Circulating cell-free tumor-DNA (cfDNA) testing (‘liquid biopsy’) is increasingly being employed both in clinical trials as well as clinical practice. With respect to colon cancer, the trends in cfDNA are associated with the responses observed (1). Furthermore, it is not only helpful in identifying inherent mutations conferring resistance to targeted therapies e.g., RAS/RAF mutations, it is also useful in identifying acquired mechanisms of resistance e.g., MET-amplifications as a mechanism of resistance to anti-epidermal growth factor receptor (anti-EGFR) monoclonal antibodies (2,3).
While knowledge of the mutational status of a given patient with colorectal cancer is adequately gained through cfDNA for the fair majority of patients with metastatic disease, the one thing that it lacks in providing is information about mismatch repair deficiency status of the tumor. Patients with mismatch repair defects (dMMR) colorectal cancers are very sensitive to immune-checkpoint blockade. This is secondary to the higher mutational load leading to more “non-self” antigens. The landmark trial published in 2015 by Le and colleagues showed that while 7 out of 9 patients with dMMR colorectal cancers had a response to PD-1 blockade, 0 out of 18 patients with mismatch repair-proficient (pMMR) tumors responded (4). Patients with dMMR tumors had on average 1,782 mutations as opposed to 73 mutations per pMMR tumors on whole-exome sequencing.
Analysis of mismatch-repair status [or microsatellite instability (MSI)] without tissue sample is challenging. However, given the fact that this leads to a higher number of somatic mutations, this information can be used to predict the MMR/MSI status of the tumor; hence, predicting the potential of response to immunotherapy.
We have had two patients with dMMR tumors now whose cfDNA testing show 12 and 13 mutations versus a median of six mutations in patients with pMMR tumors (Figure 1). This is on a commercial platform (Guardant360®) that uses a 73-gene panel. Though it is simplistic, one can still potentially identify patients who may be candidates for immunotherapy by gauging the mutational burden reported. Here in this letter, an observation with representative cases is presented.
Figure 1.
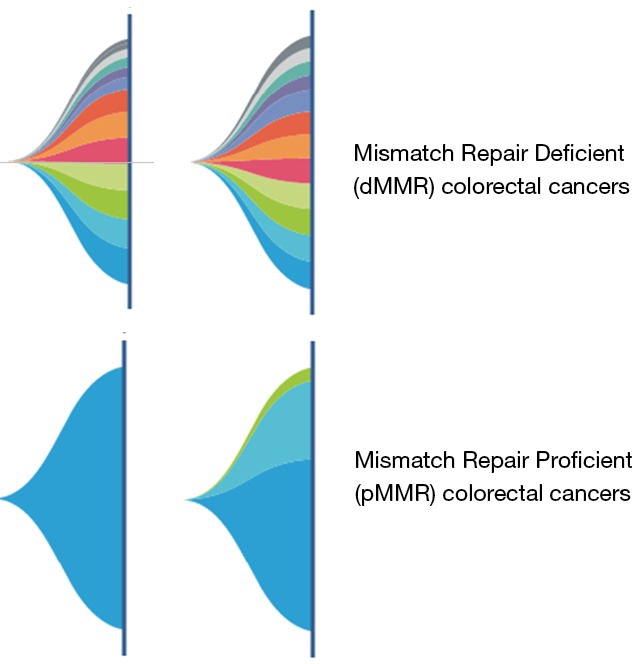
The array of somatic mutations seen in patients with mismatch repair deficient tumors (dMMR—top panel) versus the number and spectrum of mutations seen in patients with mismatch repair proficient tumors (pMMR—bottom panel); each mutation represented by a color based on the percentage composition.
Obviously, this would have to be validated on a larger cohort. However, since the results are so discernable, alongside prior knowledge of whole-exome sequencing of tissue specimens in patients with dMMR versus pMMR (1,782 mutations versus 73 mutations respectively), the observation and findings should still be significant. The difference is striking. Given the effectiveness of cfDNA testing in capturing the mutational landscape of patients with various cancers, the utility of this may go beyond colorectal cancers in identifying patients who may benefit from immunotherapy.
Acknowledgements
None.
Footnotes
Conflicts of Interest: The author has no conflicts of interest to declare.
References
- 1.Hong DS, Morris VK, El Osta B, et al. Phase IB Study of Vemurafenib in Combination with Irinotecan and Cetuximab in Patients with Metastatic Colorectal Cancer with BRAFV600E Mutation. Cancer Discov 2016;6:1352-65. 10.1158/2159-8290.CD-16-0050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Emburgh BO, Sartore-Bianchi A, Di Nicolantonio F, et al. Acquired resistance to EGFR-targeted therapies in colorectal cancer. Mol Oncol 2014;8:1084-94. 10.1016/j.molonc.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stickler JH, Banks KC, Nagy RJ, et al. Blood-based genomic profiling of circulating cell-free tumor DNA (ctDNA) in 1397 patients (pts) with colorectal cancer (CRC). J Clin Oncol 2017;35:abstract 584. [Google Scholar]
- 4.Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. 10.1056/NEJMoa1500596 [DOI] [PMC free article] [PubMed] [Google Scholar]


