Abstract
Background
Knocking down neuronal LINGO-1 using short hairpin RNAs (shRNAs) might enhance axon regeneration in the CNS. Integration-deficient lentiviral vectors have great potential as a therapeutic delivery system for CNS injuries. However, recent studies have revealed that shRNAs can induce an interferon response resulting in off-target effects and cytotoxicity.
Methods
CNS neurons were transduced with integration-deficient lentiviral vectors in vitro. The transcriptional effect of shRNA expression was analysed using qRT-PCR and northern blots were used to assess shRNA production.
Results
Integration-deficient lentiviral vectors efficiently transduced CNS neurons and knocked down LINGO-1 mRNA in vitro. However, an increase in cell death was observed when lentiviral vectors encoding an shRNA were applied or when high vector concentrations were used. We demonstrate that high doses of vector or the use of vectors encoding shRNAs can induce an up-regulation of interferon stimulated genes (OAS1 and PKR) and a down-regulation of off- target genes (including p75NTR and NgR1). Furthermore, the northern blot demonstrated that these negative consequences occur even when lentiviral vectors express low levels of shRNAs. Together, these results may explain why neurite outgrowth was not enhanced on an inhibitory substrate after transduction with lentiviral vectors encoding an shRNA targeting LINGO-1.
Conclusions
These findings highlight the importance of including appropriate controls to verify silencing specificity and the requirement to check for an interferon response when conducting RNA interference experiments. However, the potential benefits that RNA interference and viral vectors offer to gene-based therapies to CNS injuries cannot be overlooked and demand further investigation.
Keywords: RNA interference, neurons, lentiviral vector, interferon response, shRNA, CNS, Integration-deficient
Introduction
Following an injury to the adult central nervous system (CNS), neurons show a very limited regenerative response, which results in their failure to successfully form functional connections with their original targets. This is due to a number of factors including a reduced intrinsic growth state [1–6] and the presence of growth inhibitory molecules in the CNS that form a molecular and physical barrier to regeneration [7–14]. Over the past twenty years there have been steady advancements in molecular techniques that have elevated gene therapy into a promising therapeutic strategy for CNS repair.
Integration-deficient lentiviral vectors are among the most promising therapeutic delivery systems for gene therapy to the CNS. Lentiviral vectors can provide long-term expression by integration into the host cells’ genome. Although the specific integration sites are unpredictable it has been demonstrated that lentiviral vectors preferentially integrate into active gene loci [15], which can result in insertional mutagenesis, the activation of proto-oncogenes [16, 17] and the formation of cancers in both mouse models and human clinical trials [18–20]. To circumvent this, integration-deficient lentiviral vectors can be generated by mutating the integrase coding sequence, resulting in the expression of the transgene from episomes [21, 22]. Integration-deficient lentiviral vectors can be produced at high titres and express a transgene at a level comparable to vectors incorporating the wild-type integrase [21]. Robust and stable expression of enhanced green fluorescent protein (eGFP) has been observed for at least nine months in post-mitotic retinal pigment epithelium, and for at least one month in striatal and hippocampal neurons in vivo [21]. Both integration proficient and deficient lentiviral vectors have been shown to efficiently transduce CNS neurons in vitro [23, 24] and in the brain and spinal cord in vivo [21, 25–30]. Furthermore, integration-proficient lentiviral vectors have been shown to successfully mediate RNA interference in neurons in vitro [31] and in vivo where they have been successfully used to reverse the neurodegenerative and behavioural deficits in animal models of Amyotrophic lateral sclerosis and Alzheimer’s disease [32, 33]. We have produced the first integration-deficient lentiviral vectors capable of RNA interference (see also Peluffo et al., manuscript in preparation), and the present study aims to characterise in more detail the cellular response to such vectors.
RNA interference is a process of post-transcriptional gene regulation, which results in the knockdown of a target gene [34, 35]. DNA plasmids can be designed to express short hairpin RNAs (shRNAs), which mimic the structure of endogenous pre-miRNAs and are usually transcribed from Pol III promoters [36, 37]. Once expressed, the shRNAs are processed by the endogenous micro RNA (miRNA) biogenesis pathway resulting in the generation of functional small interfering RNA (siRNA) that can silence the targeted mRNA [38]. RNA interference has the potential to attenuate the production of proteins that are inhibitory to neurite outgrowth thereby enhancing neurite outgrowth in vitro and regeneration in vivo. Previous studies have demonstrated that siRNA-mediated knockdown of NgR1, p75NTR and RhoA can enhance the neurite outgrowth of dorsal root ganglion (DRG) neurons and retinal ganglion cells (RGCs) when cultured in the presence of myelin extract in vitro [39, 40]. Furthermore RNA silencing of the ubiquitin ligase Cdh1-APC has been shown to increase neurite outgrowth of postnatal cerebellar granule neurons (CGNs) on permissive and inhibitory myelin substrates in vitro [41–43].
Over the past decade there have seen an increasing number of studies reporting side effects of RNA interference and it has now been extensively demonstrated that shRNAs and siRNAs can induce a cellular innate immune response resulting in the production of interferons (IFNs) that can activate interferon stimulated genes (ISGs), induce a global inhibition in gene expression and result in cell death [40, 44–50]. IFNs are inducible cytokines produced and secreted by multiple cell types including neurons in response to double stranded RNA (dsRNA), a common indicator of viral infection [51–53]. Both viral and non-viral mediated RNA interference has been reported to augment the expression of three key ISGs; 2’,5’-oligoadenylate synthase 1 (OAS1) [44–47, 54], protein kinase R (PKR) [45, 49] and myxovirus resistance 1 (Mx1) [40, 49].
The aim of this study was to enhance the neurite outgrowth of postnatal CNS neurons cultured on a growth inhibitory substrate by constructing an integration-deficient lentiviral vector capable of mediating RNA interference. LINGO-1 was selected as the target for knockdown as it has been shown to be an essential component of the neuronally expressed Nogo receptor (NgR) complex [55], which restricts axon growth. Several myelin associated inhibitory proteins, including myelin associated glycoprotein (MAG), oligodendrocyte-myelin glycoprotein (OMgp) and the Nogo-66 domain of Nogo-A bind and signal through the NgR complex [13, 56–59]. Furthermore inhibition of LINGO-1 using a LINGO-1-Fc fusion protein has previously been shown to enhance neurite outgrowth in vitro and sprouting and functional recovery in vivo [55, 60]. To date, it was not known whether RNA interference can reduce levels of LINGO-1 in neurons and whether or not this promotes axon growth.
Using integration-deficient lentiviral vectors expressing shRNAs, primary postnatal neurons were efficiently transduced and LINGO-1 expression was significantly knocked down in vitro. However, an increase in cell death was observed when lentiviral vectors encoding an shRNA were applied (relative to those that did not encode shRNAs). Moreover, an increase in cell death was observed for all the lentiviral vectors (with or without shRNAs) when high viral concentrations were used. Furthermore, following transduction and LINGO-1 knockdown the neurite outgrowth of CGNs was not enhanced on an inhibitory MAG substrate. Subsequent experiments demonstrated that high concentrations of lentiviral vectors and low level shRNA expression generated an IFN response that may have been responsible for a component of the observed knockdown, cytotoxicity and failure to promote neurite outgrowth.
Materials and Methods
Cell lines
HEK 293T and HeLa cells were maintained in Dulbecco’s modified eagle’s medium (D-MEM) supplemented with 10% fetal bovine serum (FBS) and 100 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen, UK). CHO-MAG and CHO-R2 cells (kind gift from Professor Marie Filbin) and were maintained in a humidified incubator at 37°C, 5% CO2 in DMEM supplemented with 10% FBS (Invitrogen, UK), 2mM L-glutamine (Sigma, UK), 0.04 mg/ml L-Proline (Sigma, UK), 7.5 µg/ml Glycine (Sigma, UK), 0.73 µg/ml Thymidine (Sigma, UK), 100 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen, UK).
Cerebellar granule neuron (CGN) dissociation
Postnatal day 7-9 (P7-9) Lister hooded rat pups were killed by decapitation, the cerebellum was dissected and the meninges were removed in 3 ml Calcium and Magnesium free media (CMF) (sterile water supplemented with 0.4 mg/ml KCl (Sigma, UK), 0.06 mg/ml KH2PO4 (Sigma, UK), 7.65 mg/ml NaCl (Sigma, UK), 0.35 mg/ml NaHCO3 (Sigma, UK), 0.048 mg/ml Na2HPO4 (Sigma, UK), 2.38 mg/ml Hepes (Sigma, UK), pH 7.2). The dissected cerebellum was then finely diced with a razor blade before being incubated in 5 ml 0.05% trypsin EDTA (Invitrogen, UK) in CMF for 15 minutes at 37°C. The trypsin EDTA (Invitrogen, UK) was deactivated using an equal volume of 10% FBS (Invitrogen, UK) in CMF and the cell pellet was transferred to a fresh tube containing 2 ml CMF and 0.5 ml 5 mg/ml DNase I (Sigma, UK). The cell pellet was mechanically triturated 8 times using a 5 ml pipette and 4 times using a 2 ml pipette and left to settle for 5 minutes. 1.5 ml of supernatant was removed and pelleted by centrifugation at 100 x g for 5 minutes and then resuspended in 5 ml SATO media (D-MEM supplemented with 2% FBS (Invitrogen, UK), 1% N2 supplement (Invitrogen, UK) and 100 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen, UK)). Cells were counted using a haemocytometer.
Plasmid construction
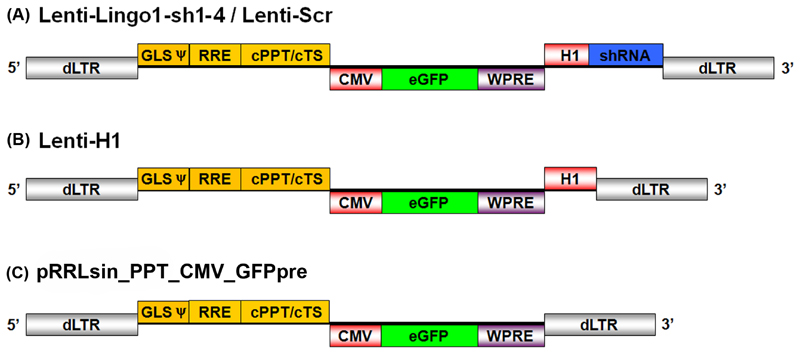
Plasmid Lenti-H1 (Figure 1B) was generated by amplifying the H1 promoter from the pSUPER_gfp_neo plasmid (kind gift from Dr. Thijn R. Brummelkamp, Dr. Rene Bernards and Dr. Reuven Agami) using PCR and cloning it into the pRRLsin_PPT_CMV_GFPpre [61] lentiviral transfer plasmid (Figure 1C). The PCR primers added a unique MCS consisting of four restriction enzyme sites (MluI, SbfI, PstI and XmaI) after the H1 promoter. PCR primer sequences were as follows: H1 cloning Forward 5’-GCG-CGC-GAA-TTC-GAA-CGC-TGA-CGT-CAT-CAA-3’ and Reverse 5’-GCG-CGC-GGT-ACC-CCC-GGG-ATA-CCT-GCA-GGA-CGC-GTG-TGG-TCT-CAT-ACA-GAA-CTT-ATA-AGA-3’. The primers were reconstituted to 100 µM using 1xTE (10 mM Tris (Sigma, UK), 1 mM EDTA (Sigma, UK), pH 8.0) and a published set of PCR conditions were used [62]. The PCR product and the pRRLsin_PPT_CMV_GFPpre plasmid were cleaved overnight at 37°C using EcoRI and Acc65I (New England BioLabs, USA). The PCR product was then ligated into the linearised pRRLsin_PPT_CMV_GFPpre plasmid downstream of an existing expression cassette encoding eGFP under the control of a CMV promoter and a woodchuck hepatitis virus post-transcriptional regulatory element (WPRE) generating Lenti-H1 plasmid (Figure 1).
Figure 1.
Schematic representation of the integration-deficient lentiviral vectors. Schematic depicts linear form of double stranded DNA after completion of reverse transcription. All vectors contain an eGFP expression cassette under the control of a CMV promoter and including a WPRE. The cassettes are flanked by the self-inactivating 5’ deleted LTR. (A) Lenti-LINGO1-sh1-4/Lenti-Scr contain an shRNA expression cassette driven by the H1 promoter placed downstream of the eGFP expression cassette. (B) Lenti-H1 contains the H1 promoter but no shRNA or Pol III termination sequence. dLTR, deleted long terminal repeat; GLS, gag leader sequence incorporating the packaging element (y); RRE, rev response element; cPPT/cTS, central polypurine tract and central termination sequence; CMV, cytomegalovirus immediate early promoter; eGFP, enhancer green fluorescent protein; H1, H1-RNA promoter; shRNA, short hairpin RNA; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element.
shRNA design
Four shRNA constructs (LINGO1-sh1-4) were designed according to the pSUPER protocol (OligoEngine, USA) (Table 1). The antisense sequences targeting LINGO-1 were designed using the Dharmacon siDESIGN® centre algorithm with the exception of LINGO1-sh4, which was based on a previously published sequence [63]. Each shRNA was 64 nucleotides in length consisting of a 19 nucleotide sense and antisense sequence separated by a 9 nucleotide non-complementary loop sequence. This was flanked by a 5’ mutated MluI restriction site and a 3’ RNA polymerase III termination signal followed by a PstI restriction site. The antisense sequences were designed to target the ORF of the rat LINGO-1 gene (GeneID: 315691) and to exclude sequences with a GC content lower than 30% and higher than 64%. Sequences with a predicted high score for successful silencing, low seed frequency, high number of mismatches to sequence records other than the target gene were preferentially selected. A scrambled non-targeting shRNA sequence (Lenti-Scr) was used as a negative control (Table 1). The shRNA sequences were cloned in to the Lenti-H1 plasmid downstream of the H1 promoter (Figure 1A).
Table 1. shRNA sequences.
| Name | Sense | Loop | Antisense |
|---|---|---|---|
| LINGO1-sh1 | CTACATGTTCCAAGACCTA | TTCAAGAGA | TAGGTCTTGGAACATGTAG |
| LINGO1-sh2 | GCTCAATGTTTCTGGCAAC | TTCAAGAGA | GTTGCCAGAAACATTGAGC |
| LINGO1-sh3 | ACAAAGCACAACATCGAAA | TTCAAGAGA | TTTCGATGTTGTGCTTTGT |
| LINGO1-sh4 | GATCGTCATCCTGCTAGAC | TTCAAGAGA | GTCTAGCAGGATGACGATC |
| Lenti-Scr | TGGTTTACATGTCGACTAA | TTCAAGAGA | TTAGTCGACATGTAAACCA |
Lentiviral production and titration
Third generation, self-inactivating, integration-deficient lentiviral vectors pseudotyped with the VSV-G envelope glycoprotein were produced as previously described [21, 64]. Briefly, lentiviral vectors were generated by transient transfection of HEK293T cells with the pMD2.VSV-G, pRSV.REV and pMDLg/pRREintD64V packaging plasmids and the relevant transfer plasmid. The harvested HEK293T cell medium was centrifuged at 690 x g for 10 minutes at room temperature and then filtered through a 0.22 µm filter (Nalgene, USA) to remove cell debris. The filtered medium was then harvested and transferred to high speed polyallomer centrifuge tubes (Beckman, USA) and centrifuged at 50 000 x g in a SW32Ti rotor (Beckman, USA) for 2 hours at 4°C. The vector was then resuspended in DMEM, centrifuged at 1400 x g for 10 minutes and incubated with 5 u/ml DNaseI (Promega, USA) and 10 mM MgCl2 (Sigma, UK) for 30 minutes. The vector was then aliquoted and stored at -80°C. The lentiviral titre was determined by serial dilution and transduction of HeLa cells followed by flow cytometry as previously described [21]. Prior to use, all the lentiviral vectors were titre matched to 1x108 transducing units/ml.
Quantitative RT-PCR analysis of mRNA
Total cellular RNA was isolated using TRIZOL reagent (Invitrogen, UK) and treated with DNase I (Qiagen, UK). For first strand cDNA synthesis, 100 units of Superscript II reverse transcriptase (Invitrogen, UK) was used with 250 ng random primers (Invitrogen, UK), 10 mM dNTP mix (Invitrogen, UK) and 500 ng total RNA. Quantitative RT-PCR (qRT-PCR) was performed using the RotorGene-3000 (Corbett Life Science, Australia). Each PCR reaction contained 20 ng of cDNA, 25 ng/μl of the relevant forward and reverse primers and 4 μl SYBR Green PCR Master Mix (Roche, USA). Forward and reverse primers for LINGO-1 were designed using Primer3, sequences were as follows: Forward 5’-TGG-ACA-TCA-GCG-AGA-ACA-AG-3’ and Reverse 5’-ATG-CAA-TCT-GAC-CTC-CAT-CC-3’. Forward and reverse primers for GAPDH, p75NTR, NgR1, OAS1, PKR and Mx1 were designed using NCBI primer-BLAST tool, sequences were as follows: GAPDH forward 5’-ATG-GGA-AGC-TGG-TCA-TCA-AC-3’ and reverse 5’-CCA-CAG-TCT-TCT-GAG-TGG-CA-3’. p75NTR forward 5’-GAG-GTG-GGC-TCG-GGA-CTC-GT-3’ and reverse 5’-CGG-GGG-CGT-AGA-CCT-TGG-GA-3’. NgR1 forward 5’-CGG-CTG-CCG-ACA-TGG-GTG-TT-3’ and reverse 5’-ATA-GGC-CAG-GCC-CCA-GCT-CC-3’. OAS1 forward 5’-ACA-GCA-ATC-CTG-ATC-CCA-AG-3’ and reverse 5’-ACC-AGT-TCC-AAG-ATT-GTC-CG-3’. PKR forward 5’-AAC-AGC-CCT-GGA-AAA-TGA-TG-3’ and reverse 5’-TTA-CGG-GTT-GTC-AAT-GCT-TTC-3’. Mx1 forward 5’-AGA-GGA-GCC-ATG-GAG-AGT-CA-3’ and reverse 5’-AAA-GCC-AGG-AGA-CAT-CCC-TT-3’. Standard curves were obtained for each of the target genes (LINGO-1, GAPDH, p75NTR, NgR1, OAS1, PKR and Mx1) using three-fold serial dilutions of E15 rat head cDNA. A single peak on the melt curve analysis confirmed the specificity of the PCR primers.
Transduction efficiency and viability assay
24-well assay plates (Nunc, UK) were coated with 0.01% poly-L-lysine (PLL) (Sigma, UK) in PBS (Invitrogen, UK) for 2 hours and washed twice with PBS (Invitrogen, UK). CGNs were dissociated from postnatal day 7-9 rat pups and 5x104 cells were plated per well and cultured at 37°C, 5% CO2 for 24 hours in 300 µl SATO media. Integration-deficient lentiviral vectors were added to the wells at a range of viral concentrations (MOI 1, 5, 10, 20 or 50). For the NVC, un-supplemented D-MEM was added to the wells instead of virus. The plates were then incubated for 72 hours at 37°C, 5% CO2
Neurite outgrowth assay
96-well assay plates (Nunc, UK) were pre-coated with 30 µl 0.01% poly-L-Lysine (Sigma, UK) in PBS (Invitrogen, UK) for 2 hours and washed with PBS (Invitrogen, UK). The 96-well plate was then coated with 30 µl 10 µg/ml fibronectin from bovine plasma (Sigma, UK) in DMEM (Invitrogen, UK) for 2 hours at 37°C and washed once with DMEM (Invitrogen, UK). The CHO-MAG and CHO-R2 cells were plated at a density of 4x104 cells per well in 100 µl CHO cell media and incubated overnight at 37°C, 5% CO2. 24 hours later P7-9 CGNs were dissociated and 1x104 plated onto the confluent monolayer of CHO-MAG or CHO-R2 cells in 100 µl SATO media. 2 hours after plating, the CGNs were transduced with the integration-deficient lentiviral vectors at an MOI 10. A NVC was included whereby un-supplemented DMEM was added instead of virus. A positive control was also included where NVC cells were given 50 µM of the synthetic ROCK inhibitor Y-27632 (Sigma, UK). After incubating for 24 hours an additional 100µl SATO media was added to the wells, which were then incubated for a further 48 hours.
Immunocytochemistry
CGN cultures were fixed using cold 4% paraformaldehyde in PBS (pH 7) for 20 minutes and then washed once with PBS (Invitrogen, UK). Neurons were stained for beta-III tubulin using a mouse monoclonal antibody (1:2000 Promega, USA) in PBS (Invitrogen, UK) containing 10% normal goat serum (NGS) (Invitrogen, UK) and 0.3% Triton X (Sigma, UK) and incubated on a shaker for 1 hour at room temperature. The cultures were then washed 3 times with PBS (Invitrogen, UK) and incubated with goat anti-mouse Alexa Fluor 546 conjugated secondary antibody (1:1000, Molecular Probes) and DAPI to visualise the nucleus in PBS (Invitrogen, UK) with 0.3% Triton X-100 (Sigma, UK) on a shaker for 45 minutes at room temperature. The cultures were then washed a further 3 times with PBS (Invitrogen, UK) and left in PBS containing 0.02% sodium azide (Sigma, UK).
Northern blot
Northern blotting was used to detect the expression of the LINGO1-sh4 shRNA and siRNA following transduction with Lenti-LINGO1-sh4 or transfection with the LINGO1-sh4 DNA plasmid. The oligonucleotide probe was designed to be complementary to the LINGO1-sh4 antisense siRNA sequence. The probes sequence was: 5’-GAT-CGT-CAT-CCT-GCT-AGA-C-3’. DNA oligonucleotide sequences that were identical to the shRNA and siRNA sequences that should be expressed by LINGO-sh4 were designed and included as positive controls. The sequence of the shRNA positive control was: 5’-GAT-CGT-CAT-CCT-GCT-AGA-CTT-CAA-GAG-AGT-CTA-GCA-GGA-TGA-CGA-TCT-T-3’ and the siRNA positive control was: 5’-GTC-TAG-CAG-GAT-GAC-GAT-C-3’. HEK 293T cells were either transduced with Lenti-LINGO1-sh4 at an MOI 10 or transfected with 3 µg of the LINGO1-sh4 plasmid using PEI. 72 hours after transduction/transfection, the total cellular RNA was isolated using TRIZOL reagent (Invitrogen, UK). End labelling of the oligonucleotide probe and ladder was performed using the Decade labelling kit (Ambion, USA). Briefly, 1 µl of 100 pmol oligonucleotide probe or 100 ng Decade ladder, 1 µl 10x forward labelling buffer, 1 µl Diethylpyrocarbonate (DEPC)-pretreated water, 1 µl γ-32P-ATP 250 µCi (Perkin Elmer, UK) and 1 µl T4 polynucleotide kinase were combined and incubated at 37°C for 60 minutes. The probe and ladder were then purified using G25 Illustra Micropin purification cartridges (GE Healthcare Life Sciences, UK). 20 μg of cellular RNA, 0.5 µl of γ-32P-labelled ladder and 100 ng of the positive control oligonucleotides in DEPC water and 2x Novex loading buffer (Invitrogen, UK) were separated on a 15% acrylamide gel. The RNA was then transferred to a nitrocellulose membrane (Invitrogen, UK) and UV-crosslinked. The membrane was pre-hybridized with UltraHyb-Oligo (Ambion USA) and then probed with the γ-32P-labelled probe overnight at 35°C. The membrane was then washed in 2x standard sodium citrate (SSC) and 0.1% sodium dodecyl sulfate (SDS) for 5 minutes at 35°C and exposed to Biomax maximum resolution autoradiography film (Sigma, UK) for 21 days. Transcript sizes were determined using the Decade ladder (Ambion, USA).
Imaging and analysis
8 fields of view were taken per well using the IN Cell Analyzer 1000 semi-automated cell imager with a 10x Nikon ApoPlan objective (GE Healthcare Life Sciences, UK). Cell counts were obtained using the semi-automated IN Cell Developer Toolbox software (GE Healthcare Life Sciences, UK) and custom-written algorithms. Transduction efficiency was determined by counting the number of eGFP and beta-III tubulin positive cells. Neuronal viability was assessed by counting the total number of beta-III tubulin immunoreactive cells. Percentage transduction was then calculated by dividing the number of transduced neurons by the total number of neurons multiplied by one hundred. The data represents 3 independent experiments using 1 well per condition, (n = 3/group). The relative levels of LINGO-1 mRNA were determined by cross-referencing with the standard curve and the fold change was determined by normalising to GAPDH: the knockdown experiment using an MOI of 10 was carried out once using three wells per condition (n = 3/group), while the knockdown experiment using an MOI of 50 was carried out once using four wells per condition (n = 4/group). The relative levels of p75NTR and NgR1 were determined by cross-referencing with a standard curve and the fold change by normalising to GAPDH. Both experiments (MOI 10 and 50) were carried out once using four wells per condition (n = 4/group). The mean neurite length per neuron was determined by measuring the neurite length of beta-III tubulin positive cells divided by the total number of neurons: this experiment was carried out once using 8 wells per condition, (n = 8/group). The relative levels of OAS1, PKR and Mx1 mRNA were determined by cross-referencing with a standard curve and the fold change was determined by normalising to GAPDH. The ISG qRT-PCR experiments at an MOI 10 was carried out once using three wells per condition (n = 3/group) and the ISG qRT-PCR experiments using an MOI 50 were carried out once using four wells per condition (n = 4/group).
Statistical analysis
Statistical analysis was carried out using SPSS 17.0 (SPSS Inc, USA). A threshold level of significance (α) of 0.05 was selected. Graphs show means ± standard errors of the mean. The Kolmogorov-Smirnov and Levene’s tests were used to test for normality and the equality of variances. To statistically analyse the transduction efficiency and viability a one-way ANOVA was used to test for differences among the groups followed by Tukey post-hoc testing to analyse the effects between groups. LINGO-1 knockdown was analysed using a one-way ANOVA to test for differences among the groups followed by Tukey post-hoc testing to analyse the effects between groups. To analyse the expression level of p75NTR and NgR1 a one-way ANOVA was used to test for differences among the groups followed by Dunnett’s post hoc tests to compare the groups to the no virus control. To analyse the expression of ISGs at an MOI of 10 a one-way ANOVA was used to test for differences among the groups followed by Tukey post-hoc tests to analyse the effects between groups. The data for an MOI of 50 was not normally distributed so a Kruskal-Wallis was performed to test for differences among the groups followed by Mann-Whitney post-hoc tests to analyse the effects between groups. We report exact P values for the Mann-Whitney comparisons: these were not adjusted for multiple comparisons. To analyse the neurite outgrowth a one-way ANOVA was used to test for differences among the groups followed by Dunnett’s post hoc tests to compare groups to the negative control.
Results
The generation of shRNA encoding integration-deficient lentiviral vectors
The H1-RNA promoter (H1 promoter) was cloned into a lentiviral transfer plasmid downstream of an eGFP expression cassette as described in the methods, generating Lenti-H1, which also contains cloning sites for shRNAs downstream of the H1 promoter. The H1 promoter has previously been demonstrated to efficiently drive the expression of the shRNAs [36]. We then generated integration-deficient lentiviral vectors expressing shRNAs targeting LINGO-1 (Lenti-LINGO1-sh1, Lenti-LINGO1-sh2, Lenti-LINGO1-sh3, Lenti-LINGO1-sh4), a vector expressing a scrambled non-targeting shRNA that had been microarray-tested by Dharmacon to have minimal targeting of known genes in the rat genome (Lenti-Scr) and a vector containing the H1 promoter and multiple cloning site (MCS) but not expressing an shRNA (Lenti-H1). A schematic of the different lentiviral vectors can be seen in Figure 1.
Integration-deficient lentiviral vectors efficiently transduce postnatal CGNs in vitro
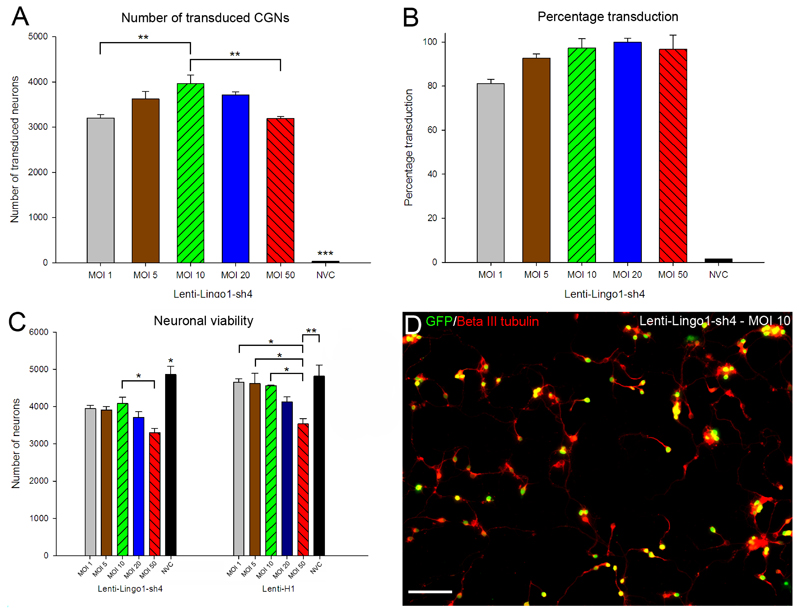
P7-9 rat CGNs were transduced in 96 well plates using a range of viral doses (multiplicity of infection (MOI) 1, 5, 10, 20 or 50). Importantly, a no virus control (NVC) was also included where the cells were left untreated. Cultures were fixed after seventy two hours and immunolabelled for beta-III tubulin. An automated image capture and analysis system was used for unbiased measurement of transduction efficiency and cell survival. Lentiviral vectors transduced neurons with high efficiency at all MOIs (Figure 2). Transduction efficiency was similar for all the lentiviral vectors tested. Using Lenti-LINGO1-sh4 as an example, the number of transduced neurons was affected by MOI (df = 5(12), F = 167, P < 0.001, one-way ANOVA, n = 3/group) (Figure 2A). The number of transduced neurons increased between an MOI 1 and 10, before decreasing at higher viral concentrations. Tukey post-hoc tests indicated a difference in the number of neurons transduced between all the concentrations used and the NVC (P values < 0.001). Significantly more neurons were transduced when using an MOI 10 compared to using an MOI 1 (P < 0.01) and MOI 50 (P < 0.01) but not an MOI 5 and 20 (P > 0.05). The percentage of neurons that were transduced, however, generally did not change with viral concentrations higher than MOI 10 where the transduction efficiency was already above 90 % (Figure 2B, D). MOI 10 was therefore optimum for CGN transduction.
Figure 2.
CGNs are efficiently transduced in vitro, although viability was affected by shRNAs and viral concentration. (A) Quantification of the mean number of transduced CGNs using Lenti-LINGO1-sh4. The number of eGFP positive CGNs was counted and the mean number of transduced CGNs plotted for each MOI. There were significantly more CGNs transduced using an MOI 10 compared to MOI 1 or 50 (** P < 0.01). Values represent mean and SEM; analysis was performed using one way ANOVA with Tukey post-hoc tests, n = 3. (B) Quantification of the percentage transduction efficiency, which was calculated by dividing the number of transduced CGNs by the total number CGNs multiplied by one hundred. Percentage transduction did not change with viral concentrations higher than MOI 10 where the transduction efficiency was already above 90%. Values represent mean and SEM, n = 3. (C) Quantification of the mean number of CGNs after transduction with Lenti-LINGO1-sh4 or Lenti-H1. The number of CGNs was counted and the mean number plotted for each MOI. Neuronal viability was affected by MOI and there was a decrease in neuronal viability at higher viral concentrations. Neuronal viability was also differentially affected by the viral vectors. There were different patterns of neuronal viability between the lentiviral vectors that encoded shRNAs and the viral vectors that did not encode an shRNA. Values represent mean and SEM, analysis was performed using one way ANOVA with Tukey post-hoc tests, n = 3/group. (D) CGNs are efficiently transduced by Lenti LINGO1-sh4 at an MOI of 10. Transduced CGNs expressing eGFP (green) and stained for beta-III tubulin (red) appear yellow. Scale bar: 100 μm.
CGN viability is reduced by both lentiviral concentration and shRNA expression in vitro
We investigated whether lentiviral vector concentration affected neuronal viability. The pattern of neuronal viability was similar for all the shRNA expressing lentiviral vectors. For Lenti-LINGO1-sh4 a difference in neuronal viability was detected between the different viral concentrations, indicating that MOI contributes to cell death (df = 5(12), F = 12.40, P < 0.001, one-way ANOVA, n = 3/group) (Figure 2C). Tukey post-hoc tests indicated that there was a difference in neuronal viability between all the MOI’s used and the NVC (P values < 0.05). There was also a significant reduction in neuronal viability between MOI 10 and MOI 50 (P < 0.05). A different pattern of viability was observed when the lentiviral vectors did not express an shRNA. Using Lenti-H1, neuronal viability was affected by viral concentrations (df = 5(12), F = 6.41, P < 0.01, one-way ANOVA, n = 3/group) (Figure 2C). However, unlike lentiviral vectors that encode an shRNA, Tukey post-hoc tests indicated that there was no difference in neuronal viability between MOI 1-20 and the NVC (P values > 0.05). Only at high viral concentrations was a statistically significantly difference in viability detected: Using MOI 50 significantly reduced viability compared to MOI 1-10 (P < 0.05) or the NVC (P < 0.01). Thus neuronal viability appears to be affected by both viral concentration and the expression of an shRNA. As there was a significant reduction in viability between MOI 10 and 50 using Lenti-LINGO1-sh4 and MOI 10 achieved almost complete transduction, we conclude that MOI 10 is the optimal concentration for CGN transduction.
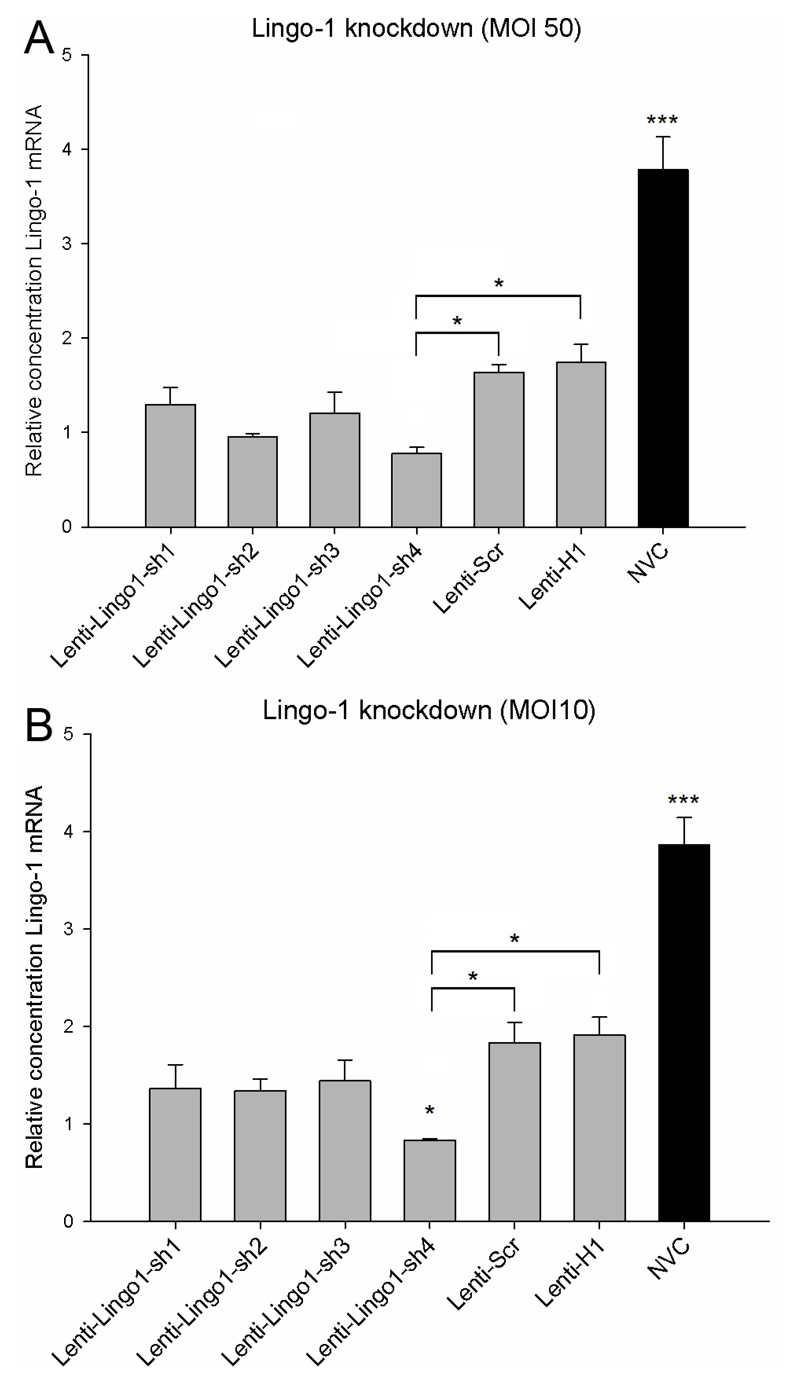
Lenti-LINGO1-sh4 significantly knocks down LINGO-1 mRNA in vitro at an MOI 10 and 50, although knockdown is also observed with the control vectors
Using an MOI of 10 or 50, P7-9 CGNs were transduced with either Lenti-LINGO1-sh1, Lenti-LINGO1-sh2, Lenti-LINGO1-sh3, Lenti-LINGO1-sh4, Lenti-Scr or Lenti-H1. A NVC was also included. Seventy two hours after transduction, knockdown of LINGO-1 mRNA was assessed using qRT-PCR. LINGO-1 mRNA levels were significantly affected by the different lentiviral vectors at an MOI of 50 (df = 6(21), F = 29.0, P < 0.001, one-way ANOVA, n = 4/group) (Figure 3A). Tukey post-hoc tests revealed that Lenti-LINGO1-sh4 significantly knocked down LINGO-1 expression 2.1 fold, 2.2 fold and 4.8 fold respectively compared to Lenti-Scr (P < 0.05), Lenti-H1 (P < 0.05) and NVC (P < 0.001). Lenti-LINGO1-sh1, Lenti-LINGO1-sh2 and Lenti-LINGO1-sh3 did not significantly knockdown the level of LINGO-1 mRNA compared to the Lenti-Scr and Lenti-H1 controls (P values > 0.05). However, unexpectedly all the lentiviral vectors including the Lenti-Scr and Lenti-H1 controls significantly knocked down the level of LINGO-1 mRNA compared to the NVC (P < 0.001). Due to concerns with cytotoxicity at high viral concentrations we investigated whether using an MOI 10 would also significantly knockdown LINGO-1. A statistically significant difference in LINGO-1 mRNA levels was detected between the different lentiviral vectors (df = 6(14), F = 24.1, P < 0.001, one-way ANOVA, n = 3/group) (Figure 3B). Tukey post-hoc tests revealed that Lenti-LINGO1-sh4 significantly knocked down LINGO-1 expression 2.2 fold, 2.3 fold and 4.6 fold respectively compared to Lenti-Scr (P < 0.05), Lenti-H1 (P < 0.05) and NVC (P < 0.001). Lenti-LINGO1-sh1, Lenti-LINGO1-sh2 and Lenti-LINGO1-sh3 did not significantly knockdown the level of LINGO-1 mRNA compared to the Lenti-Scr and Lenti-H1 controls (P values > 0.05). Again surprisingly, all the lentiviral vectors including the Lenti-Scr and Lenti-H1 controls significantly knocked down the level of LINGO-1 mRNA compared to the NVC (P < 0.001).
Figure 3.
Lenti-LINGO1-sh4 knocks down LINGO-1 mRNA, although non-specific silencing was also observed with the control vectors. (A) Quantification of the relative level of LINGO-1 mRNA in CGNs transduced with lentiviral vectors at an MOI 50. Lenti-LINGO1-sh4 significantly knocked down LINGO-1 expression 2.1 fold and 2.2 fold respectively compared to Lenti-Scr and Lenti-H1 (* P < 0.05). In addition, significantly lower levels of LINGO-1 were detected in CGNs transduced with any of the lentiviral vectors compared to the NVC (*** P < 0.001). Values represent mean and SEM, analysis was performed using one way ANOVA with Tukey post-hoc tests, n = 4/group. (B) Quantification of the relative level of LINGO-1 mRNA in CGNs after transduction at an MOI 10. Lenti-LINGO1-sh4 significantly knocked down LINGO-1 expression 2.2 fold and 2.3 fold respectively compared to Lenti-Scr, Lenti-H1 (* P < 0.05). However, significantly lower levels of LINGO-1 were detected in CGNs transduced with any of the lentiviral vectors compared to the NVC (*** P < 0.001). Values represent mean and SEM, analysis was performed using one way ANOVA with Tukey post-hoc tests, n = 3/group.
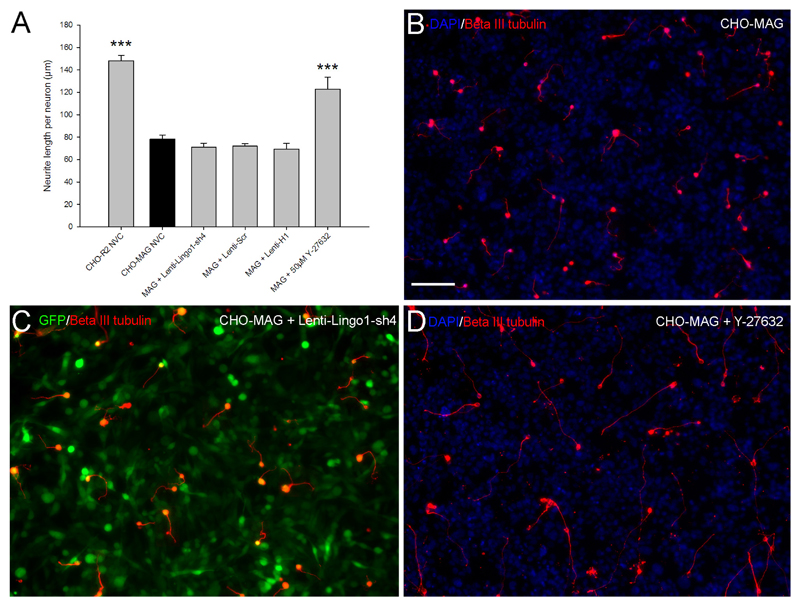
Transduction of CGNs with Lenti-LINGO1-sh4 at an MOI of 10 does not enhance neurite outgrowth on an inhibitory MAG substrate
CGNs were plated onto either a confluent layer of MAG expressing CHO cells (CHO-MAG) or CHO cells that express the reverse peptide sequence of MAG (CHO-R2) as a control. The cells were subsequently transduced with integration-deficient lentiviral vectors expressing either Lenti-LINGO1-sh4, Lenti-Scr or Lenti-H1 at an MOI 10. A NVC was included as a negative control and 50 µM Y-27632, a synthetic Rho-kinase inhibitor was added to other NVCs as a positive control. Seventy two hours after transduction the CGNs were fixed, stained and the mean neurite length per transduced neuron was measured. Statistical analysis revealed a significant difference between the groups (df =5(18), F = 33.61, P < 0.001, one-way ANOVA, n = 4/group) (Figure 4A). Dunnett’s post-hoc test indicated that CGNs plated on the CHO-R2 cells had significantly longer neurites compared to the CGNs plated on the CHO-MAG cells (CHO-R2, 148±5 µm; CHO-MAG, 78.3±3.6 µm; values represent mean ± SEM, n = 4/group, *** P < 0.001) (Figure 4A and B). Addition of 50 µM Y-27632 to NVC CGNs cultured on the CHO-MAG cells resulted in a significant increase in the neurite outgrowth (Y-27632, 122.7±10.9 µm; CHO-MAG, 78.3±3.6 µm; values represent mean ± SEM, n = 4/group, *** P < 0.001) (Figure 4A, D). In contrast, there was no significant difference in the neurite outgrowth of CGNs transduced with lentiviral vectors expressing an shRNA targeting LINGO-1 compared to CGNs transduced with any of the control lentiviral vectors or the NVC (Lenti-LINGO1-sh4, 71±3.6 µm; Lenti-Scr, 71.9±2.4 µm; Lenti-H1, 69.5±4.9 µm; NVC, 78.3±3.6 µm; values represent mean ± SEM, n = 4/group, P > 0.05) (Figure 4A, C). Thus despite substantial knockdown of LINGO-1 mRNA by lentiviral vectors in general, and by Lenti-LINGO1-sh4 in particular (Figure 3), this did not promote neurite growth in vitro.
Figure 4.
Integration-deficient lentiviral vectors encoding an shRNA targeting LINGO-1 do not enhance neurite outgrowth. (A) The neurite length of beta-III tubulin positive CGNs was measured, divided by the total number of neurons and the mean neurite length per neuron plotted. CGNs plated on the CHO-R2 cells had significantly longer neurites compared to CGNs plated on the inhibitory CHO-MAG cells (*** P < 0.001). CHO-MAG inhibition could be partially reversed with using 50 μM Y 27632 (*** P < 0.001). Lenti-LINGO1-sh4 did not significantly increase neurite outgrowth of CGNs plated on an inhibitory MAG substrate compared to the control lentiviral vectors or the NVC (P > 0.05). Values represent mean and SEM, analysis was performed using one way ANOVA, with Dunnett’s post-hoc tests comparing to CHO-MAG cells, P < 0.001, n = 8/group. (B) CGN neurite outgrowth is inhibited by CHO-MAG cells. (C) Transduction with Lenti-LINGO1-sh4 does not alleviate the MAG inhibition and enhance CGN neurite outgrowth. (D) The ROCK inhibitor Y-27632 partially reverses the MAG inhibition and promotes neurite outgrowth. Scale bar: 100 μm.
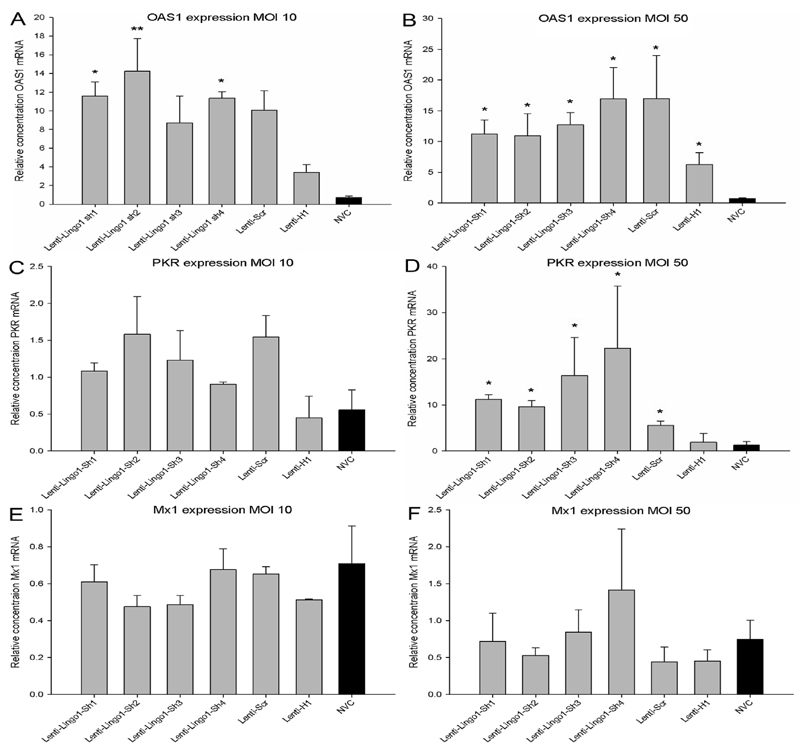
Interferon stimulated genes OAS1 and PKR but not Mx1 are up-regulated following transduction with lentiviral vectors encoding shRNAs
Due to the non-specific knockdown of LINGO-1 and the cytotoxicity observed after transduction with high concentrations of lentiviral vector, the expression of three typical ISGs (OAS1, PKR and Mx1) was examined seventy two hours after transduction using qRT-PCR. OAS1 expression was significantly affected after transduction with lentiviral vectors at an MOI of 10 (df = 6(14), F = 5.83, P < 0.01, one-way ANOVA, n = 3/group) (Figure 5A). Tukey post-hoc tests indicated that there was a significant increase in OAS1 expression between Lenti-LINGO1-sh1 (P < 0.05), Lenti-LINGO1-sh2 (P < 0.01), Lenti-LINGO1-sh4 (P < 0.05) and the NVC. There was a trend for increased OAS1 expression between Lenti-Scr and NVC (P = 0.06). However, there was no significant difference in OAS1 expression between either Lenti-LINGO1-sh3 or Lenti-H1 and the NVC (P values > 0.05). CGNs were also transduced with lentiviral vectors at an MOI 50, which also led to a significant difference in OAS1 expression between the vectors (df = 6, Chi-square = 13.31, P < 0.05, Kruskal-Wallis, n = 4/group) (Figure 5B). Mann-Whitney post-hoc tests revealed that all lentiviral vectors significantly increased OAS1 expression compared to the NVC (P values = 0.03). There was no significant difference in OAS1 expression between the shRNA expressing viruses and the non-shRNA expressing Lenti-H1 (P values > 0.05). There was also no significant difference in OAS1 expression between the two different MOI used (df = 1(35), F = 1.04, P > 0.05, two-way ANOVA, n = 3/group).
Figure 5.
OAS1 and PKR induction following transduction with integration-deficient lentiviral vectors encoding shRNAs. (A) Quantification of the relative level of OAS1 mRNA in CGNs transduced using an MOI 10. There was a significant increase in OAS1 expression with Lenti-LINGO1-sh1, Lenti-LINGO1-sh2 and Lenti-LINGO1-sh4 compared to the NVC. Values represent mean and SEM, analysis was performed using one way ANOVA with Tukey post-hoc tests, * P < 0.05, ** P < 0.01, n = 3/group. (B) Quantification of the relative level of OAS1 mRNA in CGNs transduced using an MOI 50. All the lentiviral vectors significantly increased OAS1 expression compared to the NVC. Values represent mean and SEM, analysis was performed using Kruskal-Wallis with Mann-Whitney post-hoc tests, * P < 0.05, n = 4/group. (C) Quantification of the relative level of PKR mRNA in CGNs transduced using an MOI 10. PKR expression was not significantly affected compared to the NVC. Values represent mean and SEM, analysis was performed using one way ANOVA, P > 0.05, n = 3/group. (D) Quantification of the relative level of PKR mRNA in CGNs transduced using an MOI 50. PKR expression was significantly increased in CGNs transduced with any of the shRNA expressing lentiviral vectors compared to the NVC. Values represent mean and SEM, analysis was performed using Kruskal-Wallis with Mann Whitney post-hoc tests, * P < 0.05, n = 4/group. (E) Quantification of the relative level of Mx1 mRNA in CGNs transduced at an MOI 10. No significant difference in Mx1 expression was observed compared to the NVC. Values represent mean and SEM, analysis was performed using one way ANOVA, P > 0.05, n = 3/group. (F) Quantification of the relative level of Mx1 mRNA in CGNs transduced using an MOI 50. Mx1 expression was not significantly affected compared to the NVC. Values represent mean and SEM, analysis was performed using Kruskal-Wallis, P > 0.05, n = 4/group.
Any apparent differences in PKR expression did not reach statistical significance following transduction of CGNs with lentiviral vectors at an MOI 10 (df = 6(14), F = 1.934, P > 0.05, one-way ANOVA) (Figure 5C). In contrast, at an MOI of 50 a statistically significant difference in PKR expression was observed between the groups (df = 6, Chi-square = 16.94, P < 0.05, Kruskal-Wallis, n = 4/group) (Figure 5D). Mann-Whitney post-hoc tests revealed that PKR expression was significantly increased in CGNs transduced with any of the shRNA expressing lentiviral vectors compared to the NVC (P = 0.03) but no significant difference in PKR expression was detected between the non-shRNA expressing Lenti-H1 and the NVC (P = 0.49). There was also a significant increase in PKR expression between CGNs transduced with Lenti-LINGO1-sh1 (P values = 0.03) and Lenti-H1 and a trend for Lenti-LINGO1-sh2 and Lenti-LINGO1-sh3 (P = 0.06). However, there was no significant difference in PKR expression between Lenti-LINGO1-sh4 and Lenti-H1 (P = 0.11) or Lenti-Scr and Lenti-H1 (P = 0.34). There was also a significant increase in PKR expression between CGNs transduced with lentiviral vectors at an MOI 10 and 50 (df = 1(35), F = 7.47, P < 0.01, two-way ANOVA, n = 3/group).
Mx1 expression was not significantly affected following transduction by any of the viral vectors at an MOI of 10 (df = 6(14), F = 0.936, P > 0.05, one-way ANOVA) (Figure 5E) or at an MOI of 50 (df = 6, Chi-square = 2.68, P > 0.05, Kruskal-Wallis) (Figure 5F). There was also no significant difference in Mx1 expression between the two different viral concentrations used (df = 1(35), F = 0.48, P > 0.05, two-way ANOVA, n = 3/group).
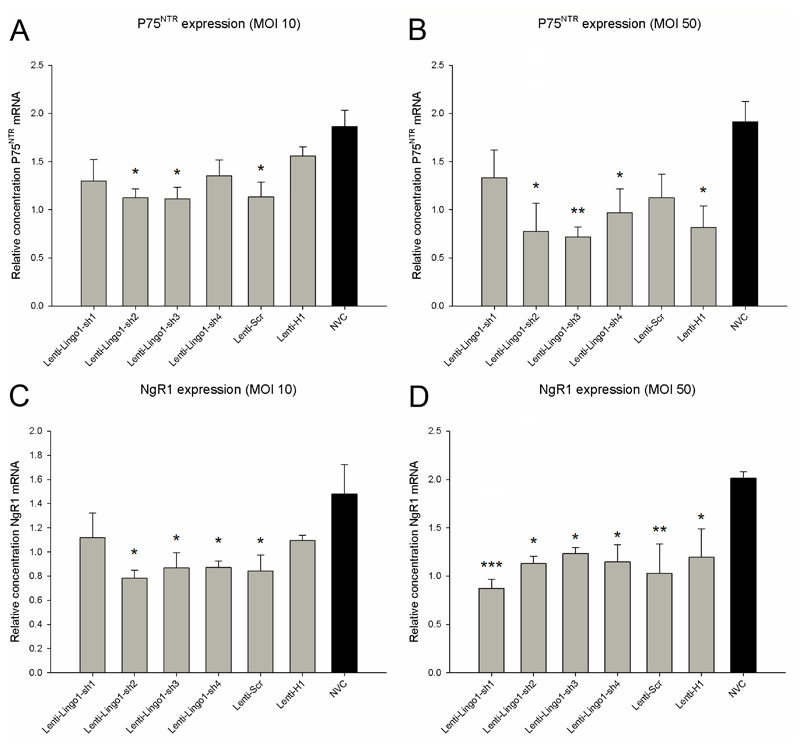
The expression of off-target genes p75NTR and NgR1 are reduced following lentiviral vector transduction
To determine whether lentiviral vector transduction reduces the expression of off-target genes, we compared the expression of two other genes (p75NTR and NgR1) in transduced and untransduced CGNs using qRT-PCR. p75NTR expression was significantly affected after transduction with lentiviral vectors at an MOI 10 (df = 6(21), F = 3.33, P < 0.05, one-way ANOVA, n = 4/group) (Figure 6A). Dunnett’s post-hoc tests demonstrated that p75NTR expression was significantly attenuated in CGNs transduced with Lenti-LINGO1-sh2 (P < 0.05), Lenti-LINGO1-sh3 (P < 0.05) or Lenti-Scr (P < 0.05) compared to the NVC, but no differences in p75NTR expression were detected between the NVC and Lenti-LINGO1-sh1, Lenti-LINGO1-sh4 or Lenti-H1 (P values > 0.05). Transduction using an MOI 50 also resulted in significant differences in p75NTR expression between the groups (df = 6(21), F = 3.13, P < 0.05, one-way ANOVA, n = 4/group) (Figure 6B). Dunnett’s post-hoc tests indicated that compared to the NVC p75NTR expression was significantly reduced in CGNs transduced with Lenti-LINGO1-sh2 (P < 0.05), Lenti-LINGO1-sh3 (P < 0.01), Lenti-LINGO1-sh4 (P < 0.05) and Lenti-H1 (P < 0.05). Lenti-LINGO1-sh1 and Lenti-Scr did not affect p75NTR expression compared to the NVC (P values > 0.05).
Figure 6.
Off-target genes p75NTR and NgR1 are reduced following lentiviral vector transduction. (A) Quantification of the relative level of p75NTR mRNA in CGNs transduced using an MOI 10. There was a significant decrease in p75NTR expression with Lenti-LINGO1-sh2, Lenti-LINGO1-sh3 and Lenti-Scr compared to the NVC. Values represent mean and SEM, analysis was performed using one way ANOVA with Dunnett’s post-hoc tests comparing to the NVC, * P < 0.05, n = 4/group. (B) Quantification of the relative level of p75NTR mRNA in CGNs transduced using an MOI 50. p75NTR expression was significantly reduced in CGNs transduced with Lenti-LINGO1-sh2, Lenti-LINGO1-sh3, Lenti-LINGO1-sh4 and Lenti-H1 compared to the NVC. Values represent mean and SEM, analysis was performed using one way ANOVA with Dunnett’s post-hoc tests comparing to the NVC, * P < 0.05, ** P < 0.01 n = 4/group. (C) Quantification of the relative level of NgR1 mRNA in CGNs transduced using an MOI 10. NgR1 expression was significantly decreased in CGNs transduced with Lenti-LINGO1-sh2, Lenti-LINGO1-sh3, Lenti-LINGO1-sh4 and Lenti-Scr compared to the NVC. Values represent mean and SEM, analysis was performed using one way ANOVA with Dunnett’s post-hoc tests comparing to the NVC, * P < 0.05, n = 4/group (D) Quantification of the relative level of NgR1 mRNA in CGNs transduced using an MOI 50. There was a significant decrease in NgR1 expression with any of the lentiviral vectors compared to the NVC. Values represent mean and SEM, analysis was performed using one way ANOVA with Dunnett’s post-hoc tests comparing to the NVC, * P < 0.05, ** P < 0.01, *** P < 0.001, n = 4/group.
NgR1 expression was also assessed. NgR1 mRNA was significantly affected when CGNs were transduced using an MOI of 10 (df = 6(21), F = 2.91, P < 0.05, one-way ANOVA, n = 4/group) (Figure 6C). Dunnett’s post-hoc tests revealed that NgR1 expression was significantly decreased in CGNs transduced with Lenti-LINGO1-sh2 (P < 0.05), Lenti-LINGO1-sh3 (P < 0.05), Lenti-LINGO1-sh4 (P < 0.05) or Lenti-Scr (P < 0.05) compared to the NVC. However no difference in NgR1 expression was detected between Lenti-LINGO1-sh1 or Lenti-H1 and the NVC (P values > 0.05). Transduction of CGNs using an MOI of 50 resulted in significant differences between the groups (df = 6(21), F = 4.04, P < 0.01, one-way ANOVA, n = 4/group) (Figure 6D). Dunnett’s post-hoc tests demonstrated that all the lentiviral vectors significantly attenuated NgR1 expression compared to the NVC (P values < 0.05).
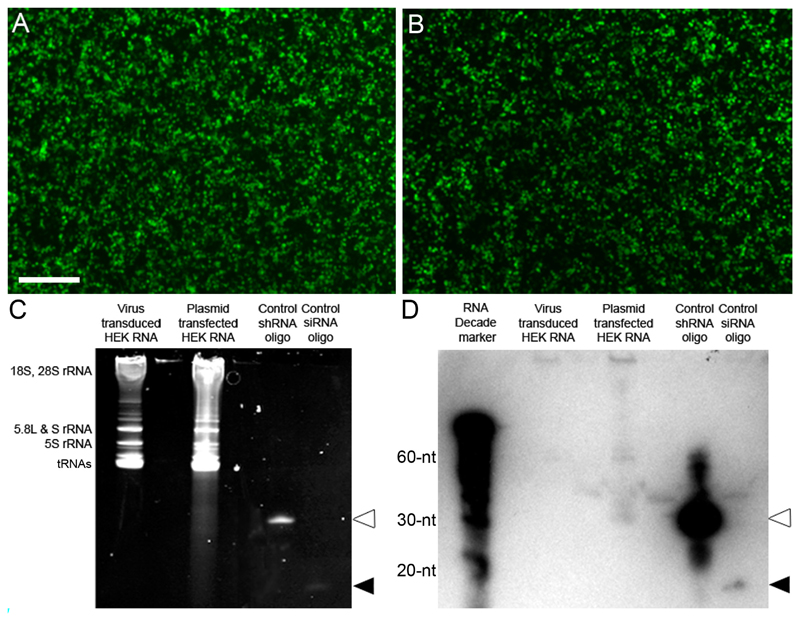
Northern blots detect low level expression of shRNA after plasmid transfection but do not confirm the expression of the shRNA or siRNA following lentiviral transduction
Cellular RNA was extracted from HEK cells that had either been transfected with the DNA plasmid expressing LINGO1-sh4 using polyethylenimine (PEI) or transduced with integration-deficient lentiviral vectors expressing Lenti-LINGO1-sh4 at an MOI 10. Both viral and non-viral methods of transfection were effective, giving high transfection and transduction efficiencies that were visually assessed using eGFP expression (Figure 7A and B). RNA samples were loaded onto a polyacrylamide gel, separated using electrophoresis and stained using ethidium bromide (thereby confirming good quality RNA; Figure 7C) prior to transferring onto a membrane for hybridisation. Expression of the shRNA and siRNA from LINGO1-sh4 was examined using a radioactively labelled DNA probe complementary to the siRNA antisense sequence. In addition, two DNA oligonucleotides were designed; one identical to the LINGO-1-sh4 shRNA and the other identical to the LINGO-1-sh4 siRNA. These were used as positive controls to confirm the northern blot had worked. These probes were loaded into gel lanes in parallel with the samples of RNA. The northern blot did not display bands for the LINGO1-sh4 shRNA or the siRNA in RNA extracted from the viral transduced HEK cells (Figure 7D). In the lane containing RNA from the DNA transfected HEK cells, a faint band representing the shRNA was detected. However, no band was present for the siRNA (Figure 7D). Bands representing the positive control shRNA and siRNA were present, confirming that the probe was correctly designed and labelled and that the northern blot had been carried out correctly (Figure 7D, white arrowhead = shRNA, black arrowhead = siRNA).
Figure 7.
Northern blots do not detect a high level of shRNA expression. (A) Image of eGFP positive HEK cells that were efficiently transduced using Lenti-LINGO1-sh4 at an MOI 10. Scale bar: 500 μm. (B) Image of eGFP positive HEK cells that were efficiently transfected with the LINGO1-sh4 plasmid using PEI. (C) Image of the resolved RNA gel, the bands correspond to the different ribosomal RNA (rRNA) species or transfer RNAs (tRNAs) as labelled on the left side. The RNA resolved from the plasmid transfected HEK cells shows some smearing. The shRNA (white arrowhead) and siRNA (black arrowhead) positive controls can be seen, confirming the design and sensitivity of the probe (D) Image of the small transcript northern blot. Neither the shRNA nor siRNA was detected from the virus transduced HEK cell RNA. A faint band representing the shRNA was detected from the plasmid transfected HEK cell RNA but a band for the siRNA was not detected. Bands representing the shRNA (white arrowhead) and siRNA (black arrowhead) positive controls were detected.
Discussion
The aim of this study was to enhance the neurite outgrowth of postnatal CNS neurons by attenuating their response to myelin associated proteins. To achieve this, an integration-deficient lentiviral vector capable of mediating RNA interference were designed and generated to knockdown the expression of LINGO-1, an essential component of the NgR that has previously been shown to be involved in the inhibition of neurite outgrowth [55]. However, while conducting these experiments we unexpectedly detected several negative side effects of transduction with integration-deficient lentiviral vectors encoding shRNAs.
LINGO-1 mRNA was significantly knocked down in CGNs by a lentiviral vector expressing LINGO1-sh4 compared to the control lentiviral vectors. This is consistent with a previous study that demonstrated knockdown of the LINGO-1 mRNA level in oligodendrocytes following transduction with lentiviral vectors expressing an identical siRNA sequence [63]. The three other shRNA sequences targeting LINGO-1 did not mediate significant mRNA knockdown compared to the control lentiviral vectors, demonstrating the requirement to test multiple shRNA sequences to determine the optimal shRNA. Unexpectedly, LINGO-1 mRNA was significantly reduced following transduction with any of the lentiviral vectors when compared to the NVC. This included the control lentiviral vectors, one of which encoded the H1 promoter without an shRNA and the other a scrambled non-targeting shRNA. It is possible that the lentiviral vector encoding just the H1 promoter did lead to expression of some downstream RNA sequence because this vector lacked an RNA polymerase III termination sequence immediately downstream of the H1 promoter. This data suggests that transduction with lentiviral vectors that do not encode an shRNA targeting LINGO-1 can knockdown the LINGO-1 transcript level via a non-specific mechanism. This may be due to the induction of an IFN response, leading to the production of IFNs that activate ISGs and result in a non-specific global inhibition in gene expression and a subsequent reduction in LINGO-1. Since this effect was observed using an MOI 10 it raises concerns that even relatively standard concentrations of lentiviral vector can generate non-specific knockdown. The time scale and cellular system under study may also be of relevance. We have also shown reduced protein levels of the channel transient receptor potential cation channel subfamily V member 1 (TRPV1) in cultured DRG neurons, using an integrating and non-integrating lentiviral vector of similar architecture to the one in the current study (Peluffo et al., manuscript in preparation). In this study the control H1 lentiviral vector lacking an shRNA did not reduce TRPV1 protein levels after five days compared to the NVC, even though the study was carried out at MOI 50 (Peluffo et al., manuscript in preparation). However, in a separate study investigating knockdown of the voltage gated calcium channel subunit α2δ1 in DRGs, two western blots demonstrated a reduction in α2δ1 protein level using the control H1 lentiviral vector compared to the NVC after 8 days in culture (Supplementary figure 1). Taken together these studies highlight the importance of using appropriate controls (e.g., no virus and scrambled controls) and monitoring off-target effects, including the interferon response, in studies involving lentiviral vectors and RNAi.
Transduction with Lenti-LINGO1-sh4 did not significantly enhance the neurite outgrowth of CGNs on a growth inhibitory MAG substrate. This was inconsistent with previous studies that have demonstrated that inhibiting the function of LINGO-1 using DN-LINGO-1 or LINGO-1-Fc in CGNs attenuated RhoA activation and the inhibitory effects of myelin inhibitors resulting in enhanced neurite outgrowth in vitro [55] and functional recovery in vivo [60]. There are several possible explanations for this disparity. Firstly, the neurite outgrowth assay used in this experiment may not be sensitive enough to detect the enhanced neurite outgrowth, although this seems unlikely as enhanced CGN neurite outgrowth was detected with Y-27632 (a synthetic ROCK inhibitor), which was used as a positive control. Secondly, the residual expression of LINGO-1 left after knockdown may be enough to maintain the neuron's responsiveness to myelin associated inhibitors, although the majority of LINGO-1 mRNA was reduced by this vector (relative to the NVC). Thirdly, de novo gene expression has been shown to be necessary for some phases of axon growth [65] and the induction of ISGs and an IFN response, which was shown to accompany transduction with these lentiviral vectors, may initiate a cellular shift away from a potential neurite growth state via the widespread down-regulation of gene expression, thereby masking any growth promoting effects of LINGO-1 knockdown. Fourthly, the lentiviral vectors may not be expressing the shRNA at sufficiently high levels, which would mean the previously detected LINGO-1 knockdown was non-specific and solely the result of an IFN response.
To address the latter possibility, northern blots were used to evaluate expression of the shRNA and siRNA from the lentiviral vectors and the DNA transfer plasmid. To our surprise, the northern blots did not detect expression of the shRNA or siRNA from the lentiviral vector in HEK cells. However, after non-viral transfection of the LINGO1-sh4 plasmid in to HEK cells, a low intensity band was detected, which represented the shRNA. However, no subsequent siRNA band was detected and the RNA in this lane had smeared during separation potentially making the shRNA band a false positive. The failure to detect the shRNA or siRNA was not a consequence of poor transduction/transfection as the HEK cells demonstrated high transfection and transduction efficiency (assessed via eGFP fluorescence). In addition, the positive controls built into the experiment were detected, confirming that the northern blot was executed successfully. However, a large amount of siRNA positive control oligonucleotide was loaded into the gel and yet only a faint band was observed even with a long exposure, potentially suggesting that the blot was not sensitive enough to detect the siRNA and that this assay requires further optimisation. The result indicates that the shLENTImax plasmid expresses only low levels of shRNA and undetectable levels of siRNA. However, our other experiments do provide multiple lines of indirect evidence for expression of the shRNAs by the lentiviral vectors: namely, increased cell death, knockdown of LINGO-1 and the up-regulation of ISGs in cells transduced with lentiviral vectors encoding shRNAs relative to the lentiviral vectors that did not encode an shRNA. The failure to express shRNAs at high levels could be due to the relative positioning of the promoter to the shRNA, which has been demonstrated to be important for the production of a functional shRNA [66, 67]. Alternatively, it may be a result of promoter interference between the two heterologous promoters: The Pol II CMV promoter driving expression of eGFP may be interfering with the expression of the shRNA by the Pol III H1 promoter. Promoter interference between the CMV promoter and elongation factor-1α (EF1α) promoter in late generation lentiviral constructs has previously been demonstrated [68].
We assessed the expression of three well characterised ISGs (OAS1, PKR and Mx1) following transduction of CGNs. OAS1 expression is induced by IFNs and once expressed OAS1 is activated by double stranded RNA. This leads to the synthesis of 2’,5’-oligoadenylates, which bind and activate the endoribonuclease RNase-L. RNase-L non-specifically degrades cellular and viral RNA, resulting in a global non-specific inhibition in gene expression and cell death [51]. There have been multiple studies showing OAS1 up-regulation following lentiviral vector mediated expression of shRNAs [44, 46, 47, 54] or transfection with siRNAs [45]. Following transduction of CGNs using an MOI 10, OAS1 expression was only up-regulated in CGNs transduced with lentiviral vectors encoding an shRNA. However, following transduction using an MOI 50, OAS1 expression was up-regulated in CGNs transduced with any of the lentiviral vectors, including the lentiviral vector that did not encode an shRNA. This indicates that at lower vector concentrations low level shRNA expression is required to stimulate up-regulation of OAS1 expression but at higher viral concentrations lentiviral transduction alone is sufficient to induce OAS1 expression. PKR is an IFN inducible protein that is autophosphorylated and activated by dsRNA. Following activation, PKR can phosphorylate and activate eukaryotic protein synthesis initiation factor 2 (eIF-2) that results in a global inhibition of mRNA translation. PKR also induces the further production of IFNs leading to enhanced expression of ISGs and induces cellular apoptosis through eIF-2 and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kappa B) mediated signalling [48, 51, 69]. PKR has previously been shown to be up-regulated following addition of siRNAs [45, 49]. In the present study PKR expression was not significantly up-regulated following transduction with any of the lentiviral vectors at an MOI 10. However, at an MOI 50, PKR expression had increased in CGNs following transduction with any of the lentiviral vectors encoding shRNAs. Mx1 is also an IFN-induced gene and is part of the Mx GTPase family of proteins that inhibits RNA synthesis in response to viral infection and double stranded RNA [51, 70]. Several studies have shown that siRNAs increase the expression of Mx1 in a dose dependent manner in CNS neurons [40, 49]. However, in the present study Mx1 expression was not affected by any of the lentiviral vectors at either concentration. This suggests that Mx1 is not involved in the lentiviral vector induced cell death or the non-specific knockdown observed in the present study. These results confirm the findings from previous studies which demonstrated that lentiviral vectors and shRNAs can induce an IFN response. This data emphasises that it is crucial to assess ISG levels relative to cells that have not been transduced or transfected at all.
Due to the observed interferon response we investigated whether transduction with lentiviral vectors results in a reduction in the expression of off-target genes. We measured the expression of p75NTR and NgR1 because these genes are expressed by CGNs [55], can bind to LINGO-1 and form a functional NgR complex [55] but are not targeted for knockdown by the LINGO-1 shRNAs. Using an MOI of 10, p75NTR and NgR1 expression were only attenuated in CGNs transduced with lentiviral vectors expressing an shRNA. However after transduction at an MOI of 50 both p75NTR and NgR1 expression levels were significantly reduced by both lentiviral vectors encoding an shRNA as well as those not encoding an shRNA. This data suggests that at low viral vector concentrations, shRNA expression stimulates an interferon response leading to a non-specific down-regulation of both target and off-target gene expression. However at higher viral concentrations lentiviral transduction alone is able to stimulate an interferon response, resulting in the non-specific down-regulation of both target and off-target genes and conceivably induce an even more widespread reduction in gene expression.
Both CGN transduction and viability were affected by viral concentration and shRNA expression. This is consistent with previous studies showing that shRNA expression by Pol III promoters can result in cytotoxicity in vitro and fatality in vivo [40, 44–47, 49, 71, 72]. These studies have extensively demonstrated that high levels of shRNA expression can activate an IFN response leading to global changes in gene expression and cell death. In contrast, the results from the present study suggest that even at very low levels of expression, shRNAs can induce an IFN response leading to an increased incidence of cell death. Lentiviral vectors have generally been demonstrated to be safe and well tolerated in the CNS [23, 29]. However, our data also suggests that the integration-deficient lentiviral vectors themselves may contribute to the induction of an IFN response, as has been previously reported for integrating lentiviral vectors. Lentiviral vectors have previously been shown to rapidly induce a type I IFN response in hepatocytes, which interfered with cell transduction and viability [73, 74]. In addition, a recent study by Bauer et al showed that transduction of cortical neurons with integration-proficient lentiviral vectors resulted in elevated levels of cleaved caspase-3 protein and increased expression of the ISG OAS1, which was then further increased by shRNA expression [54]. Our data is in agreement with these studies and demonstrates that at viral concentrations above MOI 10, integration-deficient lentiviral vectors themselves can induce the expression of OAS1 and an IFN response in CNS neurons resulting in a down-regulation of off-target genes and decreased viability. Furthermore, when the integration-deficient lentiviral vectors encode an shRNA we saw additional effects on cytotoxicity and ISG expression. These results strongly suggest that the viral concentration should be titrated to determine the lowest MOI that can be used to produce effective knockdown and the required functional effect.
Our data emphasises the need for appropriate controls to confirm sequence-specific RNA silencing and assess the level of cytotoxicity. What constitutes an appropriate control has recently come under scrutiny [75] as each siRNA sequence has a unique immunostimulatory response, and it must be ensured that the control siRNA has a similar immunostimulatory response to the therapeutic siRNA sequence. This problem was recently highlighted by a set of studies that had used an siRNA to induce the silencing of vascular endothelial growth factor (VEGF) to generate a therapeutic effect, which was not observed when using a commonly used negative control siRNA that targeted eGFP [75]. However, it was later discovered that the control siRNA had an unusually low immunostimulatory response and that the observed therapeutic effect was actually due to the induction of an IFN response by the VEGF siRNA [50, 76]. These studies raise concerns over how many other RNA interference induced therapeutic effects are the result of an induced IFN response rather than the sequence-specific silencing of the targeted gene. Measures can be taken when designing an siRNA to minimise its IFN response [77]. Specific immunostimulatory sequence motifs such as GU-rich sequences and 5’-UGU-3’ have been identified that should be avoided [78], it has also shown that the U6 promoter produces a larger IFN response compared to the H1 promoter [79]. It has been reported that specific modifications to the siRNA, such as the “Stealth” modification [40] or the transfer of the shRNA into a shRNA-miR structure can reduce the induction of ISGs and the IFN response [54, 80].
In conclusion, this study has demonstrated that although integration-deficient lentiviral vectors can efficiently transduce postnatal CGNs in vitro, transduction with lentiviral vectors at low concentrations when encoding shRNAs or at high concentrations without an shRNA can induce the up-regulation of ISGs that result in an IFN response leading to the non-specific down-regulation of both target and off-target genes and ultimately cell death. In addition, it was demonstrated via northern blot that the lentiviral transfer plasmid only expressed low levels of shRNA, which may be a result of promoter interference. Together, these results may explain why CGN neurite outgrowth was not enhanced on an inhibitory MAG substrate following transduction with lentiviral vectors expressing a shRNA targeting LINGO-1 despite substantial knockdown.
While there are still concerns with using lentiviral vectors and shRNAs for RNA interference, they still present a powerful tool that can be effective as long as the appropriate design criteria, modifications and controls are used. Alternatively, AAV vectors have been reported to display negligible cytotoxicity and immunogenicity and provide efficient CNS transduction [81–85]; however cytotoxicity and lethality associated with shRNA expression has still been reported for AAV vectors [86–90]. shRNA-miRs that mimic the structure of endogenous miRNAs seem to be capable of bypassing the majority of the silencing side effects and may therefore represent more promising therapeutic mediators of RNA interference [54, 79, 91].
Acknowledgements
This study was supported by a Research Councils UK Academic Fellowship (L.M.), the British Pharmacological Society’s Integrative Pharmacology Fund (L.M), Friends of Guy’s Hospital Research Grants (L.M. and R.J.Y.-M.), a Biotechnology and Biological Sciences Research Council’s Doctoral Training Grant (T.H.), financial support from the 7th EU Framework Programme (PERSIST project, grant agreement n°222878, R.J.Y-M.) and a grant from Genoma España (R.J.Y.-M.). We would also like to thank Dr. Thijn R. Brummelkamp, Dr. Rene Bernards and Dr. Reuven Agami for the pSUPER plasmid and Professor Marie Filbin for the CHO-MAG and CHO-R2 cells.
Footnotes
Conflict of Interest Statement
The authors declare no conflict of interest.
References
- 1.Goldberg JL. Intrinsic neuronal regulation of axon and dendrite growth. Curr Opin Neurobiol. 2004;14(5):551–557. doi: 10.1016/j.conb.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 2.Park KK, Liu K, Hu Y, et al. Science. 5903. Vol. 322. New York, NY: 2008. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway; pp. 963–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore DL, Blackmore MG, Hu Y, et al. Science. 5950. Vol. 326. New York, NY: 2009. KLF family members regulate intrinsic axon regeneration ability; pp. 298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mason MR, Lieberman AR, Anderson PN. Corticospinal neurons up-regulate a range of growth-associated genes following intracortical, but not spinal, axotomy. The European journal of neuroscience. 2003;18(4):789–802. doi: 10.1046/j.1460-9568.2003.02809.x. [DOI] [PubMed] [Google Scholar]
- 5.Bulsara KR, Iskandar BJ, Villavicencio AT, Skene JH. A new millenium for spinal cord regeneration: growth-associated genes. Spine. 2002;27(17):1946–1949. doi: 10.1097/00007632-200209010-00030. [DOI] [PubMed] [Google Scholar]
- 6.Plunet W, Kwon BK, Tetzlaff W. Promoting axonal regeneration in the central nervous system by enhancing the cell body response to axotomy. Journal of neuroscience research. 2002;68(1):1–6. doi: 10.1002/jnr.10176. [DOI] [PubMed] [Google Scholar]
- 7.Fawcett JW, Asher RA. The glial scar and central nervous system repair. Brain research bulletin. 1999;49(6):377–391. doi: 10.1016/s0361-9230(99)00072-6. [DOI] [PubMed] [Google Scholar]
- 8.Silver J, Miller JH. Regeneration beyond the glial scar. Nat Rev Neurosci. 2004;5(2):146–156. doi: 10.1038/nrn1326. [DOI] [PubMed] [Google Scholar]
- 9.Xie F, Zheng B. White matter inhibitors in CNS axon regeneration failure. Exp Neurol. 2008;209(2):302–312. doi: 10.1016/j.expneurol.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moon LD, Asher RA, Rhodes KE, Fawcett JW. Regeneration of CNS axons back to their target following treatment of adult rat brain with chondroitinase ABC. Nat Neurosci. 2001;4(5):465–466. doi: 10.1038/87415. [DOI] [PubMed] [Google Scholar]
- 11.Bradbury EJ, Moon LD, Popat RJ, et al. Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature. 2002;416(6881):636–640. doi: 10.1038/416636a. [DOI] [PubMed] [Google Scholar]
- 12.Mukhopadhyay G, Doherty P, Walsh FS, Crocker PR, Filbin MT. A novel role for myelin-associated glycoprotein as an inhibitor of axonal regeneration. Neuron. 1994;13:757–767. doi: 10.1016/0896-6273(94)90042-6. [DOI] [PubMed] [Google Scholar]
- 13.Wang KC, Koprivica V, Kim JA, et al. Oligodendrocyte-myelin glycoprotein is a Nogo receptor ligand that inhibits neurite outgrowth. Nature. 2002;417(6892):941–944. doi: 10.1038/nature00867. [DOI] [PubMed] [Google Scholar]
- 14.Chen MS, Huber AB, van der Haar ME, et al. Nogo-A is a myelin-associated neurite outgrowth inhibitor and an antigen for monoclonal antibody IN-1. Nature. 2000;403:434–439. doi: 10.1038/35000219. [DOI] [PubMed] [Google Scholar]
- 15.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110(4):521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 16.Baum C, Fehse B. Mutagenesis by retroviral transgene insertion: risk assessment and potential alternatives. Current opinion in molecular therapeutics. 2003;5(5):458–462. [PubMed] [Google Scholar]
- 17.Baum C, Kustikova O, Modlich U, Li Z, Fehse B. Mutagenesis and oncogenesis by chromosomal insertion of gene transfer vectors. Human gene therapy. 2006;17(3):253–263. doi: 10.1089/hum.2006.17.253. [DOI] [PubMed] [Google Scholar]
- 18.Li Z, Dullmann J, Schiedlmeier B, et al. Science. 5567. Vol. 296. New York, NY: 2002. Murine leukemia induced by retroviral gene marking; p. 497. [DOI] [PubMed] [Google Scholar]
- 19.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, et al. Science. 5644. Vol. 302. New York, NY: 2003. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1; pp. 415–419. [DOI] [PubMed] [Google Scholar]
- 20.Themis M, Waddington SN, Schmidt M, et al. Oncogenesis Following Delivery of a Nonprimate Lentiviral Gene Therapy Vector to Fetal and Neonatal Mice. Mol Ther. 2005;12(4):763–771. doi: 10.1016/j.ymthe.2005.07.358. [DOI] [PubMed] [Google Scholar]
- 21.Yanez-Munoz RJ, Balaggan KS, MacNeil A, et al. Effective gene therapy with nonintegrating lentiviral vectors. Nature medicine. 2006;12(3):348–353. doi: 10.1038/nm1365. [DOI] [PubMed] [Google Scholar]
- 22.Philpott NJ, Thrasher AJ. Use of nonintegrating lentiviral vectors for gene therapy. Human gene therapy. 2007;18(6):483–489. doi: 10.1089/hum.2007.013. [DOI] [PubMed] [Google Scholar]
- 23.Blomer U, Naldini L, Kafri T, Trono D, Verma IM, Gage FH. Highly efficient and sustained gene transfer in adult neurons with a lentivirus vector. Journal of virology. 1997;71(9):6641–6649. doi: 10.1128/jvi.71.9.6641-6649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Philippe S, Sarkis C, Barkats M, et al. Lentiviral vectors with a defective integrase allow efficient and sustained transgene expression in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(47):17684–17689. doi: 10.1073/pnas.0606197103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yip PK, Wong LF, Pattinson D, et al. Lentiviral vector expressing retinoic acid receptor β2 promotes recovery of function after corticospinal tract injury in the adult rat spinal cord. Human molecular genetics. 2006;15(21):3107–3118. doi: 10.1093/hmg/ddl251. [DOI] [PubMed] [Google Scholar]
- 26.Yip PK, Wong LF, Sears TA, Yanez-Munoz RJ, McMahon SB. Cortical overexpression of neuronal calcium sensor-1 induces functional plasticity in spinal cord following unilateral pyramidal tract injury in rat. PLoS biology. 2010;8(6):e1000399. doi: 10.1371/journal.pbio.1000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rahim AA, Wong AMS, Howe SJ, et al. Efficient gene delivery to the adult and fetal CNS using pseudotyped non-integrating lentiviral vectors. Gene Ther. 2009;16(4):509–520. doi: 10.1038/gt.2008.186. [DOI] [PubMed] [Google Scholar]
- 28.Low K, Blesch A, Herrmann J, Tuszynski MH. A dual promoter lentiviral vector for the in vivo evaluation of gene therapeutic approaches to axon regeneration after spinal cord injury. Gene Ther. 2010 doi: 10.1038/gt.2010.14. [DOI] [PubMed] [Google Scholar]
- 29.Naldini L, Blomer U, Gage FH, Trono D, Verma IM. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(21):11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naldini L, Blomer U, Gallay P, et al. Science. 5259. Vol. 272. New York, NY: 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector; pp. 263–267. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen TT, Marion I, Hasholt L, Lundberg C. Neuron-specific RNA interference using lentiviral vectors. The journal of gene medicine. 2009;11(7):559–569. doi: 10.1002/jgm.1333. [DOI] [PubMed] [Google Scholar]
- 32.Singer O, Marr RA, Rockenstein E, et al. Targeting BACE1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nat Neurosci. 2005;8(10):1343–1349. doi: 10.1038/nn1531. [DOI] [PubMed] [Google Scholar]
- 33.Raoul C, Abbas-Terki T, Bensadoun JC, et al. Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nature medicine. 2005;11(4):423–428. doi: 10.1038/nm1207. [DOI] [PubMed] [Google Scholar]
- 34.Zamore PD. RNA interference: listening to the sound of silence. Nature structural biology. 2001;8(9):746–750. doi: 10.1038/nsb0901-746. [DOI] [PubMed] [Google Scholar]
- 35.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 36.Brummelkamp TR, Bernards R, Agami R. Science. 5567. Vol. 296. New York, NY: 2002. A system for stable expression of short interfering RNAs in mammalian cells; pp. 550–553. [DOI] [PubMed] [Google Scholar]
- 37.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes & development. 2002;16(8):948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cullen BR. RNAi the natural way. Nature genetics. 2005;37(11):1163–1165. doi: 10.1038/ng1105-1163. [DOI] [PubMed] [Google Scholar]
- 39.Ahmed Z, Dent RG, Suggate EL, et al. Disinhibition of neurotrophin-induced dorsal root ganglion cell neurite outgrowth on CNS myelin by siRNA-mediated knockdown of NgR, p75NTR and Rho-A. Molecular and cellular neurosciences. 2005;28(3):509–523. doi: 10.1016/j.mcn.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 40.Suggate EL, Ahmed Z, Read ML, et al. Optimisation of siRNA-mediated RhoA silencing in neuronal cultures. Molecular and Cellular Neuroscience. 2009;40(4):451–462. doi: 10.1016/j.mcn.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 41.Konishi Y, Stegmuller J, Matsuda T, Bonni S, Bonni A. Science. 5660. Vol. 303. New York, NY: 2004. Cdh1-APC controls axonal growth and patterning in the mammalian brain; pp. 1026–1030. [DOI] [PubMed] [Google Scholar]
- 42.Stegmuller J, Konishi Y, Huynh MA, Yuan Z, DiBacco S, Bonni A. Cell-Intrinsic Regulation of Axonal Morphogenesis by the Cdh1-APC Target SnoN. Neuron. 2006;50(3):389–400. doi: 10.1016/j.neuron.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 43.Kim AH, Bonni A. Thinking within the D box: initial identification of Cdh1-APC substrates in the nervous system. Molecular and cellular neurosciences. 2007;34(3):281–287. doi: 10.1016/j.mcn.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 44.Bridge AJ, Pebernard S, Ducraux A, Nicoulaz A-L, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nature genetics. 2003;34(3):263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- 45.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nature cell biology. 2003;5(9):834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 46.Fish RJ, Kruithof EK. Short-term cytotoxic effects and long-term instability of RNAi delivered using lentiviral vectors. BMC molecular biology. 2004;5:9. doi: 10.1186/1471-2199-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pebernard S, Iggo RD. Determinants of interferon-stimulated gene induction by RNAi vectors. Differentiation; research in biological diversity. 2004;72(2–3):103–111. doi: 10.1111/j.1432-0436.2004.07202001.x. [DOI] [PubMed] [Google Scholar]
- 48.Sledz CA, Williams BR. RNA interference and double-stranded-RNA-activated pathways. Biochemical Society transactions. 2004;32(Pt 6):952–956. doi: 10.1042/BST0320952. [DOI] [PubMed] [Google Scholar]
- 49.Read ML, Mir S, Spice R, et al. Profiling RNA interference (RNAi)-mediated toxicity in neural cultures for effective short interfering RNA design. The journal of gene medicine. 2009;11(6):523–534. doi: 10.1002/jgm.1321. [DOI] [PubMed] [Google Scholar]
- 50.Rossi JJ. Innate Immunity confounds the clinical efficacy of small interfering RNAs (siRNAs) Gene Ther. 2009;16(5):579–580. doi: 10.1038/gt.2009.26. [DOI] [PubMed] [Google Scholar]
- 51.Samuel CE. Antiviral Actions of Interferons. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dafny N, Yang PB. Interferon and the central nervous system. European journal of pharmacology. 2005;523(1–3):1–15. doi: 10.1016/j.ejphar.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 53.Delhaye S, Paul S, Blakqori G, et al. Neurons produce type I interferon during viral encephalitis. 2006;103:7835–7840. doi: 10.1073/pnas.0602460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bauer M, Kinkl N, Meixner A, et al. Prevention of interferon-stimulated gene expression using microRNA-designed hairpins. Gene Ther. 2008;16(1):142–147. doi: 10.1038/gt.2008.123. [DOI] [PubMed] [Google Scholar]
- 55.Mi S, Lee X, Shao Z, et al. LINGO-1 is a component of the Nogo-66 receptor/p75 signaling complex. Nat Neurosci. 2004;7(3):221–228. doi: 10.1038/nn1188. [DOI] [PubMed] [Google Scholar]
- 56.Fournier AE, GrandPre T, Strittmatter SM. Identification of a receptor mediating Nogo-66 inhibition of axonal regeneration. Nature. 2001;409(6818):341–346. doi: 10.1038/35053072. [DOI] [PubMed] [Google Scholar]
- 57.Domeniconi M, Cao Z, Spencer T, et al. Myelin-associated glycoprotein interacts with the Nogo66 receptor to inhibit neurite outgrowth. Neuron. 2002;35(2):283–290. doi: 10.1016/s0896-6273(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 58.Liu BP, Fournier A, GrandPre T, Strittmatter SM. Science. 5584. Vol. 297. New York, NY: 2002. Myelin-associated glycoprotein as a functional ligand for the Nogo-66 receptor; pp. 1190–1193. [DOI] [PubMed] [Google Scholar]
- 59.McGee AW, Strittmatter SM. The Nogo-66 receptor: focusing myelin inhibition of axon regeneration. Trends in neurosciences. 2003;26(4):193–198. doi: 10.1016/S0166-2236(03)00062-6. [DOI] [PubMed] [Google Scholar]
- 60.Ji B, Li M, Wu WT, et al. LINGO-1 antagonist promotes functional recovery and axonal sprouting after spinal cord injury. Molecular and cellular neurosciences. 2006;33(3):311–320. doi: 10.1016/j.mcn.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 61.Follenzi A, Ailles LE, Bakovic S, Geuna M, Naldini L. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nature genetics. 2000;25(2):217–222. doi: 10.1038/76095. [DOI] [PubMed] [Google Scholar]
- 62.Paddison PJ, Cleary M, Silva JM, et al. Cloning of short hairpin RNAs for gene knockdown in mammalian cells. Nature methods. 2004;1(2):163–167. doi: 10.1038/nmeth1104-163. [DOI] [PubMed] [Google Scholar]
- 63.Mi S, Miller RH, Lee X, et al. LINGO-1 negatively regulates myelination by oligodendrocytes. Nat Neurosci. 2005;8(6):745–751. doi: 10.1038/nn1460. [DOI] [PubMed] [Google Scholar]
- 64.Demaison C, Parsley K, Brouns G, et al. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency [correction of imunodeficiency] virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Human gene therapy. 2002;13(7):803–813. doi: 10.1089/10430340252898984. [DOI] [PubMed] [Google Scholar]
- 65.Smith DS, Skene JH. A transcription-dependent switch controls competence of adult neurons for distinct modes of axon growth. J Neurosci. 1997;17(2):646–658. doi: 10.1523/JNEUROSCI.17-02-00646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xia H, Mao Q, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nature biotechnology. 2002;20(10):1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- 67.Davidson BL, Harper SQ. Viral delivery of recombinant short hairpin RNAs. Methods in enzymology. 2005;392:145–173. doi: 10.1016/S0076-6879(04)92009-5. [DOI] [PubMed] [Google Scholar]
- 68.Curtin JA, Dane AP, Swanson A, Alexander IE, Ginn SL. Bidirectional promoter interference between two widely used internal heterologous promoters in a late-generation lentiviral construct. Gene Ther. 2008;15(5):384–390. doi: 10.1038/sj.gt.3303105. [DOI] [PubMed] [Google Scholar]
- 69.Kumar A, Haque J, Lacoste J, Hiscott J, Williams BR. Double-stranded RNA-dependent protein kinase activates transcription factor NF-kappa B by phosphorylating I kappa B. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(14):6288–6292. doi: 10.1073/pnas.91.14.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pitossi F, Blank A, Schroder A, et al. A functional GTP-binding motif is necessary for antiviral activity of Mx proteins. 1993;67:6726–6732. doi: 10.1128/jvi.67.11.6726-6732.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grimm D, Streetz KL, Jopling CL, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441(7092):537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 72.Davidson BL, Boudreau RL. RNA interference: a tool for querying nervous system function and an emerging therapy. Neuron. 2007;53(6):781–788. doi: 10.1016/j.neuron.2007.02.020. [DOI] [PubMed] [Google Scholar]
- 73.Brown BD, Sitia G, Annoni A, et al. In vivo administration of lentiviral vectors triggers a type I interferon response that restricts hepatocyte gene transfer and promotes vector clearance. Blood. 2007;109(7):2797–2805. doi: 10.1182/blood-2006-10-049312. [DOI] [PubMed] [Google Scholar]
- 74.Nayak S, Herzog RW. Progress and prospects: immune responses to viral vectors. Gene Ther. 2009 doi: 10.1038/gt.2009.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Michels S, Schmidt-Erfurth U, Rosenfeld PJ. Promising new treatments for neovascular age-related macular degeneration. Expert opinion on investigational drugs. 2006;15(7):779–793. doi: 10.1517/13543784.15.7.779. [DOI] [PubMed] [Google Scholar]
- 76.Kleinman ME, Yamada K, Takeda A, et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452(7187):591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Judge A, MacLachlan I. Overcoming the innate immune response to small interfering RNA. Human gene therapy. 2008;19(2):111–124. doi: 10.1089/hum.2007.179. [DOI] [PubMed] [Google Scholar]
- 78.Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nature biotechnology. 2005;23(4):457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 79.An DS, Qin FX-F, Auyeung VC, et al. Optimization and Functional Effects of Stable Short Hairpin RNA Expression in Primary Human Lymphocytes via Lentiviral Vectors. Mol Ther. 2006;14(4):494–504. doi: 10.1016/j.ymthe.2006.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boudreau RL, Martins I, Davidson BL. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol Ther. 2009;17(1):169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Papale A, Cerovic M, Brambilla R. Viral vector approaches to modify gene expression in the brain. Journal of neuroscience methods. 2009;185(1):1–14. doi: 10.1016/j.jneumeth.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 82.Kay MA, Glorioso JC, Naldini L. Viral vectors for gene therapy: the art of turning infectious agents into vehicles of therapeutics. Nature medicine. 2001;7(1):33–40. doi: 10.1038/83324. [DOI] [PubMed] [Google Scholar]
- 83.Burger C, Nash K, Mandel RJ. Recombinant adeno-associated viral vectors in the nervous system. Human gene therapy. 2005;16(7):781–791. doi: 10.1089/hum.2005.16.781. [DOI] [PubMed] [Google Scholar]
- 84.McCown TJ. Adeno-associated virus (AAV) vectors in the CNS. Current gene therapy. 2005;5(3):333–338. doi: 10.2174/1566523054064995. [DOI] [PubMed] [Google Scholar]
- 85.Hutson TH, Verhaagen J, Yáñez-Muñoz RJ, Moon LD. Corticospinal tract transduction: a comparison of seven adeno-associated viral vector serotypes and a non-integrating lentiviral vector. Gene Ther. 2012 Jan;19(1):49–60. doi: 10.1038/gt.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ehlert EM, Eggers R, Niclou SP, Verhaagen J. Cellular toxicity following application of adeno-associated viral vector-mediated RNA interference in the nervous system. BMC neuroscience. 2010;11(1):20. doi: 10.1186/1471-2202-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Grimm D, Wang L, Lee JS, Schürmann N, Gu S, Börner K, Storm TA, Kay MA. Argonaute proteins are key determinants of RNAi efficacy, toxicity, and persistence in the adult mouse liver. J Clin Invest. 2010 Sep 1;120(9):3106–19. doi: 10.1172/JCI43565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ulusoy A, Sahin G, Björklund T, Aebischer P, Kirik D. Dose optimization for long-term rAAV-mediated RNA interference in the nigrostriatal projection neurons. Mol Ther. 2009 Sep;17(9):1574–84. doi: 10.1038/mt.2009.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Khodr CE, Sapru MK, Pedapati J, Han Y, West NC, Kells AP, Bankiewicz KS, Bohn MC. An alpha-synuclein AAV gene silencing vector ameliorates a behavioral deficit in a rat model of Parkinson's disease, but displays toxicity in dopamine neurons. Brain Res. 2011 Jun 13;1395:94–107. doi: 10.1016/j.brainres.2011.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Martin JN, Wolken N, Brown T, Dauer WT, Ehrlich ME, Gonzalez-Alegre P. Lethal toxicity caused by expression of shRNA in the mouse striatum: implications for therapeutic design. Gene Ther. 2011 Jul;18(7):666–73. doi: 10.1038/gt.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McBride JL, Boudreau RL, Harper SQ, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(15):5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]