Abstract
The mechanism underlying increased concentrations of cancer stem cell (CSC)-associated factors in non-small cell lung cancer (NSCLC) cells treated with transforming growth factor β1 (TGFβ1) and tumor necrosis factor α (TNFα), is still not clear. The purpose of this study was to investigate the possible role of CD44 in the regulation of CSC-associated genes, by analyzing the effect of CD44 knockdown on their expression. A549, a NSCLC cell line that expresses CD44 antigen, was treated with TGFβ1 and TNFα. Small-interfering ribonucleic acid (siRNA) that specifically targets the CD44 gene was used to knockdown the expression of CD44 in A549. The gene expressions of CD44, CXCR4, POU5F1 (octamer-binding transcription factor 4 [Oct4]), PROM1, NANOG, c-Myc, KLF4, and SOX2, as well as of CDH1 (E-cadherin), CDH2 (N-cadherin), VIM (vimentin), and FN1 (fibronectin) were analyzed in A549 cells by quantitative reverse transcription polymerase chain reaction (RT-qPCR). Cell morphology was observed using light microscopy. After TGFβ1/TNFα treatment, increased expressions of CXCR4 and POU5F1 were detected. Silencing of CD44 gene expression was confirmed by RT-qPCR. The knockdown of CD44 decreased the CXCR4 and POU5F1 gene expressions in TGFβ1/TNFα-treated A549 cells. However, the silencing of CD44 did not affect the morphology of TGFβ1/TNFα-treated A549 cells nor it reversed epithelial-mesenchymal transition (EMT) gene signature induced by TGFβ1/TNFα in A549 cells. Our preliminary findings suggest that the CD44 gene may have a role in regulating CXCR4 and POU5F1 gene expressions, independently of the EMT signaling pathway.
KEY WORDS: CD44, CXCR4 receptor, Oct4, POU5F1, non-small cell lung cancer cells, NSCLC, C-X-C chemokine receptor type 4, octamer-binding transcription factor 4
INTRODUCTION
Cancer stem cells (CSCs) are a minor population of tumor cells that have the ability to self-renew. Dysregulation of this stem cell self-renewal process is likely a basis for the development of cancer [1]. In addition, CSCs have been identified to play a role in cancer resistance and progression [2,3]. Therefore, CSC-targeted therapy provides the opportunity to significantly improve cancer patient outcomes. CSC markers have been reported in different cancer types, including CD44 [4-6], C-X-C chemokine receptor type 4 (CXCR4) [7], and octamer-binding transcription factor 4 (Oct4) [8].
CD44 was first reported as a CSC marker by Al-Hajj et al. in 2003 [6], and since then, has been extensively investigated in cancers of various organs. For example, in non-small cell lung cancer (NSCLC) it was reported that the cells expressing CD44 have stem cell-like characteristics [9]. We previously reported that the interaction between CD44 and its ligand hyaluronate in NSCLC cells in which CD44 was reintroduced induces chemoresistance and restores the cell susceptibility to the macrophage cytotoxicity [10,11].
Accumulating evidence also suggests that tumor growth factor β1 (TGFβ1) induces stem-cell characteristics in non-small lung cancer cells and breast cancer cells [12,13]. Another study reported that TGFβ1 increased the expression levels of Yamanaka factors (i.e., Oct3/4, sex determining region Y-box 2 [SOX2], Kruppel-like factor 4 [KLF4], and Myc proto-oncogene protein [Myc]) in a dose-dependent manner [14]. Moreover, CD44 overexpression was associated with TGFβ-mediated mesenchymal phenotype in hepatocellular carcinoma [15]. However the role of CD44 in the regulation of other CSC markers is still unclear.
In this study, we examined the role of CD44 in the regulation of CXCR4 and Oct4. A549 NSCLC cell line was treated with TGFβ1 and TNFα. The treatment with TGFβ1 and TNFα increased the expression of several CSC genes, including Oct4 gene POU5F1 and CXCR4 gene. The role of CD44 in the regulation of CSC gene expression was investigated.
MATERIALS AND METHODS
Cell culture and reagents
The lung adenocarcinoma cell line A549 was used in this study. A549 cells, predominantly expressing CD44 standard isoform, were purchased from the American Type Culture Collection (Manassas, VA, USA). The cell line was verified to be mycoplasma-free. The cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS), penicillin, and streptomycin (100 U/mL and 100 μg/mL, respectively), and were grown in a humidified 5% CO2 atmosphere at 37°C in an incubator where the oxygen tension was held at 21%. Recombinant soluble human TGFβ1 was obtained from Peprotech (Rocky Hill, NJ, USA) and recombinant soluble human TNF-α was obtained from eBioscience (San Diego, CA, USA). For TGFβ1/TNFα treatment, 10 μM of TGFβ1 and 100 μM of TNFα were applied to A549 cells for 48 hours.
Cell morphology analysis
A549 cells were plated in six-well dishes at a density of 1.5 × 105 cells per well and allowed to adhere for 24 hours. The cells were then treated with TGFβ1/TNFα for 24 hours. Representative images of A549 cells were captured by phase-contrast microscopy (Olympus, Tokyo, Japan).
Quantitative reverse transcription polymerase chain reaction (RT-qPCR)
RNA extraction and complementary DNA (cDNA) synthesis
The total RNA was extracted from A549 cell cultures using miRvana miRNA Isolation Kit (Ambion, Austin, TX, USA), according to the manufacturer’s instructions. Briefly, the cells were grown to 80-90% confluence in 100-mm dishes and lysed with 600 μl of Lysis Binding Buffer (Ambion). RNA was extracted using acid phenol-chloroform (Life Technologies, Frederick, MD, USA) and RNA-rich layers were separated by centrifugation. Next, RNA molecules were precipitated with ethanol 99.5%. Then, RNA was rinsed with Wash Solution 1 and 2/3 and dissolved in RNase-free UltraPure Distilled Water (Invitrogen, Grand Island, NY, USA). The concentration of RNA was measured by Nanodrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Five hundred nanograms of total RNA was reverse-transcribed into cDNA using ReverTra Ace® cDNA synthesis kit (Toyobo, Osaka, Japan), according to the manufacturer’s instructions.
Real-time PCR
Real-time PCR was performed using SYBR Green Master Mix (Toyobo) and StepOnePlus™ Real-Time PCR System (Applied Biosystem, CA, USA), according to the manufacturer’s instructions. The cycling conditions were as follows: initial denaturation at 95°C for 20 seconds and 40 cycles of amplification (denaturation at 95°C for 3 seconds, and annealing and extension at 60°C for 30 seconds). Real-time PCR was performed in triplicate and β-actin expression was used as internal control. The mRNA expression of the following genes was analyzed: CD44, CXCR4, POU5F1, PROM1 (prominin-1, CD133), NANOG, c-Myc, KLF4, and SOX2, as well as the mRNA expression of epithelial marker CDH1 (E-cadherin) and several mesenchymal markers including CDH2 (N-cadherin), VIM (vimentin), and FN1 (fibronectin). The primers used for real-time PCR are provided in Table 1.
TABLE 1.
Forward and backward primers used for quantitative reverse transcription polymerase chain reaction (RT-qPCR)

RNA interference
Small interfering RNAs (siRNAs) targeting CD44 (Stealth Select RNAi siRNA) were custom synthesized by Invitrogen. We used Stealth™ RNAi siRNA Negative Control Duplexes from Invitrogen (Cat. No. 12935-100) as negative control. To exclude off-target effect, A549 cells were transfected with two different specific siRNAs (siCD44 #1 and #2 groups) and one non-specific control using Lipofectamine® RNAiMAX (Invitrogen) [siControl group], according to the manufacturer’s instructions. The cells were detached and diluted in complete growth medium without antibiotics and then plated in each of the wells. RNAi duplex and Lipofectamine RNAiMAX were mixed in Opti-MEM I reduced serum medium (Gibco, Massachusetts, USA) and incubated for 15 minutes at room temperature. RNAi duplex-Lipofectamine™ RNAiMAX complexes were added to the cell containing wells. The cells were then incubated for 48 hours at 37°C. The sequences of the siRNA against CD44 were as follows:
Stealth CD44 oligo #1: 5’-GCAAGUCUCAGGAAAUGGUGCAUUU-3’;
Stealth CD44 oligo #2: 5’-GCUGACCUCUGCAAGGCUUUCAAUA-3’.
Statistical analysis
Differences between the two groups were analyzed using the unpaired Student’s t-test. The differences were considered significant at p < 0.05. Statistical analyses were performed using GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA).
RESULTS
TGFβ1/TNFα treatment increased the expression of stem cell-related factors in A549 cells
Following 48 hours of TGFβ1/TNFα treatment, we measured mRNA expression of CD44, CXCR4, POU5F1, CD133, NANOG, c-Myc, KLF4, and SOX2 in A549 cells, using RT-qPCR. Our results showed upregulated expression of CXCR4 and POU5F1 in TGFβ1/TNFα-treated A549 cells (Figure 1), however, no increase was observed in the expression of CD133, NANOG, c-Myc, KLF4, and SOX2 genes (data not shown).
FIGURE 1.
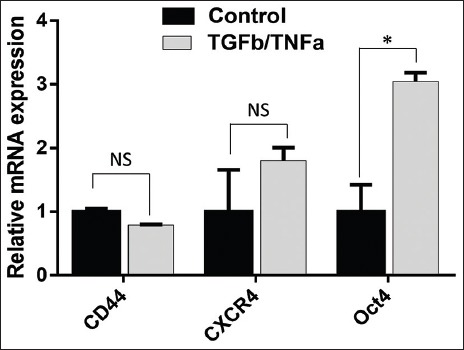
The expression of the stem-cell related genes in A549 cell line treated with TGFβ1/TNFα. A549 cells were treated with TGFβ1/TNFα for 24 hours. Quantitative reverse transcription polymerase chain reaction (RT-qPCR) was performed in triplicate with primers specific for CD44, CXCR4, and POU5F1 (Oct4) genes, and the results were normalized to actin expression. Data are expressed as mean ± standard deviation. *p<0.05 indicates a significant difference, while NS indicates a non-significant difference.
Knockdown of CD44 reduced the expression of CXCR4 and POU5F1 in TGFβ1/TNFα-treated A549 cells
Parental A549 cells showed moderate expression of CD44, which was not markedly changed after treating the cells with TGFβ1/TNFα for 48 hours (Figure 2A). The silencing of CD44 was performed in TGFβ1/TNFα-treated A549 cells using siRNA oligos that target CD44 (siCD44 #1 and #2 groups). CD44 mRNA level was less than 5% in both siCD44 groups (Figure 2A), suggesting that the siRNA successfully suppressed the CD44 expression.
FIGURE 2.
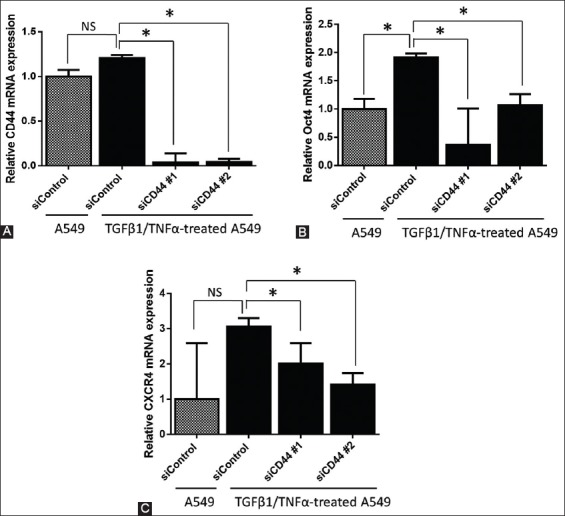
Effects of CD44 silencing on the expression of CXCR4 and POU5F1 (Oct4). CD44 gene was knocked down in TGFβ1/TNFα-treated A549 cells by small interfering RNA (siRNA) against CD44, in a 6-well plate. Next, quantitative reverse transcription polymerase chain reaction (RT-qPCR) was performed to determine the relative mRNA levels of (A) CD44, (B) POU5F1 (Oct4), and (C) CXCR4 and the results were normalized to actin expression. Two specific siRNAs for CD44 (siCD44#1 and #2) and one non-specific siRNA as a control (siControl) were used. Data are expressed as mean ± standard deviation. *p<0.05 indicates a significant difference, while NS indicates a non-significant difference.
To investigate the effect of CD44 on the expression of stem-related factors, we analyzed POU5F1 and CXCR4 mRNA expression using RT-qPCR (Figure 2B and 2C, respectively). The knockdown of CD44 using both siRNA oligos significantly reduced the mRNA expression of POU5F1 and CXCR4 in TGFβ1/TNFα-treated A549 cells (p < 0.05).
Knockdown of CD44 did not reverse TGFβ1/TNFα-induced epithelial-mesenchymal transition (EMT) in A549 cells
We also measured the mRNA expression of epithelial marker E-cadherin and several mesenchymal markers, i.e., N-cadherin, vimentin, and fibronectin, using RT-qPCR. As shown in Figure 3A, the mRNA expression of E-cadherin was significantly lower in siCD44 groups compared to siControl group. Similarly, the mRNA expression of N-cadherin (Figure 3B), vimentin (Figure 3C), and fibronectin (Figure 3D) was not suppressed by CD44 gene silencing in TGFβ1/TNFα-treated group. Moreover, TGFβ1/TNFα-treated A549 cells were irregularly spindle-shaped (Figure 3E), and the knockdown of CD44 did not reverse these morphological changes to regular, cuboidal cell shape.
FIGURE 3.
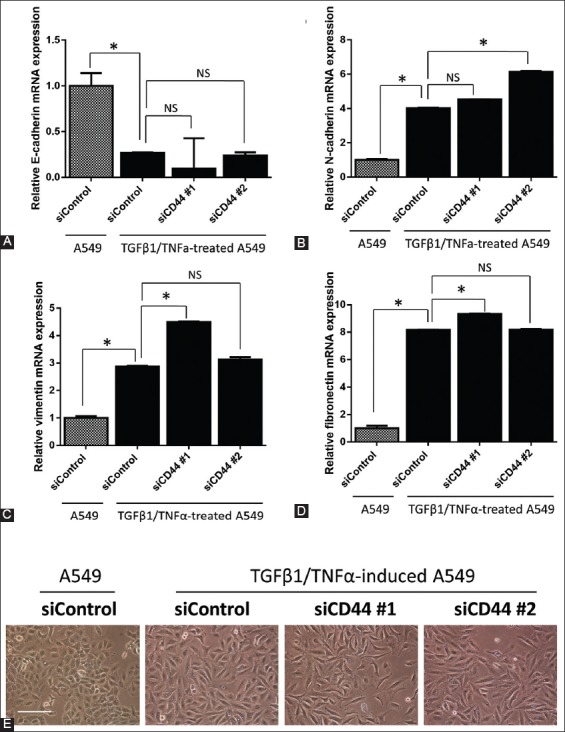
The mRNA expression of (A) E-cadherin, (B) N-cadherin, (C) vimentin, and (D) fibronectin in CD44 knockdown A549 cells, after TGFβ1/TNFα treatment. Data were normalized to actin expression. (E) A549 cells (1.5 × 105) in 6-well plates were reverse transfected with control siRNA oligo (siControl) and two siRNA oligos against CD44 (siCD44 #1 and #2) and allowed to adhere for 24 hours. The cells were then treated with TGFβ1/TNFα for 24 hours. Representative images of A549 cells were photographed. The bar represents 200 μM. The results are expressed as mean ± standard deviation. *p<0.05 indicates a significant difference, while NS indicates a non-significant difference.
DISCUSSION
Here, we demonstrated that treating A549 cells with TGFβ1/TNFα increased the mRNA expression of several stem-related factors, such as CXCR4 and Oct4. Furthermore, we showed that CD44 was associated with the upregulation of TGFβ1/TNFα-induced stem-related factors in the lung cancer cells. Similarly, a previous report suggested that TGFβ1 treatment upregulates CXCR4 expression in primary human monocyte-derived macrophages (MDMs) and rat microglia [16]. Using human basal cell carcinoma cells as an in vitro model, it was demonstrated that the regulation of CXCR4 expression by TGFβ1 is mediated by the phosphorylation of extracellular signal-regulated protein kinases 1 and 2 (ERK1/2)-ETS1 pathway [17]. In addition, the transcription factor Oct4, which is able to reprogram mouse and human somatic cells to undifferentiated, pluripotent stem cells, was found to be significantly increased in A549 cells treated with TGFβ1 compared to untreated controls [18]. The results demonstrated in the current study are consistent with the above-mentioned research.
CD44 is a ubiquitous, multi-structural and multi-functional, cell surface adhesion molecule involved in cell-cell and cell-matrix interactions [19]. Moreover, CD44 is a receptor for hyaluronic acid (HA), an anionic, nonsulfated glycosaminoglycan [20]. As a member of single-span transmembrane glycoprotein family, CD44 is involved in pathological processes, particularly in cancer pathogenesis [21]. Different methods have been developed for targeting CD44, including liposome-encapsulated doxorubicin [20], lipid-based nanovectors [22], and polypropylenimine (PPI) dendrimer-siRNA complex [23]. The efficient delivery of siRNA to target cells has been a challenge for therapeutic application of siRNA, due to limited intake of siRNA for gene silencing by tissue-constructing cells [24]. In this study, the silencing of CD44 by siRNA was successful, i.e., 0.036560 relative mRNA level was detected for siRNA oligo number 1 and 0.042390 for oligo number 2, compared to siControl group.
Recently, the interaction between CD44 and CXCR4 in the presence of C-X-C motif chemokine 12 (CXCL12) was demonstrated, and this association was inhibited by a low-molecular weight HA [25]. In gastric cancer cells, together with human epidermal growth factor receptor 2 (HER2), CD44 upregulated CXCR4 via epigenetic silencing of microRNA-139. However, until now, no study has reported the effect of CD44 knockdown on the CXCR4 expression. To the best of our knowledge, this preliminary study is the first to report decreased expression of CXCR4 and POU5F1 after CD44 gene knockdown by siRNA.
Co-stimulation with TGFβ/TNFα could induce EMT in various cancer models. However, in our study, the knockdown of CD44 did not restore the mRNA expression of epithelial marker E-cadherin, nor it downregulated the expression of mesenchymal markers fibronectin, vimentin, and N-cadherin. Also, the CD44 knockdown did not cause changes in the morphology of A549 cells. These findings suggest that the regulation of POU5F1 and CXCR4 expression was independent of EMT signaling pathway in our TGFβ1/TNFα-induced EMT model.
One of the limitations of our in vitro study is that it did not explain if the CD44-mediated regulation of CXCR4 and POU5F1 expression was due to a direct or indirect molecular mechanism. In addition, in this preliminary study, we did not confirm the suppression of CD44 at the protein level, which should be assessed using Western blot analysis. Moreover, the biological significance of CD44 silencing in terms of self-renewal ability of cells, as well as tumorigenicity of TGFβ1/TNFα-induced lung cancer cells in vivo, should be further explored. Future studies should also clarify intracellular signaling cascades that are associated with the expression of CD44-mediated stem cell-related factors.
CONCLUSION
Overall, our results indicate that CD44 possibly have a role in regulating other CSC markers, particularly CXCR4 and Oct4, and that this processes is independent of EMT signaling pathway. It is important to investigate the implication of CD44 in the self-renewal ability of lung cancer cells using both in vitro and in vivo models.
DECLARATION OF INTERESTS
The authors declare no conflict of interests.
ACKNOWLEDGMENTS
This study was supported by Grants-in-Aid for Scientific Research No. 23591906 (Fumiyuki Takahashi) and by Post-graduate Scholarship No. 91006006265 (Fariz Nurwidya) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. We received important advice and expertise from Ms. Takako Ikegami and Ms. Tomomi Ikeda, Laboratory of Molecular and Biochemical Research, Research Support Center, Juntendo University Graduate School of Medicine.
REFERENCES
- [1].Al-Hajj M, Becker MW, Wicha M, Weissman I, Clarke MF. Therapeutic implications of cancer stem cells. Curr Opin Genet Dev. 2004;14(1):43–7. doi: 10.1016/j.gde.2003.11.007. https://doi.org/10.1016/j.gde.2003.11.007. [DOI] [PubMed] [Google Scholar]
- [2].Murakami A, Takahashi F, Nurwidya F, Kobayashi I, Minakata K, Hashimoto M, et al. Hypoxia increases gefitinib-resistant lung cancer stem cells through the activation of insulin-like growth factor 1 receptor. PLoS One. 2014;9(1):e86459. doi: 10.1371/journal.pone.0086459. https://doi.org/10.1371/journal.pone.0086459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nurwidya F, Murakami A, Takahashi F, Takahashi K. Lung cancer stem cells: Tumor biology and clinical implications. Asia Pac J Clin Oncol. 2012;8(3):217–22. doi: 10.1111/j.1743-7563.2012.01550.x. https://doi.org/10.1111/j.1743-7563.2012.01550.x. [DOI] [PubMed] [Google Scholar]
- [4].Fillmore C, Kuperwasser C. Human breast cancer stem cell markers CD44 and CD24: Enriching for cells with functional properties in mice or in man? Breast Cancer Res. 2007;9(3):303. doi: 10.1186/bcr1673. https://doi.org/10.1186/bcr1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].de Beca FF, Caetano P, Gerhard R, Alvarenga CA, Gomes M, Paredes J, et al. Cancer stem cells markers CD44, CD24 and ALDH1 in breast cancer special histological types. J Clin Pathol. 2013;66(3):187–91. doi: 10.1136/jclinpath-2012-201169. https://doi.org/10.1136/jclinpath-2012-201169. [DOI] [PubMed] [Google Scholar]
- [6].Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–8. doi: 10.1073/pnas.0530291100. https://doi.org/10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jung MJ, Rho JK, Kim YM, Jung JE, Jin YB, Ko YG, et al. Upregulation of CXCR4 is functionally crucial for maintenance of stemness in drug-resistant non-small cell lung cancer cells. Oncogene. 2013;32(2):209–21. doi: 10.1038/onc.2012.37. https://doi.org/10.1038/onc.2012.37. [DOI] [PubMed] [Google Scholar]
- [8].Kim RJ, Nam JS. Oct4 expression enhances features of cancer stem cells in a mouse model of breast cancer. Lab Anim Res. 2011;27(2):147–52. doi: 10.5625/lar.2011.27.2.147. https://doi.org/10.5625/lar.2011.27.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Leung EL, Fiscus RR, Tung JW, Tin VP, Cheng LC, Sihoe AD, et al. Non-small cell lung cancer cells expressing CD44 are enriched for stem cell-like properties. PLoS One. 2010;5(11):e14062. doi: 10.1371/journal.pone.0014062. https://doi.org/10.1371/journal.pone.0014062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ohashi R, Takahashi F, Cui R, Yoshioka M, Gu T, Sasaki S, et al. Interaction between CD44 and hyaluronate induces chemoresistance in non-small cell lung cancer cell. Cancer Lett. 2007;252(2):225–34. doi: 10.1016/j.canlet.2006.12.025. https://doi.org/10.1016/j.canlet.2006.12.025. [DOI] [PubMed] [Google Scholar]
- [11].Takahashi K, Takahashi F, Hirama M, Tanabe KK, Fukuchi Y. Restoration of CD44S in non-small cell lung cancer cells enhanced their susceptibility to the macrophage cytotoxicity. Lung Cancer. 2003;41(2):145–53. doi: 10.1016/s0169-5002(03)00224-1. https://doi.org/10.1016/S0169-5002(03)00224-1. [DOI] [PubMed] [Google Scholar]
- [12].Pirozzi G, Tirino V, Camerlingo R, Franco R, La Rocca A, Liguori E, et al. Epithelial to mesenchymal transition by TGFβ-1 induction increases stemness characteristics in primary non small cell lung cancer cell line. PLoS One. 2011;6(6):e21548. doi: 10.1371/journal.pone.0021548. https://doi.org/10.1371/journal.pone.0021548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cufi S, Vazquez-Martin A, Oliveras-Ferraros C, Martin-Castillo B, Joven J, Menendez JA. Metformin against TGFbeta-induced epithelial-to-mesenchymal transition (EMT): From cancer stem cells to aging-associated fibrosis. Cell Cycle. 2010;9(22):4461–8. doi: 10.4161/cc.9.22.14048. https://doi.org/10.4161/cc.9.22.14048. [DOI] [PubMed] [Google Scholar]
- [14].Yamasaki S, Taguchi Y, Shimamoto A, Mukasa H, Tahara H, Okamoto T. Generation of human induced pluripotent stem (Ips) cells in serum- and feeder-free defined culture and TGF-B1 regulation of pluripotency. PLoS One. 2014;9(1):e87151. doi: 10.1371/journal.pone.0087151. https://doi.org/10.1371/journal.pone.0087151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mima K, Okabe H, Ishimoto T, Hayashi H, Nakagawa S, Kuroki H, et al. CD44s regulates the TGF-β-mediated mesenchymal phenotype and is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Res. 2012;72(13):3414–23. doi: 10.1158/0008-5472.CAN-12-0299. https://doi.org/10.1158/0008-5472-CAN-12-0299. [DOI] [PubMed] [Google Scholar]
- [16].Chen S, Tuttle DL, Oshier JT, Knot HJ, Streit WJ, Goodenow MM, et al. Transforming growth factor-beta1 increases CXCR4 expression, stromal-derived factor-1alpha-stimulated signalling and human immunodeficiency virus-1 entry in human monocyte-derived macrophages. Immunology. 2005;114(4):565–74. doi: 10.1111/j.1365-2567.2004.02110.x. https://doi.org/10.1111/j.1365-2567.2004.02110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chu CY, Sheen YS, Cha ST, Hu YF, Tan CT, Chiu HC, et al. Induction of chemokine receptor CXCR4 expression by transforming growth factor-β1 in human basal cell carcinoma cells. J Dermatol Sci. 2013;72(2):123–33. doi: 10.1016/j.jdermsci.2013.06.011. https://doi.org/10.1016/j.jdermsci.2013.06.011. [DOI] [PubMed] [Google Scholar]
- [18].Tirino V, Camerlingo R, Bifulco K, Irollo E, Montella R, Paino F, et al. TGF-β1 exposure induces epithelial to mesenchymal transition both in CSCs and non-CSCs of the A549 cell line, leading to an increase of migration ability in the CD133+A549 cell fraction. Cell Death Dis. 2013;4:e620. doi: 10.1038/cddis.2013.144. https://doi.org/10.1038/cddis.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Naor D, Sionov RV, Ish-Shalom D. CD44: Structure, function, and association with the malignant process. Adv Cancer Res. 1997;71:241–319. doi: 10.1016/s0065-230x(08)60101-3. https://doi.org/10.1016/S0065-230X(08)60101-3. [DOI] [PubMed] [Google Scholar]
- [20].Eliaz RE, Szoka FC., Jr Liposome-encapsulated doxorubicin targeted to CD44: A strategy to kill CD44-overexpressing tumor cells. Cancer Res. 2001;61(6):2592–601. [PubMed] [Google Scholar]
- [21].Orian-Rousseau V, Ponta H. Perspectives of CD44 targeting therapies. Arch Toxicol. 2015;89(1):3–14. doi: 10.1007/s00204-014-1424-2. DOI: 10.1007/s00204-014-1424-2. [DOI] [PubMed] [Google Scholar]
- [22].Arpicco S, De Rosa G, Fattal E. Lipid-based nanovectors for targeting of CD44-overexpressing tumor cells. J Drug Deliv. 2013;2013:860780. doi: 10.1155/2013/860780. https://doi.org/10.1155/2013/860780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Shah V, Taratula O, Garbuzenko OB, Taratula OR, Rodriguez-Rodriguez L, Minko T. Targeted nanomedicine for suppression of CD44 and simultaneous cell death induction in ovarian cancer: An optimal delivery of siRNA and anticancer drug. Clin Cancer Res. 2013;19(22):6193–204. doi: 10.1158/1078-0432.CCR-13-1536. https://doi.org/10.1158/1078-0432-CCR-13-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Asai-Tajiri Y, Matsumoto K, Fukuyama S, Kan OK, Nakano T, Tonai K, et al. Small interfering RNA against CD86 during allergen challenge blocks experimental allergic asthma. Respir Res. 2014;15:132. doi: 10.1186/s12931-014-0132-z. https://doi.org/10.1186/s12931-014-0132-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fuchs K, Hippe A, Schmaus A, Homey B, Sleeman JP, Orian-Rousseau V. Opposing effects of high- and low-molecular weight hyaluronan on CXCL12-induced CXCR4 signaling depend on CD44. Cell Death Dis. 2013;4:e819. doi: 10.1038/cddis.2013.364. https://doi.org/10.1038/cddis.2013.364. [DOI] [PMC free article] [PubMed] [Google Scholar]


