Abstract
Originally discovered as part of C1, the initiation component of the classical complement pathway, it is now appreciated that C1q regulates a variety of cellular processes independent of complement activation. C1q is a complex glycoprotein assembled from 18 polypeptide chains, with a C-terminal globular head region that mediates recognition of diverse molecular structures, and an N-terminal collagen-like tail that mediates immune effector mechanisms. C1q mediates a variety of immunoregulatory functions considered important in the prevention of autoimmunity such as the enhancement of phagocytosis, regulation of cytokine production by antigen presenting cells, and subsequent alteration in T-lymphocyte maturation. Furthermore, recent advances indicate additional roles for C1q in diverse physiologic and pathologic processes including pregnancy, tissue repair, and cancer. Finally, C1q is emerging as a critical component of neuronal network refinement and homeostatic regulation within the central nervous system. This review summarizes the classical functions of C1q and reviews novel discoveries within the field.
Keywords: complement C1q, autoimmunity, pregnancy, wound healing, tumor, neurobiology
1. The complement C1q molecule: from early appearance in evolutionary time to recent recombinant expression
At the time of its discovery at the end of the 19th century complement was described as a heat-labile factor able to cooperate with antibodies to eliminate bacteria. By the mid-1920s complement was recognized as consisting of four components (C1 to C4, with names assigned in order of their discovery), and their isolation from serum was achieved in the early 1940s (Sim et al., 2016). The macromolecular nature of C1 as a complex of three protein entities, namely C1q, the recognition subunit, and its associated proteases C1r and C1s, was defined in the early 1960s, i.e. more than half a century ago (Lepow et al., 1963; Naff et al., 1964). The fact that C1q and the initiating proteins of the classical complement pathway were first discovered is likely related to the high concentrations of these proteins in serum, by comparison with the corresponding recognition proteins (mannan-binding lectin (MBL), ficolins) and associated proteases (MBL-associated serine proteases) of the lectin complement pathway. From this point of view C1q can be considered undoubtedly as an old molecule.
In addition, C1q is also “old” in being present early evolutionary time. An orthologue of vertebrate C1q proteins has been identified in amphioxus, the modern survivors of the ancient basal chordate lineage (Gao et al., 2014), well before the appearance of jaw vertebrates and adaptive immunity. C1q from Branchiostoma japonicum (BjC1q) possibly activates a primitive C1q–mediated complement system, through the binding of Ig-domain-containing amphioxus proteins as well as C4-activating proteases. BjC1q can also bind mammalian C1r and C1s proteases, as well as human IgG, and thus can replace human C1q to activate the human classical pathway. The C1q molecule further evolved with three different A, B and C subunits, possibly during the diversification of cartilaginous fishes, with clear sequence signatures characterizing their functional evolutionary diversification (Gao et al., 2014; Tariq et al., 2015; Goshima et al., 2016). Although only one C1q, C1r, C1s protein sequence has been obtained from the liver transcriptome of a hammerhead shark, the presence of three C1q chains in the nurse shark has been reported at the protein level, suggesting that C1q could be expressed in extra-hepatic sites in sharks as in mammals (Goshima et al., 2016). A recent study details the C1q sequences in the Chinese goose and the corresponding evolutionary relationships of the three C1q subunits in duck, chicken, bird and alligator (Tariq et al., 2015).
1.1. C1q ultrastructure and polypeptide chain composition
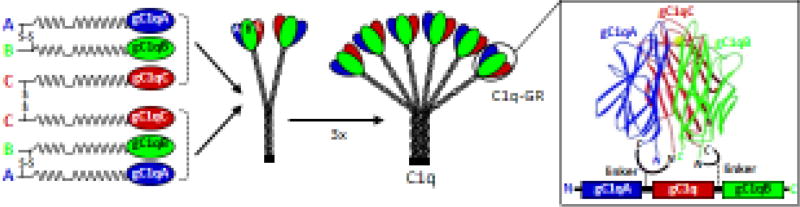
Electron microscopy studies in the 1970s revealed the typical shape of C1q as resembling a bunch of flowers, with six peripheral globular regions each connected by fibrillar strands to a central bundle of fibers (Shelton et al., 1972; Svehag et al., 1972). In parallel, detailed biochemical studies revealed that C1q is a complex glycoprotein assembled from 18 polypeptide chains of three different types named A, B, and C of 29, 27, and 23 kDa, respectively. Each chain comprises an N-terminal collagen-like sequence and a C-terminal globular gC1q module, with disulfide bridges linking the N-terminal ends of the A and B chains and of two C chains (Reid and Porter, 1976). Each A-B dimer associates with a C chain, resulting in a basic subunit comprised of two disulfide-linked heterotrimeric collagen-like triple helices prolonged by globular domains (Fig. 1). Biochemical characterization of the C1q protein culminated in the determination of the amino acid sequences of the entire A and B chains and that of the collagen-like region of the C chain by N-terminal Edman sequencing (Reid, 1979; Reid et al., 1982). Both cDNA and genomic clones were isolated shortly afterwards for the A and B chains (Reid, 1985), and the isolation of the cDNA sequence of the C chain allowed completion of the entire derived amino acid sequence of the C1q molecule (Sellar et al., 1991). The three genes were found to be aligned 5’-3’ in the same orientation and in the order A-C-B on human chromosome 1p.
Fig. 1.
Schematic representation of the assembly of the C1q molecule and of the recombinant single chain globular region. C1q is assembled from three polypeptide chains (A, B and C) each containing an N-terminal collagen-like sequence and a C-terminal globular gC1q module. A particular inter-chain disulfide pattern results in a basic subunit comprised of two heterotrimeric collagen-like stalks prolonged by globular domains (C1q–GR). Three subunits associate to yield the full-length protein with a typical shape of a bouquet of six flowers. The crystal structure of the trimeric C1q–GR shows that the N- and C- terminal ends of the 3 chains are in close proximity which allowed insertion of short linkers between the gC1qA-gC1qC and gC1qC-gC1qB modules to yield a recombinant single chain construct.
1.2. C1q functional domains
The presence of two distinct types of structures in the C1q molecule, i.e. the globular heads and the collagen-like fragments, allowed their isolation by limited proteolysis of C1q with collagenase and pepsin, respectively (Brodsky-Doyle et al., 1976; Hughes-Jones and Gardner, 1979). Functional studies showed that the globular regions are mainly responsible for target recognition, including binding to the Fc region of immunoglobulins (Hughes-Jones and Gardner, 1979), but also to bacterial and viral surface proteins, as well as altered self elements (apoptotic cells, amyloid and prion proteins) (Gaboriaud et al., 2011). The collagen-like regions mediate immune effector mechanisms, including complement activation through interaction with the C1r and C1s proteases (Siegel and Schumaker, 1983) and enhancement of phagocytosis through interaction with cell receptors (Bobak et al., 1987).
Isolation of the globular region of C1q allowed determination of its X-ray crystal structure, revealing a compact heterotrimeric assembly with a calcium ion bound at the top (Gaboriaud et al., 2003). Each of the three gC1q modules exhibits distinct surface patterns of charged and hydrophobic residues, which likely endow them with a diversity of specific recognition properties. In addition, the compact structure of the trimer allows ligand binding through residues contributed by two or three chains, thereby broadening the recognition spectrum of C1q (Gaboriaud et al., 2003). This structure confirmed the structural homology between tumor necrosis factor (TNF) and gC1q–containing proteins, previously revealed in the crystal structures of the homotrimeric gC1q domain of mouse adiponectin (Shapiro and Scherer, 1998) and collagen X (Bogin et al., 2002).
1.3. Recombinant C1q and site-directed mutagenesis studies
Detailed investigation of the structure-function relationships of C1q required its production in a recombinant form. The first achievements consisted in bacterial expression of soluble fusion proteins of the individual gC1q domains (A, B and C) with maltose binding protein (MBP), which were purified by affinity chromatography on an amylose resin. Characterization of their interaction properties revealed that these domains have likely evolved as functionally autonomous entities with differential ligand-binding properties (Kishore et al., 2003). Site-directed mutagenesis revealed that the binding sites for a number of C1q target molecules, including IgG, IgM, pentraxins and bacterial components such as Salmonella lipopolysaccharides and Klebsiella outer membrane protein K36, are closely overlapping (Kojouharova et al., 2003; Kojouharova et al., 2004; Roumenina et al., 2006; Zlatarova et al., 2006; Roumenina et al., 2008), but differ from the binding sites for the viral glycoproteins gp41 of HIV-1 and gp21 of HTLV-1 (Kojouharova et al., 1998; Kishore et al., 2003).
Production of full-length recombinant C1q has been a major technical bottleneck for a long time, owing to the three chain structure, particular inter-chain disulfide pattern and numerous post-translational modifications of the molecule. For example, the collagen-like sequences of C1q contain the repeating Gly-X-Y triplet where X is often a proline residue and Y a hydroxyproline or hydroxylysine residue, the latter being frequently modified with glucosylgalactosyl disaccharide units (Reid, 1979; Pflieger et al., 2010). However, this challenge was achieved in 2013 with the production of the full-length recombinant protein in stably transfected mammalian 293-F cells (Bally et al., 2013). The recombinant C1q molecule is similar to serum-derived C1q, as judged from biochemical analysis and electron microscopy imaging, and retains the ability to associate with the C1r and C1s proteases, to recognize physiological C1q ligands including IgG and pentraxin 3, and to trigger complement activation. The production of recombinant C1q opened the way for deciphering the interaction properties of this protein by site-directed mutagenesis, as illustrated by the identification of the C1s–C1r–C1r–C1s–binding residues in the C1q collagen-like stems (Bally et al., 2013). The availability of recombinant C1q should allow engineering of C1q molecules lacking specific functions (i.e. complement activation or binding to a protein partner) which will represent unique tools to study the role of C1q in various cellular contexts.
The latest progress in recombinant C1q production was the generation of a single-polypeptide form of the human C1q globular region (C1q–scGR) (Moreau et al., 2016), based on a strategy previously used to produce a single-chain form of the homotrimeric globular domain of adiponectin, a protein structurally related to C1q (Ge et al., 2010). Indeed the crystal structures of the globular domains of mouse adiponectin (Shapiro and Scherer, 1998) and of C1q (Gaboriaud et al., 2003) revealed the close proximity of the N- and C-termini of the three gC1q modules at the base of the trimer (Fig. 1), allowing connection of adjacent monomers by short linkers. Physicochemical, structural, and functional characterization of the recombinant single chain fragment, produced at high yield in stably transfected 293-F cells, showed that its 3D–structure and binding properties were similar to those of the three-chain fragment generated by collagenase digestion of serum-derived C1q (Moreau et al., 2016).
This novel recombinant tool will allow more reliable assessment of the effects of single residue mutations in a heterotrimeric context as present in native C1q and the ability to test the hypothesis that recognition of certain ligands involves residues contributed by several subunits.
2. Classical functions: complement, phagocytosis and cytokine regulation
2.1. The classical complement pathway
Originally discovered as a part of C1, the classical complement pathway initiation complex C1qC1r2C1s2, C1q was quickly shown to have the recognition motif for the Fc portion of IgM and IgG. Appropriate binding of the C1q globular head domains to multiple Fc regions of antibody in an immune complex alters the conformation of the serine protease C1r, enabling autoactivation of C1r and then cleavage of C1s. C1r activation somehow involves conformational changes, since it is produced in a proenzyme form, with a collapsed binding site conformation (Gohara and Di Cera, 2011). Activated C1s, also a serine protease, then cleaves C4 and C2 to generate the classical complement C3 convertase, C4b2b. As such, this leads to the generation of chemotactic peptides C3a and C5a and thus recruitment of immune effector cells to the site of infection, and initiates antibody mediated killing of pathogens by the downstream complement components via the generation of membranolytic C5b-9 and/or opsonization of the pathogen by C3b (also iC3b, C4b and C1q) followed by complement receptor mediated ingestion and degradation by phagocytes. Evidence has since accumulated of antibody independent activation of C1 via C1q binding to other exogenous molecules such as lipid A and other constituents of bacteria and viruses indicating a primary role of C1 in protection from pathogens. In addition, endogenous substances such as apoptotic cells, beta sheet amyloid fibrils, pentraxins and others have been shown to bind C1q and activate C1 (reviewed in (Bohlson et al., 2007)). This diversity of possible interactions is derived from the 3 independently folded A, B, and C globular domains and combination of sites as indicated above. The outcome of these interactions depends on both the complement proteins present at the site as well as the ability of the activating substance to bind complement activation inhibitors (reviewed in (Sjoberg et al., 2009)).
2.2. Enhancement of phagocytosis
In the late 1980s, a role for C1q that was independent of complement activation emerged with the observation that immobilized C1q, in the absence of C1r and C1s, triggered an enhancement of FcγR-mediated phagocytosis when targets were coated with a sub-optimal concentration of antibody (Bobak et al., 1987). This observation suggested that C1q may be beneficial during early stages of an immune response, or in the immunocompromised, conditions that may correspond to limited antibody production. The activity required the collagen-like tail of C1q (Bobak et al., 1987), and extended to enhancement of CR1-mediated phagocytosis (Bobak et al., 1988). Moreover, the enhancement of phagocytic function was shared with structurally similar “defense collagens” including SP-A and MBL (Tenner et al., 1989; Tenner et al., 1995). C1q stimulated enhanced phagocytic function when it was either immobilized on a substrate, or within a soluble immune-complex (IC) (Webster et al., 2001).
Systemic lupus erythematosus (SLE) is an autoimmune disease characterized by the generation of autoantibodies (reviewed in (Beurskens et al., 2015)) and IC deposition leading to chronic inflammation and tissue/organ destruction. C1q–deficiency in humans results in development of SLE or an SLE-like syndrome (Manderson et al., 2004; Stegert et al., 2015) consistent with a role for C1q in clearance of IC. In addition, either complete genetic C1q deficiency in mice, or a lower production of C1q by murine peritoneal macrophages, has been linked to the presence of autoantibodies and murine lupus nephritis (Botto et al., 1998; Miura-Shimura et al., 2002). Therefore, C1q may be beneficial in facilitating clearance of IC in vivo thus limiting pathology associated with SLE both indirectly, via activation of the classical complement pathway and deposition of C3b (the ligand for CR1), or directly via enhanced phagocytosis of IC. However, an additional role of C1q in the clearance of apoptotic cells was highlighted when C1q knock out mice displayed a deficit in clearance of peritoneally injected apoptotic cells and enhanced glomerulonephritis (Botto et al., 1998). Apoptotic cells are considered the source of auto-antigen in SLE, and C1q binds to surface blebs on apoptotic human keratinocytes (Korb and Ahearn, 1997) via the globular head region (Navratil et al., 2001). Studies over the last 15 years confirmed that C1q enhances phagocytosis of a variety of apoptotic cells by multiple cell types both in vitro and in vivo (reviewed in (Galvan et al., 2012a)). Interestingly, most recently Elkon and colleagues, using an inhibitor of C1s, demonstrated that C1q facilitates apoptotic cell engulfment independent of classical pathway activation (Colonna et al., 2016).
2.3. Programmed engulfment of apoptotic cells
C1q triggers immediate responses from phagocytes resulting in enhanced phagocytosis of a variety of targets (e.g. IC, antibody and complement- opsonized targets and apoptotic cells) as discussed above, and this phenomenon has been the subject of much investigation. However, over the last decade we have begun to appreciate the role of C1q in programming responses from phagocytic cells resulting in enhanced phagocytic capacity and alternation in pro-inflammatory cytokine synthesis (reviewed in (Bohlson et al., 2014)). Analysis of gene expression profiles in C1q–stimulated mouse bone marrow derived macrophages (BMDM) identified multiple pro-phagocytic genes that were upregulated by C1q. Among these genes, Mer tyrosine kinase and its ligand Gas6 were upregulated in C1q–stimulated BMDM, and C1q–dependent enhanced engulfment of apoptotic cells required de novo Mer expression in vitro since Mer-deficient macrophages failed to respond to C1q with enhanced engulfment of apoptotic cells (Galvan et al., 2012b). Unlike the immediate signal for enhanced phagocytosis which is mediated by the collagen-like tails of C1q, and conserved with MBL, programmed efferocytosis required the full-length C1q molecule and was not shared with MBL. While primary human macrophages failed to upregulate Mer expression following prolonged stimulation with C1q alone (Hulsebus et al., 2016), human monocytes and dendritic cells cultured with the combination of C1q and HMGB1 upregulated Mer, suggesting with a conserved pathway in mouse and human cells (Son et al., 2016). These data are consistent with the observation that there is a reduction in the expression of pro-engulfment genes in macrophages from patients with SLE (Majai et al., 2014). Future studies should reveal the contribution of C1q to expression of pro-engulfment genes in vivo.
2.4. Anti-inflammatory role of C1q parallels clearance of immune complexes and apoptotic cells
As discussed above, C1q has been shown to play a role in enhancing phagocytosis of IC and apoptotic cells, likely to be critical in the prevention of chronic inflammation and autoimmunity. In tissue where C1q can be synthesized in the absence of C1r and Cls in response to injury (Bensa et al., 1983; Fonseca et al., 2011), this would facilitate rapid clearance of proinflammatory IC or cellular debris. In addition, however, C1q decreases proinflammatory cytokine release and promotes production of anti-inflammatory mediators in macrophages, dendritic cells and microglia (Bohlson et al., 2014), consistent with silent clearance of cell debris, and provides a checkpoint for induction of an immune response to cell antigens. Specifically, while enhancing clearance, C1q bound to apoptotic cells directed human monocyte-derived macrophages (HMDM) ingesting these apoptotic cells towards a less inflammatory state, characterized by induction of gene expression and protein secretion of the anti-inflammatory cytokines IL-27 and IL-10, as well as decreasing inflammasome activity and the secretion of mature IL-1β (Benoit et al., 2012). This anti-inflammatory function of C1q skews the adaptive immune system towards a more regulatory state and a limited induction of an immune response to the autoantigens being ingested. Importantly, in a mixed-leukocyte reaction, human macrophages ingesting C1q–coated apoptotic cells added to allogeneic or autologous T cells significantly and substantially decreased Th17 and Th1 subset proliferation, and demonstrated a trend towards increased Treg proliferation relative to macrophages ingesting apoptotic cells without C1q. Similarly, C1q–polarized dendritic cells decreased autologous Th17 and Th1 proliferation relative to dendritic cells ingesting apoptotic cells in the absence of C1q (Clarke et al., 2015). These data are consistent with the suppressive effect of C1q on dendritic cell maturation (Castellano et al., 2007) and that of Lu and colleagues (Teh et al., 2011) demonstrating that primary human monocytes differentiated to dendritic cells in the presence of immobilized C1q showed reduced induction of allogeneic Th1 and Th17 cells. Thus, C1q–polarized phagocytic cells regulate T effector cell activation, essentially “sculpting” the adaptive immune response to avoid autoimmunity while clearing dying cells. While there have been some studies suggesting the possibility of a direct C1q interaction with T cells, more definitive investigations are needed (reviewed in (Clarke and Tenner, 2014)). The recent confirmation of the predominant role of C1q (rather than C1 activation and C3b deposition) on clearance and immunosuppression of apoptotic cells should direct therapeutic intervention in SLE and other autoimmune diseases (Colonna et al., 2016).
C1q–dependent immunoregulatory functions have also been observed in IC clearance via different mechanisms. Interestingly, in the human system, C1q–bound to ICs markedly shifted IC binding to monocytes and away from plasmacytoid DCs (pDCs), thereby reducing the expression of the majority of IFNα-response genes induced by ICs (Santer et al., 2010; Santer et al., 2012) and characteristic of the IFN -signature associated with SLE progression (Baechler et al., 2003). In the mouse, Ossendorp and colleagues have reported that C1q enhances uptake of IC in splenic dendritic cells, and thereby promotes cross presentation to CD8+ T cells (Ho et al., 2017). Further studies are needed to determine the contributions of these activities to regulating the immune response.
3. Interaction with cell surface proteins: old and new receptors
The interaction of C1q with the lymphocyte surface was suggested in the early 1970s (Dickler and Kunkel, 1972) and subsequently demonstrated for most cells of the peripheral blood under various conditions, as well as cells involved in tissue repair and wound healing including platelets, fibroblasts, endothelial cells, mast cells, and epithelial cells (reviewed by (Kouser et al., 2015)). Deciphering which are the key molecules involved in these different contexts remains under investigation. Rather than “one C1q receptor or even receptor complex”, it is now proposed that different complexes of surface molecules act in concert upon engagement of C1q to induce function relevant to the microenvironment, either to maintain homeostasis, or to respond to infection or other challenges in tissue. Apart from historical C1q–binding proteins, cC1qR (CRT) and gC1qR, which lack transmembrane domains, known C1q receptors might be grouped according to the nature of their extracellular domains: (i) large multmodular ectodomains involved in interaction with multiple ligands (CR1, LRP1 and the recently identified scavenger receptors SR-F1 and SR-F3); (ii) integrins (α2-β1 and CR3/αM-β2); (iii) Ig-like receptors (RAGE, LAIR-1 and CD33) and (iv) the lectin receptors DC-SIGN and DC-SIGNR. Another important feature of C1q receptors lies in the existence of soluble counterparts, arising from gene alternative splicing (soluble RAGE and DC-SIGN isoforms), gene duplication (LAIR-2), or proteolytic ectodomain shedding (as reported for CR1 and LRP1). These soluble proteins might act as decoy receptors by competing with cell surface receptors for ligand binding and preventing subsequent signal transmission, thereby playing a role in regulation of C1q–mediated cellular functions. The main features of C1q receptors are summarized in Table 1 and we will address a few of these below.
Table 1.
Main features of C1q receptors
| Names (CD number) Family |
Cellular expression |
Ectodomain composition |
C1q region recognized |
Main protein ligands /partners |
C1q–mediated biological response |
|---|---|---|---|---|---|
|
| |||||
| gC1qR, C1qBP, p33, p32 | Mitochondrial protein | “doughnut-like” trimer | gC1q | DC-SIGN, CRT β1 integrin | Regulation of DC differentiation (trimolecular complex with DC-SIGN) |
|
| |||||
| cC1qR, calreticulin, CRT | ER lectin-like chaperone | Lectin-like + “arm” | cC1q | MBL, ficolins, LRP1, CD59 | Eat-me signal for apoptotic cells phagocytosis |
| gC1q | |||||
|
| |||||
| CR1 (CD35) Regulator of complement activation | Erythrocytes, leukocytes, DCs | 4 long homologous repeats of CCP modules | cC1q | C3b, C4b MBL, ficolins | Possible role in the clearance of immune complexes |
|
| |||||
| LRP1,α2M receptor (CD91) LDL receptor | MФs, DCs, nonprofessional phagocytes | 4 clusters of LA, WTD and EGF-like modules | cC1q | Lipoproteins, ECM proteins, CRT, CR3, protease-inhibitor complexes | Uptake of apoptotic cells |
| gC1q | |||||
|
| |||||
| α2β1 β1 integrin | leukocytes, platelets | α-β heterodimer | cC1q | Collagen, laminin MBL, SP-A, CRT | Activation of peritoneal mast cells |
|
| |||||
| CR3, Mac1, αMβ2 (CD11bCD18) β2 integrin | Specific MФ populations, DCs | α-β heterodimer | ? | iC3b, sRAGE, LRP1, collagen | Apoptotic cells clearance (trimolecular complex with RAGE) |
|
| |||||
| SREC-I, SCARF1, SR-F1 Scavenger receptor | Macrophages, DCs, endothelial cells | 7 EGF-like modules | ? | oxLDL, HSP, CRT, SRF-2 ectodomain | Apoptotic cells clearance |
|
| |||||
| Megf10, SR-F3 Scavenger receptor | Astrocytes, myosatellite cells | EMI + 15 EGF-like modules | ? | amyloid-β peptide (Aβ42) | Apoptotic cells clearance by astrocytes |
|
| |||||
| RAGE, SR-J Ig superfamily receptor | Leukocytes, DCs, endothelial cells, neurons | 3 IgG-like modules (V-C1-C2) | gC1q | HMGB1, AGE, PS, CR3, | Apoptotic cells clearance (trimolecular complex with CR3) Differentiation of monocytes to M2-like macrophages (tetramolecular complex with LAIR-1 and HMGB1) |
|
| |||||
| LAIR-1 (CD305) Inhibitory Ig-like immune receptor | Myeloid and lymphoid immune cells | Ig-like module (C2) | cC1q | MBL, SP-D, collagens) | Restriction of DC differentiation and activation Differentiation of monocytes to anti-inflammatory M2-like MФs (tetramolecular complex with RAGE and HMGB1) |
|
| |||||
| Siglec-3 (CD33) Inhibitory Ig-like immune receptor | Myeloid immune cells | 2 IgG-like modules (V-C2) | gC1q | CD14 | Restriction of DC differentiation and activation (trimolecular complex with LAIR-1) |
|
| |||||
| DC-SIGN (CD209) C-type lectin receptor | Most immune cells, Macrophages, DCs, PMNs | Tetramer; neck + C-lectin domain | gC1q | ICAM-3, gC1qR | Regulation of DC differentiation (trimolecular complex with gC1qR) |
3.1. cC1q and gC1q receptors
Paradoxically, the first two C1q binding proteins that were described as receptors for the collagen-like and the globular regions of C1q (cC1qR/collectin receptor/calreticulin (CRT) and gC1qR, respectively) (Malhotra et al., 1993; Ghebrehiwet et al., 1994) turned out to be multifunctional, primarily intracellular proteins located in the endoplasmic reticulum (cC1qR/CRT) and in mitochondria (gC1qR/p33), but which were detected at the surface of a wide variety of cells. Since both proteins lack a transmembrane domain or lipid anchor, they need to function in association with other surface molecules. (Ogden et al., 2001; Vandivier et al., 2002; Ghiran et al., 2003) (Hosszu et al., 2012). There are however contradictory reports and definitive experiments remain to be done to establish functional correlates of these protein interactions (see below). In addition, the term cC1qR, although historical, is probably no longer appropriate since it has been demonstrated that cC1qR is identical to CRT and also binds to the globular regions of C1q (Kovacs et al., 1998; Paidassi et al., 2011).
3.2. Receptors involved in C1q enhancement of phagocytosis
Identification of the receptor(s) required for C1q–dependent phagocytosis has remained enigmatic despite years of investigation. Monoclonal antibodies reactive with CD93, a transmembrane glycoprotein expressed on phagocytic cells, inhibited C1q–dependent enhanced phagocytosis (Guan et al., 1994; Steinberger et al., 2002). However, CD93 did not bind directly to C1q (McGreal et al., 2002), and mice deficient in CD93, though deficient in clearance of apoptotic cells in vivo, responded to C1q with enhanced phagocytic function in vitro (Norsworthy et al., 2004). Similarly, antibodies against CRT inhibited C1q–dependent engulfment (Ogden et al., 2001). CRT has been localized to the plasma membrane of multiple cell types, including apoptotic cells and cancer cells, where it functions as an “eat me” signal. While CRT-dependent engulfment of apoptotic cells required expression of LRP1/CD91 on the phagocyte (Gardai et al., 2005), LRP1/CD91 was not required for C1q–dependent enhancement of FcγR-mediated phagocytosis, nor was it required for C1q–dependent engulfment of apoptotic cells in the presence of serum (Lillis et al., 2008). These data suggest that multiple receptors or multi-molecular complexes may regulate C1q–dependent phagocytic function, as discussed further below.
3.3. Multimodular endocytic and scavenger receptors
CR1 is known as a receptor for complement fragments C4b and C3b (immune adherence receptor) and a regulator of complement activation, (reviewed in (Krych-Goldberg and Atkinson, 2001) ). C1q has been shown to interact with CR1 LHR-D in vitro (Klickstein et al., 1997), but the contribution of the C1q–CR1 interaction in immune adherence remains to be fully elucidated.
LRP1, a large, 600-kDa multifunctional endocytic receptor of the LDL receptor family, has been shown to interact with C1q in vitro (Duus et al., 2010a) but as mentioned above, at least the ability of C1q to enhance engulfment of apoptotic cells or antibody-coated targets is not impaired by the lack of CD91/LRP (Lillis et al., 2008). Both CR1 and LRP1 have been shown to interact with other defense collagens such as MBL and ficolins in vitro (Duus et al., 2010b; Jacquet et al., 2013), suggesting binding of these receptors to the conserved collagen-like regions of the defense collagens.
The class F scavenger receptors SR-F1 (alias SCREC-I/SCARF1) and SR-F3 (alias Megf10) have been suggested to play a crucial role in the C1q–dependent engulfment of apoptotic cells and in preventing autoimmunity (Ramirez-Ortiz et al., 2013; Iram et al., 2016). SR-F1 has been described as an endocytic receptor for CRT (Berwin et al., 2004), but no interaction was detected using isolated proteins (Ramirez-Ortiz et al., 2013). It has also been shown to interact through its ectodomain with the related scavenger receptor SRF-2 (Ishii et al., 2002), but the possible role of this interaction in the context of C1q binding remains to be investigated. Recessive mutations in Megf10/SR- F3 result in a rare disease characterized by early-onset myopathy, respiratory distress and dysphagia (EMARDD), and cells expressing EMARDD mutations are defective at engulfment of apoptotic cells and internalization of C1q (Iram et al., 2016). However, the role of C1q in progression of EMARDD and the protein domains involved in C1q–SR-F1 or -SR-F3 interaction remain to be determined.
3.4. Integrins α2β1 and αMβ2
Integrin α2β1 is known as a receptor for extracellular matrix components, including collagen and laminin. It has been shown to interact with CRT at the surface of various cells, including platelets (Elton et al., 2002) and with C1q and other members of the collectin family such as MBL and SP-A. The C1q-α2β1 interaction has been suggested to play a role in mast cell activation and cytokine secretion and to involve the collagen-like part of C1q and the integrin inserted (I) domain, similar to that involved in collagen binding (Edelson et al., 2006).
Integrin αMβ2 (or CR3) is a phagocytic integrin that plays an essential role in the uptake of target particles (e.g. microorganisms and apoptotic cells) opsonized with complement C3 fragments (reviewed in (Ehlers, 2000)), as well as is essential for the generation of superoxide by human neutrophils triggered by immobilized C1q (Goodman et al., 1995). CR3 has been shown to interact extracellularly with C1q receptor components including LRP1 (Ranganathan et al., 2011) and RAGE, and directly with C1q (Ma et al., 2012). Since the collagen domain of C1q is sufficient for the CD18-dependent induction of superoxide in neutrophils, and CR3 has recently been reported to bind collagens in vitro (Lahti et al., 2013), it is possible that the interaction of CR3 involves the collagen-like region of C1q, but this has not yet been biochemically determined.
3.5. Ig-like receptors RAGE, LAIR-1 and CD33
RAGE is a multi-ligand receptor for endogenous danger signals such as AGEs, HMGB1 and DNA, and participates in apoptotic cell clearance by acting as a phosphatidylserine (PS) receptor (reviewed in (Sorci et al., 2013). It was shown recently to bind to the globular regions of C1q and to enhance C1q–mediated phagocytosis of apoptotic cells, a process proposed to involve a ternary complex with the CR3 receptor (Ma et al., 2012).
LAIR-1 is an inhibitory collagen receptor involved in the control of various phases of the immune response (reviewed by (Meyaard, 2008)). LAIR-1 and its soluble homologue LAIR-2 have been shown to interact with C1q and related proteins, including MBL and surfactant protein D, through their collagen-like regions (Olde Nordkamp et al., 2014b; Olde Nordkamp et al., 2014a). LAIR-1 engagement by C1q inhibits DC differentiation and activation either during steady state or inflammation and is suggested to help maintain monocyte tolerance (Son et al., 2012; Son and Diamond, 2015). In addition, C1q was reported recently to suppress the HMGB1-polarization of monocytes and maintain an anti-inflammatory M2-like macrophage, a pathway mediated through a complex with both RAGE and LAIR-1 and depending on the relative levels of C1q and HMGB1 (Son et al., 2016).
CD33 is an inhibitory immunoreceptor of the sialic acid Ig-like lectin (Siglec) family of receptors able to promote sialic acid-dependent cell adhesion (Crocker et al., 2007). C1q was reported recently to crosslink CD33 and LAIR-1, thereby triggering phosphorylation of CD33/LAIR-1 inhibitory motifs (Son et al., 2017). While LAIR-1 interacted with the collagenlike tail of C1q, CD33 bound to the globular head region. Taken together, these data suggest that full length C1q concurrently crosslinks multiple inhibitory receptors to regulate myeloid cell activation.
3.6. C-type lectin receptors DC-SIGN and DC-SIGNR
DC-SIGN is a C-type lectin receptor involved in multiple functions including adhesion, pathogen recognition and antigen presentation (Garcia-Vallejo and van Kooyk, 2013). Interactions between neutrophils and DCs are mediated through interaction of DC-SIGN with CR3 (Ludwig et al., 2006). DC-SIGN and the related protein DC-SIGNR were reported recently to bind to C1q globular heads (Hosszu et al., 2012; Pednekar et al., 2016). In addition, C1q and gC1qR were shown to associate with DC-SIGN on the surface of immature DCs and to regulate DC differentiation and function. Interestingly, SIGN-R1, a DC-SIGN homolog expressed on mouse splenic marginal zone macrophages, was shown to enhance apoptotic cells clearance through interaction with C1q and subsequent complement activation (Prabagar et al., 2013). The SIGNR1-C1q interaction has been proposed to involve the primary carbohydrate binding site in SIGN-R1 and the polysaccharides ending in a 2,6 sialic acid present in the globular domain of the C1q A chain (Silva-Martin et al., 2014), whereas binding of C1q to DC-SIGN was Ca2+-dependent and inhibited by mannan, suggesting the involvement of C1q mannose residues (Hosszu et al., 2012).
4. C1q- Recent advances and novel findings
4.1 Expression of C1q at tissue level
While plasma C1q can be produced by the Kupffer cells (macrophages of the liver), it is now known that the bone marrow myeloid cells can completely rescue C1q deficiency (Petry et al., 2001) and that many tissue myeloid cells can be induced to produce C1q, including mast cells and chondrocytes (Bradley et al., 1996; van Schaarenburg et al., 2016). As described above, C1q has long been recognized as initiator of the classical pathway of complement (C) activation and its tissue deposition when co-localized with C4 and C3 was usually regarded as an indication of local C activation (Ricklin et al., 2010; Merle et al., 2015). However, increasing evidence collected over recent years suggests that local presence of C1q should not necessarily be considered a marker of C activation as it is not always associated with C4 deposition (Kouser et al., 2015). In addition, while plasma containing circulating C1q as part of C1 complex (Ziccardi and Tschopp, 1982) may represent a possible source of tissue-bound C1q in cases of breached vasculature, it is increasing evident that C1q can be synthesized and secreted locally by various cell types, including macrophages, dendritic cells, fibroblasts, and mast cells that are ubiquitously distributed throughout the body (Ghebrehiwet et al., 2012). In addition, other cells selectively localized in specific tissues and organs such as microglial cells, glomerular and tubular cells, osteoclasts, and trophoblasts contribute to local production of C1q (Fonseca et al., 2017; Xavier et al., 2017). Although C1q is likely to be secreted in limited amount at the extravascular sites, recent evidence suggests that locally synthesized C1q is involved in the regulation of multiple cellular functions in addition to the cellular control of innate and adaptive immunity, as described above.
4.2. Role of C1q in the pathophysiology of pregnancy
Interest in placenta as site of C involvement was prompted several years ago by the recognition that this newly formed tissue connecting the developing embryo to the mother undergoes profound changes to allow successful embryo implantation and regular progression of pregnancy. These changes include extensive tissue remodeling that occurs primarily in the maternal decidua and is associated with an inflammatory process. The inflammation that develops during embryo implantation is sustained by cytokines and chemokines released by decidual cells and hormonal changes and is compatible with a successful pregnancy (Bulla et al., 2012). The cell debris formed as a result of tissue remodeling and the inflammatory process contributes to trigger C activation. Analysis of term and preterm placenta performed in the eighties and the early nineties revealed the presence of both early and late C components in the stroma of villi around the vessels, in the perivillous deposits of fibrin. C1q was also detected in maternal decidua in areas of fibrinoid necrosis localized in the basal plate and fibrinogen deposits in the wall of uteroplacental arteries (Girardi et al., 2006). More recent studies have highlighted the contribution of C1q to tissue remodeling in placenta acting predominantly on extravillous trophoblasts that depart from the cell columns of the anchoring villi and invade the maternal decidua reaching the inner third of the myometrium. A large number of these cells surrounds the uterine spiral arteries forming cell cuffs and contribute together with decidual NK cells to the extensive vascular remodeling characterized by loss of the musculoelastic structure of the vessel wall replaced by fibrinoid material (Bulla et al., 2012). As a result of these structural changes, the arteries transform into large vessels with markedly reduced resistance that favors unrestricted blood flow in the intervillous space (Bulla et al., 2005; Pijnenborg et al., 2006). The extravillous trophoblasts were found to express the messages for the three chains of C1q when they start invading the decidua (Agostinis et al., 2010). The molecule secreted from these cells, which are widely distributed in decidual tissue, binds to the extracellular matrix and, by interacting with cell surface-expressed receptor for the globular head of C1q and the associated α4β1 integrin, delivers an activation signal and promotes trophoblast migration. The relevance of these findings to pregnancy outcome is supported by the impaired labyrinth development and the vessel remodeling observed in pregnant C1q–deficient mice compared to wild-type mice (Agostinis et al., 2010). Moreover, the absence of C1q predisposes these mice to manifest clinical and laboratory signs of pre-eclampsia including hypertension, albuminuria, reduced expression of VEGF, increased level of circulating VEGF receptor1 (sFlt-1), and increased fetal loss (Singh et al., 2011). Interestingly, trophoblasts surrounding unremodeled spiral arteries found in a substantial number in pre-eclamptic placentae and considered as a characteristic feature of this clinical condition, fail to express C1q (Agostinis et al., 2011).
A distinct group of extravillous trophoblasts enters the lumen of the spiral arteries and migrates upward like metastatic cells acquiring an endothelial cell phenotype that allows the endovascular trophoblasts to adhere and partially replace the endothelium. (Bulla et al., 2005). C1q plays an important role in this process acting as a molecular bridge between endovascular trophoblasts and decidual endothelial cells. These are the only endothelial cells that synthesize and express C1q on the cell surface under physiological conditions (Bulla et al., 2008). The ability of both cells to produce C1q and to express the receptors for C1q favors a cross interaction between the two cell types.
4.3. Contribution of C1q to tissue repair
Wound healing is probably the best studied example of tissue regeneration and proceeds through three successive phases comprising blood clotting and inflammation, proliferation characterized by granuloma tissue formation and angiogenesis, and tissue remodeling (Sun et al., 2014). C1q has been shown to act at a later stage of the regeneration process and to favor wound healing stimulating angiogenesis in a C activation-independent manner (Bossi et al., 2014). Immunohistochemical analysis of the granulation tissue formed in the proliferation phase revealed strong C1q staining of the vessel endothelium and the surrounding stroma in the lesional skin in the absence of C4 and C3. The endothelial cells of newly formed vessels, unlike the cells of normal skin, were found to express the messages for the three chains of C1q and to synthesize the molecule detected on the cell surface. By interacting with the receptor for the globular head (gC1qR), C1q proved to be effective in stimulating proliferation and migration of endothelial cells and in inducing vessel formation evaluated both in vitro by the tube formation assay and ex-vivo by the rat aortic ring assay. The proangiogenic activity of C1q was further supported by the observation that the mean vascular density in the wound was significantly reduced in C1q−/− mice compared to wild-type animals and was restored to normal after topical application of C1q (Bossi et al., 2014).
C1q has been shown to have a different effect on skeletal muscle regeneration after injury causing impaired tissue repair in aged wild-type mice that was not seen in C1q deficient mice and that this effect was due to C1q–dependent activation of the canonical Wnt pathway involved in the regulation of mammalian aging. (Naito et al., 2012). Although the contribution of the classical pathway downstream of C1 activation was excluded by the failure of C3-deficiency to prevent the impairment of C1q–induced muscle regeneration, the finding that the activation of Wnt signaling was inhibited by C1-inhibitor or a neutralizing antibody to C1s suggest that C1s is also involved in this process (Naito et al., 2012). The same group has subsequently shown that inhibition of the angiotensin II receptor, suppresses C1q synthesis in injured muscles and enhanced recovery of muscle function (Yabumoto et al., 2015), providing potential for therapeutic application if these results translate to humans.
4.4. Effect of C1q on cancer development
Tumor growth is largely dependent on the intrinsic properties of cancer cells as well as on several factors, including the C system, that may either control or promote its progression (Markiewski et al., 2008; Macor et al., 2015). C1q has recently attracted special attention for its potential contribution to regulation of tumor growth following the detection of substantial amounts of this C component in the tumor microenvironment (Bulla et al., 2016). Analysis of tissue specimens obtained from solid tumors including colon, lung, breast, pancreatic carcinoma and melanoma revealed C1q deposition on the vascular endothelium, and in the stroma, mesenchymal cells, and mononuclear cells surrounding the tumor mass at the edge of tumor invasion. Interestingly, the cancer cells failed to express C1q in all tumors examined. A syngeneic model of melanoma developed in C57BL/6 mice showed prolonged survival of tumor-bearing mice and slower tumor growth in C1q–deficient mice compared to wild-type mice. The slower cancer progression observed in the absence of C1q was associated with reduced vascular density and was unrelated to local recruitment of suppressor cells that favor tumor expansion by inhibiting the immune response (Bulla et al., 2016). Similar results were obtained using a syngeneic mouse model of lung carcinoma. In vitro experiments showed that surface-bound C1q was able to promote adhesion, proliferation and migration of the melanoma cells used for the tumor model and to protect them from apoptosis. Although the ligands for C1q on cancer cells that mediate its biological effects were not identified, gC1qR was hyper-expressed in several human tumors (Dembitzer et al., 2012), and is likely to be one of them.
It is important to emphasize that not all cancer cells respond to C1q in the same way. Hong et al. (Hong et al., 2009) showed that C1q induced apoptosis and growth suppression of the human prostate DU145 cells as a result of direct activation of the tumor suppressor WW-domain containing oxidoreductase (WOX1). Interestingly, analysis of prostate tissue revealed that C1q was present in normal prostate while it was hypo-expressed in benign prostatic hyperplasia (BPH) and prostate cancer further supporting the contribution of C1q to the control of cell proliferation. In addition, C1q was reported to induce a pro-apoptotic effect on an ovarian cell line, SKOV3, acting via a TNF-α induced apoptosis pathway that involves upregulation of Bax and Fas (Kaur et al., 2016). Although these in vitro data were obtained analyzing cell lines under experimental conditions that do not reflect the complex tumor microenvironment, recent in vivo findings confirmed the protective activity of C1q against the mammary carcinoma that develops spontaneously in BALB-neuT mice (Bandini et al., 2016). C1q–deficient BALB-neuT mice manifested an accelerated tumor growth associated with an increased number of intra-tumoral vessels compared to wild-type neuT mice. The difference in tumor progression between C1q–deficient and WT mice was attributed to a reduced activation of WW domain containing oxidoreductase (WWOX) in C1q–deficient mice. Unfortunately, no attempt was made to reverse the accelerated progression of tumor in C1q–deficient mice by administration of C1q or bone marrow transplantation.
These contradictory data on the effect of C1q on tumor growth in the in vivo studies may be explained by differences in mouse strains, type of tumors and tumor models used. However, two important points should be considered when translating the murine data to cancer patients. One is that C1q has been detected in the microenvironment of growing solid tumors and its expression has been associated with cancer invasion and metastatic foci (Bulla et al., 2016). More importantly, analysis of gene expression in human breast cancer by Winslow and colleagues (Winslow et al., 2015) revealed an increased expression of the genes for the three chains of C1q associated with a poor prognosis suggesting that C1q was not protective and was able to promote tumor growth. Given the conflicting results obtained by different groups, further studies are warranted to define the role of C1q in cancer development.
4.5. C1q in the nervous system
With exciting new discoveries of critical roles for classical complement pathway in pruning of neuronal synapses, the influence of the complement system in central nervous system has been substantially expanded (Stephan et al., 2012). How complement impacts disease progression is clearly complex, with evidence of both complement-dependent detrimental and protective effects (reviewed in (Wyss-Coray and Rogers, 2012)).
C1q expression is upregulated following neuronal injury, during aging and early in neurodegenerative disorders such as Alzheimer’s disease (reviewed in (Veerhuis et al., 2011)). Similar to its activities in the periphery, C1q enhances the uptake of apoptotic neurons and neuronal blebs by microglia, the primary phagocytic cell of the central nervous system. More rapid ingestion by microglia of apoptotic neurons or neuronal blebs should prevent toxic intracellular contents (such as high concentrations of glutamate) from being released thereby preventing excitotoxic damage to surrounding neurons. C1q either immobilized or bound to apoptotic neurons also suppresses the LPS-induced production of proinflammatory cytokines IL-1α, IL-1β, IL-6 and TNF-α, all suggesting anti-inflammatory/homoeostatic role for C1q in the brain (Fraser et al., 2010), and predicting neuroprotection.
In an unexpected discovery, it was observed that in vitro C1q, in the absence of other complement components, induces gene expression critical for neuronal survival and neurite outgrowth (Pisalyaput and Tenner, 2008; Benoit and Tenner, 2011) and protection against oligomeric and fibrillar Aβ-induced neuronal death (Benoit et al., 2013). Microarray analysis demonstrated that C1q up-regulates genes involved in membrane and cytoskeleton processes, in growth of neurites and cholesterol metabolism, and in the regulation of the expression of neurotrophic factors (Benoit and Tenner, 2011). The transcriptional programs stimulated by C1q in nutrient stressed or Aβ-injured neurons share some pathways (Benoit and Tenner, 2011; Benoit et al., 2013). A common signaling event is the phosphorylation and nuclear translocation of the transcription factor CREB. CREB is a transcription factor vital for long-term memory and synaptic plasticity, neurogenesis (Tchantchou et al., 2009; Li et al., 2011) and induction of neurotrophic factors in the CNS (Colangelo et al., 1998), thus suggesting mechanisms by which the C1q–induced neuroprotection may occur.
However, as injury increases, synthesis of most other complement factors is also increased. Interestingly, in the presence of other complement components the removal or loss of sialic acid on neuronal membranes results in binding of C1 via C1q, activation of complement, deposition of C3b on the neurons and subsequent ingestion of those membranes by microglia via CR3, the receptor for C3b/iC3b (Linnartz et al., 2012). How CD33, a sialic acid binding negative regulator of the immune system that is expressed on human microglia and that can interact with C1q (as described above), influences this activity, if at all, remains to be defined (reviewed in (Linnartz-Gerlach et al., 2014)). It is firmly established both in vitro and in vivo that fibrillar (beta sheet fibrils) Aβ (fAβ) which accumulate in Alzheimer’s disease (AD) can activate both the alternative and the classical complement pathways in the absence of antibody. Thus, in the AD brain both activators and the complement proteins are present leading to the colocalization of C1q, C3b/iC3b and C4b to nearly 100% of the fibrillar amyloid plaques (vs. diffuse plaques) in AD in humans and in mouse models of AD. In addition, activated microglia and astrocytes and the complement membranolytic complex C5b-9 have been detected in the areas containing the fibrillar plaques in AD (Webster et al., 1997) as reviewed in (Veerhuis et al., 2011), suggesting a detrimental outcome of complement activation in neurodegenerative diseases.
Innovative studies by Stevens, Barres and colleagues, beautifully demonstrated that activation of the early components of the classical complement pathway, C1, C4 and C3 during normal development contributes to synaptic pruning esssential for proper neuron circuit construction (Stevens et al., 2007). In contrast to the predominant synthesis of C1q in the adult brain by microglia (Fonseca et al., 2017), here C1q was produced by the neurons in a TGFβ and activity dependent fashion (Bialas and Stevens, 2013). Subsequently, these investigators and others have further uncovered that excessive complement-mediated synapse elimination is associated with neuronal degeneration (Williams et al., 2016) and with cognitive loss in diverse mouse models of aging and neurological disorders (Shi et al., 2015; Hong et al., 2016; Lui et al., 2016; Vasek et al., 2016). Substantial suggestions of correlates in the human diseases including not only neurodegenerative diseases such as AD and frontal temporal dementia (Hong et al., 2016; Lui et al., 2016), but also cognitive loss experienced by some after West Nile Virus infection (Vasek et al., 2016)and perhaps psychiatric disorders such as schizophrenia (Sekar et al., 2016), suggest contributions of these processes to human disease.
It remains to be determined what are the contributions of complement mediated excessive synaptic pruning, C5a induced microglial polarization, and other complement mediated functions to the cognitive loss seen in these diseases as well as the neuroprotective pathways that appear to be modulated by complement and/or C1q alone. This should enable selective targeting and/or modulation of complement activation products or their receptors as an effective strategy for retaining the neuroprotective, anti-inflammatory and phagocytic functions of complement, while dampening proinflammatory induced damage.
In summary, the C1q molecule continues to surprise and intrigue complement biologists as novel activities are discovered. Being evolutionarily ancient, it has been selected to be structurally diverse and capable of complexing with multiple binding partners in solution and on cell membranes to influence local and systemic activities. While there are many disorders where C1q–targeted therapies may be possible, a comprehensive understanding of downstream consequences of modulating this multifunctional protein will be critical for success in the clinic.
Highlights.
Classical and new functions of complement component C1q are reviewed
Recombinant C1q provides a new tool for structure-function analysis
C1q immunoregulatory functions influence innate and adaptive immunity
Novel unexpected roles for C1q in pregnancy, wound repair and cancer were identified
C1q is a multifunctional homeostatic regulator in the CNS
Acknowledgments
The work included in this review was partly supported by the French National Research Agency grants ANR-09-PIRI-0021 and ANR-16-CE11-0019-01 (CG,NMT), the NIH AG00536 (AJT), and NIH NIAID 1R15AI117474 (SSB).
Abbreviations
- Aβ
amyloid peptide
- AD
Alzheimer’s disease
- AGE
Advanced Glycation End-product
- C
complement
- cC1qR
receptor for the collagen tail of C1q
- CCP
complement control protein
- CNS
central nervous system
- CR
complement receptor
- CRT
calreticulin
- DC
dendritic cell
- DC-SIGN
DC-specific intercellular adhesion molecule (ICAM)-3 grabbing non integrin
- DC-SIGNR
DC-SIGN-related protein
- ECM
extracellular matrix
- EGF
epidermal growth factor
- EMI
domain present in proteins of the EMILIN family
- ER
endoplasmic reticulum
- gC1qR
receptor for the globular “heads” of C1q
- HMGB1
high mobility group protein B1
- HSP
heat-shock protein
- IC
immune complexes
- ICAM
intercellular adhesion molecule
- LA
LDL receptor-class A
- LAIR-1
leukocyte associated immunoglobulin like receptor 1
- LHR
long homologous repeat
- LPS
lipopolysaccharide
- LRP1
low density lipoprotein receptor related protein 1
- LTA
lipoteichoic acid
- MBL
mannose-binding lectin
- PS
phosphatidylserine
- RAGE
AGE receptor
- SLE
systemic lupus erythematosus
- SP
surfactant protein
- SR
scavenger receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors report no conflict of interest.
References
- Agostinis C, Bulla R, Tripodo C, Guarnotta C, De Seta F, Tonon M, Spessotto P, Botto M, Tedesco F. Preeclampsia is associated withdefective production of C1qby invasive trophoblast. Molecular Immunology. 2011;48:1678–1679. [Google Scholar]
- Agostinis C, Bulla R, Tripodo C, Gismondi A, Stabile H, Bossi F, Guarnotta C, Garlanda C, De Seta F, Spessotto P, Santoni A, Ghebrehiwet B, Girardi G, Tedesco F. An alternative role of C1q in cell migration and tissue remodeling: contribution to trophoblast invasion and placental development. Journal of Immunology. 2010;185:4420–4429. doi: 10.4049/jimmunol.0903215. [DOI] [PubMed] [Google Scholar]
- Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, Grande WJ, Hughes KM, Kapur V, Gregersen PK, Behrens TW. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bally I, Ancelet S, Moriscot C, Gonnet F, Mantovani A, Daniel R, Schoehn G, Arlaud GJ, Thielens NM. Expression of recombinant human complement C1q allows identification of the C1r/C1s–binding sites. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:8650–8655. doi: 10.1073/pnas.1304894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandini S, Macagno M, Hysi A, Lanzardo S, Conti L, Bello A, Riccardo F, Ruiu R, Merighi IF, Forni G, Iezzi M, Quaglino E, Cavallo F. The non-inflammatory role of C1q during Her2/neu-driven mammary carcinogenesis. Oncoimmunology. 2016;5:e1253653. doi: 10.1080/2162402X.2016.1253653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit ME, Tenner AJ. Complement protein C1q–mediated neuroprotection is correlated with regulation of neuronal gene and microRNA expression. J Neurosci. 2011;31:3459–3469. doi: 10.1523/JNEUROSCI.3932-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit ME, Clarke EV, Morgado P, Fraser DA, Tenner AJ. Complement protein C1q directs macrophage polarization and limits inflammasome activity during the uptake of apoptotic cells. J Immunol. 2012;188:5682–5693. doi: 10.4049/jimmunol.1103760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit ME, Hernandez MX, Dinh ML, Benavente F, Vasquez O, Tenner AJ. C1q–induced LRP1B and GPR6 Proteins Expressed Early in Alzheimer Disease Mouse Models, Are Essential for the C1q–mediated Protection against Amyloid-beta Neurotoxicity. J BiolChem. 2013;288:654–665. doi: 10.1074/jbc.M112.400168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensa JC, Reboul A, Colomb MG. Biosynthesis in vitro of complement subcomponents C1q, C1s and C1 inhibitor by resting and stimulated human monocytes. BiochemJ. 1983;216:385–392. doi: 10.1042/bj2160385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berwin B, Delneste Y, Lovingood RV, Post SR, Pizzo SV. SREC-I, a type F scavenger receptor, is an endocytic receptor for calreticulin. J Biol Chem. 2004;279:51250–51257. doi: 10.1074/jbc.M406202200. [DOI] [PubMed] [Google Scholar]
- Beurskens FJ, van Schaarenburg RA, Trouw LA. C1q, antibodies and anti-C1q autoantibodies. Mol Immunol. 2015;68:6–13. doi: 10.1016/j.molimm.2015.05.010. [DOI] [PubMed] [Google Scholar]
- Bialas AR, Stevens B. TGF-beta signaling regulates neuronal C1q expression and developmental synaptic refinement. NatNeurosci. 2013;16:1773–1782. doi: 10.1038/nn.3560. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bobak DA, Frank MM, Tenner AJ. C1q acts synergistically with phorbol dibutyrate to activate CR1-mediated phagocytosis by human mononuclear phagocytes. EurJImmunol. 1988;18:2001–2007. doi: 10.1002/eji.1830181220. [DOI] [PubMed] [Google Scholar]
- Bobak DA, Gaither TA, Frank MM, Tenner AJ. Modulation of FcR function by complement: subcomponent C1q enhances the phagocytosis of IgG-opsonized targets by human monocytes and culture-derived macrophages. Journal of Immunology. 1987;138:1150–1156. [PubMed] [Google Scholar]
- Bogin O, Kvansakul M, Rom E, Singer J, Yayon A, Hohenester E. Insight into Schmid metaphyseal chondrodysplasia from the crystal structure of the collagen X NC1 domain trimer. Structure. 2002;10:165–173. doi: 10.1016/s0969-2126(02)00697-4. [DOI] [PubMed] [Google Scholar]
- Bohlson SS, Fraser DA, Tenner AJ. Complement proteins C1q and MBL are pattern recognition molecules that signal immediate and long-term protective immune functions. Mol Immunol. 2007;44:33–43. doi: 10.1016/j.molimm.2006.06.021. [DOI] [PubMed] [Google Scholar]
- Bohlson SS, O’Conner SD, Hulsebus HJ, Ho MM, Fraser DA. Complement, c1q, and c1q–related molecules regulate macrophage polarization. Front Immunol. 2014;5:402. doi: 10.3389/fimmu.2014.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi F, Tripodo C, Rizzi L, Bulla R, Agostinis C, Guarnotta C, Munaut C, Baldassarre G, Papa G, Zorzet S, Ghebrehiwet B, Ling GS, Botto M, Tedesco F. C1q as a unique player in angiogenesis with therapeutic implication in wound healing. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:4209–4214. doi: 10.1073/pnas.1311968111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto M, Dell’agnola C, Bygrave AE, Thompson EM, Cook HT, Petry F, Loos M, Pandolfi PP, Walport MJ. Homozygous C1q deficiency causes glomerulonephritis associated with multiple apoptotic bodies. NatGenet. 1998;19:56–59. doi: 10.1038/ng0598-56. [DOI] [PubMed] [Google Scholar]
- Bradley K, North J, Saunders D, Schwaeble W, Jeziorska M, Woolley DE, Whaley K. Synthesis of classical pathway complement components by chondrocytes. Immunology. 1996;88:648–656. [PMC free article] [PubMed] [Google Scholar]
- Brodsky-Doyle B, Leonard KR, Reid KB. Circular-dichroism and electron-microscopy studies of human subcomponent C1q before and after limited proteolysis by pepsin. Biochem J. 1976;159:279–286. doi: 10.1042/bj1590279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulla R, Bossi F, Tedesco F. The complement system at the embryo implantation site: friend or foe? Front Immunol. 2012;3:55. doi: 10.3389/fimmu.2012.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulla R, Villa A, Bossi F, Cassetti A, Radillo O, Spessotto P, De Seta F, Guaschino S, Tedesco F. VE-cadherin is a critical molecule for trophoblast-endothelial cell interaction in decidual spiral arteries. Experimental Cell Research. 2005;303:101–113. doi: 10.1016/j.yexcr.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Bulla R, Agostinis C, Bossi F, Rizzi L, Debeus A, Tripodo C, Radillo O, De Seta F, Ghebrehiwet B, Tedesco F. Decidual endothelial cells express surface-bound C1q as a molecular bridge between endovascular trophoblast and decidual endothelium. Molecular Immunology. 2008;45:2629–2640. doi: 10.1016/j.molimm.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulla R, Tripodo C, Rami D, Ling GS, Agostinis C, Guarnotta C, Zorzet S, Durigutto P, Botto M, Tedesco F. C1q acts in the tumour microenvironment as a cancer-promoting factor independently of complement activation. Nat Commun. 2016;7:10346. doi: 10.1038/ncomms10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano G, Woltman AM, Schlagwein N, Xu W, Schena FP, Daha MR, van KC. Immune modulation of human dendritic cells by complement. EurJ Immunol. 2007;37:2803–2811. doi: 10.1002/eji.200636845. [DOI] [PubMed] [Google Scholar]
- Clarke EV, Tenner AJ. Complement modulation of T cell immune responses during homeostasis and disease. JLeukocBiol. 2014;96:745–756. doi: 10.1189/jlb.3MR0214-109R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke EV, Weist BM, Walsh CM, Tenner AJ. Complement protein C1q bound to apoptotic cells suppresses human macrophage and dendritic cell-mediated Th17 and Th1 T cell subset proliferation. JLeukocBiol. 2015;97:147–160. doi: 10.1189/jlb.3A0614-278R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colangelo AM, Johnson PF, Mocchetti I. beta-adrenergic receptor-induced activation of nerve growth factor gene transcription in rat cerebral cortex involves CCAAT/enhancer-binding protein delta. ProcNatlAcadSciUSA. 1998;95:10920–10925. doi: 10.1073/pnas.95.18.10920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna L, Parry GC, Panicker S, Elkon KB. Uncoupling complement C1s activation from C1q binding in apoptotic cell phagocytosis and immunosuppressive capacity. Clin Immunol. 2016;163:84–90. doi: 10.1016/j.clim.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- Dembitzer FR, Kinoshita Y, Burstein D, Phelps RG, Beasley MB, Garcia R, Harpaz N, Jaffer S, Thung SN, Unger PD, Ghebrehiwet B, Peerschke EI. gC1qR expression in normal and pathologic human tissues: differential expression in tissues of epithelial and mesenchymal origin. Journal of Histochemistry and Cytochemistry. 2012;60:467–474. doi: 10.1369/0022155412440882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickler HB, Kunkel HG. Interaction of aggregated -globulin with B lymphocytes. Journal of Experimental Medicine. 1972;136:191–196. doi: 10.1084/jem.136.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duus K, Hansen EW, Tacnet P, Frachet P, Arlaud GJ, Thielens NM, Houen G. Direct interaction between CD91 and C1q. FEBS J. 2010a;277:3526–3537. doi: 10.1111/j.1742-4658.2010.07762.x. [DOI] [PubMed] [Google Scholar]
- Duus K, Thielens NM, Lacroix M, Tacnet P, Frachet P, Holmskov U, Houen G. CD91 interacts with mannan-binding lectin (MBL) through the MBL-associated serine protease-binding site. FEBS J. 2010b;277:4956–4964. doi: 10.1111/j.1742-4658.2010.07901.x. [DOI] [PubMed] [Google Scholar]
- Edelson BT, Stricker TP, Li Z, Dickeson SK, Shepherd VL, Santoro SA, Zutter MM. Novel collectin/C1q receptor mediates mast cell activation and innate immunity. Blood. 2006;107:143–150. doi: 10.1182/blood-2005-06-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MR. CR3: a general purpose adhesion-recognition receptor essential for innate immunity. Microbes Infect. 2000;2:289–294. doi: 10.1016/s1286-4579(00)00299-9. [DOI] [PubMed] [Google Scholar]
- Elton CM, Smethurst PA, Eggleton P, Farndale RW. Physical and functional interaction between cell-surface calreticulin and the collagen receptors integrin alpha2beta1 and glycoprotein VI in human platelets. Thromb Haemost. 2002;88:648–654. [PubMed] [Google Scholar]
- Fonseca MI, Chu SH, Berci AM, Benoit ME, Peters DG, Kimura Y, Tenner AJ. Contribution of complement activation pathways to neuropathology differs among mouse models of Alzheimer’s disease. J Neuroinflammation. 2011;8:4. doi: 10.1186/1742-2094-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca MI, Chu SH, Hernandez MX, Fang MJ, Modarresi L, Selvan P, MacGregor GR, Tenner AJ. Cell-specific deletion of C1qa identifies microglia as the dominant source of C1q in mouse brain. J Neuroinflammation. 2017;14:48. doi: 10.1186/s12974-017-0814-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser DA, Pisalyaput K, Tenner AJ. C1q enhances microglial clearance of apoptotic neurons and neuronal blebs, and modulates subsequent inflammatory cytokine production. J Neurochem. 2010;112:733–743. doi: 10.1111/j.1471-4159.2009.06494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriaud C, Frachet P, Thielens NM, Arlaud GJ. The human c1q globular domain: structure and recognition of non-immune self ligands. Front Immunol. 2011;2:92. doi: 10.3389/fimmu.2011.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriaud C, Juanhuix J, Gruez A, Lacroix M, Darnault C, Pignol D, Verger D, Fontecilla-Camps JC, Arlaud GJ. The crystal structure of the globular head of complement protein C1q provides a basis for its versatile recognition properties. J Biol Chem. 2003;278:46974–46982. doi: 10.1074/jbc.M307764200. [DOI] [PubMed] [Google Scholar]
- Galvan MD, Greenlee-Wacker MC, Bohlson SS. C1q and phagocytosis: the perfect complement to a good meal. JLeukocBiol. 2012a;92:489–497. doi: 10.1189/jlb.0212099. [DOI] [PubMed] [Google Scholar]
- Galvan MD, Foreman DB, Zeng E, Tan JC, Bohlson SS. Complement component C1q regulates macrophage expression of Mer tyrosine kinase to promote clearance of apoptotic cells. Journal of Immunology. 2012b;188:3716–3723. doi: 10.4049/jimmunol.1102920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Li M, Ma J, Zhang S. An amphioxus gC1q protein binds human IgG and initiates the classical pathway: Implications for a C1q–mediated complement system in the basal chordate. Eur J Immunol. 2014;44:3680–3695. doi: 10.1002/eji.201444734. [DOI] [PubMed] [Google Scholar]
- Garcia-Vallejo JJ, van Kooyk Y. The physiological role of DC-SIGN: a tale of mice and men. Trends Immunol. 2013;34:482–486. doi: 10.1016/j.it.2013.03.001. [DOI] [PubMed] [Google Scholar]
- Gardai SJ, McPhillips KA, Frasch SC, Janssen WJ, Starefeldt A, Murphy-Ullrich JE, Bratton DL, Oldenborg PA, Michalak M, Henson PM. Cell-Surface Calreticulin Initiates Clearance of Viable or Apoptotic Cells through trans-Activation of LRP on the Phagocyte. Cell. 2005;123:321–334. doi: 10.1016/j.cell.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Ge H, Xiong Y, Lemon B, Lee KJ, Tang J, Wang P, Weiszmann J, Hawkins N, Laudemann J, Min X, Penny D, Wolfe T, Liu Q, Zhang R, Yeh WC, Shen W, Lindberg R, Wang Z, Sheng J, Li Y. Generation of novel long-acting globular adiponectin molecules. J Mol Biol. 2010;399:113–119. doi: 10.1016/j.jmb.2010.03.062. [DOI] [PubMed] [Google Scholar]
- Ghebrehiwet B, Hosszu KK, Valentino A, Peerschke EI. The C1q family of proteins: insights into the emerging non-traditional functions. Front Immunol. 2012:3. doi: 10.3389/fimmu.2012.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghebrehiwet B, Lim BL, Peerschke EI, Willis AC, Reid KB. Isolation, cDNA cloning, and overexpression of a 33-kD cell surface glycoprotein that binds to the globular “heads” of C1q. J Exp Med. 1994;179:1809–1821. doi: 10.1084/jem.179.6.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiran I, Klickstein LB, Nicholson-Weller A. Calreticulin is at the surface of circulating neutrophils and uses CD59 as an adaptor molecule. J Biol Chem. 2003;278:21024–21031. doi: 10.1074/jbc.M302306200. [DOI] [PubMed] [Google Scholar]
- Girardi G, Bulla R, Salmon JE, Tedesco F. The complement system in the pathophysiology of pregnancy. Molecular Immunology. 2006;43:68–77. doi: 10.1016/j.molimm.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Gohara DW, Di Cera E. Allostery in trypsin-like proteases suggests new therapeutic strategies. Trends Biotechnol. 2011;29:577–585. doi: 10.1016/j.tibtech.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman EB, Anderson DC, Tenner AJ. C1q triggers neutrophil superoxide production by a unique CD18-dependent mechanism. JLeukocBiol. 1995;58:168–176. doi: 10.1002/jlb.58.2.168. [DOI] [PubMed] [Google Scholar]
- Goshima M, Sekiguchi R, Matsushita M, Nonaka M. The complement system of elasmobranches revealed by liver transcriptome analysis of a hammerhead shark, Sphyrna zygaena. Dev Comp Immunol. 2016;61:13–24. doi: 10.1016/j.dci.2016.03.009. [DOI] [PubMed] [Google Scholar]
- Guan E, Robinson SL, Goodman EB, Tenner AJ. Cell surface protein identified on phagocytic cells modulates the C1q–mediated enhancement of phagocytosis. Journal of Immunology. 1994;152:4005–4016. [PubMed] [Google Scholar]
- Ho NI, Camps MGM, de Haas EFE, Trouw LA, Verbeek JS, Ossendorp F. C1q–Dependent Dendritic Cell Cross-Presentation of In Vivo-Formed Antigen-Antibody Complexes. J Immunol. 2017;198:4235–4243. doi: 10.4049/jimmunol.1602169. [DOI] [PubMed] [Google Scholar]
- Hong Q, Sze CI, Lin SR, Lee MH, He RY, Schultz L, Chang JY, Chen SJ, Boackle RJ, Hsu LJ, Chang NS. Complement C1q activates tumor suppressor WWOX to induce apoptosis in prostate cancer cells. PLoS One. 2009;4:e5755. doi: 10.1371/journal.pone.0005755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S, Merry KM, Shi Q, Rosenthal A, Barres BA, Lemere CA, Selkoe DJ, Stevens B. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 2016;352:712–716. doi: 10.1126/science.aad8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosszu KK, Valentino A, Vinayagasundaram U, Vinayagasundaram R, Joyce MG, Ji Y, Peerschke EI, Ghebrehiwet B. DC-SIGN, C1q, and gC1qR form a trimolecular receptor complex on the surface of monocyte-derived immature dendritic cells. Blood. 2012;120:1228–1236. doi: 10.1182/blood-2011-07-369728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes-Jones NC, Gardner B. Reaction between the isolated globular sub-units of the complement component C1q and IgG-complexes. Mol Immunol. 1979;16:697–701. doi: 10.1016/0161-5890(79)90010-5. [DOI] [PubMed] [Google Scholar]
- Hulsebus HJ, O’Conner SD, Smith EM, Jie C, Bohlson SS. Complement Component C1q Programs a Pro-Efferocytic Phenotype while Limiting TNFalpha Production in Primary Mouse and Human Macrophages. Front Immunol. 2016;7:230. doi: 10.3389/fimmu.2016.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iram T, Ramirez-Ortiz Z, Byrne MH, Coleman UA, Kingery ND, Means TK, Frenkel D, El Khoury J. Megf10 Is a Receptor for C1Q That Mediates Clearance of Apoptotic Cells by Astrocytes. J Neurosci. 2016;36:5185–5192. doi: 10.1523/JNEUROSCI.3850-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii J, Adachi H, Aoki J, Koizumi H, Tomita S, Suzuki T, Tsujimoto M, Inoue K, Arai H. SREC-II, a new member of the scavenger receptor type F family, trans-interacts with SREC-I through its extracellular domain. J Biol Chem. 2002;277:39696–39702. doi: 10.1074/jbc.M206140200. [DOI] [PubMed] [Google Scholar]
- Jacquet M, Lacroix M, Ancelet S, Gout E, Gaboriaud C, Thielens NM, Rossi V. Deciphering complement receptor type 1 interactions with recognition proteins of the lectin complement pathway. J Immunol. 2013;190:3721–3731. doi: 10.4049/jimmunol.1202451. [DOI] [PubMed] [Google Scholar]
- Kaur A, Sultan SH, Murugaiah V, Pathan AA, Alhamlan FS, Karteris E, Kishore U. Human C1q Induces Apoptosis in an Ovarian Cancer Cell Line via Tumor Necrosis Factor Pathway. Front Immunol. 2016;7:599. doi: 10.3389/fimmu.2016.00599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore U, Gupta SK, Perdikoulis MV, Kojouharova MS, Urban BC, Reid KB. Modular organization of the carboxyl-terminal, globular head region of human C1q A, B, and C chains. j immunol. 2003;171:812–820. doi: 10.4049/jimmunol.171.2.812. [DOI] [PubMed] [Google Scholar]
- Klickstein LB, Barbashov SF, Liu T, Jack RM, Nicholson-Weller A. Complement receptor type 1 (CR1, CD35) is a receptor for C1q. Immunity. 1997;7:345–355. doi: 10.1016/s1074-7613(00)80356-8. [DOI] [PubMed] [Google Scholar]
- Kojouharova MS, Panchev ID, Tchorbadjieva MI, Reid KB, Hoppe HJ. Differential binding of IgG and of a HIV gp41 peptide by the B chain and A chain globular head sequences of C1q, respectively. Journal of Immunology. 1998;161:4325–4331. [PubMed] [Google Scholar]
- Kojouharova MS, Tsacheva IG, Tchorbadjieva MI, Reid KB, Kishore U. Localization of ligand-binding sites on human C1q globular head region using recombinant globular head fragments and single-chain antibodies. Biochimica et Biophysica Acta. 2003;1652:64–74. doi: 10.1016/j.bbapap.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Kojouharova MS, Gadjeva MG, Tsacheva IG, Zlatarova A, Roumenina LT, Tchorbadjieva MI, Atanasov BP, Waters P, Urban BC, Sim RB, Reid KB, Kishore U. Mutational analyses of the recombinant globular regions of human C1q A, B, and C chains suggest an essential role for arginine and histidine residues in the C1q–IgG interaction. Journal of Immunology. 2004;172:4351–4358. doi: 10.4049/jimmunol.172.7.4351. [DOI] [PubMed] [Google Scholar]
- Korb LC, Ahearn JM. C1q binds directly and specifically to surface blebs of apoptotic human keratinocytes: complement deficiency and systemic lupus erythematosus revisited. J Immunol. 1997;158:4525–4528. [PubMed] [Google Scholar]
- Kouser L, Madhukaran SP, Shastri A, Saraon A, Ferluga J, Al-Mozaini M, Kishore U. Emerging and Novel Functions of Complement Protein C1q. Front Immunol. 2015;6:317. doi: 10.3389/fimmu.2015.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs H, Campbell ID, Strong P, Johnson S, Ward FJ, Reid KB, Eggleton P. Evidence that C1q binds specifically to CH2-like immunoglobulin gamma motifs present in the autoantigen calreticulin and interferes with complement activation. Biochemistry. 1998;37:17865–17874. doi: 10.1021/bi973197p. [DOI] [PubMed] [Google Scholar]
- Krych-Goldberg M, Atkinson JP. Structure-function relationships of complement receptor type 1. ImmunolRev. 2001;180:112–122. doi: 10.1034/j.1600-065x.2001.1800110.x. [DOI] [PubMed] [Google Scholar]
- Lahti M, Heino J, Kapyla J. Leukocyte integrins alphaLbeta2, alphaMbeta2 and alphaXbeta2 as collagen receptors--receptor activation and recognition of GFOGER motif. Int J Biochem Cell Biol. 2013;45:1204–1211. doi: 10.1016/j.biocel.2013.03.016. [DOI] [PubMed] [Google Scholar]
- Lepow IH, Naff GB, Todd EW, Pensky J, Hinz CF. Chromatographic resolution of the first component of human complement into three activities. J Exp Med. 1963;117:983–1008. doi: 10.1084/jem.117.6.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YF, Cheng YF, Huang Y, Conti M, Wilson SP, O’Donnell JM, Zhang HT. Phosphodiesterase-4D knock-out and RNA interference-mediated knock-down enhance memory and increase hippocampal neurogenesis via increased cAMP signaling. JNeurosci. 2011;31:172–183. doi: 10.1523/JNEUROSCI.5236-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillis AP, Greenlee MC, Mikhailenko I, Pizzo SV, Tenner AJ, Strickland DK, Bohlson SS. Murine low-density lipoprotein receptor-related protein 1 (LRP) is required for phagocytosis of targets bearing LRP ligands but is not required for C1q–triggered enhancement of phagocytosis. Journal of Immunology. 2008;181:364–373. doi: 10.4049/jimmunol.181.1.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnartz-Gerlach B, Kopatz J, Neumann H. Siglec functions of microglia. Glycobiology. 2014;24:794–799. doi: 10.1093/glycob/cwu044. [DOI] [PubMed] [Google Scholar]
- Linnartz B, Kopatz J, Tenner AJ, Neumann H. Sialic acid on the neuronal glycocalyx prevents complement C1 binding and complement receptor-3-mediated removal by microglia. J Neurosci. 2012;32:946–952. doi: 10.1523/JNEUROSCI.3830-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig IS, Geijtenbeek TB, van Kooyk Y. Two way communication between neutrophils and dendritic cells. Curr Opin Pharmacol. 2006;6:408–413. doi: 10.1016/j.coph.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Lui H, et al. Progranulin Deficiency Promotes Circuit-Specific Synaptic Pruning by Microglia via Complement Activation. Cell. 2016;165:921–935. doi: 10.1016/j.cell.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Rai V, Hudson BI, Song F, Schmidt AM, Barile GR. RAGE binds C1q and enhances C1q–mediated phagocytosis. Cell Immunol. 2012;274:72–82. doi: 10.1016/j.cellimm.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Macor P, Secco E, Mezzaroba N, Zorzet S, Durigutto P, Gaiotto T, De Maso L, Biffi S, Garrovo C, Capolla S, Tripodo C, Gattei V, Marzari R, Tedesco F, Sblattero D. Bispecific antibodies targeting tumor-associated antigens and neutralizing complement regulators increase the efficacy of antibody-based immunotherapy in mice. Leukemia. 2015;29:406–414. doi: 10.1038/leu.2014.185. [DOI] [PubMed] [Google Scholar]
- Majai G, Kiss E, Tarr T, Zahuczky G, Hartman Z, Szegedi G, Fesus L. Decreased apopto-phagocytic gene expression in the macrophages of systemic lupus erythematosus patients. Lupus. 2014;23:133–145. doi: 10.1177/0961203313511557. [DOI] [PubMed] [Google Scholar]
- Malhotra R, Willis AC, Jensenius JC, Jackson J, Sim RB. Structure and homology of human C1q receptor (collectin receptor) Immunology. 1993;78:341–348. [PMC free article] [PubMed] [Google Scholar]
- Manderson AP, Botto M, Walport MJ. The role of complement in the development of systemic lupus erythematosus. AnnuRevImmunol. 2004;22:431–456. doi: 10.1146/annurev.immunol.22.012703.104549. [DOI] [PubMed] [Google Scholar]
- Markiewski MM, DeAngelis RA, Benencia F, Ricklin-Lichtsteiner SK, Koutoulaki A, Gerard C, Coukos G, Lambris JD. Modulation of the antitumor immune response by complement. Nat Immunol. 2008;9:1225–1235. doi: 10.1038/ni.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGreal EP, Ikewaki N, Akatsu H, Morgan BP, Gasque P. Human C1qRp is identical with CD93 and the mNI-11 antigen but does not bind C1q. Journal of Immunology. 2002;168:5222–5232. doi: 10.4049/jimmunol.168.10.5222. [DOI] [PubMed] [Google Scholar]
- Merle NS, Church SE, Fremeaux-Bacchi V, Roumenina LT. Complement System Part I - Molecular Mechanisms of Activation and Regulation. Front Immunol. 2015;6:262. doi: 10.3389/fimmu.2015.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyaard L. The inhibitory collagen receptor LAIR-1 (CD305) J Leukoc Biol. 2008;83:799–803. doi: 10.1189/jlb.0907609. [DOI] [PubMed] [Google Scholar]
- Miura-Shimura Y, Nakamura K, Ohtsuji M, Tomita H, Jiang Y, Abe M, Zhang D, Hamano Y, Tsuda H, Hashimoto H, Nishimura H, Taki S, Shirai T, Hirose S. C1q regulatory region polymorphism down-regulating murine C1q protein levels with linkage to lupus nephritis. Journal of Immunology. 2002;169:1334–1339. doi: 10.4049/jimmunol.169.3.1334. [DOI] [PubMed] [Google Scholar]
- Moreau C, Bally I, Chouquet A, Bottazzi B, Ghebrehiwet B, Gaboriaud C, Thielens N. Structural and Functional Characterization of a Single-Chain Form of the Recognition Domain of Complement Protein C1q. Front Immunol. 2016;7:79. doi: 10.3389/fimmu.2016.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naff GB, Pensky J, Lepow IH. The Macromolecular Nature of the First Component of Human Complement. J Exp Med. 1964;119:593–613. doi: 10.1084/jem.119.4.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito AT, et al. Complement C1q activates canonical Wnt signaling and promotes aging-related phenotypes. Cell. 2012;149:1298–1313. doi: 10.1016/j.cell.2012.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navratil JS, Watkins SC, Wisnieski JJ, Ahearn JM. The globular heads of C1q specifically recognize surface blebs of apoptotic vascular endothelial cells. Journal of Immunology. 2001;166:3231–3239. doi: 10.4049/jimmunol.166.5.3231. [DOI] [PubMed] [Google Scholar]
- Norsworthy PJ, Fossati-Jimack L, Cortes-Hernandez J, Taylor PR, Bygrave AE, Thompson RD, Nourshargh S, Walport MJ, Botto M. Murine CD93 (C1qRp) contributes to the removal of apoptotic cells in vivo but is not required for C1q–mediated enhancement of phagocytosis. J Immunol. 2004;172:3406–3414. doi: 10.4049/jimmunol.172.6.3406. [DOI] [PubMed] [Google Scholar]
- Ogden CA, deCathelineau A, Hoffmann PR, Bratton D, Ghebrehiwet B, Fadok VA, Henson PM. C1q and mannose binding lectin engagement of cell surface calreticulin and CD91 initiates macropinocytosis and uptake of apoptotic cells. J Exp Med. 2001;194:781–795. doi: 10.1084/jem.194.6.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olde Nordkamp MJ, van Eijk M, Urbanus RT, Bont L, Haagsman HP, Meyaard L. Leukocyte-associated Ig-like receptor-1 is a novel inhibitory receptor for surfactant protein D. J Leukoc Biol. 2014a;96:105–111. doi: 10.1189/jlb.3AB0213-092RR. [DOI] [PubMed] [Google Scholar]
- Olde Nordkamp MJ, Boross P, Yildiz C, Jansen JH, Leusen JH, Wouters D, Urbanus RT, Hack CE, Meyaard L. Inhibition of the classical and lectin pathway of the complement system by recombinant LAIR-2. J Innate Immun. 2014b;6:284–292. doi: 10.1159/000354976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paidassi H, Tacnet-Delorme P, Verneret M, Gaboriaud C, Houen G, Duus K, Ling WL, Arlaud GJ, Frachet P. Investigations on the C1q–calreticulin-phosphatidylserine interactions yield new insights into apoptotic cell recognition. J Mol Biol. 2011;408:277–290. doi: 10.1016/j.jmb.2011.02.029. [DOI] [PubMed] [Google Scholar]
- Pednekar L, Pandit H, Paudyal B, Kaur A, Al-Mozaini MA, Kouser L, Ghebrehiwet B, Mitchell DA, Madan T, Kishore U. Complement Protein C1q Interacts with DC-SIGN via Its Globular Domain and Thus May Interfere with HIV-1 Transmission. Front Immunol. 2016;7:600. doi: 10.3389/fimmu.2016.00600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry F, Botto M, Holtappels R, Walport MJ, Loos M. Reconstitution of the complement function in C1q–deficient (C1qa−/−) mice with wild-type bone marrow cells. Journal of Immunology. 2001;167:4033–4037. doi: 10.4049/jimmunol.167.7.4033. [DOI] [PubMed] [Google Scholar]
- Pflieger D, Przybylski C, Gonnet F, Le Caer JP, Lunardi T, Arlaud GJ, Daniel R. Analysis of human C1q by combined bottom-up and top-down mass spectrometry: detailed mapping of post-translational modifications and insights into the C1r/C1s binding sites. Mol Cell Proteomics. 2010;9:593–610. doi: 10.1074/mcp.M900350-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijnenborg R, Vercruysse L, Hanssens M. The uterine spiral arteries in human pregnancy: facts and controversies. Placenta. 2006;27:939–958. doi: 10.1016/j.placenta.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Pisalyaput K, Tenner AJ. Complement component C1q inhibits beta-amyloid- and serum amyloid P-induced neurotoxicity via caspase- and calpain-independent mechanisms. JNeurochem. 2008;104:696–707. doi: 10.1111/j.1471-4159.2007.05012.x. [DOI] [PubMed] [Google Scholar]
- Prabagar MG, Do Y, Ryu S, Park JY, Choi HJ, Choi WS, Yun TJ, Moon J, Choi IS, Ko K, Young Shin C, Cheong C, Kang YS. SIGN-R1, a C-type lectin, enhances apoptotic cell clearance through the complement deposition pathway by interacting with C1q in the spleen. Cell Death Differ. 2013;20:535–545. doi: 10.1038/cdd.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Ortiz ZG, Pendergraft WF, 3rd, Prasad A, Byrne MH, Iram T, Blanchette CJ, Luster AD, Hacohen N, El Khoury J, Means TK. The scavenger receptor SCARF1 mediates the clearance of apoptotic cells and prevents autoimmunity. Nat Immunol. 2013;14:917–926. doi: 10.1038/ni.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan S, Cao C, Catania J, Migliorini M, Zhang L, Strickland DK. Molecular basis for the interaction of low density lipoprotein receptor-related protein 1 (LRP1) with integrin alphaMbeta2: identification of binding sites within alphaMbeta2 for LRP1. J Biol Chem. 2011;286:30535–30541. doi: 10.1074/jbc.M111.265413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid KB. Complete amino acid sequences of the three collagen-like regions present in subcomponent C1q of the first component of human complement. Biochem J. 1979;179:367–371. doi: 10.1042/bj1790367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid KB. Molecular cloning and characterization of the complementary DNA and gene coding for the B-chain of subcomponent C1q of the human complement system. Biochem J. 1985;231:729–735. doi: 10.1042/bj2310729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid KB, Porter RR. Subunit composition and structure of subcomponent C1q of the first component of human complement. Biochem J. 1976;155:19–23. doi: 10.1042/bj1550019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid KB, Gagnon J, Frampton J. Completion of the amino acid sequences of the A and B chains of subcomponent C1q of the first component of human complement. Biochem J. 1982;203:559–569. doi: 10.1042/bj2030559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumenina LT, Popov KT, Bureeva SV, Kojouharova M, Gadjeva M, Rabheru S, Thakrar R, Kaplun A, Kishore U. Interaction of the globular domain of human C1q with Salmonella typhimurium lipopolysaccharide. Biochimica et Biophysica Acta. 2008;1784:1271–1276. doi: 10.1016/j.bbapap.2008.04.029. [DOI] [PubMed] [Google Scholar]
- Roumenina LT, Ruseva MM, Zlatarova A, Ghai R, Kolev M, Olova N, Gadjeva M, Agrawal A, Bottazzi B, Mantovani A, Reid KB, Kishore U, Kojouharova MS. Interaction of C1q with IgG1, C-reactive Protein and Pentraxin 3: Mutational Studies Using Recombinant Globular Head Modules of Human C1q A, B, and C Chains. Biochemistry. 2006;45:4093–4104. doi: 10.1021/bi052646f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santer DM, Wiedeman AE, Teal TH, Ghosh P, Elkon KB. Plasmacytoid dendritic cells and C1q differentially regulate inflammatory gene induction by lupus immune complexes. Journal of Immunology. 2012;188:902–915. doi: 10.4049/jimmunol.1102797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santer DM, Hall BE, George TC, Tangsombatvisit S, Liu CL, Arkwright PD, Elkon KB. C1q deficiency leads to the defective suppression of IFN-alpha in response to nucleoprotein containing immune complexes. Journal of Immunology. 2010;185:4738–4749. doi: 10.4049/jimmunol.1001731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar A, Bialas AR, de RH, Davis A, Hammond TR, Kamitaki N, Tooley K, Presumey J, Baum M, Van DV, Genovese G, Rose SA, Handsaker RE, Daly MJ, Carroll MC, Stevens B, McCarroll SA. Schizophrenia risk from complex variation of complement component 4. Nature. 2016 doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellar GC, Blake DJ, Reid KB. Characterization and organization of the genes encoding the A-, Band C-chains of human complement subcomponent C1q. The complete derived amino acid sequence of human C1q. Biochem J. 1991;274(Pt 2):481–490. doi: 10.1042/bj2740481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L, Scherer PE. The crystal structure of a complement-1q family protein suggests an evolutionary link to tumor necrosis factor. Curr Biol. 1998;8:335–338. doi: 10.1016/s0960-9822(98)70133-2. [DOI] [PubMed] [Google Scholar]
- Shelton E, Yonemasu K, Stroud RM. Ultrastructure of the human complement component, Clq (negative staining-glutamine synthetase-biologically active Clq) Proc Natl Acad Sci U S A. 1972;69:65–68. doi: 10.1073/pnas.69.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Colodner KJ, Matousek SB, Merry K, Hong S, Kenison JE, Frost JL, Le KX, Li S, Dodart JC, Caldarone BJ, Stevens B, Lemere CA. Complement C3-Deficient Mice Fail to Display Age-Related Hippocampal Decline. JNeurosci. 2015;35:13029–13042. doi: 10.1523/JNEUROSCI.1698-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RC, Schumaker VN. Measurement of the association constants of the complexes formed between intact C1q or pepsin-treated C1q stalks and the unactivated or activated C1r2C1s2 tetramers. Mol Immunol. 1983;20:53–66. doi: 10.1016/0161-5890(83)90105-0. [DOI] [PubMed] [Google Scholar]
- Silva-Martin N, Bartual SG, Ramirez-Aportela E, Chacon P, Park CG, Hermoso JA. Structural basis for selective recognition of endogenous and microbial polysaccharides by macrophage receptor SIGN-R1. Structure. 2014;22:1595–1606. doi: 10.1016/j.str.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Sim RB, Schwaeble W, Fujita T. Complement research in the 18th-21st centuries: Progress comes with new technology. Immunobiology. 2016;221:1037–1045. doi: 10.1016/j.imbio.2016.06.011. [DOI] [PubMed] [Google Scholar]
- Singh J, Ahmed A, Girardi G. Role of complement component C1q in the onset of preeclampsia in mice. Hypertension. 2011;58:716–724. doi: 10.1161/HYPERTENSIONAHA.111.175919. [DOI] [PubMed] [Google Scholar]
- Sjoberg AP, Trouw LA, Blom AM. Complement activation and inhibition: a delicate balance. Trends Immunol. 2009;30:83–90. doi: 10.1016/j.it.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Son M, Diamond B. C1q–mediated repression of human monocytes is regulated by leukocyte-associated Ig-like receptor 1 (LAIR-1) Mol Med. 2015;20:559–568. doi: 10.2119/molmed.2014.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son M, Santiago-Schwarz F, Al-Abed Y, Diamond B. C1q limits dendritic cell differentiation and activation by engaging LAIR-1. Proc Natl Acad Sci U S A. 2012;109:E3160–3167. doi: 10.1073/pnas.1212753109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son M, Diamond B, Volpe BT, Aranow CB, Mackay MC, Santiago-Schwarz F. Evidence for C1q–mediated crosslinking of CD33/LAIR-1 inhibitory immunoreceptors and biological control of CD33/LAIR-1 expression. Sci Rep. 2017;7:270. doi: 10.1038/s41598-017-00290-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son M, Porat A, He M, Suurmond J, Santiago-Schwarz F, Andersson U, Coleman TR, Volpe BT, Tracey KJ, Al-Abed Y, Diamond B. C1q and HMGB1 reciprocally regulate human macrophage polarization. Blood. 2016 doi: 10.1182/blood-2016-05-719757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorci G, Riuzzi F, Giambanco I, Donato R. RAGE in tissue homeostasis, repair and regeneration. Biochim Biophys Acta. 2013;1833:101–109. doi: 10.1016/j.bbamcr.2012.10.021. [DOI] [PubMed] [Google Scholar]
- Stegert M, Bock M, Trendelenburg M. Clinical presentation of human C1q deficiency: How much of a lupus? Mol Immunol. 2015;67:3–11. doi: 10.1016/j.molimm.2015.03.007. [DOI] [PubMed] [Google Scholar]
- Steinberger P, Szekeres A, Wille S, Stockl J, Selenko N, Prager E, Staffler G, Madic O, Stockinger H, Knapp W. Identification of human CD93 as the phagocytic C1q receptor (C1qRp) by expression cloning. J LeukocBiol. 2002;71:133–140. [PubMed] [Google Scholar]
- Stephan AH, Barres BA, Stevens B. The complement system: an unexpected role in synaptic pruning during development and disease. AnnuRevNeurosci. 2012;35:369–389. doi: 10.1146/annurev-neuro-061010-113810. [DOI] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, Sher A, Litke AM, Lambris JD, Smith SJ, John SW, Barres BA. The classical complement cascade mediates CNS synapse elimination. Cell. 2007;131:1164–1178. doi: 10.1016/j.cell.2007.10.036. [DOI] [PubMed] [Google Scholar]
- Sun BK, Siprashvili Z, Khavari PA. Advances in skin grafting and treatment of cutaneous wounds. Science. 2014;346:941–945. doi: 10.1126/science.1253836. [DOI] [PubMed] [Google Scholar]
- Svehag SE, Manhem L, Bloth B. Ultrastructure of human C1q protein. Nat New Biol. 1972;238:117–118. doi: 10.1038/newbio238117a0. [DOI] [PubMed] [Google Scholar]
- Tariq M, Chen R, Yuan H, Liu Y, Wu Y, Wang J, Xia C. De novo transcriptomic analysis of peripheral blood lymphocytes from the Chinese goose: gene discovery and immune system pathway description. PloS one. 2015;10:e0121015. doi: 10.1371/journal.pone.0121015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchantchou F, Lacor PN, Cao Z, Lao L, Hou Y, Cui C, Klein WL, Luo Y. Stimulation of neurogenesis and synaptogenesis by bilobalide and quercetin via common final pathway in hippocampal neurons. JAlzheimersDis. 2009;18:787–798. doi: 10.3233/JAD-2009-1189. [DOI] [PubMed] [Google Scholar]
- Teh BK, Yeo JG, Chern LM, Lu J. C1q regulation of dendritic cell development from monocytes with distinct cytokine production and T cell stimulation. Mol Immunol. 2011;48:1128–1138. doi: 10.1016/j.molimm.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Tenner AJ, Robinson SL, Ezekowitz RA. Mannose binding protein (MBP) enhances mononuclear phagocyte function via a receptor that contains the 126,000 M(r) component of the C1q receptor. Immunity. 1995;3:485–493. doi: 10.1016/1074-7613(95)90177-9. [DOI] [PubMed] [Google Scholar]
- Tenner AJ, Robinson SL, Borchelt J, Wright JR. Human Pulmonary Surfactant Protein (SP-A), a Protein Structurally Homologous to C1q, Can Enhance FcR- and CR1-mediated Phagocytosis. The Journal of Biological Chemistry. 1989;264:13923–13928. [PubMed] [Google Scholar]
- van Schaarenburg RA, Suurmond J, Habets KL, Brouwer MC, Wouters D, Kurreeman FA, Huizinga TW, Toes RE, Trouw LA. The production and secretion of complement component C1q by human mast cells. Mol Immunol. 2016;78:164–170. doi: 10.1016/j.molimm.2016.09.001. [DOI] [PubMed] [Google Scholar]
- Vandivier RW, Ogden CA, Fadok VA, Hoffmann PR, Brown KK, Botto M, Walport MJ, Fisher JH, Henson PM, Greene KE. Role of surfactant proteins A, D, and C1q in the clearance of apoptotic cells in vivo and in vitro: calreticulin and CD91 as a common collectin receptor complex. J Immunol. 2002;169:3978–3986. doi: 10.4049/jimmunol.169.7.3978. [DOI] [PubMed] [Google Scholar]
- Vasek MJ, et al. A complement-microglial axis drives synapse loss during virus-induced memory impairment. Nature. 2016;534:538–543. doi: 10.1038/nature18283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerhuis R, Nielsen HM, Tenner AJ. Complement in the brain. Mol Immunol. 2011;48:1592–1603. doi: 10.1016/j.molimm.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster S, Lue LF, Brachova L, Tenner AJ, McGeer PL, Terai K, Walker DG, Bradt B, Cooper NR, Rogers J. Molecular and cellular characterization of the membrane attack complex, C5b-9, in Alzheimer’s disease. NeurobiolAging. 1997;18:415–421. doi: 10.1016/s0197-4580(97)00042-0. [DOI] [PubMed] [Google Scholar]
- Webster SD, Galvan MD, Ferran E, Garzon-Rodriguez W, Glabe CG, Tenner AJ. Antibody-mediated phagocytosis of the amyloid beta-peptide in microglia is differentially modulated by C1q. Journal of Immunology. 2001;166:7496–7503. doi: 10.4049/jimmunol.166.12.7496. [DOI] [PubMed] [Google Scholar]
- Williams PA, Tribble JR, Pepper KW, Cross SD, Morgan BP, Morgan JE, John SW, Howell GR. Inhibition of the classical pathway of the complement cascade prevents early dendritic and synaptic degeneration in glaucoma. MolNeurodegener. 2016;11:26. doi: 10.1186/s13024-016-0091-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winslow S, Leandersson K, Edsjo A, Larsson C. Prognostic stromal gene signatures in breast cancer. Breast Cancer Res. 2015;17:23. doi: 10.1186/s13058-015-0530-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss-Coray T, Rogers J. Inflammation in Alzheimer disease-a brief review of the basic science and clinical literature. Cold Spring HarbPerspectMed. 2012;2:a006346. doi: 10.1101/cshperspect.a006346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier S, Sahu RK, Landes SG, Yu J, Taylor RP, Ayyadevara S, Megyesi J, Stallcup WB, Duffield JS, Reis ES, Lambris JD, Portilla D. Pericytes and immune cells contribute to complement activation in tubulointerstitial fibrosis. Am J Physiol Renal Physiol. 2017;312:F516–F532. doi: 10.1152/ajprenal.00604.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabumoto C, Akazawa H, Yamamoto R, Yano M, Kudo-Sakamoto Y, Sumida T, Kamo T, Yagi H, Shimizu Y, Saga-Kamo A, Naito AT, Oka T, Lee JK, Suzuki J, Sakata Y, Uejima E, Komuro I. Angiotensin II receptor blockade promotes repair of skeletal muscle through down-regulation of aging-promoting C1q expression. SciRep. 2015;5:14453. doi: 10.1038/srep14453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziccardi RJ, Tschopp J. The dissociation properties of native C1. Biochemical and Biophysical Research Communications. 1982;107:618–623. doi: 10.1016/0006-291x(82)91536-4. [DOI] [PubMed] [Google Scholar]
- Zlatarova AS, Rouseva M, Roumenina LT, Gadjeva M, Kolev M, Dobrev I, Olova N, Ghai R, Jensenius JC, Reid KB, Kishore U, Kojouharova MS. Existence of different but overlapping IgG- and IgM-binding sites on the globular domain of human C1q. Biochemistry. 2006;45:9979–9988. doi: 10.1021/bi060539v. [DOI] [PubMed] [Google Scholar]