Abstract
The EcoRV DNA-(adenine-N6)-methyltransferase (M.EcoRV) specifically modifies the first adenine residue within GATATC sequences. During catalysis, the enzyme flips its target base out of the DNA helix and binds it into a target base binding pocket which is formed in part by Lys16 and Tyr196. A cytosine residue is accepted by wild-type M.EcoRV as a substrate at a 31-fold reduced efficiency with respect to the kcat/KM values if it is located in a CT mismatch substrate (GCTATC/GATATC). Cytosine residues positioned in a CG base pair (GCTATC/GATAGC) are modified at much more reduced rates, because flipping out the target base is much more difficult in this case. We intended to change the target base specificity of M.EcoRV from adenine-N6 to cytosine-N4. To this end we generated, purified and characterized 15 variants of the enzyme, containing single, double and triple amino acid exchanges following different design approaches. One concept was to reduce the size of the target base binding pocket by site-directed mutagenesis. The K16R variant showed an altered specificity, with a 22-fold preference for cytosine as the target base in a mismatch substrate. This corresponds to a 680-fold change in specificity, which was accompanied by only a small loss in catalytic activity with the cytosine substrate. The K16R/Y196W variant no longer methylated adenine residues at all and its activity towards cytosine was reduced only 17-fold. Therefore, we have changed the target base specificity of M.EcoRV from adenine to cytosine by rational protein design. Because there are no natural paragons for the variants described here, a change of the target base specificity of a DNA interacting enzyme was possible by rational de novo design of its active site.
INTRODUCTION
DNA methylation plays important roles in the control of gene expression, DNA replication and DNA repair (1–4). In eukaryotes it is also involved in development and epigenetic processes such as parental imprinting and X-inactivation (1,5). So far, three functional cytosine-C5 methyltransferases (MTases) have been identified in mammals, which all are essential in mice (6,7), and more than 1000 DNA MTases of different specificity have been found in bacteria (8). Prokaryotic DNA MTases recognize short DNA sequences and using S-adenosylmethionine (AdoMet) as the methyl group donor they specifically modify one base within the recognition sequence at a defined position yielding 6-methyladenine, 4-methylcytosine or 5-methylcytosine.
It is the goal of molecular enzymology to understand the chemical mechanism of enzymatic catalysis and regulation to provide a detailed understanding of the cellular metabolism. Based on the advances of molecular enzymology, it is often attempted to design enzyme inhibitors or to improve the properties of enzymes for biotechnical applications. In particular, a rational redesign of the specificity of an enzyme can be viewed as the ultimate challenge to our understanding of the principles and details of enzyme action. Several issues make DNA MTases to an ideal model system for design projects aiming to change the specificity of enzymes. (i) All DNA MTases whose structures are known share one common fold (reviewed in 9) that consists of two domains; one large domain containing the binding sites for the cofactor and the catalytic center and one smaller domain that participates in DNA binding and recognition. (ii) Structures are available for three adenine-N6 MTases, M.TaqI, DpnM and M.RsrI (10–12) and one cytosine-N4 MTase, M.PvuII (13). Since DpnM is closely related to M.EcoRV (11,14,15), this work mainly refers to the DpnM structure. (iii) Structural and biochemical studies have shown that all DNA MTases share one mechanistic feature, namely that methylation of the target base is always preceded by flipping the base out of the DNA helix (16–18). The flipped base is bound into a hydrophobic binding pocket within the large domain of the DNA MTase where catalysis takes place (reviewed in 19). (iv) The sequence specificity of DNA MTases has been successfully altered by domain swapping experiments at least in some cases (20–26).
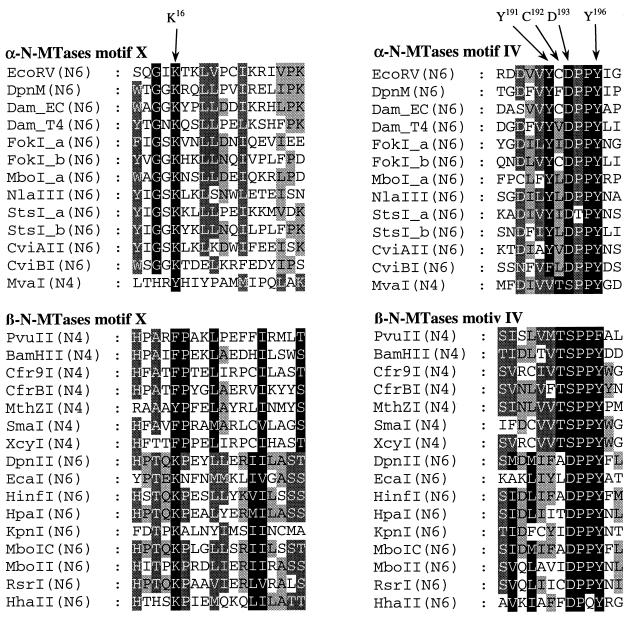
In this study, we attempted to change the substrate specificity of the EcoRV DNA-(adenine-N6)-methyltransferase (E.C. 2.1.1.72) to a DNA-(cytosine-N4)-methyltransferase (E.C. 2.1.1.113) using rational de novo protein-design. We followed two approaches: first we employed multiple sequence alignments of adenine-N6 MTases and of cytosine-N4 MTases (11,13,15,27) and systematically searched for residues that are conserved within but dissimilar between these groups (Fig. 1). Using this strategy we selected six amino acid residues for analysis and generated 12 single, double and triple mutants: we exchanged Asp193 of M.EcoRV to Ser because most cytosine MTases have an SPP(Y/F) tetrapeptide in their active site motif IV whereas a (D/N)PP(Y/F) sequence is found in adenine-N6 MTases (the corresponding sequence in M.EcoRV is: Asp193, Pro194, Pro195, Tyr196) (Fig. 1). There are few cytosine-N4 MTases with a DPPY motif in the active center, like M.BamHI, which is not included in the alignment shown in Figure 1. Thus, the SPP(Y/F) motif is not required for cytosine-N4 methylation. So far, no SPP(Y/F) enzyme has been characterized as an adenine-N6 Mtase. Ser229 was exchanged to aspartic acid and glutamic acid because in the cytosine-N4 MTase M.PvuII the SPPF-serine is part of a charge relay system with Asp96 of motif VI which might be relevant for catalysis (13). In adenine-N6 MTases, a serine is located at the corresponding position in motif VI (Ser239 in DpnM), such that the SPPF..D arrangement of M.PvuII could correspond to a DPPY..S in adenine-N6 MTases. In fact, Ser229 in M.EcoRV, the residue corresponding to Ser239 in DpnM (15) has been implicated previously in target base recognition by M.EcoRV (15,28). A K16Y mutant of M.EcoRV was prepared, because it has been suggested that Lys21 which corresponds to Lys16 of M.EcoRV is involved in target base recognition of DpnM (11). A lysine residue is conserved in all adenine-N6 MTases at this position whereas all cytosine-N4 MTases with an SPP(Y/F) motif carry a phenylalanine or tyrosine (Fig. 1). In the context of the D193S exchange we have also prepared Y196F, Y191V and C192T variants, because (i) M.EcoRV has a tyrosine at position 196, where M.PvuII has a phenylalanine, (ii) Tyr191 in M.EcoRV is strongly conserved among α-adenine-N6 MTases, but cytosine-N4 MTases carry a valine at the equivalent location and (iii) a threonine is highly conserved in cytosine-N4 MTases at the position of Cys192 in M.EcoRV (Fig. 1).
Figure 1.
Alignments of conserved regions of adenine and cytosine-specific DNA MTases belonging to the α- and β-group of MTases (11,13,15,27). The amino acid residues of M.EcoRV that were subjected to mutagenesis in this study are marked with arrows. Shading is according to the Blosum 62 scoring matrix, with black shading for 100%, dark grey for 80% and light grey for 60% conserved amino acid residues.
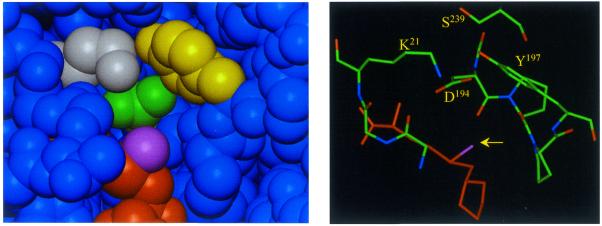
In an alternative approach that does not rely on amino acid sequence alignments but on structural data, we have inspected the model of the adenine binding pocket of M.EcoRV for suitable candidates to change specificity (Fig. 2). In this approach we did not intend to transplant some residues from the active site of a cytosine-N4 MTases to M.EcoRV but to make the target base binding pocket smaller, such that it could only accommodate cytosine and not adenine. In the structure of the M.TaqI–DNA complex, the extrahelical base is contacted by residues from the motifs IV, VIII and X (18). Since M.TaqI is from the γ-type of N-MTases and M.EcoRV is from the α-type, which are related by a circular permutation of the large domain and a different position of insertion of the small domain into the framework of the large domain (27,29), it is difficult to assign the corresponding amino acid residues of M.EcoRV and M.TaqI. Therefore, we used the structure of the DpnM MTase (11), which has 27% amino acid sequence identity to M.EcoRV and can be used as a template structure to obtain a structural model for M.EcoRV. We selected two target residues, Lys16 (from motif X) and Tyr196 (from motif IV) for mutagenesis. These residues were changed in a conservative fashion to arginine and tryptophan with the intention to decrease the size of the target base binding pocket with minimal change of the chemical properties of the corresponding residues. Therefore, the K16R, Y196W and K16R/Y196W double mutant were prepared.
Figure 2.
Adenine binding pocket of the DpnM methyltransferase (11). The left panel displays a surface view into the binding pocket. The side chains of Lys21, Asp194 and Tyr197 are colored white, green and yellow, respectively. AdoMet is shown in orange with the activated methyl group highlighted in violet. The right panel shows a similar view, with the activated methyl group marked by an arrow. The labeled residues Lys21, Asp194, Tyr197 and Ser239 of DpnM correspond to Lys16, Asp193, Tyr196 and Ser229 in M.EcoRV.
MATERIALS AND METHODS
Synthetic oligonucleotides
HPLC-purified oligodeoxyribonucleotides were purchased from Interactiva (Ulm, Germany). Methylation of adenine residues was determined using RV_ME. This substrate contains a hemimethylated EcoRV site (GATATC/GmATATC) that can only be modified in the upper DNA strand. Methylation of cytosine residues was measured using RV_C/A, which is identical to RV_ME apart from an exchange of the target base from adenine to cytosine. Methylation of cytosine residues not located in a base mismatch was investigated using the RV_CG substrate, which contains a GCTATC/GmATAGC-site. To investigate the importance of the G in the fifth position of the lower strand in RV_CG we also used RV_AT and RV_AG, which contain a canonical GATATC/GmATATC site and a GATATC/GmATAGC site, respectively. With each of the variants, methylation kinetics were also carried out with the double methylated oligonucleotide RV_MM as a negative control.
The oligodeoxyribonucleotide sequences are shown below. RV_ME, d(GATCGTAGATATCGCATCGA)/d(Bt-TTTTCGATGCGmATATCTACGATC); RV_C/A, d(GATCGTAGCTATCGCATCGA)/d(Bt-TTTTCGATGCGmATATCTACGATC); RV_CG, (CGCGGCCGCTATCCCGGGC)/d(Bt-TTTTTTGCCCGGGmATAGCGGCCGCG); RV_MM, d(CGCGGCCGmATATCCCGGGC)/d(Bt-TTTTTTGCCCGGGmATATCGGCCGCG); RV_AT, d(CGCGGCCGATATCCCGGGC)/d(Bt-TTTTTTGCCCGGGmATATCGGCCGCG) and RV_AG, d(CGCGGCCGATATCCCGGGC)/d(Bt-TTTTTTGCCCGG-GmATAGCGGCCGCG).
Mutagenesis and protein purification
Mutagenesis was carried out following a protocol described by Kirsch and Joly (30). M.EcoRV and M.EcoRV variants were purified using a GST-tag at the N-terminal end of the protein as described (15,28).
Kinetic analyses using the biotin/avidin microplate assay
Details of the avidin/biotin MTase assay have been described (31). Methylation reactions with M.EcoRV were carried out in 50 mM HEPES, pH 7.5, 50 mM NaCl, 1 mM EDTA, 50 ng/µl BSA in the presence of labeled [methyl-3H]AdoMet (specific activity: 2.68 × 1015 Bq/mol; NEN) at ambient temperature. After defined times, aliquots containing 2 pmol DNA were removed and the incorporation of radioactivity analyzed as described (31). Unless otherwise stated, 0.5 µM oligonucleotide and 0.76 µM AdoMet were used. To measure the steady-state kinetic parameters, the concentrations of the oligonucleotides and the cofactor 10 were varied between 0.1–2 µM and 0.76–10 µM, respectively. The slopes of the steady-state phase of the individual reaction progress curves were determined by linear regression and the rates obtained fitted to the Michaelis–Menten model.
Under the conditions of our experiments, biphasic time courses were observed with highly active variants and good substrates in which a fast exponential phase is followed by a slower linear phase. To derive an initial slope, the data were fitted to equation 1 (32).
CPMtheo = BL + f × (1 – exp–k1 × t) + k2 × t 1
with BL [counts per minute (CPM)], baseline of radioactivity; f [CPM], size of the exponential phase; k1 [s–1], rate constant of DNA methylation during the exponential phase; k2 [CPM/s], rate constant of DNA methylation during the linear phase.
To obtain the initial rate of DNA methylation (kmet) in CPM/s, equation 1 was differentiated at t = 0 which results in equation 2. The CPM/s values obtained with equation 2 can easily be converted into turnover numbers per enzyme molecule per second by considering the specific activity of the AdoMet (2.68 × 1015 Bq/mol), the amount of DNA transferred into each well of the microplate (2 pmol) and the concentration of the enzyme (as indicated).
kmet = f × k1 + k2 2
Structural modeling of M.EcoRV
Using the DpnM structure (PDB code 2DPM) as template and a multiple sequence alignment of M.EcoRV and several dam MTases, the structure of M.EcoRV could be modeled by the SWISS-Model server (http://www.expasy.ch/swissmod/) (33,34). The model shows a very good superposition of the structural model for M.EcoRV with the DpnM structure in all parts relevant for this work.
RESULTS
Screening of the variants for an altered specificity
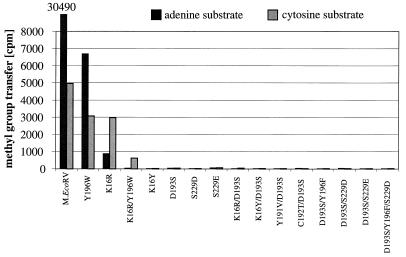
We have investigated the M.EcoRV enzyme, a DNA-(adenine-N6)-MTase that modifies the first adenine residue within GATATC sequences. It was the aim of this work to characterize the target base specificity of M.EcoRV and alter it from adenine to cytosine. The chemical reactions catalyzed by adenine-N6 and cytosine-N4 DNA MTases are very similar because in each case the exocyclic amino groups of heterocyclic aromatic ring systems are methylated. Both families of enzymes have similar active sites and show a partial functional overlap, because several adenine-DNA MTases, including M.EcoRV, also show residual catalytic activity with cytosine residues as target base (35) and M.PvuII, a cytosine-N4 MTase, also modifies adenine residues (36). Thus, the question arose whether an enzyme of one class could be converted into an enzyme of the other class by protein design, which is investigated here for M.EcoRV as the target enzyme. Fifteen M.EcoRV variants containing single, double and triple amino acid exchanges were prepared by site-directed mutagenesis. The proteins were expressed in Escherichia coli and purified as GST fusion proteins. To screen for an altered specificity, the enzymes were incubated with the adenine (RV_ME, recognition site, GATATC/GmATATC) and cytosine substrates (RV_C/A, recognition site, GCTATC/GmATATC) and the methylation of the DNA was detected. Note that in RV_C/A, only the target adenine is exchanged by cytosine. As shown in Figure 3, 9 out of 15 variants were inactive or only showed residual activity. This result underscores the important role of Asp193 and Ser229 in M.EcoRV (15,28). As we intended to design an active MTase with an altered specificity, these variants were not investigated further.
Figure 3.
Screening of all variants for an altered specificity. Either the adenine (RV_ME, black bars) or cytosine substrate (RV_C/A, gray bars) (0.5 µM) was incubated with 1 µl protein preparation of M.EcoRV or M.EcoRV mutants (2–40 µM) in 10 µl methylation buffer containing 0.76 µM [methyl-3H]-AdoMet for 1 h at 37°C.
Only three of the variants displayed considerable catalytic activity in vitro, namely K16R, Y196W and K16R/Y196W. Like wild-type M.EcoRV, all the variants showed a significant activity with the cytosine substrate as well. Previously we have shown by HPLC analysis that in this reaction 4-methylcytosine is formed by wild-type M.EcoRV (35). Here, as an additional control we carried out methylation reactions with a substrate that contains a fully methylated EcoRV site (GmATATC/GmATATC). No significant methylation was detectable with any of the variants (data not shown) confirming that with the C/A substrate cytosine methylation was being observed.
Interestingly, the target base preferences of the variants were significantly altered with respect to wild-type M.EcoRV. Whereas the wild-type enzyme shows a clear preference for adenine as target base, the discrimination between these two bases is significantly weaker in the case of the Y196W variant; and the K16R variant even prefers methylation of cytosine residues. In the case of the K16R/Y196W double mutant, catalytic activity was only detectable with the cytosine substrate.
Target base specificity of wild-type M.EcoRV
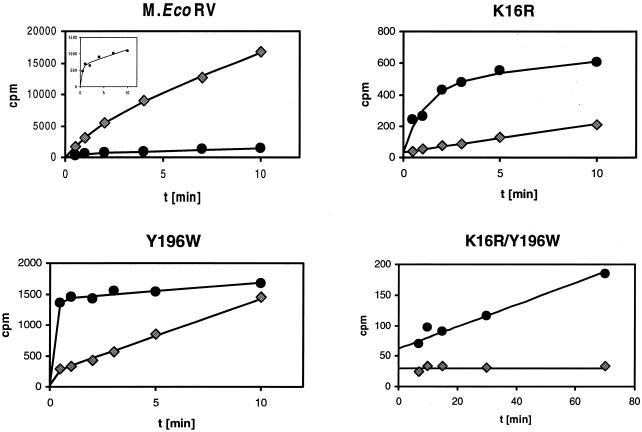
To characterize quantitatively the target base specificity of wild-type M.EcoRV and the variants that show significant catalytic activities, the kinetics of the methylation reactions of the adenine and cytosine substrates were determined. These experiments were carried out by mixing preincubated enzyme-AdoMet solutions with oligonucleotide. Under these conditions, wild-type M.EcoRV shows a biphasic methylation reaction in which a fast expontential phase is followed by a slower linear phase with the adenine substrate, because the first turnover of the enzyme is approximately five times faster than the turnover under steady-state conditions (32). This shape of the reaction progress curve is observed despite the concentration of the enzyme being higher than the concentrations of the AdoMet and the DNA because, under the experimental conditions, M.EcoRV is not saturated with either substrate. Therefore, not all DNA molecules are bound and many non-productive enzyme–DNA complexes that do not contain AdoMet are formed such that only few enzyme molecules can modify their substrates in a fast reaction that is followed by a slower linear phase.
We have previously shown that the M.EcoRV–AdoMet–DNA complex exists in an open state where DNA binding and release is possible and in a closed state where methyl group transfer can take place (32,37). The catalytic rate of the exponential phase is limited by the rate constant of the transition from the open to the closed complex. The multiple turnover rate constant is limited by the reverse reaction, namely opening of the closed complex which allows product dissociation. As shown in Figure 4, the amount of substrate that is modified during the exponential phase is strongly reduced with the cytosine substrate, indicating that fewer complexes adopt the catalytically active conformation in a fast reaction because the cytosine does not optimally fit into the binding pocket. To determine the steady-state kinetic parameters of EcoRV, kinetics of methylation of the adenine and cytosine substrates were carried out at different concentrations of AdoMet and oligonucleotide. The results obtained (Table 1) are in close agreement with data published for the His6-tagged enzyme (31,37). They show that neither KMAdoMet nor KMDNA are important for target base specificity (Table 1). Overall, for wild-type M.EcoRV, the kcat/KMDNA value was 31 ± 3 times higher with the adenine than with the cytosine substrate.
Figure 4.
Time courses of methylation of the adenine (RV_ME, diamonds) and the cytosine substrates (RV_C/A, circles) by M.EcoRV and three M.EcoRV variants. In these experiments, 0.5 µM oligonucleotide was incubated with 0.9 µM M.EcoRV or M.EcoRV mutant in methylation buffer containing 0.76 µM [methyl-3H]AdoMet. The insert in the panel showing the wild-type data displays an enlarged view on the reaction progress curve obtained with the cytosine substrate. All data points are averages of two to four individual measurements with errors always <10%. The lines show fits of the data points to equation 1 or linear regressions. The initial rates of DNA methylation of the K16R variant are 1.9 × 10–3 h–1 (adenine substrate) and 4.2 × 10–2 h–1 (cytosine substrate). The rate of methylation of the cytosine substrate by the K16R/Y196W variant is 2.0 × 10–4 h–1, whereas wild-type M.EcoRV modifies the cytosine substrate with a rate of 3.4 × 10–3 h–1 under these conditions (Table 1).
Table 1. Steady-state kinetic parameters of wild-type M.EcoRV and the K16R variant with the adenine (RV_ME) and cytosine substrates (RV_C/A)a.
| M.EcoRV variant | Adenine substrate | Cytosine substrate | ||||||
| KMAdoMet | KMDNA | kcat | kcat/KMDNA | KMAdoMet | KMDNA | kcat | kcat/KMDNA | |
| |
[µM] |
[µM] |
[h–1] |
[µM–1 h–1] |
[µM] |
[µM] |
[h–1] |
[µM–1 h–1] |
| Wildtype | 9.9 | 0.46 | 2.3 | 5.0 | 8.6 | 0.56 | 0.09 | 0.16 |
| K16R | 10 | 1.0 | 0.10 | 0.1 | 8.5 | 1.1 | 0.14 | 0.13 |
aKM and kcat values are valid ± 20% and ± 30%, respectively.
Target base specificity of the K16R variant
A detailed analysis of the kinetics of methylation of the adenine and cytosine substrates was also carried out with the K16R variant. As shown in Figure 4, this variant shows an exponential phase of the methylation reaction only with the cytosine substrate. The rate of methylation of the cytosine substrate by K16R is similar to that by wild-type M.EcoRV as illustrated by the similar amount of substrate that is modified during the exponential phase of the methylation reaction (K16R, 450 CPM; wild-type M.EcoRV, 510 CPM) and virtually identical steady-state parameters (Table 1). [500 CPM correspond to the methylation of ∼0.2% of the oligonucleotides during the exponential phase of the reaction]. However, with the adenine substrate no exponential phase was observed, indicating that now the methyl group transfers itself or an earlier step has become rate limiting. This suggests that the smaller target base binding pocket of the K16R variant only accommodates adenine poorly.
The change in the reaction mechanism of the methylation of the adenine and cytosine substrates raises problems for the quantitative comparison of the data obtained with both substrates. The rate constant of the exponential phase of methylation of the cytosine substrate by the K16R variant is ∼1 min–1. Comparison with the kcat value for the adenine substrate of 0.1 h–1 (where no exponential phase is observed) would lead to a 600-fold preference for cytosine over adenine. However, this number does not take into consideration the fact that only a small fraction of the DNA molecules are modified with the fast rate constant either because they are not bound in a ternary enzyme–DNA–AdoMet complex or because the complex does not adopt the conformation required for fast methylation. To use a more realistic model, we considered the reaction mixture to consist of two populations of enzyme molecules, one reacting with a fast rate for one first turnover and a second reacting with a slower rate. This behavior is described by equation 2, which consequently can be used to derive the initial slopes of the reaction progress curves with both the substrates. The initial rate of methylation of the cytosine substrate was about 22 times higher than the rate of methylation of the adenine substrate (Fig. 4) demonstrating that the K16R variant has a 22 ± 2-fold preference for methylation of cytosine. This ratio has been reproducibly obtained with different enzyme preparations.
Target base specificity of the Y196W variant
The Y196W variant showed biphasic reaction progress curves with both substrates (Fig. 4). With the cytosine substrate, large amounts of the DNA were modified during the exponential phase of the reaction, even more than with wild-type M.EcoRV, and a slow linear phase was observed. In contrast, the adenine substrate showed a very small amplitude of the exponential phase but a fast linear phase. Because of this complex behavior, we refrain from the definition of a simple number that characterizes the specificity of this variant. Because the variant prefers adenine over cytosine in the second phase of the reaction, we did not carry out a complete Michaelis–Menten analysis with this variant.
The results obtained with the Y196W variant suggest that the altered target base binding pocket strongly interacts with cytosine, which should result in a large amplitude of the exponential phase of the methylation reaction. This strong interaction, however, interferes with the extrusion of the methylated base and, thereby, causes a low multiple turnover rate. In contrast, adenine only fits poorly into the binding pocket. Thus, only few complexes adopt a catalytically active conformation and only a small exponential phase of the methylation reaction is observed. However, in this case, release of the modified base is easy allowing a relatively fast multiple turnover.
Target base specificity of the K16R/Y196W variant
The results obtained with the K16R/Y196W double mutant show that the effects of the individual exchanges of Lys16 to arginine and Tyr196 to tryptophan operate synergistically. The double mutant no longer modifies adenine residues. Even with cytosine, only a linear time course of methylation was observed, indicating that base flipping has become an inefficient process. However, a significant steady-state rate of methylation was observed, which was 17 ± 5-fold lower than the steady-state rate of methylation of the cytosine substrate with wild-type M.EcoRV under the same conditions (Fig. 4). Due to the reduced catalytic activity of this variant, DNA methylation experiments were not possible under multiple turnover conditions, such that a Michaelis–Menten analysis of this variant was not possible.
Methylation of cytosine residues in a non-mismatch environment
Given the results obtained so far, we were interested to determine if wild-type M.EcoRV and the mutants also were able to modify cytosine residues located in a non-mismatch site (GCTATC/GmATAGC). We therefore also tested methylation of the RV_CG substrate. In agreement with earlier results (35), M.EcoRV showed a 10–100-fold lower activity with the RV_CG substrate than with the RC_C/A substrate that carries the target cytosine in a base mismatch (Table 2 and Fig. 4). This means that either M.EcoRV is not able to flip out a cytosine that is located in a CG base pair or that the G present in the recognition sequence of RV_CG at the fifth position of the lower strand impedes DNA methylation. To distinguish between these two alternatives, we determined the kinetics of methylation of RV_AT (GATATC/GmATATC)and RV_AG (GATATC/GmATAGC). Both substrates carry an adenine residue at the target position, which is in a normal AT base pair in RV_AT but in an AG mismatch in RV_AG. The kinetics of methylation of both these substrates by wild-type M.EcoRV were virtually indistinguishable (data not shown), demonstrating that a G at the fifth position in the lower strand of the recognition sequence does not affect methylation of an adenine residue. This result implies that the low rate of methylation of the RV_CG substrate is due to a low efficiency of base flipping of the target cytosine that is base paired to guanine.
Table 2. Methylation of cytosine residues located in a GCTATC/GmATAGC site by M.EcoRV and the K16R, Y196W and K16R/Y196W variants.
| M.EcoRV variant |
DNA methylation [CPM] |
| Wild-type | 306 ± 20 |
| K16R | 78 ± 5 |
| Y196W | 333 ± 5 |
| K16R/Y196W | 27 ± 2 |
| No enzyme | 25 ± 5 |
In each case, 0.5 µM RV_CG was incubated for 1 h at ambient temperature with 1 µM of the variants.
As shown in Table 2, none of the variants shows an increased level of methylation of the RV_CG substrate. This result is not very surprising, since we altered neither the residues that contact the partner base of the target base nor those responsible for base flipping. It was not expected, therefore, to obtain a variant which forms better contacts to the G and/or has an increased capability to flip out a cytosine located in a CG base pair.
DISCUSSION
An elaborate substrate specificity is one of the most fascinating properties of enzymes. Therefore, many important questions of molecular enzymology deal with the understanding of the molecular basis of specificity, such as the mechanism of substrate recognition, and it is a big challenge to change this property by rational design. This investigation addresses the target base specificity of M.EcoRV, an enzyme which specifically modifies the first adenine within GATATC sequences. We used a rational design approach to change the target base specificity of this enzyme from adenine to cytosine by reducing the size of its target base binding pocket, changing Lys16 to arginine and Tyr196 to tryptophan, respectively. It should be noticed that there are no natural paragons for these variants, because cytosine-N4 MTases with an arginine at the position equivalent to Lys16 are unknown and the known MTases that carry a tryptophan in the active site (such as M.MunI) are adenine-N6 MTases. We generated three variants with a dramatically altered target base specificity. One of these (K16R) no longer preferred adenine, but instead showed a 22-fold preference for cytosine as target base. This means that the overall specificity was altered 680-fold by just one single mutation. This impressive change in specificity was only accompanied by a very small loss of activity, because the mutant has a similar activity towards cytosine as the target base as the wild-type enzyme. Thus, the change in specificity was accomplished by a large decrease in activity towards adenine without affecting cytosine methylation. A second variant (K16R/Y196W) did not modify adenine at all. Even in this case, the catalytic activity of the variant towards cytosine was reduced only 17-fold with respect to the wild-type enzyme. Thus, a bona fide specificity for cytosine was achieved while maintaining a reasonable catalytic activity.
The changes in specificity reported here are not unique because large changes of enzyme properties, substrate preferences and reaction specificities have been achieved by directed evolution where a detailed understanding of the transition state complex is not a prerequisite for successful design (reviewed in 38). Moreover, there are some examples of successful protein design by rational approaches, e.g. it was possible to change the cofactor specificity of dehydrogenases from NAD to NADP and vice versa (39–41) and the reaction specificity of aspartate aminotransferase was altered (42). However, so far, there are no examples where the specificity of an enzyme that specifically interacts with DNA has been completely changed by rational exchange of one or two amino acid residues. Interestingly our initial approach to alter target base specificity by transplanting critical amino acid residues from cytosine-N4 MTases to M.EcoRV completely failed, as did similar attempts in other systems. This lack of success in rational enzyme design is probably due to our limited knowledge of the structures and critical interactions, in the transition state of both the catalyzed reactions and the second shell buttressing interactions that also had to be optimized to design a new transition state. Later we used a rational de novo design approach which proved successful, demonstrating that at least in this case, it was much easier to redesign the active site of an enzyme de novo instead of trying to copy nature by introducing elements from another enzyme.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (JE 252/2-3) and by a grant of the European Community (BIO-CT98-0328).
References
- 1.Bestor T.H. and Verdine,G.L. (1994) DNA methyltransferases. Curr. Opin. Cell Biol., 6, 380–389. [DOI] [PubMed] [Google Scholar]
- 2.Cheng X. (1995) DNA modification by methyltransferases. Curr. Opin. Struct. Biol., 5, 4–10. [DOI] [PubMed] [Google Scholar]
- 3.Cheng X. (1995) Structure and function of DNA methyltransferases. Annu. Rev. Biophys. Biomol. Struct., 24, 293–318. [DOI] [PubMed] [Google Scholar]
- 4.Dryden D.T.F. (1999) Bacterial DNA methyltransferases. In Cheng,X. and Blumenthal,R.M. (eds), S-Adenosylmethionine-Dependent Methyltransferases: Structures and Functions. World Scientific Publishing, Singapore, pp. 283–340.
- 5.Robertson K.D. and Wolffe,A.P. (2000) DNA methylation in health and disease. Nature Rev. Genet., 1, 11–19. [DOI] [PubMed] [Google Scholar]
- 6.Li E., Bestor,T.H. and Jaenisch,R. (1992) Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell, 69, 915–926. [DOI] [PubMed] [Google Scholar]
- 7.Okano M., Bell,D.W., Haber,D.A. and Li,E. (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell, 99, 247–257. [DOI] [PubMed] [Google Scholar]
- 8.Roberts R.J. and Macelis,D. (2000) REBASE - restriction enzymes and methylases. Nucleic Acids Res., 28, 306–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faumann E.B., Blumenthal,R.M. and Cheng,X. (1999) Structure and evolution of AdoMet-dependent methyltransferases. In Cheng,X. and Blumenthal,R.M. (eds), S-Adenosylmethionine-Dependent Methyltransferases: Structures and Functions. World Scientific Publishing, Singapore, pp. 1–38.
- 10.Labahn J., Granzin,J., Schluckebier,G., Robinson,D.P., Jack,W.E., Schildkraut,I. and Saenger,W. (1994) Three-dimensional structure of the adenine-specific DNA methyltransferase M.TaqI in complex with the cofactor S-adenosylmethionine. Proc. Natl Acad. Sci. USA, 91, 10957–10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tran P.H., Korszun,Z.R., Cerritelli,S., Springhorn,S.S. and Lacks,S.A. (1998) Crystal structure of the DpnM DNA adenine methyltransferase from the DpnII restriction system of Streptococcus pneumoniae bound to S-adenosylmethionine. Structure, 6, 1563–1575. [DOI] [PubMed] [Google Scholar]
- 12.Scavetta R.D., Thomas,C.B., Walsh,M.A., Szegedi,S., Joachimiak,A., Gumport,R.I. and Churchill,M.E. (2000) Structure of RsrI methyltransferase, a member of the N6–adenine beta class of DNA methyltransferases. Nucleic Acids Res., 28, 3950–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong W., O’Gara,M., Blumenthal,R.M. and Cheng,X. (1997) Structure of PvuII DNA-(cytosine N4) methyltransferase, an example of domain permutation and protein fold assignment. Nucleic Acids Res., 25, 2702–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeltsch A., Sobotta,T. and Pingoud,A. (1996) Structure prediction of the EcoRV DNA methyltransferase based on mutant profiling, secondary structure analysis, comparison with known structures of methyltransferases and isolation of catalytically inactive single mutants. Protein Eng., 9, 413–423. [DOI] [PubMed] [Google Scholar]
- 15.Roth M., Helm-Kruse,S., Friedrich,T. and Jeltsch,A. (1998) Functional roles of conserved amino acid residues in DNA methyltransferases investigated by site-directed mutagenesis of the EcoRV adenine-N6-methyltransferase. J. Biol. Chem., 273, 17333–17342. [DOI] [PubMed] [Google Scholar]
- 16.Klimasauskas S., Kumar,S., Roberts,R.J. and Cheng,X. (1994) HhaI methyltransferase flips its target base out of the DNA helix. Cell, 76, 357–369. [DOI] [PubMed] [Google Scholar]
- 17.Reinisch K.M., Chen,L., Verdine,G.L. and Lipscomb,W.N. (1995) The crystal structure of HaeIII methyltransferase covalently complexed to DNA: an extrahelical cytosine and rearranged base pairing. Cell, 82, 143–153. [DOI] [PubMed] [Google Scholar]
- 18.Goedecke K., Pignot,M., Goody,R.S., Scheidig,A.J. and Weinhold,E. (2001) Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog. Nature Struct. Biol., 8, 121–125. [DOI] [PubMed] [Google Scholar]
- 19.Roberts R.J. and Cheng,X. (1998) Base flipping. Annu. Rev. Biochem., 67, 181–198. [DOI] [PubMed] [Google Scholar]
- 20.Lange C., Wild,C. and Trautner,T.A. (1996) Identification of a subdomain within DNA-(cytosine-C5)-methyltransferases responsible for the recognition of the 5′ part of their DNA target. EMBO J., 15, 1443–1450. [PMC free article] [PubMed] [Google Scholar]
- 21.Walter J., Trautner,T.A. and Noyer-Weidner,M. (1992) High plasticity of multispecific DNA methyltransferases in the region carrying DNA target recognizing enzyme modules. EMBO J., 11, 4445–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trautner T.A., Pawlek,B., Behrens,B. and Willert,J. (1996) Exact size and organization of DNA target-recognizing domains of multispecific DNA-(cytosine-C5)-methyltransferases. EMBO J., 15, 1434–1442. [PMC free article] [PubMed] [Google Scholar]
- 23.Mi S. and Roberts,R.J. (1992) How M.MspI and M.HpaII decide which base to methylate. Nucleic Acids Res., 20, 4811–4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klimasauskas S., Nelson,J.L. and Roberts,R.J. (1991) The sequence specificity domain of cytosine-C5 methylases. Nucleic Acids Res., 19, 6183–6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gann A.A.F., Campbell,A.J.B., Collins,J.F., Coulson,A.F.W. and Murray,N.E. (1987) Reassortment of DNA recognition domains and the evolution of new specificities. Mol. Microbiol., 1, 13–22. [DOI] [PubMed] [Google Scholar]
- 26.Gubler M., Braguglia,D., Meyer,J., Piekarowicz,A. and Bickle,T.A. (1992) Recombination of constant and variable modules alters DNA sequence recognition by type IC restriction-modification enzymes. EMBO J., 11, 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malone T., Blumenthal,R.M. and Cheng,X. (1995) Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyl-transferases and suggests a catalytic mechanism for these enzymes. J. Mol. Biol., 253, 618–632. [DOI] [PubMed] [Google Scholar]
- 28.Jeltsch A., Roth,M. and Friedrich,T. (1999) Mutational analysis of target base flipping by the EcoRV adenine-N6 DNA methyltransferase. J. Mol. Biol., 285, 1121–1130. [DOI] [PubMed] [Google Scholar]
- 29.Jeltsch A. (1999) Circular permutations in the molecular evolution of DNA methyltransferase. J. Mol. Evol., 49, 161–164. [DOI] [PubMed] [Google Scholar]
- 30.Kirsch R.D. and Joly,E. (1998) An improved PCR-mutagenesis strategy for two-site mutagenesis or sequence swapping between related genes. Nucleic Acids Res., 26, 1848–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roth M. and Jeltsch,A. (2000) Biotin-avidin microplate assay for the quantitative analysis of enzymatic methylation of DNA by DNA methyltransferases. Biol. Chem., 381, 269–272. [DOI] [PubMed] [Google Scholar]
- 32.Gowher H. and Jeltsch,A. (2000) Molecular enzymology of the EcoRV DNA-(adenine-N6)-methyltransferase: Kinetics of DNA binding and bending, kinetic mechanism and linear diffusion of the enzyme on DNA. J. Mol. Biol., 303, 93–110. [DOI] [PubMed] [Google Scholar]
- 33.Guex N., Diemand,A. and Peitsch,M.C. (1999) Protein modelling for all. Trends Biochem. Sci., 24, 364–367. [DOI] [PubMed] [Google Scholar]
- 34.Guex N. and Peitsch,M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modelling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- 35.Jeltsch A., Christ,F., Fatemi,M. and Roth,M. (1999) On the substrate specificity of DNA methyltransferases. J. Biol. Chem., 274, 19538–19544. [DOI] [PubMed] [Google Scholar]
- 36.Jeltsch A. (2001) The cytosine N4-methyltransferase M.PvuII also modifies adenine residues. Biol. Chem., 382, 707–710. [DOI] [PubMed] [Google Scholar]
- 37.Jeltsch A., Friedrich,T. and Roth,M. (1998) Kinetics of methylation and binding of DNA by the EcoRV adenine-N6 methyltransferase. J. Mol. Biol., 275, 747–758. [DOI] [PubMed] [Google Scholar]
- 38.Arnold F.H. and Volkov,A.A. (1999) Directed evolution of biocatalysts. Curr. Opin. Chem. Biol., 3, 54–59. [DOI] [PubMed] [Google Scholar]
- 39.Chen R., Greer,A. and Dean,A.M. (1996) Redesigning secondary structure to invert coenzyme specificity in isopropylmalate dehydrogenase. Proc. Natl Acad. Sci. USA, 93, 12171–12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen R., Greer,A. and Dean,A.M. (1995) A highly active decarboxylating dehydrogenase with rationally inverted coenzyme specificity. Proc. Natl Acad. Sci. USA, 92, 11666–11670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scrutton N.S., Berry,A. and Perham,R.N. (1990) Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature, 343, 38–43. [DOI] [PubMed] [Google Scholar]
- 42.Graber R., Kasper,P., Malashkevich,V.N., Strop,P., Gehring,H., Jansonius,J.N. and Christen,P. (1999) Conversion of aspartate aminotransferase into an L-aspartate β-decarboxylase by a triple active-site mutation. J. Biol. Chem., 274, 31203–31208. [DOI] [PubMed] [Google Scholar]