Abstract
Pirfenidone recently received FDA approval as one of the first two drugs designed to treat idiopathic pulmonary fibrosis. While the clinical data continues to support the efficacy of pirfenidone, the specific molecular mechanism of action of this drug has not been fully defined. From a chemical perspective the comparatively simple and lipophilic structure of pirfenidone combined with its administration at high doses, both experimentally and clinically, complicates some of the basic tenants of drug action and drug design. Our objective here was to identify a commercially available structural mimic of pirfenidone which retains key aspects of its physical chemical properties but does not display any of its antifibrotic effects. We tested these molecules using lung fibroblasts derived from patients with idiopathic pulmonary fibrosis and found phenylpyrrolidine based analogs of pirfenidone that were non-toxic and lacked antifibrotic activity even when applied at millimolar concentrations. Based on our findings, these molecules represent pharmacological tools for future studies delineating pirfenidone’s mechanism of action.
Keywords: Pirfenidone, Fibrosis, Antifibrotic, IPF, Idiopathic Pulmonary Fibrosis, Interstitial Lung Disease, p38
1. INTRODUCTION
Idiopathic pulmonary fibrosis (IPF) is a chronic disease characteristically defined by excessive deposition of extracellular matrix and destruction of the lung’s normal architecture. Over time this progressive scarring and alteration in tissue leads to declining lung function and often death. Despite the scope and severity of the disease burden associated with IPF, it has only recently become the target of clinically approved therapeutics, with the approval of nintedanib and pirfenidone. Interest in the latter began with the finding that phenylamine derivative substituted pyridones produce analgesic properties (Scudi et al., 1960), leading to the synthesis of 5-Methyl-1-phenyl-2-(1H)-pyridone (initially referred to as AMR-69, later referred to as pirfenidone) and description of its analgesic, antipyretic, and anti-inflammatory effects (Shreekrishna Manmohan Gadekar, 1972). Decade’s later the antifibrotic properties of pirfenidone began to emerge (Iyer et al., 1995; Kehrer and Margolin, 1997; Schelegle et al., 1997).
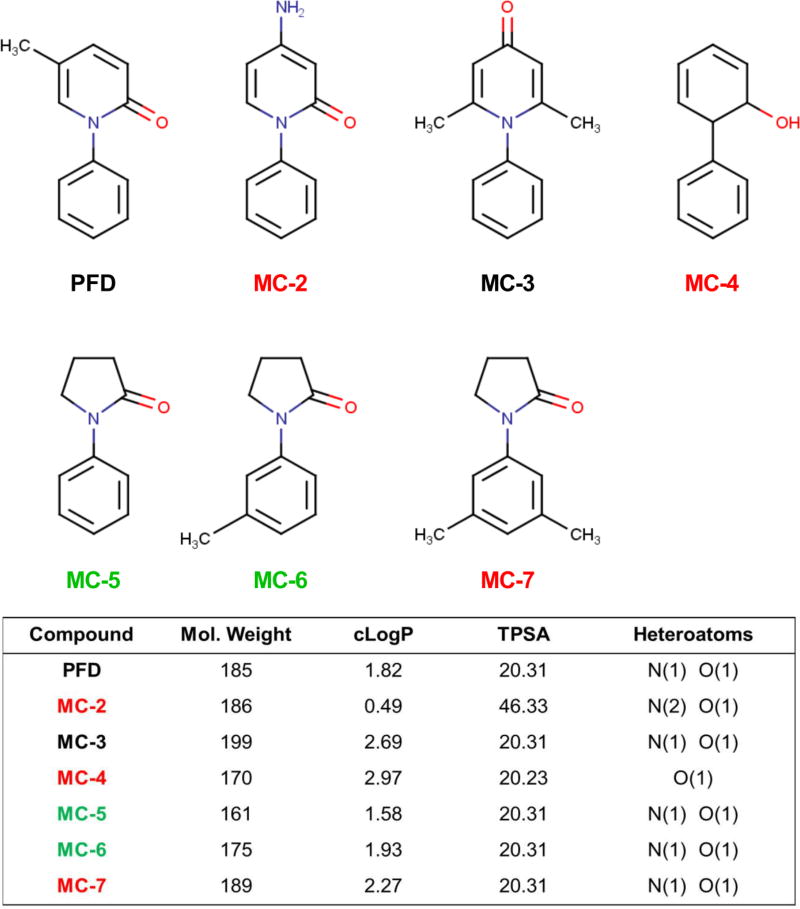
The molecular mechanism of action of pirfenidone is still not well defined. In pulmonary fibrosis, activated resident fibroblasts are thought to be the main cellular contributor to matrix deposition and scarring (Barkauskas and Noble, 2014). In an effort to identify the mechanism of action of pirfenidone, its in vitro antifibrotic efficacy was established in isolated fibroblasts (Conte et al., 2014; Hewitson et al., 2001; Lehtonen et al., 2016a). A major limitation of pirfenidone is the compound’s potency. In order to produce antifibrotic activity in vitro, concentrations between 1–5mM are required (Conte et al., 2014; Lehtonen et al., 2016b; Nakayama et al., 2008). Pirfenidone’s lack of potency, lipophilic nature, and minimal number of heteroatoms (Fig. 1) has led to debate about the potential for pirfenidone to bind to a selective target or pocket, and has stimulated an alternative hypothesis that pirfenidone simply works as a free radical scavenging antioxidant (Mitani et al., 2008; Salazar-Montes et al., 2008). Some very recent work has suggested pirfenidone is an inhibitor of the p38 mitogen-activated protein kinase (p38 MAPK) pathway (Li et al., 2016; Ma et al., 2014; Neri et al., 2016; Yin et al., 2016). A major barrier to further delineating pirfenidone’s mechanism of action is the absence of structurally similar compounds which lack antifibrotic efficacy. Inactive structural analogs are extremely valuable tools when attempting to further define the mechanism of action or molecular target of small molecules (Dancy et al., 2012; Fu et al., 2008; Lim et al., 2004).
Fig. 1.
Pirfenidone (PFD) and structural mimics: physical chemical properties. Molecular weight (Mol. Weight), calculated logarithm of the partition coefficient (cLogP), topological polar surface area (tPSA), and the non-carbon heteroatoms are compared. Based on the data presented here the compounds are color-coded to summarize whether they were antifibrotic (black), acutely toxic (red) or inactive structural mimics of pirfenidone (green).
We set out to identify a chemical mimic of pirfenidone which retains its low molecular weight and lipophilic nature but does not display any antifibrotic activity against pulmonary fibroblasts. Our criteria for an inactive mimic thus included commercial availability and non-toxicity at the very high doses (millimolar) at which pirfenidone shows antifibrotic activity. For our initial testing we identified a diverse pilot set of compounds with physical chemical properties consistent with pirfenidone (To ease communication the compounds are abbreviated MC-2 through MC-7) (Fig. 1). We tested these compounds in primary human pulmonary fibroblasts obtained from patients with IPF. As a proof of concept demonstrating the utility of these compounds, we used the inactive analogs identified here to address the role of p38 MAPK in mediating antifibrotic effects of pirfenidone in cultured lung fibroblasts.
2. MATERIALS AND METHODS
2.1. Compounds
Pirfenidone (PFD) (PubChem CID:40632) was purchased from Sigma Aldrich, St. Louis, MO. 4-amino-1-phenyl-1,2-dihydropyridin-2-one (MC-2) (PubChem CID:664174); 4,6-dimethyl-1-phenyl-1,2-dihydropyridin-2-one (MC-3) (PubChem CID:N/A); 2-phenylphenol (MC-4) (PubChem CID: 24938931); 1-phenylpyrrolidin-2-one (MC-5) (PubChem CID: 78375); 1-(3-methylphenyl)pyrrolidin-2-one (MC-6) (PubChem CID: 314213); and 1-(3,5-dimethylphenyl)pyrrolidin-2-one (MC-7) (PubChem CID: 847897) were purchased from MolPort, Riga, Latvia. All compounds were dissolved in DMSO for a stock concentration of 600mM. TPSA and cLogP were calculated using the OSIRIS Property Explorer available through the Organic Chemistry Portal: http://www.organic-chemistry.org/prog/peo/.
2.2. Cell Culture
Primary human lung fibroblasts (generously provided by Peter Bitterman and Craig Henke at the University of Minnesota) were isolated by explant culture from the lungs of subjects diagnosed with IPF who underwent lung transplantation, under a protocol approved by the University of Minnesota Institutional Review Board. Primary fibroblasts were maintained in EMEM (ATCC, Manassas, VA) containing 10% FBS, unless otherwise noted. All primary cell culture experiments were performed with cells at passage six or less.
2.3. Viability Assay
Lung fibroblasts were plated into 96-well plates in EMEM containing 10% FBS and allowed to attach for 6 h. Media was then exchanged with EMEM containing 0.1% FBS + the indicated compound concentration. All wells were treated with a final concentration of 0.5% DMSO (We have previously found these cells tolerate up to 1% DMSO without any effects on viability). After 24 h the cellular viability/toxicity was analyzed using the WST-1 reagent (Sigma Aldrich) following the manufacturer’s protocol. Results are expressed as the absorbance at 450nm relative to DMSO control.
2.4. Immuno-ECM Assay
Adapting from previously published methods (Jones et al., 2010; Vogel et al., 2014), lung fibroblasts were plated to confluence in clear-bottom 96-well plates in EMEM containing 10% FBS and allowed to attach for 6 h. Media was then exchanged with EMEM containing 0.1% FBS plus the indicated compound concentration. All wells were treated with a final concentration of 0.5% DMSO. After 72 h cells were fixed with 4% formalin, then treated with 0.25% Triton X-100 and blocked with Li-Cor Odyssey Blocking Buffer (Li-Cor Biosciences, Lincoln, NE) before overnight incubation in a polyclonal rabbit antibody for collagen I (Novus NB600-408) diluted 1:150 in blocking buffer. Wells where then incubated with IR-dye-conjugated secondary antibody (Li-Cor #926-32211) diluted 1:500. Plates were imaged via a Li-Cor OdysseyXL system with quantification performed via densitometry. Results are represented as collagen I signal intensity relative to DMSO control.
2.5. qPCR Analysis
Cells were plated and treated as indicated prior to RNA isolation using RNeasy Plus Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Isolated RNA (250ng) was then used to synthesize cDNA using SuperScript VILO (Invitrogen, Carlsbad, CA). Quantitative PCR was performed using FastStart Essential DNA Green Master (Roche Applied Science, Penzberg, Germany) and analyzed using a LightCycler 96 (Roche Applied Science). Results are expressed as a fold change by ΔΔCt relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Primers: GAPDH; F: GGAAGGGCTCATGACCACAG, R: ACAGTCTTCTGGGTGGCAGTG. ACTA2; F: GTGAAGAAGAGGACAGCACTG, R: CCCATTCCCACCATC ACC. COL1A1; F: AAGGGACACAGAGGTTTCAGTGG, R: CAGCACCAGTAGCACCATCATTTC. COL1A2; F: CTTGCAGTAACCTTATGCCTAGCA, R:CCCATCTAACCTCTCTACCCAGTCT
2.6. Western Blot Analysis
Cells were plated in EMEM containing 10% FBS and allowed to attach for 6 h. Cells were then starved for 24 h in EMEM containing 0.1% FBS prior to stimulation with 2ng/mL TGFβ with or without the indicated concentration of compound for 30 min. Total protein was isolated using RIPA buffer (Thermo Scientific, Rockford, IL) and quantified with a BCA protein assay kit (Thermo Scientific, Rockford, IL) according to manufacturer’s instructions. Membranes were probed for phospho-p38 (Cell Signaling, Danvers, MA #9211) and total p38 (Cell Signaling, Danvers, MA #9212) followed by IR conjugated secondary antibodies (Li-Cor Biosciences, Lincoln, NE). Bands were visualized and quantified using a Li-Cor OdysseyXL.
2.7. Statistical Analysis
Groups were compared by one-way ANOVA with Tukey’s multiple comparison’s test. All statistical tests were carried out using GraphPad Prism 6 with statistical significance defined as p < 0.05. Results are expressed throughout as the mean ± standard error of the mean (S.E.M.).
3. RESULTS
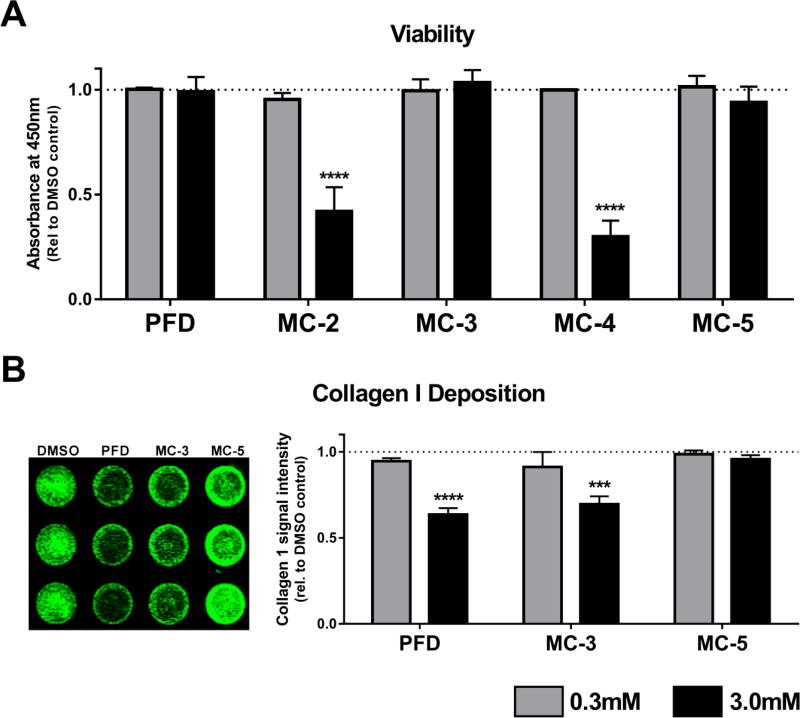
We began this campaign through molport.com searching for compounds which have a similarity of 0.65 or greater (Tanimoto metrics) to PFD. (Fig. 1). Compounds MC-2 and MC-3 retain the phenylpyridine core structure of PFD with moderate substitutions, and the phenylphenol, MC-4 and the phenylpyrrolidine MC-5 offer alternative backbones but retain the basic size and atom representation of PFD. Previous work has shown that pirfenidone can be given at millimolar concentrations in vitro without causing any cellular toxicity (Conte et al., 2014; Nakayama et al., 2008). To screen out any toxic compounds we treated IPF lung fibroblasts with 0.3 and 3.0mM concentrations of each compound for 24 h and then measured cellular viability with WST-1 reagent, a tetrazolium salt that is cleaved into a formazan dye in metabolically active, viable cells (Fig. 2A). Consistent with prior observations, PFD did not reduce cellular viability. However 3.0mM treatment with MC-2 and MC-4 was toxic to the cells. At 3.0mM MC-4 became insoluable and fell out of solution, likely due to the lipopholic nature of this compound (cLogP = 2.97).
Fig. 2.
Effects of high concentration pirfenidone and initial test compounds on cellular viability and collagen I protein deposition. A. Toxicity. Pulmonary fibroblasts derived from patients with IPF were treated for 24 h at 0.3 and 3.0mM to initially rule out any acutely toxic compounds. Data represent the mean (±S.E.M.) from three patient samples each run in duplicate (**** P < 0.0001 vs. DMSO control). B. Collagen I Deposition. One of the hallmarks of pulmonary fibrosis is activated fibroblasts which synthesize excessive extracellular matrix proteins; principally collagen I. Pulmonary fibroblasts derived from patients with IPF were treated for 72 h with PFD, MC-3, and MC-5 at 0.3 and 3.0mM to measure the effect on collagen deposition. Shown on the left is a representative image from one patient sample run in triplicate for each compound at 3.0mM. Data represent the mean (±S.E.M.) from three patient samples each run in triplicate (*** P < 0.001, **** P < 0.0001 vs. DMSO control). Pirfenidone was not acutely toxic to the cells but did reduce matrix deposition; in contrast MC-5 did not affect the cells in either assay.
In previous in vitro work (Conte et al., 2014) with pulmonary fibroblasts PFD has been shown to reduce gene expression of COL1A1 (which codes for collagen 1α1 protein, a major fibrillar collagen component in the extracellular matrix). To translate this readout into a higher-througput assay we measured collagen I protein levels using an in-cell Western blot technique. In this experiment we plated IPF lung fibroblasts at confluence in a 96-well plate and allowed them to synthesize and deposit matrix for 72 h in the presense of PFD, MC-3, MC-5 (non-toxic from Fig. 2A) or DMSO control. We then immunostained the whole well for collagen I protein and visualized and quantitatively analyzed using infrared-emitting secondary antibody. PFD reduced collagen protein levels at the higher concentration, and interestingly MC-3 appeared to perform as an active structural mimic of PFD, as it was both non-toxic to the fibroblasts and reproduced the antifibrotic phenotype of reducing collagen deposition. The phenylpyrrolidine-based backbone MC-5 appeared to be a good framework to move forward as an inactive structural mimic of pirfenidone based on the combination of low toxicity and absence of antifibrotic effect on collagen 1 expression.
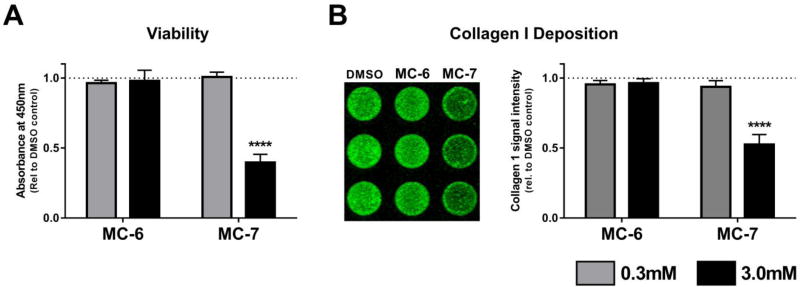
Based on the results from Fig. 2 we identified a second round of commercially availible pirfenidone mimics based off of the phenylpyrrolidine backbone. Aside from the number of hydrogens; MC-5 and PFD only differed by two carbon atoms so we searched for phenylpyrridone anologs with aditional carbon substitutions. We also reasoned that inactivity would be associated with compounds which were substituted on the phenyl ring rather than the pyridine as in PFD. Methyl, MC-6 and dimethyl, MC-7 derivatives of PFD were thus obtained and tested in the same assays as in Fig. 2. Interstingly, the dimethyl substituted analog, MC-7 was toxic to the cells (and also reduced collagen deposition), whereas MC-6, with a single methyl substitution onto MC-5, was both non-toxic and inactive in the collagen deposition assay (Fig. 3).
Fig. 3.
Methyl substituted phenylpyrrolidines effect on cellular viability and collagen I protein deposition. A. Toxicity. Pulmonary fibroblasts derived from patients with IPF were treated for 24 h with MC-6 and MC-7 at 0.3 and 3.0mM to measure acute toxicity. Data represent the mean (±S.E.M.) from three patient samples each run in duplicate (**** P < 0.0001 vs. DMSO control). B. Collagen I Deposition. Pulmonary fibroblasts derived from patients with IPF were treated for 72 h with MC-6 and MC-7 at 0.3 and 3.0mM to measure the effect on collagen deposition. Shown on the left is a representative image from one patient sample run in triplicate for each compound at 3.0mM. Data represent the mean (±S.E.M.) from three patient samples each run in triplicate (**** P < 0.0001 vs. DMSO control). Similar to MC-5 in Fig. 2, MC-6 is nontoxic and has no effect on collagen deposition.
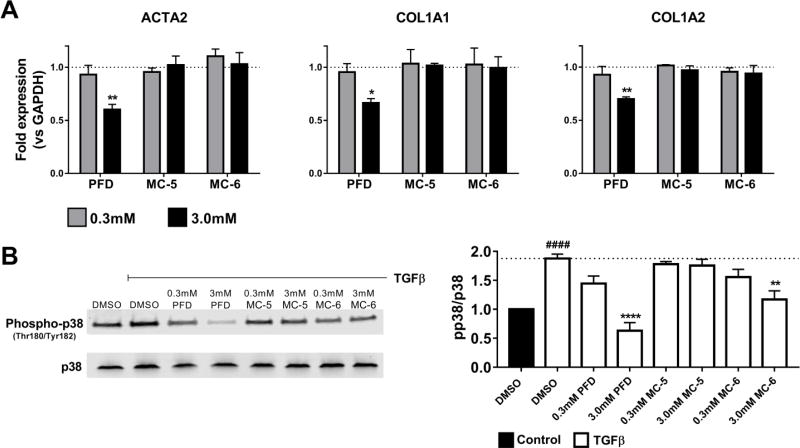
For our final analysis testing MC-5 and MC-6 against pirfenidone we measured expression of three fibrosis-associated genes: ACTA2 (coding for α-smooth muscle actin, a hallmark of activated and contractile myofibroblasts), and COL1A1 and COL1A2 (both genes required for mature collagen I protein). IPF lung fibroblasts were treated for 48 h with compounds or DMSO control and gene expression was determined by qPCR relative to the housekeeping gene GAPDH. As predicted, pirfenidone reduced expression of all three profibrotic genes at the higher concetration; in contrast, MC-5 and MC-6 exhibited no effect in this highly sensitive assay (Fig. 4A).
Fig. 4.
Pirfenidone and phenylpyrrolidine mimics effects on profibrotic gene expression and inhibition of p38 phosphorylation. A. IPF lung fibroblasts were treated for 48 h with PFD, MC-5, and MC-6 at 0.3 and 3.0 mM, followed by RNA isolation and qPCR analysis. Data represent the mean (±S.E.M.) from three patient samples (* P < 0.05, ** P < 0.01 vs. DMSO control). The approved antifibrotic drug pirfenidone reduced expression of all three genes but the inactive structural mimics had no effect. B. IPF lung fibroblasts were starved for 24 h in media containing 0.1% FBS and then stimulated with 2ng/mL TGFβ +/− PFD, MC-5, and MC-6 for 30 min. A representative blot from a single patient sample is shown on the left, while the plot on the right represents the mean (±S.E.M.) from three patient samples each run in triplicate (#### P < 0.0001 vs. DMSO control –TGFβ, *** P < 0.001, **** P < 0.0001 vs. DMSO control +TGFβ).
In response to recent evidence suggesting p38 MAPK pathway inhibition is the mechanism of action of pirfenidone (Li et al., 2016; Ma et al., 2014; Neri et al., 2016), we stimulated IPF lung fibroblasts with TGFβ for 30 min in the presence of pirfenidone and our inactive mimics (Fig. 4B). TGFβ has been shown to induce p38 phosphorylation through multiple pathways (Yu et al., 2002; Zhang, 2009), and consistent with these prior publications we observed a 2-fold induction in phospho-p38 under these conditions. As previosuly reported, pirfenidone was able to significantly inhibit this TGFβ-induced p38 activation. However, while one of the inactive structural mimics, MC-5 showed no effect on inhibitng p38 phosphorylation, the other MC-6 did significantly inhibit p38 at a concentration which showed no antifibrotic activity (Fig. 3 and Fig. 4), consistent with pirfenidone acting through additional or alternative mechanisms beyond p38 in mediating its antifibrotic effects in fibroblasts.
4. DISCUSSION
Approved in 2014 for the treatment of idiopathic pulmonary fibrosis, pirfenidone became one of the first in a class of new “antifibrotic drugs” which will likely expand in the future. Multiple groups are currently attempting to understand the mechanism of action that leads to the antifibrotic efficacy of pirfenidone. One of the basic challenges in interpreting pirfenidone’s clinical efficacy is the dosage of the drug required to produce any biological activity. We set out to identify one or several structural mimics of pirfenidone that can be used as a negative control which accounts for the amount of lipophilic compound delivered to the cells. Taken together, our data demonstrate that phenylpyrrolidine backbones produce adequate structural mimics of pirfenidone. Our results are also consistent with the concept that pirfenidone binds its antifibrotic target(s) in a specific manner that is highly dependent on the 3-dimensional planar shape of pirfenidone or potentially the 5-methyl substitution on the pyridine ring, not present in MC-5 and MC-6. Interestingly, F-351, a hydroxyl substituted analog of pirfenidone, is currently in clinical trials for liver and kidney fibrosis (Nanthakumar et al., 2015). This modification results in more “drug like” physical chemical properties than pirfenidone and retains the antifibrotic nature of the drug.
MC-5 and MC-6 are available from molport.com (MolPort-001-012-470 and MolPort-001-028-037) through multiple venders for less than $10/mg, making them very affordable, and based on their structural similarity to pirfenidone (Fig. 1) and inactivity in cellular assays of fibrosis (Fig. 2–4), should be considered as potentially informative inactive structural mimics for pirfenidone. The use of phenotype-based assays in drug discovery is continuing to expand, leading to the identification of promising drug candidates with no known mechanism of action or molecular target (Etzion and Muslin, 2009; Lee and Berg, 2013; Moffat et al., 2014). One of the common approaches to identifying or validating the molecular target of a small molecule is to design linkable chemical analogs to perform pull-down assays followed by proteomics analysis (Leslie and Hergenrother, 2008). In these types of assays a pull-down experiment can be performed using both the active and inactive compounds. Inevitably, highly abundant or adherent proteins will be observed in both cases, and only by comparing which protein(s) are exclusively associated with the active molecule can specific targets be identified (Bell et al., 2013; Colca et al., 2003). Even with very potent drugs nonspecific binding is always a concern when interpreting the effects of adding a compound to a biological system. In the case of pirfenidone where millimolar concentrations are required, these concerns are amplified. In recent years the growth of biologics based therapeutics has created a new standard for preclinical drug discovery where the use of inactive control molecules is customary. Antibody based therapies are compared to an isotype control, siRNA treatments are compared with scrambled nontargeting sequences and peptides are compared to an inactive derivative sequence. Although less exciting in the short term, inactive analogs play an important role in small molecule based drug discovery.
It is important to note that although we have shown MC-5 and MC-6 are inactive in fibrosis assays relative to pirfenidone, no small molecule is absolutely inactive. MC-5 and MC-6 are both in the PubChem database but only MC-5 has been tested in biological assays; reassuringly this compound showed no activity in these screens. The interesting results we observed in the p38 MAPK phosphorylation experiments serves as proof of concept of their utility, and suggest that further work is needed to define the role of p38 MAPK as a potential mechanism of action for pirfenidone antifibrotic effects in fibroblasts. The identification and characterization of these compounds should help facilitate further delineation of the molecular mechanism of action of pirfenidone, and ultimately lead to more potent and more efficacious antifibrotic drugs in the future.
Acknowledgments
This work was supported by NIH HL092961 and HL113796 (DJT), American Lung Association Senior Research Training Fellowship (AJH), and funds from the Caerus Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST:
There is no conflict of interest
References
- Barkauskas CE, Noble PW. Cellular Mechanisms of Tissue Fibrosis. 7. New insights into the cellular mechanisms of pulmonary fibrosis. Am J Physiol-Cell Ph. 2014;306:C987–C996. doi: 10.1152/ajpcell.00321.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JL, Haak AJ, Wade SM, Sun YH, Neubig RR, Larsen SD. Design and synthesis of tag-free photoprobes for the identification of the molecular target for CCG-1423, a novel inhibitor of the Rho/MKL1/SRF signaling pathway. Beilstein J Org Chem. 2013;9:966–973. doi: 10.3762/bjoc.9.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colca JR, McDonald WG, Waldon DJ, Thomasco LM, Gadwood RC, Lund ET, Cavey GS, Mathews WR, Adams LD, Cecil ET, Pearson JD, Bock JH, Mott JE, Shinabarger DL, Xiong LQ, Mankin AS. Cross-linking in the living cell locates the site of action of oxazolidinone antibiotics. J Biol Chem. 2003;278:21972–21979. doi: 10.1074/jbc.M302109200. [DOI] [PubMed] [Google Scholar]
- Conte E, Gili E, Fagone E, Fruciano M, Iemmolo M, Vancheri C. Effect of pirfenidone on proliferation, TGF-beta-induced myofibroblast differentiation and fibrogenic activity of primary human lung fibroblasts. Eur J Pharm Sci. 2014;58:13–19. doi: 10.1016/j.ejps.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Dancy BM, Crump NT, Peterson DJ, Mukherjee C, Bowers EM, Ahn YH, Yoshida M, Zhang J, Mahadevan LC, Meyers DJ, Boeke JD, Cole PA. Live-Cell Studies of p300/CBP Histone Acetyltransferase Activity and Inhibition. Chembiochem. 2012;13:2113–2121. doi: 10.1002/cbic.201200381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzion Y, Muslin AJ. The Application of Phenotypic High-Throughput Screening Techniques to Cardiovascular Research. Trends Cardiovas Med. 2009;19:207–212. doi: 10.1016/j.tcm.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Han J, Ishola T, Scerbo M, Adwanikar H, Ramsey C, Neugebauer V. PKA and ERK, but not PKC, in the amygdala contribute to pain-related synaptic plasticity and behavior. Mol Pain. 2008;4 doi: 10.1186/1744-8069-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitson TD, Kelynack KJ, Tait MG, Martic M, Jones CL, Margolin SB, Becker GJ. Pirfenidone reduces in vitro rat renal fibroblast activation and mitogenesis. J Nephrol. 2001;14:453–460. [PubMed] [Google Scholar]
- Iyer SN, Wild JS, Schiedt MJ, Hyde DM, Margolin SB, Giri SN. Dietary intake of pirfenidone ameliorates bleomycin-induced lung fibrosis in hamsters. J Lab Clin Med. 1995;125:779–785. [PubMed] [Google Scholar]
- Jones B, Bucks C, Wilkinson P, Pratta M, Farrell F, Sivakumar P. Development of cellbased immunoassays to measure type I collagen in cultured fibroblasts. Int J Biochem Cell Biol. 2010;42:1808–1815. doi: 10.1016/j.biocel.2010.07.011. [DOI] [PubMed] [Google Scholar]
- Kehrer JP, Margolin SB. Pirfenidone diminishes cyclophosphamide-induced lung fibrosis in mice. Toxicol Lett. 1997;90:125–132. doi: 10.1016/s0378-4274(96)03845-3. [DOI] [PubMed] [Google Scholar]
- Lee JA, Berg EL. Neoclassic Drug Discovery: The Case for Lead Generation Using Phenotypic and Functional Approaches. J Biomol Screen. 2013;18:1143–1155. doi: 10.1177/1087057113506118. [DOI] [PubMed] [Google Scholar]
- Lehtonen ST, Veijola A, Karvonen H, Lappi-Blanco E, Sormunen R, Korpela S, Zagai U, Skold MC, Kaarteenaho R. Pirfenidone and nintedanib modulate properties of fibroblasts and myofibroblasts in idiopathic pulmonary fibrosis. Respir Res. 2016a;17 doi: 10.1186/s12931-016-0328-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtonen ST, Veijola A, Karvonen H, Lappi-Blanco E, Sormunen R, Korpela S, Zagai U, Skold MC, Kaarteenaho R. Pirfenidone and nintedanib modulate properties of fibroblasts and myofibroblasts in idiopathic pulmonary fibrosis. Respir Res. 2016b;17:14. doi: 10.1186/s12931-016-0328-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie BJ, Hergenrother PJ. Identification of the cellular targets of bioactive small organic molecules using affinity reagents. Chem Soc Rev. 2008;37:1347–1360. doi: 10.1039/b702942j. [DOI] [PubMed] [Google Scholar]
- Li Z, Liu X, Wang B, Nie Y, Wen J, Wang Q, Gu C. Pirfenidone suppresses MAPK signaling pathway to reverse epithelial-mesenchymal transition and renal fibrosis. Nephrology (Carlton) 2016 doi: 10.1111/nep.12831. [DOI] [PubMed] [Google Scholar]
- Lim S, Kang KW, Park SY, Kim SI, Choi YS, Kim ND, Lee KU, Lee HK, Pak YK. Inhibition of lipopolysaccharide-induced inducible nitric oxide synthase expression by a novel compound, mercaptopyrazine, through suppression of nuclear factor-kappaB binding to DNA. Biochem Pharmacol. 2004;68:719–728. doi: 10.1016/j.bcp.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Ma Z, Pan YL, Huang WH, Yang YW, Wang ZY, Li Q, Zhao Y, Zhang XY, Shen ZR. Synthesis and biological evaluation of the pirfenidone derivatives as antifibrotic agents. Bioorg Med Chem Lett. 2014;24:220–223. doi: 10.1016/j.bmcl.2013.11.038. [DOI] [PubMed] [Google Scholar]
- Mitani Y, Sato K, Muramoto Y, Karakawa T, Kitamado M, Iwanaga T, Nabeshima T, Maruyama K, Nakagawa K, Ishida K, Sasamoto K. Superoxide scavenging activity of pirfenidone-iron complex. Biochem Biophys Res Commun. 2008;372:19–23. doi: 10.1016/j.bbrc.2008.04.093. [DOI] [PubMed] [Google Scholar]
- Moffat JG, Rudolph J, Bailey D. Phenotypic screening in cancer drug discovery - past, present and future. Nature Reviews Drug Discovery. 2014;13:588–602. doi: 10.1038/nrd4366. [DOI] [PubMed] [Google Scholar]
- Nakayama S, Mukae H, Sakamoto N, Kakugawa T, Yoshioka S, Soda H, Oku H, Urata Y, Kondo T, Kubota H, Nagata K, Kohno S. Pirfenidone inhibits the expression of HSP47 in TGF-beta1-stimulated human lung fibroblasts. Life Sci. 2008;82:210–217. doi: 10.1016/j.lfs.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Nanthakumar CB, Hatley RJD, Lemma S, Gauldie J, Marshall RP, Macdonald SJF. Dissecting fibrosis: therapeutic insights from the small-molecule toolbox. Nature Reviews Drug Discovery. 2015;14:693–720. doi: 10.1038/nrd4592. [DOI] [PubMed] [Google Scholar]
- Neri T, Lombardi S, Faita F, Petrini S, Balia C, Scalise V, Pedrinelli R, Paggiaro P, Celi A. Pirfenidone inhibits p38-mediated generation of procoagulant microparticles by human alveolar epithelial cells. Pulm Pharmacol Ther. 2016;39:1–6. doi: 10.1016/j.pupt.2016.05.003. [DOI] [PubMed] [Google Scholar]
- Salazar-Montes A, Ruiz-Corro L, Lopez-Reyes A, Castrejon-Gomez E, Armendariz-Borunda J. Potent antioxidant role of pirfenidone in experimental cirrhosis. Eur J Pharmacol. 2008;595:69–77. doi: 10.1016/j.ejphar.2008.06.110. [DOI] [PubMed] [Google Scholar]
- Schelegle ES, Mansoor JK, Giri S. Pirfenidone attenuates bleomycin-induced changes in pulmonary functions in hamsters. P Soc Exp Biol Med. 1997;216:392–397. doi: 10.3181/00379727-216-44187. [DOI] [PubMed] [Google Scholar]
- Scudi JV, Reisner DB, Childress SJ, Walter LA. In: Substituted 1-m-aminophenyl-2-pyridones. Office USP, editor. WALLACE & TIERNAN INC; United States: 1960. [Google Scholar]
- Shreekrishna Manmohan Gadekar . In: N-SUBSTITUTED PYRIDONE AND GENERAL METHOD FOR PREPARING PYRIDONES. Office USP, editor. United States: 1972. [Google Scholar]
- Vogel ER, VanOosten SK, Holman MA, Hohbein DD, Thompson MA, Vassallo R, Pandya HC, Prakash YS, Pabelick CM. Cigarette smoke enhances proliferation and extracellular matrix deposition by human fetal airway smooth muscle. Am J Physiol Lung Cell Mol Physiol. 2014;307:L978–986. doi: 10.1152/ajplung.00111.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin N, Qi X, Tsai S, Lu Y, Basir Z, Oshima K, Thomas JP, Myers CR, Stoner G, Chen G. p38 gamma MAPK is required for inflammation-associated colon tumorigenesis. Oncogene. 2016;35:1039–1048. doi: 10.1038/onc.2015.158. [DOI] [PubMed] [Google Scholar]
- Yu L, Hebert MC, Zhang YE. TGF-beta receptor-activated p38 MAP kinase mediates Smad-independent TGF-beta responses. Embo J. 2002;21:3749–3759. doi: 10.1093/emboj/cdf366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]