Abstract
The replicative and translational machinery utilizes the unique geometry of canonical G•C and A•T/U Watson-Crick base pairs to discriminate against DNA and RNA mismatches in order to ensure high fidelity replication, transcription, and translation. There is growing evidence that spontaneous errors occur when mismatches adopt a Watson-Crick-like geometry through tautomerization and/or ionization of the bases. Studies employing NMR relaxation dispersion recently showed that wobble dG•dT and rG•rU mismatches in DNA and RNA duplexes transiently form tautomeric and anionic species with probabilities (≈0.01–0.40%) that are in concordance with replicative and translational errors. While computational studies indicate that these exceptionally short-lived and low-abundance species form Watson-Crick like base pairs, their conformation could not be directly deduced from the experimental data, and alternative pairing geometries could not be ruled out. Here, we report direct NMR evidence that the transient tautomeric and anionic species form hydrogenbonded Watson-Crick like base pairs. A guanine-to-inosine substitution, which selectively knocks out a Watson-Crick type (G)N2H2•••O2(T) hydrogen bond, significantly destabilized the transient tautomeric and anionic species, as assessed by lack of any detectable chemical exchange by imino nitrogen rotating frame spin relaxation (R1ρ) experiments. An 15N R1ρ NMR experiments targeting the amino nitrogen of guanine (dG-N2) provides direct evidence for Watson-Crick (G)N2H2•••O2(T) hydrogen bonding in the transient tautomeric state. The strategy presented in this work can be generally applied to examine hydrogen-bonding patterns in nucleic acid transient states including in other tautomeric and anionic species that are believed to play roles in replication and translational errors.
Graphical abstract
In their paper describing the structure of the DNA double helix, Watson and Crick hypothesized that spontaneous mutations could arise when bases adopt their rare energetically less favorable tautomeric forms1 as this could allow mismatches such as dA•dC and dG•dT to pair up in a Watson-Crick (WC) like geometry. Topal and Fresco extended this idea to also encompass a wider range of tautomeric WC-like mismatches that together could account for a broader spectrum of transition and transversion mutations2 as well as miscoding events during translation.3 They also hypothesized that the probability of tautomerization could be an important determinant of misincorporation probability during replication and translation.2,3
Many decades later, it is now well-established that the replicative and translational machinery utilize the unique WC geometry (Figure 1a) to discriminate against mismatches.4–8 In addition, there is considerable evidence that tautomeric9 as well as anionic10 WC-like mismatches play roles in replication2,11–13 and translation errors.3,14,15 WC-like mismatches have been observed in crystal structures of polymerase9,11 and the ribosome8,14,15 in catalytically active conformations. Chemical modifications that stabilize the tautomeric and anionic WC-like species also result in increased misincorporation and base substitution probabilities.10,16–18 Recently, studies based on NMR relaxation dispersion (RD)19–21 targeting the imino nitrogen of guanine (G-N1) and thymidine / uridine (T-N3 / U-N3) in DNA and RNA duplexes provided evidence for spontaneous transitions from wobble (WB) G•T/U mismatches (Figure 1b) toward short-lived and low-populated WC-like tautomeric (Figure 1c) and anionic (Figure 1d) mismatches.22 Consistent with the Topal and Fresco hypothesis, these transitions toward WC-like mismatches occur with probabilities (10−3–10−5) that are comparable to the probabilities of misincorporation.22
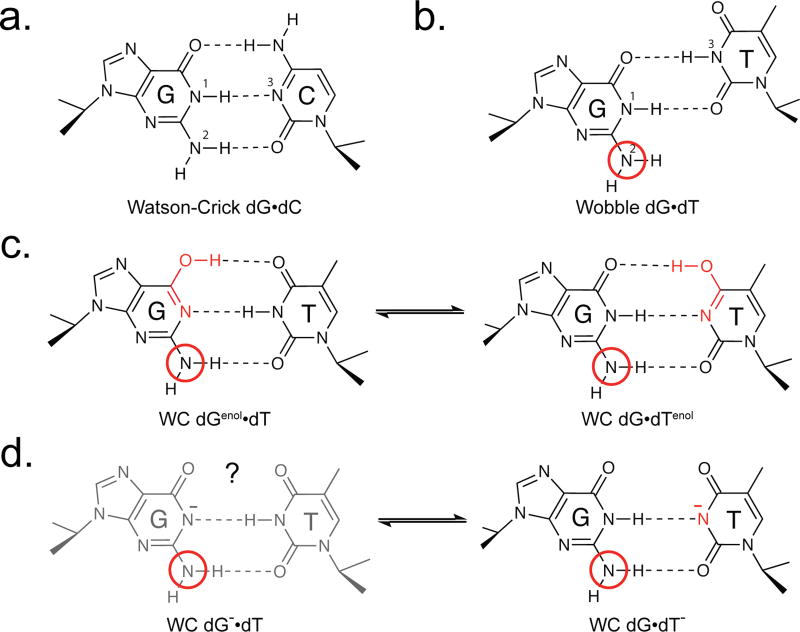
Figure 1.
Wobble and Watson-Crick like dG•dT mismatches (a) Watson-Crick dG•dC base pair. (b) Wobble dG•dT base pair with dG-N2 highlighted with a red circle. (c) Enolic ES1 features a rapid equilibrium between major dGenol•dT and minor dG•dTenol (d) Anionic ES2 dG•dT−. A dynamic equilibrium with a minor dG−•dT that is not detectable by relaxation dispersion cannot be ruled out.22
Although the NMR 15N imino RD data strongly suggest deprotonation of G-N1 or T/U-N3, whether or not the resulting enolic (Genol and Tenol/Uenol) and anionic (T−/U−) bases do indeed adopt a WC-like base pair (bp) stabilized by hydrogen-bonding (H-bonding) remains to be established (Figure 1c/d). Characterizing H-bonds in short-lived low-populated species that form though a subtle rearrangement of protons presents a challenge to existing biophysical methods (Figure 1). Here, we developed two complimentary approaches and obtained evidence for WC-like H-bonding in transient tautomeric and anionic dG•dT mismatches.
The prior 15N NMR RD study22 showed that ground state (GS) dG•dT mismatches exist in dynamic equilibrium with two short-lived and low-abundance species referred to as ‘excited states’ (ESs).19,22–25 The pH-independent ES1 is characterized by significantly downfield shifted dG-N1 (ΔωN1 = ωES1–ωGS ≈ 36–40 p.p.m.) and dT-N3 (ΔωN3 ≈ 9–19 p.p.m.) chemical shifts.11,22 This was interpreted as evidence for deprotonation of dG-N1 and T/U-N3 to form a major Genol and minor T/Uenol tautomeric species that exist in fast exchange on the NMR timescale (Figure 1c). ES2 had a population (pB) that increased with pH and featured a smaller downfield shift in G-N1 (ΔωN1 ≈ 1–10 p.p.m.) and much larger downfield shift in T/U-N3 (ΔωN3 ≈ 45–57). This was interpreted as ionization of T/U-N3 to form anionic T−/U− (Figure 1d).22
While the NMR data did not provide information regarding the conformation of these deprotonated bases, there is indirect evidence that they form WC-like bps (Figure 1c/d). First, the downfield shift in ES1 for the H-bond acceptor nitrogen (N1/3) was smaller than expected based on deprotonating isolated dGTP-N1 and dTTP-N3.22 This could in part be explained by H-bonding between N1 and N3 (Figure 1).22,26 Second, the relative energetic stability of ES1 measured by NMR RD was in good agreement with values computed using density functional theory calculations for H-bonded WC-like tautomeric species.26,27 However, without direct experimental evidence for H-bonding, alternative conformations22 could not be entirely ruled out.
In the WB dG•dT bp, the exocyclic amino group of guanine does not form a H-bond with dT though it may form a relatively weak water-mediated H-bond.28 In contrast, the same amino group is predicted to form an N-H•••O H-bond in both ES1 and ES2 (Figure 1c/d), analogous to that observed in G•C bps (Figure 1a). It correctly positions the purine N3 and pyrimidine O2 H-bond acceptors for sequence-independent recognition by polymerases.29 If the ESs do indeed adopt a WC-like geometry capable of replicative and translational errors, then they are expected to be stabilized by a WC-like N2H2•••O2 H-bond. Omission of the amino group via a guanine to inosine substitution has been shown to have little effect (≈0.1 kcal mol−1) on the stability of the GS dG•dT WB.30 In contrast, by knocking out the WC-like N2H2•••O2 H-bond, it is expected to destabilize ES1 and ES2 by at least ≈1 kcal mol−1, and to diminish their abundance to levels (<0.01%) undetectable by RD NMR (Figure 2a).2,11,22
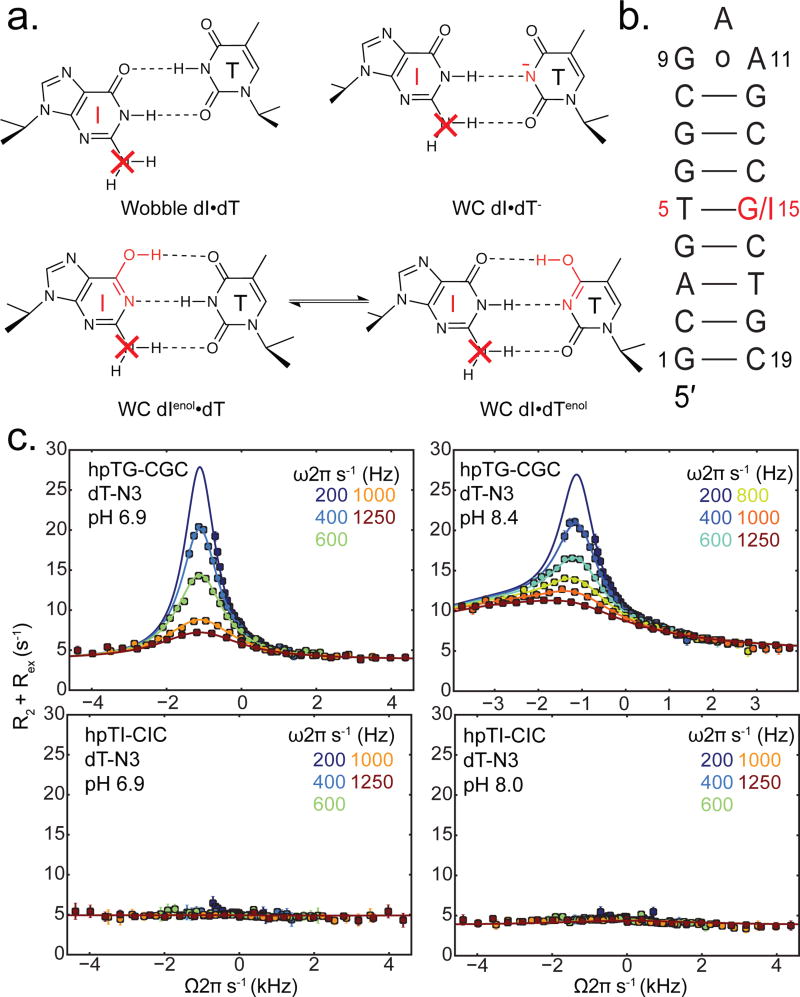
Figure 2.
Guanine (dG) to inosine (dI) substitution suppresses 15N imino dT-N3 relaxation dispersion in dG•dT. (a) Removal of the exocyclic dG-NH2 amino group selectively knocks out a H-bond in the proposed excited state Watson-Crick like mismatches (enolic dT•dIenol, dTenol•dI, and anionic dT•dI) without substantially affecting the H-bonds and stability of ground state wobble (dT•dI). (b) hpTG-CGC DNA hairpin construct used in RD measurements. (c) 15N RD profiles measured for dT-N3 in dT•dG (top) and dT•dI (bottom) at variable pH (inset). Solid lines represent best fit to RD data. Error bars represent experimental uncertainty (one s.d.).
We examined how a guanine (dG) to inosine (dI) substitution (Figure 2a) affects off-resonance R1ρ RD profiles measured for dT-N3 in a dG•dT mismatch embedded in a DNA hairpin (hpTG-CGC) used in prior studies (Figure 2b).22 Comparison of NMR spectra for the two duplexes confirms that the dG to dI substitution has little effect on the GS WB conformation (Figure S1a) consistent with previous studies.30,31 Additionally, inosine is expected to have a similar tautomeric behavior to guanine based on experimental32 and computational studies.33,34 As expected, at pH = 6.9 in which ES2 is suppressed, we observed significant dT-N3 RD in dG•dT consistent with a single ES. Fitting the RD profiles to a two-state model yields exchange parameters (pB = 0.169 ± 0.002% and kex = k1 + k−1 = 2896 ± 96 s−1, ΔωN3 = 18.4 ± 0.12 p.p.m.) that are consistent with the tautomeric ES1 (Figure 2). A second ES (pB = 0.031 ± 0.001%, kex = 17030 ± 1230 s−1, ΔωN3 = 49.4 ± 1.50 p.p.m.) is observed upon increasing the pH to 8.4 that is consistent with anionic ES2 (dG•dT−).22 In sharp contrast, the dT-N3 RD profiles measured for the dI•dT mismatch were flat at both pH 6.9 and 8.0 (Figure 2c) and over a range of temperatures (Figure S2b). These results indicate that a dG to dI substitution significantly destabilizes both the tautomeric ES1 and anionic ES2 by selectively knocking out a Watson-Crick type N2H2•••O2 H-bond as assessed by the loss of detectable chemical exchange at the dT-N3.
To more directly probe the WC-like (G)N2H2•••O2(T) H-bond, we adapted, with minor modifications (Supporting), the selective R1ρ experiment first introduced for proteins35 and then adapted for nucleic acid imino N1/3 nitrogens36 to measure dG-N2 RD (Figure 1b). The selective R1ρ experiment employs Hartmann-Hahn cross polarization37,38 (CP) to selectively excite individual resonances. In dG•dC WC bps, the amino group is involved in a (G)N2H2•••O2(C) H-bond (Figure 1a) and appears as a single NMR resonance with dG-N2 ≈ 72 p.p.m. In contrast, the WB dG•dT amino group is not H-bonded (Figure 1b) and the two equivalent protons appear as a single resonance with dG-N2 ≈ 74–76 p.p.m. Therefore, a transition from the GS WB to a WC-like ES, in which it forms a (G)N2H2•••O2(T) H-bond (Figure 1b), is predicted to induce a significant downfield shift (ΔωG-N2 >2 p.p.m.) (Figure S2a) in the dG-N2 chemical shift which should give rise to measurable 15N RD.
RD measurements targeting amino nitrogens have not yet been reported in nucleic acids. However, the utility of 15N CPMG RD measurements targeting amino NH2 groups in protein side chains has already been established.39 These studies have highlighted additional considerations in RD experiments for AX2 spin systems, including the need to account for potential contributions arising from N-H/N-H dipole-dipole (DDN2Ha/DDN2Hb) cross-correlated cross-relaxation.39,40 Fortunately, because the angle between the two N-H dipoles in amino NH2 groups is ≈120°, these contributions are predicted to be small for the constructs studied here and can safely be ignored.41 This was verified by observation of monoexponential decays and 15N multiplets with approximately 1:2:1 intensity ratios in a coupled HSQC spectrum (Figure S2c/d/S3).
As a negative control, we measured dG-N2 R1ρ in a canonical dG•dC WC bp within a hairpin duplex (Figure S2e) under high pH conditions where chemical exchange due to transient HG bps is suppressed (Figure S2f). As expected, the RD profiles were all flat with no signs of chemical exchange (Figure 3a).
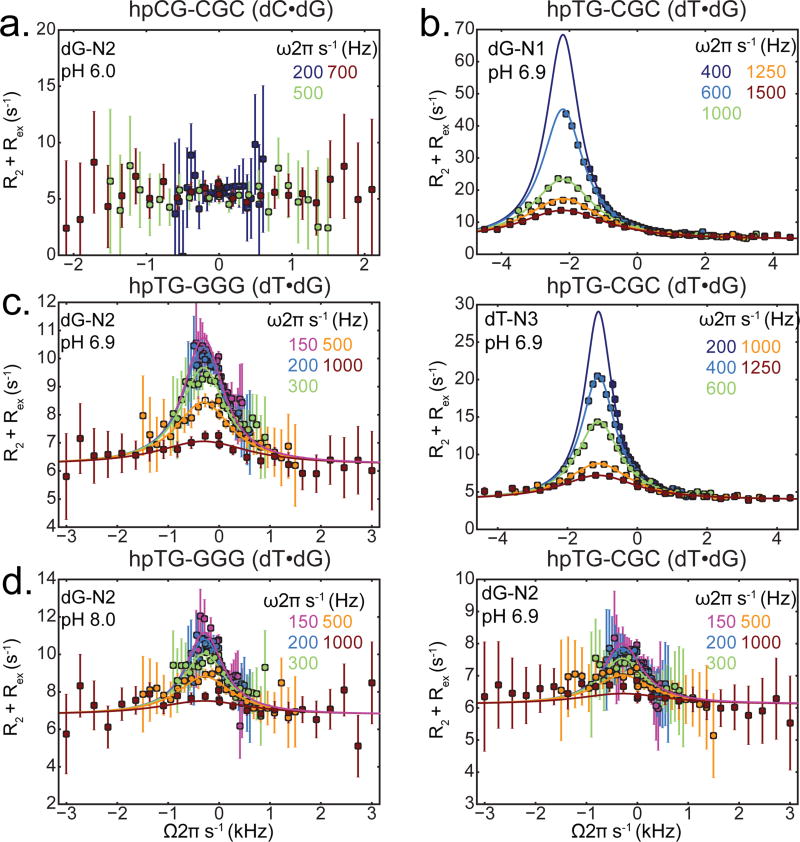
Figure 3.
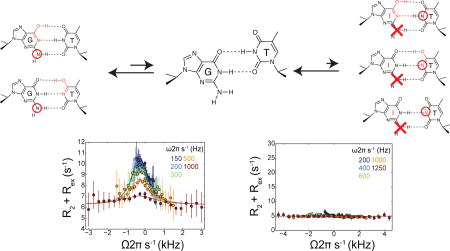
dG-N2 RD profiles. (a) dG-N2 RD profiles measured in a Watson-Crick dG•dC base pair at pH 6.0 and 25°C showing absence of chemical exchange. (b) 15N RD profiles measured in a Wobble dG•dT mismatch for dG-N1, dT-N3, and dG-N2 in hpTG-CGC (see Figure 2b) at pH 6.9 25 °C. Solid line represents a global fit of the N1/N2/N3 RD data with shared populations and exchange rates. dG-N2 RD profiles measured in a Wobble dG•dT mismatch in hpTG-GGG at (c) pH 6.9, (d) pH 8.0. Solid lines represent individual fits of the N2 RD data.
Next, we measured dG-N2 RD in a site-specifically labeled dG•dT mismatch embedded within the hpTG-CGC construct (Figure 2b). Measurements were initially carried out at neutral pH so as to suppress ES2. In contrast to the dG•dC WC bp, we observed clear signs of dG-N2 RD in the dG•dT mismatch. Two-state analysis of the dG-N2 RD profiles (Supporting) yielded exchange parameters (pB = 0.24 ± 0.04% and kex = 2486 ± 208) that are in good agreement with those obtained for ES1 (pB = 0.17 ± 0.002% and kex = 2743 ± 41) based on N1/3 RD measurements.22 Indeed, the RD data measured for dG-N2, dG-N1, and dT-N3 at pH 6.9 could be globally fitted (Figure 3b), confirming that that they are reporting on the same GS-ES1 exchange process.
Importantly, the differences in the dG-N2 chemical shift between the GS and ES (ΔωG-N2 = +4.85 ± 0.19 p.p.m.) deduced by the 2-state analysis of the dG-N2 RD data are in excellent agreement with the experimentally observed differences (ΔωG-N2 ≈ +4 p.p.m.) between the dG-N2 chemical shift in H-bonded WC dG•dC and non-H-bonded WB dG•dT bps in the same sequence context (Figure S2a). We also carried out analogues dG-N2 RD measurements for a dG•dT mismatch in a second unique sequence context and distinct ES1/ES2 exchange parameters (Figure S2g). Once again we observed dG-N2 RD profiles that yield exchange parameters (pB = 0.53 ± 0.037% and kex = 3087 ± 100) that are in good agreement with those obtained for ES1 based on N1/3 RD measurements (pB = 0.37 ± 0.004% and kex = 2797 ± 41). The value of ΔωG-N2 ≈ +5.15 p.p.m. is consistent with H-bonding and in agreement with the experimentally observed differences (ΔωG-N2 ≈ +4 p.p.m.) between the dG-N2 chemical shifts in WC dG•dC and WB dG•dT bps.
dG-N2 is also predicted to be H-bonded in ES2. However, simulations show that due to the much faster GS-ES2 exchange kinetics (kex ≈ 50,000 s−1 as compared to ≈ 2000 s−1 for ES1), the ES2 contribution to dG-N2 RD is expected to be negligible (Figure S4) even at pH = 8.0, in which the population of both ES1 (0.23%) and ES2 (0.22%) are significant.22 In accord with this expectation, we observed insignificant changes in the dG-N2 RD profiles measured in the dG•dT mismatch in hpTG-GGG when increasing to pH 8.0 (Figure 3c/d).
In conclusion, our results provide direct experimental evidence that tautomeric and anionic dG•dT ESs adopt a WC-like geometry that mimics both the shape and H-bond patterns of canonical WC bps, and therefore satisfy the key requirements for inducing replication and translation errors. The methodology described in this work provides a new means for characterizing H-bond alignments and base-pairing patterns in transient nucleic acid conformational states.
Supplementary Material
Acknowledgments
We thank Fabien Ferrage (École normale supérieure, France), Geoffrey Bodenhausen (École normale supérieure, France), Alexander Hansen (The Ohio State University, USA), Lewis Kay (University of Toronto, Canada) and Ananya Majumdar (Johns Hopkins University) for critical input. We also thank members of the Al-Hashimi lab for critical comments and the Duke University Magnetic Resonance Spectroscopy Center for technical assistance. This work was supported by an NIH grant (RO1 GM089846) awarded to H.M.A.
Footnotes
ASSOCIATED CONTENT
Details of sample preparation, NMR experiments, and NMR RD profiles. This material is available free of charge via the Internet at http://pub.acs.org.
No competing financial interests have been declared.
References
- 1.Watson JD, Crick FHC. Nature. 1953;171:964. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 2.Topal MD, Fresco JR. Nature. 1976;263:285. doi: 10.1038/263285a0. [DOI] [PubMed] [Google Scholar]
- 3.Topal MD, Fresco JR. Nature. 1976;263:289. doi: 10.1038/263289a0. [DOI] [PubMed] [Google Scholar]
- 4.Kunkel TA. Cold Spring Harb. Symp. Quat. Biol. 2009;74:91. doi: 10.1101/sqb.2009.74.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freudenthal BD, Beard WA, Shock DD, Wilson SH. Cell. 2013;154:157. doi: 10.1016/j.cell.2013.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koag MC, Nam K, Lee S. Nucleic Acids Res. 2014;17:11233. doi: 10.1093/nar/gku789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westhof E, Yusupov M, Yusupova G. F1000 Prime Rep. 2014;6:19. doi: 10.12703/P6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. Nature. 2012;484:256. doi: 10.1038/nature10913. [DOI] [PubMed] [Google Scholar]
- 9.Wang W, Hellings HW, Beese LS. Proc. Natl. Acad. Sci. U.S.A. 2011;108:17644. doi: 10.1073/pnas.1114496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu H, Eritja R, Bloom LB, Goodman MF. J. Biol. Chem. 1993;268:15935. [PubMed] [Google Scholar]
- 11.Bebenek K, Pederson LC, Kunkel TA. Proc. Natl. Acad. Sci. U.S.A. 2011;108:1862. doi: 10.1073/pnas.1012825108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris VH, Smith CL, Cummins WJ, Hamilton AL, Adams H, Dickman M, Hornby DP, Williams DM. J. Mol. Biol. 2003;326:1389. doi: 10.1016/s0022-2836(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 13.Freudenthal BD, Beard WA, Cuneo MJ, Dyrkheeva NS, Wilson SH. Nat. Struct. Mol. Biol. 2015;22:924. doi: 10.1038/nsmb.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Cell. 2002;326:1389. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 15.Rozov A, Demeshkina N, Westhof E, Yusupov M, Yusupova G. Nat. Commun. 2015;6:7251. doi: 10.1038/ncomms8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warren JJ, Forsberg LJ, Beese LS. Proc. Natl. Acad. Sci. U.S.A. 2006;103:19701. doi: 10.1073/pnas.0609580103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rozov A, Demeshkina N, Khusainov I, Westhod E, Yusupov M, Yusupova G. Nat Commun. 2016;7:10457. doi: 10.1038/ncomms10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weixlbaumer A, Murphy FV, 4th, Dziergowska A, Malkiewicz A, Vendeix FA, Agris PF, Ramakrishnan V. Nat. Struct. Mol. Biol. 2007;14:498. doi: 10.1038/nsmb1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao B, Zhang Q. Curr. Opin. Struct. Biol. 2015;30:134. doi: 10.1016/j.sbi.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen AL, Nikolova EN, Casiano-Negroni A, Al-Hashimi HM. J. Am. Chem. Soc. 2009;131:3818. doi: 10.1021/ja8091399. [DOI] [PubMed] [Google Scholar]
- 21.Palmer AG, III, Massi F. Chem. Rev. 2006;106:1700. doi: 10.1021/cr0404287. [DOI] [PubMed] [Google Scholar]
- 22.Kimsey IJ, Petzold K, Sathyamoorthy B, Stein ZW, Al-Hashimi HM. Nature. 2015;519:315. doi: 10.1038/nature14227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mulder FA, Mittermaier A, Hon B, Dahlquist FW, Kay LE. Nat. Struct. Biol. 2001;8:932. doi: 10.1038/nsb1101-932. [DOI] [PubMed] [Google Scholar]
- 24.Dethoff EA, Petzold K, Chugh J, Casiano-Negroni A, Al-Hashimi HM. Nature. 2012;491:724. doi: 10.1038/nature11498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikolova EN, Kim E, Wise AA, O’Brien PJ, Andricioaei I, Al-Hashimi HM. Nature. 2011;470:498. doi: 10.1038/nature09775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nomura K, Hoshino R, Shimizu E, Hoshiba Y, Danilov VI, Kurita N. J. Mod. Phys. 2013;04:422. [Google Scholar]
- 27.Brovarets OO, Hovorun DM. J. Biomol Struct. Dyn. 2015;33:2297. doi: 10.1080/07391102.2015.1046936. [DOI] [PubMed] [Google Scholar]
- 28.Ho PS, Frederick CA, Quigley GJ, van der Marel GA, van Boom JH, Wang AH, Rich A. EMBO J. 1985;4:3617. doi: 10.1002/j.1460-2075.1985.tb04125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiefer JR, Mao C, Braman JC, Beese LS. Nature. 1998;391:304. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- 30.Watkins NE, SantaLucia J. Nucleic Acids Res. 2005;33:6258. doi: 10.1093/nar/gki918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cruse WB, Aymani J, Kennard O, Brown T, Jack AG, Leonard GA. Nucleic Acid Res. 1989;17:55. doi: 10.1093/nar/17.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramaswamy S, Evens FE. J. Mol. Biol. 1989;89:249. [Google Scholar]
- 33.Brovarets OO, Hovorun DM. J. Biomol. Struct. Dyn. 2013;31:913. doi: 10.1080/07391102.2012.715041. [DOI] [PubMed] [Google Scholar]
- 34.Costas ME, Acevedo-Chávez R. J. Solution Chem. 2012;41:864. [Google Scholar]
- 35.Korzhnev DM, Orekhov VY, Kay LE. J. Am. Chem. Soc. 2005;127:713. doi: 10.1021/ja0446855. [DOI] [PubMed] [Google Scholar]
- 36.Nikolova EN, Gottardo FL, Al-Hashimi HM. J. Am. Chem. Soc. 2012;134:3667. doi: 10.1021/ja2117816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiarparin E, Pelupessy P, Bodenhausen G. Mol. Phys. 1998;95:759. [Google Scholar]
- 38.Pelupessy P, Chiarparin E. Concepts Magn. Reson. 2000;12:103. [Google Scholar]
- 39.Mulder FAA, Skrynnikov NR, Hon B, Dahlquist FW, Kay LE. J. Am. Chem. Soc. 2001;123:967. doi: 10.1021/ja003447g. [DOI] [PubMed] [Google Scholar]
- 40.Skrynnikov NR, Mulder FAA, Hon B, Dahlquist FW, Kay LE. J. Am. Chem. Soc. 2001;123:4556. doi: 10.1021/ja004179p. [DOI] [PubMed] [Google Scholar]
- 41.Pelupessy P, Ferrage F, Bodenhausen G. J. Chem. Phys. 2007;126:134508. doi: 10.1063/1.2715583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.