and C). Collectively, these results suggest that STAT3 regulates transcription factor HNF4α and in turn activates HBV gene expression and replication.
Figure 8.
Inhibition of signal transducer and activator of transcription 3 by AG-490 and its consequences for hepatitis B virus (HBV) infection. HBV-infected PHHs (A, B) or HepaRG cells (C, D) were treated for 24 hours either with dimethyl sulfoxide (DMSO) or with AG-490 (20 or 100 μM). (A, C) Expression levels of HBV pregenomic RNA (pgRNA) and HNF4α, HNF3α, and HNF3β genes were determined by real-time reverse transcription polymerase chain reaction. Gene expression levels in HBV-infected cells treated with DMSO were set to 100%. (B, D) Levels of hepatitis B early antigen (HBeAg) secreted into the cell culture medium were determined by as signal-to-control (S/CO) ratio by enzyme-linked immunosorbent assay at 24 hours after treatment with AG-490. All values are shown as mean ± SD, statistical significance was calculated relative to the respective DMSO-control sample (n = 3; *P < .05; **P < .01; ***P < .001; Student's t test). ns, not significant; PHH, primary human hepatocytes.
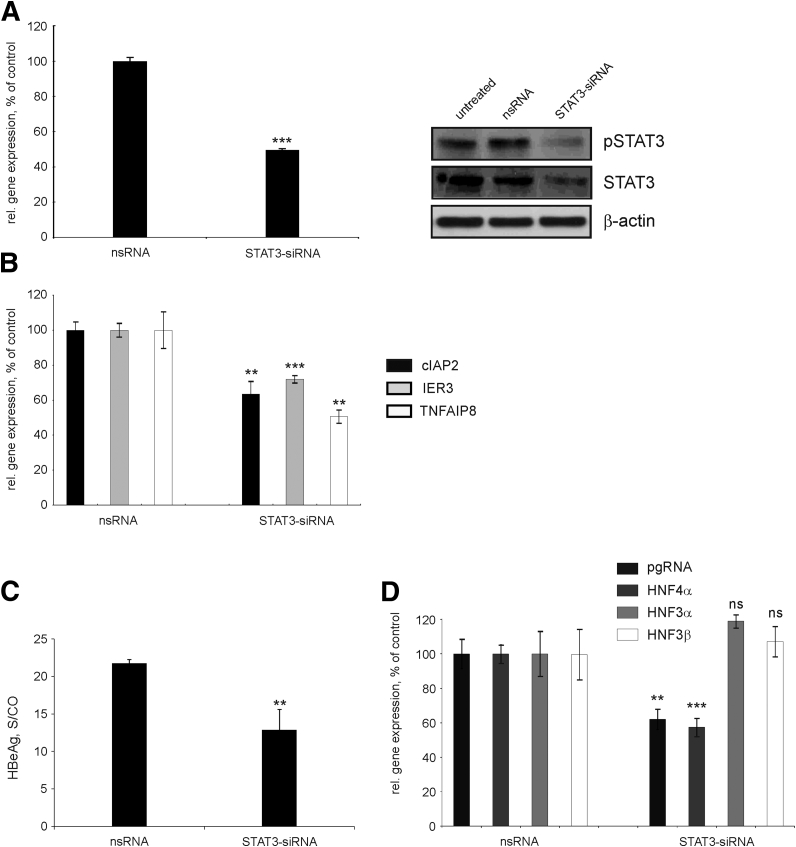
To further confirm the results obtained with AG-490, we knocked down STAT3 by STAT3-specific siRNA. As primary hepatocytes are difficult to transfect, we used HBV-replicating HepG2-H1.3 cells. Transfection of HepG2-H1.3 cells with STAT3-siRNA resulted in ∼50% reduction in STAT3 expression at the mRNA and at the protein levels (Figure 9A) and an ∼70% decrease of phosphorylated STAT3. Importantly, siRNA-mediated inhibition of STAT3 significantly reduced expression levels of cIAP2, IER3, and TNFAIP8 genes in HepG2-H-1.3 cells by 37%, 30%, and 50%, respectively (Figure 9B).
Figure 9.
Knockdown of signal transducer and activator of transcription 3 (STAT3) by STAT3-specific small interfering (siRNA) and its consequences. (A) Hepatitis B virus (HBV)–replicating HepG2-H1.3 cells were either left untreated or transfected with 5 nM of STAT3-siRNA or nonsilencing siRNA (nsRNA) as control. STAT3 messenger RNA (mRNA) levels (left panel) were determined by real-time reverse transcription polymerase chain reaction (RT-PCR). Cells transfected with nsRNA were set to 100%. Levels of phosphorylated STAT3 (pSTAT3) and STAT3 were examined by Western blot analysis of total cellular proteins (right panel) using anti-pSTAT3 and anti-STAT3 antibodies, respectively. Membranes were reprobed with anti–β actin antibodies to control equal protein loading. (B) Gene expression levels of cIAP2, IER3, and TNFAIP8 in HepG2-H1.3 cells transfected with STAT3 siRNA were determined by real-time RT-PCR relative to those transfected with nsRNA (set to 100%). (C) Levels of hepatitis B early antigen (HBeAg) secreted into the cell culture medium were determined by the HBeAg enzyme-linked immunosorbent assay and shown as signal-to-control (S/CO) ratio. (D) Expression levels of pregenomic RNA (pgRNA) and HNF4α, HNF3α, and HNF3β genes were determined by real-time RT-PCR relative to the respective nsRNA control. Expression levels in cells transfected with nsRNA were set to 100%. (A–D) Values are shown as median ± SD (n = 3; **P < .01; ***P < .001; Student's t test). ns, not significant.
To confirm the effects on HBV gene expression and replication, we examined levels of secreted HBeAg and transcription of HBV pgRNA. After STAT3 knockdown, both HBeAg levels and HBV pgRNA levels significantly decreased by 41% and 38%, respectively (Figure 9C and D, respectively). As before, we also observed a strong down-regulation of HNF4α (by 43%) but not HNF3α or HNF3β expression upon knockdown of STAT3 (Figure 9D), suggesting that HNF4α links STAT3 activation and HBV replication.
Taken together, our results strongly suggest that STAT3 serves as a proviral host factor activating HBV transcription and replication. Since inhibition of STAT3 activation resulted in induction of apoptosis, down-regulation of antiapoptotic genes, and a marked decline of HBV gene expression and replication, we concluded that the activation of STAT3 is required to prevent apoptosis and to support HBV persistence and replication.
Discussion
Our study revealed that HBV replication strongly activates STAT3 signaling in PHHs, infected HepaRG cells, HBV-replicating hepatoma cells, and mice, as well as in chronically HBV-infected human liver tissue and in HBV-related HCC. STAT3 activation led to the up-regulation of genes encoding for acute phase response proteins and proteins involved in cell survival. In addition, STAT3 activation supported HBV replication. Inhibition of JAK-kinases in HBV-infected PHH cultures prevented STAT3 activation by phosphorylation and induced apoptosis. Inhibition of STAT3 phosphorylation as well as STAT3 knockdown by siRNA led to down-regulation of antiapoptotic genes and reduction of HBV gene expression and replication. Thus, STAT3 activation is beneficial for the virus not only by supporting HBV replication but also by sustaining survival of HBV replicating hepatocytes.
Using genome-wide gene expression profiling, we identified a number of differentially expressed genes in HBV-infected primary human hepatocyte cultures, and investigated those genes in stably HBV-producing cell lines as well as in tumor and nontumorous tissue of patients chronically infected with HBV. We found that most of the regulated genes are controlled by the STAT3 signaling pathway and serve to restore cellular homeostasis or oxygen radical detoxification and favor hepatocyte survival. This suggests a link to HBV-related cancer development for which STAT3 activation seems important.15, 49 Intriguingly, our data demonstrate that activation of STAT3 is a prerequisite for efficient HBV replication in HBV-infected hepatocytes.
Microarray analysis revealed only limited differences in gene expression profiles between primary human hepatocyte cultures infected with HBV and noninfected controls. However, hepatocytes obviously responded to HBV infection by the induction of a STAT3-dependent acute phase response restoring cell homeostasis and restricting proteolytic activity and tissue damage. The fact that Mn-SOD was up-regulated and a number of CYP450 family members were down-regulated, indicated that oxidative stress contributed to this regulation.
A strong down-regulation of CYP450 enzymes in HBV-infected hepatocytes detected in our study was similar to what has been reported in HBVtg mice.50 CYP450 proteins are mono-oxygenases, which are an important source of ROS in hepatocytes. Particularly CYP3A4, the predominant CYP in the liver, was down-regulated in all analyzed PHH cultures and in HBV-expressing tumor and nontumorous tissue (Tables 3 and 4 and Figure 2). In our study, antioxidant protein Mn-SOD was up-regulated in all analyzed PHH cultures, in liver tissue from chronically infected patients actively replicating HBV as well as in HBV-related HCC (Figures 1D and 2 and Tables 3 and 4). Mn-SOD, which is regulated by STAT3, has been reported to be up-regulated by induction of HBV replication in HepAD38 hepatoma cells51 and increased in patients with acute viral hepatitis.52 It suppresses oxidative stress damage and ROS-mediated apoptosis in rat hepatocytes.35 As HBV replication caused an increase of ROS (Figure 6B–D) and oxidative stress has been shown to negatively regulate HBV gene expression,53 one could speculate that HBV activates a feedback mechanism: down-regulation of CYP450 enzymes, accompanied by induction of Mn-SOD and ROS-reducing activity proteins (IER3)54 will protect the host hepatocyte from oxidative stress–induced apoptosis and maintain virus replication despite an activated immune system. As a side effect, resistance to oxidative stress could provide a selective growth advantage for preneoplastic hepatocytes, which can also contribute to HCC development.55
Wieland et al56 found no significant change in gene regulation by microarray analysis of serial liver biopsies of experimentally infected chimpanzees before the onset of a T cell response and concluded that HBV is a stealth virus. On a first glance, this appears to be in contrast to our data. A study by Fisicaro et al57 showed that the innate immune system is activated from the beginning of HBV infection in men, indicated by the early development of natural killer and CD56+ natural killer–T cell responses. In cultured cells, proinflammatory cytokine responses, including IL-6 responses, are rapidly but transient induced after contact with HBV. HBV and its antigens even seem to inhibit subsequent recognition of other patterns.14, 58 This indicates that sustained STAT3 signaling, which we observed in the human tissue samples, requires other triggers in addition to HBV patterns (eg, inflammatory liver disease), which is lacking in chimpanzees.59
Consistent with the chimpanzee study, our microarray analyses failed to detect any activation of an IFN response during HBV replication in the absence of T cells. IRF3 protein was not activated (Figure 1D) and IFN type I– and type II–inducible genes 2’5’-oligoadenylatsynthetase and IP10 were rather down-regulated (Table 4)—most likely due to STAT3 activation which can mitigate virus induced IFN type I signaling.60
HBV protein expression has been accused to interfere with IFN type I signaling in infected hepatocytes.61 HBV S62 as well as polymerase proteins63 have been reported to interfere with host cell-autonomous immune responses within infected hepatocytes. In particular the HBx protein,64, 65, 66 seems to influence cellular immune as well as survival signaling. HBV needs to constantly express HBx, which is essential to establish and maintain HBV infection67 that otherwise is inhibited by the smc5/6 complex.68 The virus also requires S as well as polymerase proteins to produce progeny. Therefore, its replication may well actively interfere with any interferon response.
As apoptosis is an important mechanism to eliminate HBV-infected hepatocytes, primes antiviral cytokine responses in the liver via apoptotic bodies69 and is deleterious to HBV spread,22 we further analyzed the regulation of apoptosis-related genes. In all experiments, the balance between cell survival and cell death was in favor of cell survival although with some interexperimental variability. A set of antiapoptotic genes was similarly regulated upon HBV infection in at least 4 independent PHH cultures infected with HBV (Table 4) and in stably HBV-replicating cell lines (Table 5). More importantly, we observed an up-regulation of cIAP2, Mn-SOD, IER3, and TNFAIP8 in HBV-expressing tumor and nontumorous liver tissue. Of note, our data strongly suggest a pivotal role of HBV rather than hepatocarcinogenesis in up-regulation of these antiapoptotic genes, since their expression in HBV-expressing nontumorous liver tissue was significantly higher than in samples from noninfected donors (Figure 2).
A number of signaling pathways are accused to contribute to hepatocarcinogenesis. Besides NF-kb, and STAT3 activation,15 the MAPK, Akt/mTOR, WNT/b-catenin, insulin, and hepatocyte growth factor pathways are regarded to be important in this respect.70 In contrast to previously published data,71 we did not observe activation of Erk or JNK by HBV replication in HBV-replicating hepatoma cell lines (data not shown). The STAT3 pathway, in contrast, was strongly regulated by HBV replication after infection of primary human hepatocytes and HepaRG cells or in HBV-replicating hepatoma cells, and—most importantly—in all HBV-infected human liver tissue samples. Analysis of liver needle biopsies from patients with chronic hepatitis B in comparison with healthy controls in our study confirmed that HBV infection leads to a predominant STAT3 phosphorylation and its nuclear translocation in hepatocytes.
By gene expression analyses of primary human liver cell cultures infected with HBV, we also detected changes in the expression of acute phase response genes which are regulated by the IL-6/STAT3 pathway.72, 73 STAT3 is activated via phosphorylation by several tyrosine kinases in response to various intrinsic and extrinsic factors. Growth factors or a number of cytokines including IL-6 or IL-10 family members or leptin activate STAT3. Previously, we demonstrated a rapid secretion of IL-6 and other NF-kB–dependent cytokines upon contact of nonparenchymal liver cells with HBV.14 Furthermore, serum levels of IL-6 are significantly higher in hepatitis B patients than in healthy blood donors74 suggesting sustained IL-6 signaling and activation of STAT3 in HBV infection.
However, we and others detected STAT3 activation also in HBV-replicating hepatoma cell lines75, 76, 77 and in HBVtg mice78 in the absence of cytokine secretion. This requires additional, cytokine-independent intracellular activation of STAT3 by HBV replication. Generation of ROS in stably HBV-producing cell lines HepG2.2.15 and HepG2-H1.3 and upon infection of HepaRG cells with wild-type HBV (Figure 6), and oxidative stress may result in STAT3 activation as described for hepatitis C virus.79 Also, HBx protein might be responsible for intracellular STAT3 activation during HBV replication,80 as it associates with mitochondria and can cause oxidative stress by ROS induction51, 80, 81 as well as may dysregulate cellular miRNAs such as lethal-782 or microRNA-21.83 It is very likely, upon HBV replication, STAT3 is activated by multiple intra- and extracellular pathways in hepatocytes or in nonparenchymal cells. The exact contribution of each pathway in activation of STAT3 in primary hepatocytes is yet to be determined.
Inhibition of STAT3 is known to induce growth arrest and apoptosis of human HCC cells.63, 84 Interestingly, study performed by Xie et al,77 strongly suggested that some single nucleotide polymorphisms in STAT3-coding gene predispose the host with HBV mutations in enhancer or basal core promoter and precore region or in preS2 start codon mutations to hepatocarcinogenesis. By chemical inhibition of STAT3 phosphorylation and siRNA-mediated silencing of STAT3, we show that STAT3 activation is necessary to confer apoptosis resistance in primary human hepatocytes infected with HBV and in HBV-replicating HepG2-H1.3 cells. Accordingly, we detected significant down-regulation of cIAP2, Mn-SOD (PHH), IER3, and TNFAIP8 genes upon inhibition of STAT3 (Figure 7D and Figure 9B).
Interestingly, inhibition of STAT3 resulted in a marked decline of HBV gene expression, antigen secretion and replication (Figure 8, Figure 9C and D). Here, 2 possible mechanisms may become operative: (1) activated STAT3 may regulate HNF3, and together they bind to the core domain of HBV enhancer I and positively influence HBV gene expression85; alternatively, (2) HNF1,86 an important transcriptional activator of HNF4α, may up-regulate HNF4α and thus transcription of pgRNA and HBV replication.23, 87 Examining the first possible mechanism, we quantified expression of HNF3α and HNF3β mRNAs in PHHs and in HepaRG cells upon inhibition of STAT3 signaling by administration of AG-490 or specific STAT3-siRNA. In neither case, the expression of HNF3α or HNF3β was changed. In contrast, we found a strong decrease of HNF4α levels upon inhibition of STAT3 (Figure 8A and C, Figure 9D). As STAT3 is known to amplify transactivation of hepatic genes, mediated by HNF1, we speculate that this mechanism is utilized by HBV to increase transcription of viral pregenomes, which are the essential template for HBV replication.
A recent study performed by Huang et al88 demonstrates that activation of STAT3 by HBV inhibits expression of micro-RNA miR-204 acting as a tumor suppressor but also inhibiting HBV pRNA encapsidation and capsid assembly. Hence, in addition to prevention of apoptosis, attenuation of oxidative stress, and stimulation of HBV replication through HNF4α described in our study, activation of STAT3 upon HBV infection supports HBV replication and malignant transformation of infected hepatocytes by suppressing miR-204. Thus, STAT3 signaling acts at various levels in favor of HBV but also in favor of hepatocyte transformation and survival.
In summary, our data clearly demonstrate HBV-mediated activation of STAT3 signaling in different cell culture systems, in HBV-replicating mice and in HBV-replicating noncancerous liver and HCC tissue. We found that HBV activated STAT3 by intra- and extracellular signaling. Activated STAT3 is crucial to maintain efficient HBV replication. In addition, STAT3 promotes hepatocyte survival, essential for virus persistence, but in turn favors malignant transformation. Hence, inhibition of STAT3 activation represents a promising target to control HBV infection and to inhibit HBV-related HCC development.
Acknowledgments
The authors thank Uta Zedler, Andreas Untergasser, Gregor Ebert, and Diana Wagner for preparation of primary human hepatocytes; Daniel Kull and Birgit Riepl for histological and molecular analysis of hepatitis B virus–infected human liver tissue, Peter Schraml from the Institute of Surgical Pathology, University Hospital Zürich, for providing liver tissues samples; and Heike Oberwinkler, Gisela Holz, and Maureen Menning for excellent technical assistance.
Footnotes
Author contributions MH, MQ, MHW, and UP designed the study. UP, MHW, and HB acquired grants. MH, MQ, MR, SDP, MS, JHB, SA, DW, and GO performed experiments. MH, MQ, HK, SDP, AW, JLS, HB and UP analyzed the data. MH, DW, MQ, and MHW prepared the figures. MH, MQ, MHW, and UP wrote the manuscript.
Conflicts of interest The authors have declared that no relevant conflict of interest exists.
Funding The study was supported by the Deutsche Forschungsgemeinschaft (SFB 670 to U.P. and H.B., TRR 179 to U.P. and M.H.W.) and the Horizon 2020 program (HepCar consortium to U.P. and M.H.W.). M.H.W. was supported by a Helmholtz Young Investigator Award and an ERC Consolidator grant (HepatoMetaboPath). J.L.S. was supported by the Sofja Kovalevskaja Award of the Alexander von Humboldt Foundation.
References
- 1.Protzer U., Maini M.K., Knolle P.A. Living in the liver: hepatic infections. Nat Rev Immunol. 2012;12:201–213. doi: 10.1038/nri3169. [DOI] [PubMed] [Google Scholar]
- 2.Lucifora J., Xia Y., Reisinger F., Zhang K., Stadler D., Cheng X., Sprinzl M.F., Koppensteiner H., Makowska Z., Volz T., Remouchamps C., Chou W.M., Thasler W.E., Huser N., Durantel D., Liang T.J., Munk C., Heim M.H., Browning J.L., Dejardin E., Dandri M., Schindler M., Heikenwalder M., Protzer U. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 2014;343:1221–12218. doi: 10.1126/science.1243462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwon H., Lok A.S. Does antiviral therapy prevent hepatocellular carcinoma? Antivir Ther. 2011;16:787–795. doi: 10.3851/IMP1895. [DOI] [PubMed] [Google Scholar]
- 4.Ferlay J., Shin H.R., Bray F., Forman D., Mathers C., Parkin D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 5.Chen G., Luo D.Z., Liu L., Feng Z.B., Guo F., Li P. Hepatic local micro-environmental immune status in hepatocellular carcinoma and cirrhotic tissues. West Indian Med J. 2006;55:403–408. doi: 10.1590/s0043-31442006000600007. [DOI] [PubMed] [Google Scholar]
- 6.McMahon B.J. The natural history of chronic hepatitis B virus infection. Hepatology. 2009;49:S45–S55. doi: 10.1002/hep.22898. [DOI] [PubMed] [Google Scholar]
- 7.Ringelhan M., O'Connor T., Protzer U., Heikenwalder M. The direct and indirect roles of HBV in liver cancer: prospective markers for HCC screening and potential therapeutic targets. J Pathol. 2015;235:355–367. doi: 10.1002/path.4434. [DOI] [PubMed] [Google Scholar]
- 8.Neuveut C., Wei Y., Buendia M.A. Mechanisms of HBV-related hepatocarcinogenesis. J Hepatol. 2010;52:594–604. doi: 10.1016/j.jhep.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 9.Ringelhan M., Protzer U. Oncogenic potential of hepatitis B virus encoded proteins. Curr Opin Virol. 2015;14:109–115. doi: 10.1016/j.coviro.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 10.Tan Y.J. Hepatitis B virus infection and the risk of hepatocellular carcinoma. World J Gastroenterol. 2011;17:4853–4857. doi: 10.3748/wjg.v17.i44.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stauffer J.K., Scarzello A.J., Jiang Q., Wiltrout R.H. Chronic inflammation, immune escape, and oncogenesis in the liver: a unique neighborhood for novel intersections. Hepatology. 2012;56:1567–1574. doi: 10.1002/hep.25674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakamoto Y., Guidotti L.G., Kuhlen C.V., Fowler P., Chisari F.V. Immune pathogenesis of hepatocellular carcinoma. J Exp Med. 1998;188:341–350. doi: 10.1084/jem.188.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haybaeck J., Zeller N., Wolf M.J., Weber A., Wagner U., Kurrer M.O., Bremer J., Iezzi G., Graf R., Clavien P.A., Thimme R., Blum H., Nedospasov S.A., Zatloukal K., Ramzan M., Ciesek S., Pietschmann T., Marche P.N., Karin M., Kopf M., Browning J.L., Aguzzi A., Heikenwalder M. A lymphotoxin-driven pathway to hepatocellular carcinoma. Cancer Cell. 2009;16:295–308. doi: 10.1016/j.ccr.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosel M., Quasdorff M., Wiegmann K., Webb D., Zedler U., Broxtermann M., Tedjokusumo R., Esser K., Arzberger S., Kirschning C.J., Langenkamp A., Falk C., Buning H., Rose-John S., Protzer U. Not interferon, but interleukin-6 controls early gene expression in hepatitis B virus infection. Hepatology. 2009;50:1773–1782. doi: 10.1002/hep.23226. [DOI] [PubMed] [Google Scholar]
- 15.He G., Karin M. NF-kappaB and STAT3 - key players in liver inflammation and cancer. Cell Res. 2011;21:159–168. doi: 10.1038/cr.2010.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guidotti L.G., Matzke B., Schaller H., Chisari F.V. High-level hepatitis B virus replication in transgenic mice. J Virol. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang L.R., Gabel Y.A., Graf S., Arzberger S., Kurts C., Heikenwalder M., Knolle P.A., Protzer U. Transfer of HBV genomes using low doses of adenovirus vectors leads to persistent infection in immune competent mice. Gastroenterology. 2012;142:1447–1450.e3. doi: 10.1053/j.gastro.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 18.Sprinzl M.F., Oberwinkler H., Schaller H., Protzer U. Transfer of hepatitis B virus genome by adenovirus vectors into cultured cells and mice: crossing the species barrier. J Virol. 2001;75:5108–5118. doi: 10.1128/JVI.75.11.5108-5118.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Freyend M.J., Untergasser A., Arzberger S., Oberwinkler H., Drebber U., Schirmacher P., Protzer U. Sequential control of hepatitis B virus in a mouse model of acute, self-resolving hepatitis B. J Viral Hepat. 2011;18:216–226. doi: 10.1111/j.1365-2893.2010.01302.x. [DOI] [PubMed] [Google Scholar]
- 20.Schulze-Bergkamen H., Untergasser A., Dax A., Vogel H., Buchler P., Klar E., Lehnert T., Friess H., Buchler M.W., Kirschfink M., Stremmel W., Krammer P.H., Muller M., Protzer U. Primary human hepatocytes–a valuable tool for investigation of apoptosis and hepatitis B virus infection. J Hepatol. 2003;38:736–744. doi: 10.1016/s0168-8278(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 21.Sells M.A., Chen M.L., Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci U S A. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arzberger S., Hosel M., Protzer U. Apoptosis of hepatitis B virus-infected hepatocytes prevents release of infectious virus. J Virol. 2010;84:11994–12001. doi: 10.1128/JVI.00653-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quasdorff M., Hosel M., Odenthal M., Zedler U., Bohne F., Gripon P., Dienes H.P., Drebber U., Stippel D., Goeser T., Protzer U. A concerted action of HNF4αlpha and HNF1alpha links hepatitis B virus replication to hepatocyte differentiation. Cell Microbiol. 2008;10:1478–1490. doi: 10.1111/j.1462-5822.2008.01141.x. [DOI] [PubMed] [Google Scholar]
- 24.Untergasser A., Zedler U., Langenkamp A., Hosel M., Quasdorff M., Esser K., Dienes H.P., Tappertzhofen B., Kolanus W., Protzer U. Dendritic cells take up viral antigens but do not support the early steps of hepatitis B virus infection. Hepatology. 2006;43:539–547. doi: 10.1002/hep.21048. [DOI] [PubMed] [Google Scholar]
- 25.Protzer U., Seyfried S., Quasdorff M., Sass G., Svorcova M., Webb D., Bohne F., Hosel M., Schirmacher P., Tiegs G. Antiviral activity and hepatoprotection by heme oxygenase-1 in hepatitis B virus infection. Gastroenterology. 2007;133:1156–1165. doi: 10.1053/j.gastro.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 26.Birnbaum F., Nassal M. Hepatitis B virus nucleocapsid assembly: primary structure requirements in the core protein. J Virol. 1990;64:3319–3330. doi: 10.1128/jvi.64.7.3319-3330.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Omura T., Sato R., Cooper D.Y., Rosenthal O., Estabrook R.W. Function of cytochrome P-450 of microsomes. Fed Proc. 1965;24:1181–1189. [PubMed] [Google Scholar]
- 28.Ebert G., Preston S., Allison C., Cooney J., Toe J.G., Stutz M.D., Ojaimi S., Scott H.W., Baschuk N., Nachbur U., Torresi J., Chin R., Colledge D., Li X., Warner N., Revill P., Bowden S., Silke J., Begley C.G., Pellegrini M. Cellular inhibitor of apoptosis proteins prevent clearance of hepatitis B virus. Proc Natl Acad Sci U S A. 2015;112:5797–5802. doi: 10.1073/pnas.1502390112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schoemaker M.H., Ros J.E., Homan M., Trautwein C., Liston P., Poelstra K., van Goor H., Jansen P.L., Moshage H. Cytokine regulation of pro- and antiapoptotic genes in rat hepatocytes: NF-kappaB-regulated inhibitor of apoptosis protein 2 (cIAP2) prevents apoptosis. J Hepatol. 2002;36:742–750. doi: 10.1016/s0168-8278(02)00063-6. [DOI] [PubMed] [Google Scholar]
- 30.Huang Y.H., Wu J.Y., Zhang Y., Wu M.X. Synergistic and opposing regulation of the stress-responsive gene IEX-1 by p53, c-Myc, and multiple NF-kappaB/rel complexes. Oncogene. 2002;21:6819–6828. doi: 10.1038/sj.onc.1205854. [DOI] [PubMed] [Google Scholar]
- 31.Porturas T.P., Sun H., Buchlis G., Lou Y., Liang X., Cathopoulis T., Fayngerts S., Johnson D.S., Wang Z., Chen Y.H. Crucial roles of TNFAIP8 protein in regulating apoptosis and Listeria infection. J Immunol. 2015;194:5743–5750. doi: 10.4049/jimmunol.1401987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leu J.I., George D.L. Hepatic IGFBP1 is a prosurvival factor that binds to BAK, protects the liver from apoptosis, and antagonizes the proapoptotic actions of p53 at mitochondria. Genes Dev. 2007;21:3095–3109. doi: 10.1101/gad.1567107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jia S.H., Li Y., Parodo J., Kapus A., Fan L., Rotstein O.D., Marshall J.C. Pre-B cell colony-enhancing factor inhibits neutrophil apoptosis in experimental inflammation and clinical sepsis. J Clin Invest. 2004;113:1318–1327. doi: 10.1172/JCI19930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanson J.C., Bostick M.K., Campe C.B., Kodali P., Lee G., Yan J., Maher J.J. Transgenic overexpression of interleukin-8 in mouse liver protects against galactosamine/endotoxin toxicity. J Hepatol. 2006;44:359–367. doi: 10.1016/j.jhep.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 35.Terui K., Enosawa S., Haga S., Zhang H.Q., Kuroda H., Kouchi K., Matsunaga T., Yoshida H., Engelhardt J.F., Irani K., Ohnuma N., Ozaki M. Stat3 confers resistance against hypoxia/reoxygenation-induced oxidative injury in hepatocytes through upregulation of Mn-SOD. J Hepatol. 2004;41:957–965. doi: 10.1016/j.jhep.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 36.Yu H.C., Qin H.Y., He F., Wang L., Fu W., Liu D., Guo F.C., Liang L., Dou K.F., Han H. Canonical notch pathway protects hepatocytes from ischemia/reperfusion injury in mice by repressing reactive oxygen species production through JAK2/STAT3 signaling. Hepatology. 2011;54:979–988. doi: 10.1002/hep.24469. [DOI] [PubMed] [Google Scholar]
- 37.Eugenin E.A., D'Aversa T.G., Lopez L., Calderon T.M., Berman J.W. MCP-1 (CCL2) protects human neurons and astrocytes from NMDA or HIV-tat-induced apoptosis. J Neurochem. 2003;85:1299–1311. doi: 10.1046/j.1471-4159.2003.01775.x. [DOI] [PubMed] [Google Scholar]
- 38.Yang J., Wezeman M., Zhang X., Lin P., Wang M., Qian J., Wan B., Kwak L.W., Yu L., Yi Q. Human C-reactive protein binds activating Fcgamma receptors and protects myeloma tumor cells from apoptosis. Cancer Cell. 2007;12:252–265. doi: 10.1016/j.ccr.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 39.Mackiewicz A., Schooltink H., Heinrich P.C., Rose-John S. Complex of soluble human IL-6-receptor/IL-6 up-regulates expression of acute-phase proteins. J Immunol. 1992;149:2021–2027. [PubMed] [Google Scholar]
- 40.Stonans I., Stonane E., Russwurm S., Deigner H.P., Bohm K.J., Wiederhold M., Jager L., Reinhart K. HepG2 human hepatoma cells express multiple cytokine genes. Cytokine. 1999;11:151–156. doi: 10.1006/cyto.1998.0366. [DOI] [PubMed] [Google Scholar]
- 41.Garcia R., Bowman T.L., Niu G., Yu H., Minton S., Muro-Cacho C.A., Cox C.E., Falcone R., Fairclough R., Parsons S., Laudano A., Gazit A., Levitzki A., Kraker A., Jove R. Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene. 2001;20:2499–2513. doi: 10.1038/sj.onc.1204349. [DOI] [PubMed] [Google Scholar]
- 42.Meydan N., Grunberger T., Dadi H., Shahar M., Arpaia E., Lapidot Z., Leeder J.S., Freedman M., Cohen A., Gazit A., Levitzki A., Roifman C.M. Inhibition of acute lymphoblastic leukaemia by a Jak-2 inhibitor. Nature. 1996;379:645–648. doi: 10.1038/379645a0. [DOI] [PubMed] [Google Scholar]
- 43.Yang J., Chatterjee-Kishore M., Staugaitis S.M., Nguyen H., Schlessinger K., Levy D.E., Stark G.R. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res. 2005;65:939–947. [PubMed] [Google Scholar]
- 44.Lazebnik Y.A., Kaufmann S.H., Desnoyers S., Poirier G.G., Earnshaw W.C. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature. 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- 45.Raney A.K., Kline E.F., Tang H., McLachlan A. Transcription and replication of a natural hepatitis B virus nucleocapsid promoter variant is regulated in vivo by peroxisome proliferators. Virology. 2001;289:239–251. doi: 10.1006/viro.2001.1169. [DOI] [PubMed] [Google Scholar]
- 46.Chen M., Hieng S., Qian X., Costa R., Ou J.H. Regulation of hepatitis B virus ENI enhancer activity by hepatocyte-enriched transcription factor HNF3. Virology. 1994;205:127–132. doi: 10.1006/viro.1994.1627. [DOI] [PubMed] [Google Scholar]
- 47.Li M., Xie Y., Wu X., Kong Y., Wang Y. HNF3 binds and activates the second enhancer, ENII, of hepatitis B virus. Virology. 1995;214:371–378. doi: 10.1006/viro.1995.0046. [DOI] [PubMed] [Google Scholar]
- 48.Tang H., McLachlan A. Mechanisms of inhibition of nuclear hormone receptor-dependent hepatitis B virus replication by hepatocyte nuclear factor 3beta. J Virol. 2002;76:8572–8581. doi: 10.1128/JVI.76.17.8572-8581.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He G., Yu G.Y., Temkin V., Ogata H., Kuntzen C., Sakurai T., Sieghart W., Peck-Radosavljevic M., Leffert H.L., Karin M. Hepatocyte IKKbeta/NF-kappaB inhibits tumor promotion and progression by preventing oxidative stress-driven STAT3 activation. Cancer Cell. 2010;17:286–297. doi: 10.1016/j.ccr.2009.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hajjou M., Norel R., Carver R., Marion P., Cullen J., Rogler L.E., Rogler C.E. cDNA microarray analysis of HBV transgenic mouse liver identifies genes in lipid biosynthetic and growth control pathways affected by HBV. J Med Virol. 2005;77:57–65. doi: 10.1002/jmv.20427. [DOI] [PubMed] [Google Scholar]
- 51.Severi T., Ying C., Vermeesch J.R., Cassiman D., Cnops L., Verslype C., Fevery J., Arckens L., Neyts J., van Pelt J.F. Hepatitis B virus replication causes oxidative stress in HepAD38 liver cells. Mol Cell Biochem. 2006;290:79–85. doi: 10.1007/s11010-006-9167-x. [DOI] [PubMed] [Google Scholar]
- 52.Semrau F., Kuhl R.J., Ritter S., Ritter K. Manganese superoxide dismutase (MnSOD) and autoantibodies against MnSOD in acute viral infections. J Med Virol. 1998;55:161–167. [PubMed] [Google Scholar]
- 53.Zheng Y.W., Yen T.S. Negative regulation of hepatitis B virus gene expression and replication by oxidative stress. J Biol Chem. 1994;269:8857–8862. [PubMed] [Google Scholar]
- 54.Shen L., Guo J., Santos-Berrios C., Wu M.X. Distinct domains for anti- and pro-apoptotic activities of IEX-1. J Biol Chem. 2006;281:15304–15311. doi: 10.1074/jbc.M600054200. [DOI] [PubMed] [Google Scholar]
- 55.Xu X.R., Huang J., Xu Z.G., Qian B.Z., Zhu Z.D., Yan Q., Cai T., Zhang X., Xiao H.S., Qu J., Liu F., Huang Q.H., Cheng Z.H., Li N.G., Du J.J., Hu W., Shen K.T., Lu G., Fu G., Zhong M., Xu S.H., Gu W.Y., Huang W., Zhao X.T., Hu G.X., Gu J.R., Chen Z., Han Z.G. Insight into hepatocellular carcinogenesis at transcriptome level by comparing gene expression profiles of hepatocellular carcinoma with those of corresponding noncancerous liver. Proc Natl Acad Sci U S A. 2001;98:15089–15094. doi: 10.1073/pnas.241522398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wieland S., Thimme R., Purcell R.H., Chisari F.V. Genomic analysis of the host response to hepatitis B virus infection. Proc Natl Acad Sci U S A. 2004;101:6669–6674. doi: 10.1073/pnas.0401771101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fisicaro P., Valdatta C., Boni C., Massari M., Mori C., Zerbini A., Orlandini A., Sacchelli L., Missale G., Ferrari C. Early kinetics of innate and adaptive immune responses during hepatitis B virus infection. Gut. 2009;58:974–982. doi: 10.1136/gut.2008.163600. [DOI] [PubMed] [Google Scholar]
- 58.Wu J., Meng Z., Jiang M., Zhang E., Trippler M., Broering R., Bucchi A., Krux F., Dittmer U., Yang D., Roggendorf M., Gerken G., Lu M., Schlaak J.F. Toll-like receptor-induced innate immune responses in non-parenchymal liver cells are cell type-specific. Immunology. 2010;129:363–374. doi: 10.1111/j.1365-2567.2009.03179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rehermann B., Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat Rev Immunol. 2005;5:215–229. doi: 10.1038/nri1573. [DOI] [PubMed] [Google Scholar]
- 60.Wang W.B., Levy D.E., Lee C.K. STAT3 negatively regulates type I IFN-mediated antiviral response. J Immunol. 2011;187:2578–2585. doi: 10.4049/jimmunol.1004128. [DOI] [PubMed] [Google Scholar]
- 61.Lutgehetmann M., Bornscheuer T., Volz T., Allweiss L., Bockmann J.H., Pollok J.M., Lohse A.W., Petersen J., Dandri M. Hepatitis B virus limits response of human hepatocytes to interferon-alpha in chimeric mice. Gastroenterology. 2011;140:2074–2083. doi: 10.1053/j.gastro.2011.02.057. 2083.e1–2. [DOI] [PubMed] [Google Scholar]
- 62.Christen V., Duong F., Bernsmeier C., Sun D., Nassal M., Heim M.H. Inhibition of alpha interferon signaling by hepatitis B virus. J Virol. 2007;81:159–165. doi: 10.1128/JVI.01292-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun X., Zhang J., Wang L., Tian Z. Growth inhibition of human hepatocellular carcinoma cells by blocking STAT3 activation with decoy-ODN. Cancer Lett. 2008;262:201–213. doi: 10.1016/j.canlet.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 64.Hu Z., Zhang Z., Kim J.W., Huang Y., Liang T.J. Altered proteolysis and global gene expression in hepatitis B virus X transgenic mouse liver. J Virol. 2006;80:1405–1413. doi: 10.1128/JVI.80.3.1405-1413.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ng R.K., Lau C.Y., Lee S.M., Tsui S.K., Fung K.P., Waye M.M. cDNA microarray analysis of early gene expression profiles associated with hepatitis B virus X protein-mediated hepatocarcinogenesis. Biochem Biophys Res Commun. 2004;322:827–835. doi: 10.1016/j.bbrc.2004.07.188. [DOI] [PubMed] [Google Scholar]
- 66.Zhang W.Y., Xu F.Q., Shan C.L., Xiang R., Ye L.H., Zhang X.D. Gene expression profiles of human liver cells mediated by hepatitis B virus X protein. Acta Pharmacol Sin. 2009;30:424–434. doi: 10.1038/aps.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lucifora J., Durantel D., Testoni B., Hantz O., Levrero M., Zoulim F. Control of hepatitis B virus replication by innate response of HepaRG cells. Hepatology. 2010;51:63–72. doi: 10.1002/hep.23230. [DOI] [PubMed] [Google Scholar]
- 68.Decorsiere A., Mueller H., van Breugel P.C., Abdul F., Gerossier L., Beran R.K., Livingston C.M., Niu C., Fletcher S.P., Hantz O., Strubin M. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature. 2016;531:386–389. doi: 10.1038/nature17170. [DOI] [PubMed] [Google Scholar]
- 69.Canbay A., Feldstein A.E., Higuchi H., Werneburg N., Grambihler A., Bronk S.F., Gores G.J. Kupffer cell engulfment of apoptotic bodies stimulates death ligand and cytokine expression. Hepatology. 2003;38:1188–1198. doi: 10.1053/jhep.2003.50472. [DOI] [PubMed] [Google Scholar]
- 70.Whittaker S., Marais R., Zhu A.X. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29:4989–5005. doi: 10.1038/onc.2010.236. [DOI] [PubMed] [Google Scholar]
- 71.Chin R., Earnest-Silveira L., Koeberlein B., Franz S., Zentgraf H., Dong X., Gowans E., Bock C.T., Torresi J. Modulation of MAPK pathways and cell cycle by replicating hepatitis B virus: factors contributing to hepatocarcinogenesis. J Hepatol. 2007;47:325–337. doi: 10.1016/j.jhep.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 72.Trautwein C., Boker K., Manns M.P. Hepatocyte and immune system: acute phase reaction as a contribution to early defence mechanisms. Gut. 1994;35:1163–1166. doi: 10.1136/gut.35.9.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wegenka U.M., Lutticken C., Buschmann J., Yuan J., Lottspeich F., Muller-Esterl W., Schindler C., Roeb E., Heinrich P.C., Horn F. The interleukin-6-activated acute-phase response factor is antigenically and functionally related to members of the signal transducer and activator of transcription (STAT) family. Mol Cell Biol. 1994;14:3186–3196. doi: 10.1128/mcb.14.5.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang J.Y., Wang X.L., Liu P. Detection of serum TNF-alpha,IFN-beta,IL-6 and IL-8 in patients with hepatitis B. World J Gastroenterol. 1999;5:38–40. doi: 10.3748/wjg.v5.i1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Choudhari S.R., Khan M.A., Harris G., Picker D., Jacob G.S., Block T., Shailubhai K. Deactivation of Akt and STAT3 signaling promotes apoptosis, inhibits proliferation, and enhances the sensitivity of hepatocellular carcinoma cells to an anticancer agent. Atiprimod. Mol Cancer Ther. 2007;6:112–121. doi: 10.1158/1535-7163.MCT-06-0561. [DOI] [PubMed] [Google Scholar]
- 76.Koeberlein B., zur Hausen A., Bektas N., Zentgraf H., Chin R., Nguyen L.T., Kandolf R., Torresi J., Bock C.T. Hepatitis B virus overexpresses suppressor of cytokine signaling-3 (SOCS3) thereby contributing to severity of inflammation in the liver. Virus Res. 2010;148:51–59. doi: 10.1016/j.virusres.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 77.Xie J., Zhang Y., Zhang Q., Han Y., Yin J., Pu R., Shen Q., Lu W., Du Y., Zhao J., Han X., Zhang H., Cao G. Interaction of signal transducer and activator of transcription 3 polymorphisms with hepatitis B virus mutations in hepatocellular carcinoma. Hepatology. 2013;57:2369–2377. doi: 10.1002/hep.26303. [DOI] [PubMed] [Google Scholar]
- 78.Kim K., Kim K.H., Cheong J. Hepatitis B virus X protein impairs hepatic insulin signaling through degradation of IRS1 and induction of SOCS3. PLoS One. 2010;5:e8649. doi: 10.1371/journal.pone.0008649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Waris G., Turkson J., Hassanein T., Siddiqui A. Hepatitis C virus (HCV) constitutively activates STAT3 via oxidative stress: role of STAT3 in HCV replication. J Virol. 2005;79:1569–1580. doi: 10.1128/JVI.79.3.1569-1580.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 80.He P., Zhang D., Li H., Yang X., Li D., Zhai Y., Ma L., Feng G. Hepatitis B virus X protein modulates apoptosis in human renal proximal tubular epithelial cells by activating the JAK2/STAT3 signaling pathway. Int J Mol Med. 2013;31:1017–1029. doi: 10.3892/ijmm.2013.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Waris G., Huh K.W., Siddiqui A. Mitochondrially associated hepatitis B virus X protein constitutively activates transcription factors STAT3 and NF-kappa B via oxidative stress. Mol Cell Biol. 2001;21:7721–7730. doi: 10.1128/MCB.21.22.7721-7730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Y., Lu Y., Toh S.T., Sung W.K., Tan P., Chow P., Chung A.Y., Jooi L.L., Lee C.G. Lethal-7 is down-regulated by the hepatitis B virus x protein and targets signal transducer and activator of transcription 3. J Hepatol. 2010;53:57–66. doi: 10.1016/j.jhep.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 83.Li C.H., Xu F., Chow S., Feng L., Yin D., Ng T.B., Chen Y. Hepatitis B virus X protein promotes hepatocellular carcinoma transformation through interleukin-6 activation of microRNA-21 expression. Eur J Cancer. 2014;50:2560–2569. doi: 10.1016/j.ejca.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 84.Quinn C.M., Kagedal K., Terman A., Stroikin U., Brunk U.T., Jessup W., Garner B. Induction of fibroblast apolipoprotein E expression during apoptosis, starvation-induced growth arrest and mitosis. Biochem J. 2004;378:753–761. doi: 10.1042/BJ20031352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Waris G., Siddiqui A. Interaction between STAT3 and HNF-3 leads to the activation of liver-specific hepatitis B virus enhancer 1 function. J Virol. 2002;76:2721–2729. doi: 10.1128/JVI.76.6.2721-2729.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leu J.I., Crissey M.A., Leu J.P., Ciliberto G., Taub R. Interleukin-6-induced STAT3 and AP-1 amplify hepatocyte nuclear factor 1-mediated transactivation of hepatic genes, an adaptive response to liver injury. Mol Cell Biol. 2001;21:414–424. doi: 10.1128/MCB.21.2.414-424.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tang H., Banks K.E., Anderson A.L., McLachlan A. Hepatitis B virus transcription and replication. Drug News Perspect. 2001;14:325–334. [PubMed] [Google Scholar]
- 88.Huang J.Y., Chen H.L., Shih C. MicroRNA miR-204 and miR-1236 inhibit hepatitis B virus replication via 2 different mechanisms. Sci Rep. 2016;6:34740. doi: 10.1038/srep34740. [DOI] [PMC free article] [PubMed] [Google Scholar]