Abstract
DNA of replication foci attached to the nuclear matrix was isolated from Chinese hamster ovary cells and human HeLa cells synchronized at different stages of the G1 and S phases of the cell cycle. The abundance of sequences from dihydrofolate reductase ori-β and the β-globin replicator was determined in matrix-attached DNA. The results show that matrix-attached DNA isolated from cells in late G1 phase was enriched in origin sequences in comparison with matrix-attached DNA from early G1 phase cells. The concentration of the early firing ori-β in DNA attached to the matrix decreased in early S phase, while the late firing β-globin origin remained attached until late S phase. We conclude that replication origins associate with the nuclear matrix in late G1 phase and dissociate after initiation of DNA replication in S phase.
INTRODUCTION
Mammalian DNA is structurally and functionally organized into ∼104 tandemly arranged DNA domains called replicons. Within each replicon, DNA synthesis initiates from a single replication origin in a regulated way during S phase. Currently, ∼20 replication origins have been mapped in mammalian genomes, of which only a few have been characterized in detail: ori-β located downstream of the dihydrofolate reductase (DHFR) gene in Chinese hamster cells, the human β-globin replicator and the human lamin B2 origin of replication (1). Although DNA replication can be initiated in the absence of a nucleus in an in vitro replication system using Xenopus egg extract (2), thus far initiation at specific sequences has only been observed in intact nuclei (3). Furthermore, while initiation in this system occurs randomly throughout the genome in early G1 nuclei, in late G1 nuclei initiation occurs at the same replication origins normally used by the cells in vivo (4). These experiments suggest that establishment of specific initiation sites for DNA replication in the mammalian genome requires a cell cycle-dependent event and organization of the origins of replication, which includes components of the nuclear structures (5,6). A characteristic feature of mammalian DNA replication is that it occurs at a few hundred discrete foci. A typical replication focus contains a cluster of several replicons and ∼1 Mb of DNA. Recent studies demonstrate that replicon clusters are stable units of chromosomal structure and persist throughout the cell cycle and in the subsequent daughter cells. Replication foci labeled with halogenated deoxyuridines at different stages in S phase show a similar focused appearance in the next G1 phase (7–9). Furthermore, antibodies against Mcm2, a component of the initiation complex at origins of replication, co-localized with foci formed by both early and late replicating chromatin domains during G1 phase. Mcm2 was loaded gradually and cumulatively throughout G1 and excluded from the active replication foci in S phase (10). This cell cycle-dependent association of a component of the initiation complex with replication foci suggests the existence of certain temporal associations of the origins with replication foci. The replication foci exhibit specific patterns during S phase and these patterns are remarkably preserved after extractions for the nuclear matrix (11). This indicates that replication foci represent a higher order chromatin structure formed by aggregation of several 50–200 kb DNA loops attached to the nuclear matrix (12,13).
The nuclear matrix is mainly a proteinaceous structure isolated by treating nuclei with non-ionic detergents, nucleases and solutions of high and moderate ionic strength and there is a long-standing debate in the literature concerning the association of origins of replication with this structure (14). Recently we have shown that replication origins are not permanently associated with the nuclear matrix, which suggests a dynamic association during the cell cycle (15). In the present communication we have studied the cell cycle-dependent association and dissociation of two origins of replication with DNA isolated from replication foci attached to the nuclear matrix. The results show that there is an increase in origin sequences in this DNA in late G1 phase followed by a decrease in S phase after replication of the respective replicons had started.
MATERIALS AND METHODS
Cell cultures, labeling and synchronization
Chinese hamster ovary cells (CHO K1) and human HeLa cells (HeLa M) were grown in DMEM supplemented with 10% fetal bovine serum (Sigma) in an atmosphere of 96% air/4% CO2. To uniformly label DNA, exponentially growing cells (∼50% confluent) were incubated with 0.025 µCi/ml [14C]thymidine (50 mCi/mmol; Du Pont) for 24 h. Cells were synchronized in mitosis by incubating 70% confluent cultures for 4 h with 50 ng/ml nocodazole (3). Mitotic cells were collected, washed free of nocodazole and cultured under normal conditions. The fraction of cells undergoing DNA synthesis was followed by labeling of aliquots with 1 µCi/ml [3H]thymidine (70–90 Ci/mmol; Dupont) for 30 min at 37°C. For fluorescence activated cell sorting (FACS) analysis cells were collected, washed with phosphate-buffered saline, treated with 20 µg/ml RNase (Sigma) for 30 min at 37°C and stained with 20 µg/ml propidium iodide at room temperature for 30 min. Aliquots of 5 × 105 cells/sample were analyzed with a Becton Dickinson (Facscalibur) cell sorter, using ModFit software (Becton Dickinson).
Permeabilization of cells
Cells were washed twice with TBS buffer (0.15 M NaCl, 5 mM MgCl2, 10 mM Tris–HCl, pH 7.4). They were permeabilized in situ with 0.1% Triton X-100 and 0.5 mM phenylmethylsulfonyl fluoride (PMSF) in TBS at room temperature for 5 min, followed by a wash with TBS buffer.
Preparation of replication foci attached to the nuclear matrix
Replication foci attached to the nuclear matrix were prepared essentially as described by Nakayasu and Berezney (11). The chromatin of the permeabilized cells was digested in situ with 20 U/ml DNase I (Sigma) in TBS buffer at room temperature for 7 min and extracted twice with 20 mM Tris–HCl, pH 7.4, containing 0.2 M ammonium sulfate and 0.2 mM MgCl2 at room temperature for 1 min. The samples were then washed twice with TBS buffer.
Isolation and electrophoresis of DNA
For DNA isolation, cells were lysed in 0.5 M NaCl, 0.5% SDS, 20 mM EDTA, 25 mM Tris–HCl, pH 8. RNA was degraded with 200 µg/ml RNase at 37°C for 3 h. Proteins were digested with 200 µg/ml proteinase K (Sigma) overnight and, after extraction with phenol/chloroform (1:1) and with chloroform/isoamyl alcohol (24:1), DNA was precipitated with 2.5 vol ethanol. The DNA was dissolved in 1 mM EDTA, 10 mM Tris–HCl, pH 8, and the concentration was determined by reading the optical density at 260 nm. Agarose gel electrophoresis was performed in 1% agarose in 40 mM Tris–acetic acid, 2 mM EDTA, pH 8, and the gels were stained with ethidium bromide.
DNA synthesis in replication foci attached to the nuclear matrix
Cells were grown on coverslips and replication foci attached to the nuclear matrix were prepared as described above. They were washed twice with glycerol buffer (20 mM Tris–HCl, pH 7.4, 25% glycerol, 5 mM MgCl2, 0.5 mM EGTA, 0.5 mM PMSF) and then incubated with DNA synthesis medium [50 mM Tris–HCl, pH 7.4, 10 mM MgCl2, 0.5 mM EGTA, 25% glycerol, 100 µM dATP, 100 µM dCTP, 100 µM dGTP, 10 µM dTTP, 200 µM CTP, 200 µM GTP, 200 µM UTP, 4 mM ATP, 40 mM creatine phosphate, 20 µg/ml creatine phosphokinase and 1 µCi [3H]dTTP (60 Ci/mmol; DuPont)]. The amount of DNA synthesis medium was 50 µl per coverslip and incubation was for 0, 30 and 60 min at 37°C. Aphidicolin at a concentration of 100 µg/ml was added to the control reactions to inhibit chromosomal DNA replication. Following incubation, the coverslips were excessively washed with TBS, the cells collected by trypsinization (0.25% trypsin in phosphate-buffered saline) and DNA was precipitated with 10% trichloroacetic acid (TCA) at 4°C for 1 h. Precipitated material was retained on GF/C filters (Whatman), extensively washed with 5% TCA and ethanol and dried. The incorporated radioactivity was measured with a Beckman scintillation counter. To determine DNA synthesis, 3H counts resulting from [3H]dTTP incorporation in the course of the reactions were related to the 14C counts of the uniformly labeled DNA.
DNA probes
CHO genomic DNA was used as template to generate a 479 bp DNA fragment (nucleotides 888–1366 in the sequence of Leu et al.; 16) by PCR. Primers were 5′-TCGGCCTGTCTGTAATATTT-3′ and 5′-CTGTGGAGCTGCTGTGTTTT-3′. The parameters of the PCR cycle were denaturation at 96°C for 30 s, annealing at 72°C for 30 s and synthesis at 66°C for 1 min. The generated fragment was purified by electrophoresis in a 1% agarose gel and cloned in pBluescript II (KS+) according to standard procedures (17). The human β-globin origin probe represented a unique DNA sequence from the human β-globin gene. It was a 917 bp BamHI–EcoRI fragment (nucleotides 62613–63530 in the hβG locus, GenBank accession no. J00179), kindly donated by H. Cedar (Probe I in Kitsberg et al.; 18). The control probe C1 was a 1024 bp sequence from the β-globin gene locus (nucleotides 36994–38018 in the hβG locus). It was generated by PCR as described above, using the primers 5′-ACTTAACGCTTGTGTCTCCAGAC-3′ and 5′-TTACCCATGTAACAAGCCTGC-3′. The control probe C2, representing a 1.1 kb BglII–EcoRI fragment located ∼25 kb downstream of the β-globin gene, was kindly donated by H. Cedar (Probe L in Kitsberg et al.; 18). The Chinese hamster genomic DNA probe was prepared by digestion of CHO cell DNA with BamHI and HindIII and the human genomic DNA probe by digestion of HeLa cell DNA with BamHI and EcoRI.
Dot-blot hybridization
DNA probes were labeled in vitro with [32P]dCTP (3000 Ci/mmol; DuPont) using the RadPrime DNA Labeling System (Gibco BRL). For dot-blot hybridization, 10, 5, 2.5 and 1 µg matrix-attached DNA was loaded in triplicate onto Hybond-N+ membrane (Amersham) as recommended by the manufacturer using a manifold dot-blotter (Bio-Rad). Hybridization was carried out under stringent conditions (7% SDS, 0.25 M phosphate buffer, 1% bovine serum albumin, at 68°C overnight). The membranes were rinsed with 2× SSC (0.3 M NaCl, 0.03 M sodium citrate, pH 7) at room temperature (twice), washed with 2× SSC, 0.1% SDS at 68°C for 30 min (twice) and then in 0.1× SSC, 0.1% SDS at room temperature for 30 min and finally rinsed with 0.1× SSC at room temperature. For rehybridization the blots were stripped off by boiling in 0.1× SSC, 0.1% SDS for 5 min. The autoradiographs were scanned and quantified with Gel Pro Analyzer v.3 software for Windows (Media Cybernetics).
RESULTS
Characterization of the preparations of replication foci attached to the nuclear matrix
To isolate matrix-attached DNA replication foci we used the method of Nakayasu and Berezney (11). The authors have shown that it is possible to digest and extract most of the DNA to obtain residual nuclear structures containing DNA bound to the nuclear matrix that show remarkable preservation of the S phase-specific pattern of replication foci observed in vivo. Moreover, these structures were capable of synthesizing DNA without the addition of cytoplasmic extract and the replication foci visualized by labeling in vitro were indistinguishable from those visualized in intact cells by in vivo labeling. This opens the possibility to isolate and analyze DNA present in replication foci attached to the nuclear matrix during different stages of the cell cycle. The method includes permeabilization of the cells in situ with the detergent Triton X-100, followed by digestion of DNA with DNase I and extraction under mild conditions with 0.2 M ammonium sulfate.
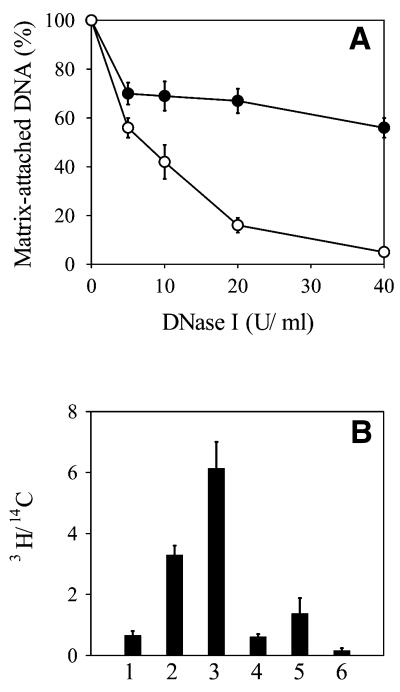
In order to calibrate the DNase I digestion, permeabilized cells were treated with increasing concentrations of the enzyme. In each case the percentage of DNA attached to the nuclear matrix was measured and its molecular weight determined by agarose gel electrophoresis (Fig. 1). It can be seen that with increasing DNase I concentration both the amount of attached DNA and its length gradually decreased. At 20 U/ml DNase I treatment ∼15% of the genomic DNA, representing heterogeneous length DNA fragments in the range 0.2–12 kb (the most abundant being ∼2 kb long), remained attached to the matrix. As our aim was to investigate the abundance of origin sequences in matrix-attached DNA, this 85% level of digestion was most suitable for our further experiments for the following considerations. First, the size of the most abundant fragments protected from nuclease digestion was comparable with the estimated size of the fragments attached to the nucleoskeleton, 1 kb (19). Second, the sizes of the fragments protected from nuclease digestion were similar to the sizes of mammalian origin regions, estimated to be at least 2 kb long, as will be seen later.
Figure 1.
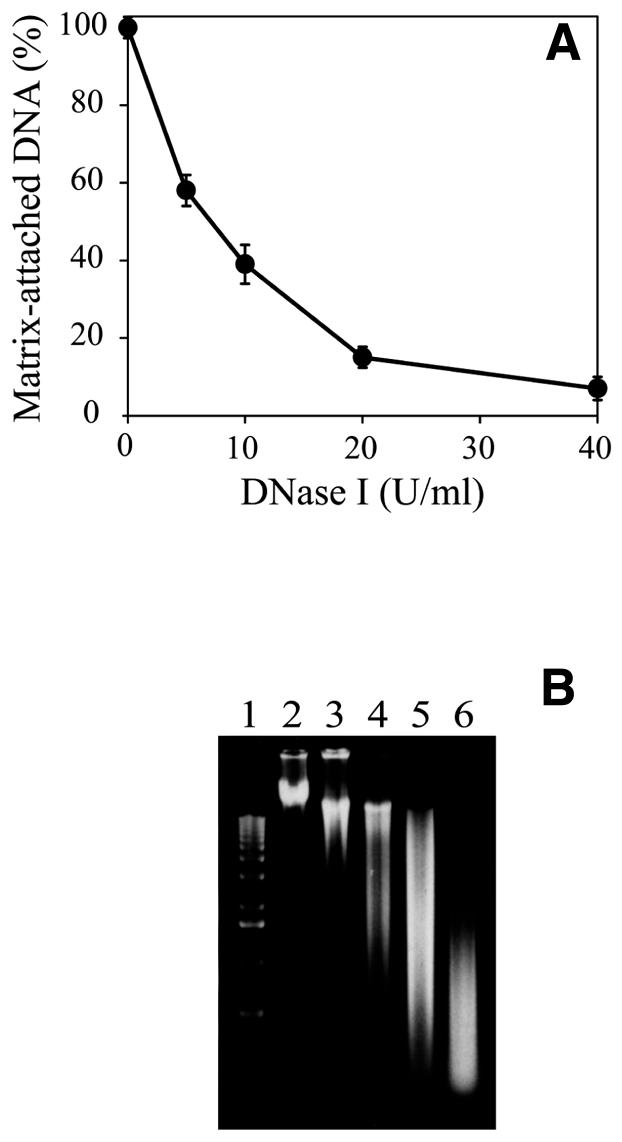
Kinetics of digestion with DNase I. (A) Percentage of matrix-attached DNA as a function of DNase I concentration. Each point is the mean of three independent experiments. Error bars show standard deviations. (B) Agarose gel electrophoresis of DNA isolated from nuclear matrix prepared with different concentrations of DNase I. Lane 1, 1 kb DNA ladder; lane 2, control DNA from permeabilized cells undigested with DNase I; lanes 3–6, DNA from cells treated with 5, 10, 20 and 40 U/ml DNase I.
To check whether the isolated residual nuclear structures represented the DNA replication sites observed in vivo, where synthesis of genomic DNA takes place, DNA of exponentially growing CHO cells was uniformly prelabeled with [14C]thymidine and then pulse labeled with 50 µCi/ml [3H]thymidine for 3 min in vivo. The matrix-attached DNA was isolated and counted to determine the percentage of bulk (14C-labeled) and newly synthesized (3H-labeled) DNA. The results presented in Figure 2A show that with increasing concentrations of DNase I the percentage of bulk DNA steadily decreased to reach ∼7%. However, even at the highest level of digestion 50% of the newly synthesized DNA remained attached to the matrix. This indicates that the matrix-attached DNA was relatively enriched in newly replicated DNA.
Figure 2.
DNA synthesis on replication foci attached to the nuclear matrix in vivo and in vitro. (A) Exponentially growing CHO cells were labeled uniformly with [14C]thymidine and then pulse labeled with 50 µCi/ml [3H]thymidine for 3 min at 37°C. Replication foci attached to the nuclear matrix were prepared with increasing concentrations of DNase I. DNA was isolated and counted. The percentage of 3H counts (filled circle) and 14C counts (open circle) were plotted versus DNase I concentration. (B) CHO cells grown on coverslips were labeled uniformly with [14C]thymidine, replication foci attached to the nuclear matrix were prepared by digestion with 20 U/ml DNase I and incubated for 0 (1), 30 (2) and 60 min (3) in DNA synthesis medium containing 1 µCi/ml [3H]dTTP; another sample was incubated for 1 h in the same medium containing 100 µg/ml aphidicolin (4). As a control, cells were permeabilized with Triton X-100 and incubated for 1 h in DNA synthesis medium containing 1 µCi/ml [3H]dTTP in the absence (5) and presence (6) of 100 µg/ml aphidicolin. Cells were collected by trypsinization, DNA was TCA precipitated and its specific radioactivity determined as the ratio of 3H to 14C counts. In both (A) and (B) each result is the mean of three independent experiments. Error bars show standard deviations.
To check whether the DNA replication machinery was preserved during the isolation procedure, CHO cells were grown on coverslips. DNA was uniformly pre-labeled with [14C]thymidine and the residual structures were prepared and incubated in DNA synthesis medium supplemented with labeled precursor for different periods. The results show that the isolated residual replication foci were able to steadily synthesize DNA for at least 60 min in an aphidicolin-sensitive manner (Fig. 2B). As a control, the same experiment was carried out with cells only permeabilized with Triton X-100 but not treated with DNase I. The specific radioactivity of DNA synthesized on the residual structures was 4.5 times higher than that of DNA isolated from permeabilized cells. This series of experiments prove that our preparations were functionally active and could be used to study the association of replication origins with DNA from replication foci attached to the nuclear matrix during the phases of the cell cycle.
Association of Chinese hamster DHFR ori-β with the nuclear matrix
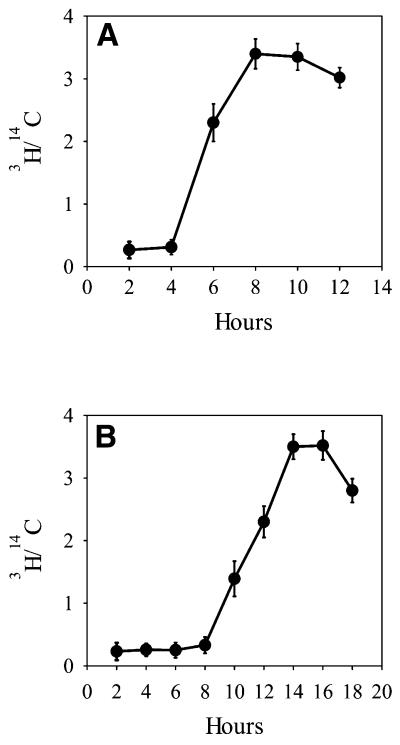
To study the dynamics of association of origins of replication with the nuclear matrix our approach was to determine the abundance of origin sequences in DNA from replication foci attached to the nuclear matrix at different time points in the G1 and S phases. CHO cells were synchronized at mitosis by incubation with nocodazole. The mitotic cells were collected and cultured for different time intervals in nocodazole-free medium. Their progression through the G1 and S phases of the cell cycle was followed by pulse labeling with radioactive DNA precursor and FACS analysis. The results show that CHO cells did not incorporate [3H]thymidine until 4 h after the nocodazole block. After this time incorporation began to increase, reaching a maximum after 8 h (Fig. 3A). FACS analysis showed that cells collected immediately after nocodazole treatment were over 90% in mitosis. With progression of these cells through the cell cycle some loss of synchronization occurred, but still 80% of CHO cells were in G1 phase 4 h after the block and ∼70% were in S phase after 6 h. The distribution of exponentially growing CHO cells in the cell cycle as determined by FACS was 34% in G0/G1, 61% in S and 5% in G2/M phase, which is consistent with the observed relatively short G1 phase of this cell line. The labeling kinetics of the synchronized cells showed an S-shaped curve with an inflection point at 5.5 h, which time was accepted as the entry point into S phase. Accordingly, hours 2 and 4 were considered as early and late G1 phase, while hours 6 and 10 were accepted as early and late S phase. Cell cycle-specific pre-replication foci in G1 and replication foci in S phase associated with the nuclear matrix from CHO cells were isolated from cells synchronized at these time points.
Figure 3.
Kinetics of DNA synthesis in synchronized CHO (A) and HeLa (B) cells. Exponentially growing cells were synchronized in mitosis by incubation with 50 ng/ml nocodazole for 4 h. Mitotic cells were collected, washed free of nocodazole and cultured. At different time intervals aliquots were incubated with 1 µCi/ml [3H]thymidine for 30 min to determine DNA synthetic activity. The 3H to 14C ratios are plotted versus time after nocodazole removal. Each point is the mean of three independent experiments. Error bars show standard deviations.
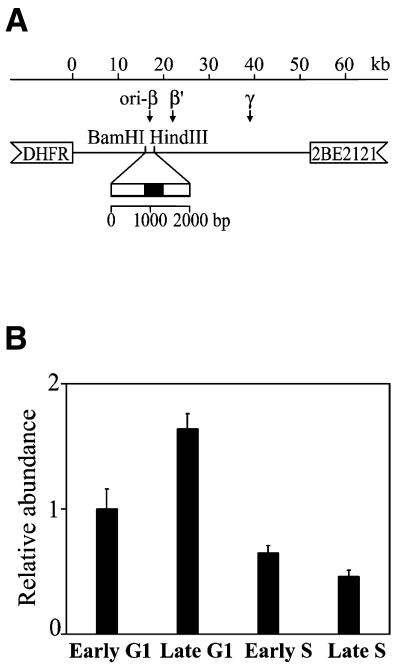
DNA was isolated and dot-blotted onto nylon membranes. These were hybridized with an in vitro labeled DNA probe containing a sequence from the DHFR ori-β, which is the most thoroughly studied mammalian origin so far. Three preferred initiation sites have been identified in the 55 kb initiation zone encompassing the non-transcribed spacer 3′ of the DHFR gene (ori-β, ori-β′ and ori-γ), of which ori-β is the primary initiation site (20); there is genetic evidence that initiation requires a specific DNA sequence (21). Several biochemical methods for mapping replication origins reveal that a 1.96 kb BamHI–HindIII fragment located ∼17 kb downstream of the DHFR gene most probably represents the core origin region of ori-β (22–29). A 479 bp unique DNA sequence (Fig. 4A) within the fragment was generated by PCR, cloned and used to probe the matrix-associated DNA isolated from CHO cells.
Figure 4.
Dynamics of association of the ori-β to the nuclear matrix during the cell cycle of CHO cells. (A) Schematic representation of the position of the ori-β probe in the non-transcribed spacer 3′ of the DHFR gene in CHO cells. A 479 bp DNA fragment (filled box) was amplified by PCR and used as a probe. (B) Relative abundance of the DHFR ori-β in the matrix-attached DNA in CHO cells. Aliquots of matrix-attached DNA isolated from CHO cells synchronized at the indicated stages of the cell cycle were dot-blotted and hybridized with DNA probe in vitro labeled with [32P]dCTP. For determination of relative abundance the blots were rehybridized with genomic DNA. The autoradiographs were scanned and quantified and the ratios of probes to genomic DNA were calculated and presented in arbitrary units, assuming the early G1 ratio as 1.0. The results are means of five independent experiments and the standard deviations are shown with error bars.
After hybridization with the origin probe the blots were stripped and rehybridized with genomic DNA from CHO cells. The autoradiographs were scanned and quantified and the signals obtained with the origin probe were related to the respective signals with genomic DNA to normalize for differences in the DNA content in the dot blots. The ratios were expressed in arbitrary units, assuming the ratio at early G1 to be 1.0. The results presented in Figure 4B show that there were differences in the intensities of the hybridization signals between matrix-attached DNA isolated at different time points in the cell cycle. DNA isolated from late G1 phase CHO cells gave an ∼1.6 times higher hybridization signal with ori-β than matrix-attached DNA from early G1 CHO cells. As the cells progressed through S phase the abundance of origin sequences in the matrix-attached active replication foci rapidly declined and reached 4 times lower values than in late G1.
Association of the human β-globin replicator with the nuclear matrix in HeLa cells
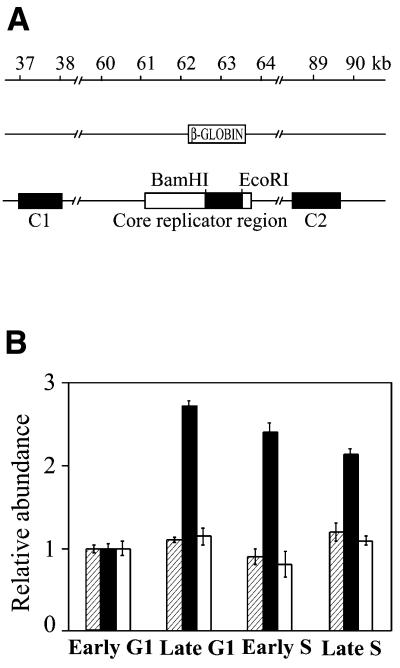
In order to determine whether these cell cycle-related dynamics of association of ori-β with the nuclear matrix are a general characteristic of mammalian origins we applied the same approach to a second origin of replication. The origin of replication located in the human β-globin gene has been mapped by two biochemical methods and there is solid genetic evidence that initiation requires a specific DNA sequence (18,30). By using a combination of site-specific recombination and a physical mapping technique Aladjem et al. (30) have shown that the β-globin origin encompasses 8 kb and consists of a core replicator region and two auxiliary elements. The fragment used as a hybridization probe was a 917 bp BamHI–EcoRI fragment derived from the core region (Fig. 5A).
Figure 5.
Dynamics of association of the β-globin origin of replication with the nuclear matrix during the cell cycle of HeLa cells. (A) Schematic representation of the positions of the probes from the human β-globin locus (filled boxes). (B) Aliquots of matrix-attached DNA isolated from HeLa cells synchronized at the indicated stages of the cell cycle were dot-blotted and hybridized with DNA probes in vitro labeled with [32P]dCTP. For determination of relative abundance the blots were rehybridized with genomic DNA. The autoradiographs were scanned and quantified and the ratios of probes to genomic DNA were calculated and presented in arbitrary units, assuming the early G1 ratio as 1.0. Shaded columns, relative abundance of the control probe C1; filled columns, relative abundance of the β-globin replicator probe; white columns, relative abundance of the control probe C2. The results are means of five independent experiments and the standard deviations are shown with error bars.
HeLa cells synchronized at mitosis did not show any significant incorporation of radioactive precursor until 8 h and incorporation was maximal at 14 h (Fig. 3B). FACS analysis confirmed that at 8 h after release from the nocodazole block ∼80% of the cells were in G1 phase and at 11 h 70% were in S phase. The distribution of non-synchronous exponentially growing HeLa cells in the cell cycle as determined by FACS was 56% in G0/G1, 28% in S and 16% in G2/M phase, confirming the greater length of the G1 phase in this case. Hours 2 and 8 were chosen as early and late G1 phase and hours 11 and 15 as early and late S phase. Accordingly, DNA from replication foci attached to the nuclear matrix was isolated from synchronized HeLa cells at these time points.
The hybridization experiments showed that the abundance of sequences from the β-globin replicator was up to 4 times higher in matrix-attached DNA isolated from late G1 phase than from early G1 phase and only slightly diminished as the cells proceeded into S phase (Fig. 5B). To see whether this cell cycle-related association of DNA sequences with the nuclear matrix is specific for origins of replication the following control experiment was done. The blots containing HeLa matrix-attached DNA were rehybridized with probe C1, located ∼25 kb upstream from the β-globin origin probe, and probe C2, located ∼25 kb downstream of the β-globin gene (Fig. 5A). In this way, the β-globin locus downstream of the locus control region was effectively covered with three probes in such a way that no overlap of the fragments to which the probes would hybridize could occur. No differences in hybridization of the control sequences were observed between early G1, late G1 and S phase (Fig. 5B).
DISCUSSION
A large body of evidence indicates that replication occurs on the nuclear matrix (31–35). This implies that any DNA sequence, including the origins of replication, will be attached to the matrix at a certain time during the cell cycle. In order to study the dynamics of association and dissociation of replication origins with the nuclear matrix we isolated the residual replication foci attached to the nuclear matrix by the method of Nakayasu and Berezney (11). The series of experiments presented in Figure 2, in which either CHO cells were in vivo pulse labeled with [3H]thymidine or isolated residual replication foci were allowed to synthesize DNA in vitro, were in perfect agreement with the results of Nakayasu and Berezney (11). They show that the in situ prepared residual replication foci represent the structures at which DNA synthesis takes place in vivo and that a functionally active DNA replication machinery anchored to the nuclear matrix was preserved during the isolation procedure. The results confirm previous studies that replication forks are associated with the nuclear matrix (31–35) and that the DNA replication machinery is immobilized by attachment to the nuclear matrix (36).
The two origin sequences we have studied showed a well-expressed tendency for preferential attachment to the matrix in late G1 phase. This stage-specific association was observed in both CHO cells and HeLa cells, although the specificity was more evident in HeLa cells, which had a longer G1 phase, permitting better discrimination between early and late G1 phase cells. Interestingly, there were differences in dissociation of the two origins of replication from the nuclear matrix as the cells proceeded into S phase and these differences were consistent with the replication timing of the two origins. While the abundance of ori-β sequences in matrix-attached DNA sharply decreased at the onset of S phase, the abundance of sequences from the β-globin origin of replication diminished only slightly, even at 15 h. The DHFR gene is a housekeeping gene and replicates in early S phase (22,24,37,38). The β-globin gene is not expressed in HeLa cells and is replicated in late S phase (18,30). The overall differences in the hybridization signals with the two origin probes during the cell cycle were ∼4-fold. This corresponded well with the percentage of DNase I digestion and the increase in specific radioactivity of matrix-attached DNA observed both in vivo and in vitro. On the other hand, the control sequences surrounding the β-globin origin did not show cell cycle-dependent variations in their association with the nuclear matrix. Thus, the conclusion can be drawn that replication origins are transiently attached to the nuclear matrix. They associate with the nuclear matrix in late G1 phase and dissociate after initiation of DNA replication in S phase.
Our results are consistent with the results of Pemov et al. (39) showing that alterations in the chromatin structure of ori-β appear at the G1/S boundary, but only in those copies of the DHFR amplicon that are attached to the nuclear matrix in CHOC 400 cells. These alterations disappeared in early S phase and were not evident in early G1 cells. Our data are also in agreement with the results of Ortega and DePamphilis (35), who showed that ori-β is not attached to the nucleoskeleton in S phase. The authors did not follow the dynamics of association of ori-β in G1 phase. They determined the association of ori-β with the nucleoskeleton only at middle G1 phase and did not observe the preferential association with the nuclear matrix described here in late G1 phase.
Recently, a third well-characterized origin of replication, that located downstream of the human lamin B2 gene, was mapped to a matrix anchorage site (40). As the authors worked with exponentially growing cells, their interpretation was that the origin was permanently associated with the nuclear matrix. This origin is localized in a 500 bp region corresponding to the 3′-non-coding end of the lamin B2 gene and the non-transcribed spacer between the gene and the 5′-end of another highly transcribed gene of unknown function and the origin includes its promoter (41). However, it is well known that transcription sites are also attached to the nuclear matrix (36). This makes the results of Lagarkova et al. (40) ambiguous because of the overlap between the cis-acting elements controlling transcription and DNA replication and they may be explained by the transcriptional activity of the region.
Our results suggest that association of origin sequences with pre-replication foci in late G1 phase may play a role in establishment of the initiation-competent state of the origins and support the hypothesis of Newport and Yan (42) that association with the replication foci regulates origin usage in mammalian cells. The question arises as to how the changes in association with the nuclear matrix occur. A number of reports in recent years have shown that chromatin moves locally as well as on a large scale during the G1 and S phases. The large-scale movements in early G1 are connected with repositioning subchromosomal regions within positionally stable territories (43,44). Several lines of evidence indicate that the subchromosomal regions are equivalent to the clustering of replicons into replication foci (7,9). In addition to the large-scale movements, small-scale refolding events within subchromosomal regions were also observed (45). It can be speculated that as a result of such small-scale refolding of subchromosomal regions mediated by protein–protein interactions, the origins of replication associate and dissociate with the nuclear matrix. This conclusion is indirectly supported by the finding that at least one protein which is a component of the initiation complex at origins of replication is loaded cumulatively on pre-replication foci in G1 and excluded from active replication foci in S phase (10).
Acknowledgments
ACKNOWLEDGEMENT
This study was supported by grant K-604/96 from the Bulgarian National Science Fund to B.A.
References
- 1.Todorovic V., Falaschi,A. and Giacca,M. (1999) Replication origins of mammalian chromosomes: the happy few. Front. Biosci., 4, d859–d868. [DOI] [PubMed] [Google Scholar]
- 2.Walter J., Sun,L. and Newport,J. (1998) Regulated chromosomal DNA replication in the absence of a nucleus. Mol. Cell, 1, 519–529. [DOI] [PubMed] [Google Scholar]
- 3.Gilbert D.M., Miyazawa,H. and DePamphilis,M.L. (1995) Site-specific initiation of DNA replication in Xenopus egg extract requires nuclear structure. Mol. Cell. Biol., 15, 2942–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu J.R. and Gilbert,D.M. (1996) A distinct G1 step required to specify the Chinese hamster DHFR replication origin. Science, 271, 1270–1272. [DOI] [PubMed] [Google Scholar]
- 5.DePamphilis M.L. (1999) Origins of DNA replication. In DePamphilis,M.L. (ed.) Concepts of Eukaryotic DNA Replication. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 45–86.
- 6.Dimitrova D.S. and Gilbert,D.M. (1999) DNA replication and nuclear organization: prospects for a soluble in vitro system. Crit. Rev. Eukaryot. Gene Expr., 9, 353–361. [DOI] [PubMed] [Google Scholar]
- 7.Jackson D.A. and Pombo,A. (1998) Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol., 140, 1285–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma H., Saramabandu,J., Devdhar,R., Acharya,R., Cheng,P., Meng,C. and Berezney,R. (1998) Spatial and temporal dynamics of DNA replication sites in mammalian cells. J. Cell Biol., 143, 1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadoni N., Linger,S., Fauth,C., Bernardi,G., Cremer,T., Turner,B.M. and Zink,D. (1999) Nuclear organization of mammalian genomes: polar chromosome territories build up functionally distinct higher order compartments. J. Cell Biol., 146, 1211–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dimitrova D.S., Todorov,I.T., Melendy,T. and Gilbert,D.M. (1999) Mcm2, but not RPA, is a component of mammalian early G1-phase prereplication complex. J. Cell Biol., 146, 709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakayasu H. and Berezney,R. (1989) Mapping replicational sites in the eukaryotic cell nucleus. J. Cell Biol., 108, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunnev D., Tsvetkov,L., Anachkova,B. and Russev,G. (1997) Clusters of replicons that fire simultaneously may be organized into superloops. DNA Cell Biol., 16, 1059–1065. [DOI] [PubMed] [Google Scholar]
- 13.Berezney R., Dubey,D.D. and Huberman,J.A. (2000) Heterogeneity of eukaryotic replicons, replicon clusters and replication foci. Chromosoma, 108, 471–484. [DOI] [PubMed] [Google Scholar]
- 14.Berezney R., Mortillaro,M.J., Ma,H., Wei,X. and Samarabandu,J. (1995) The nuclear matrix: a structural milieu for genomic function. Int. Rev. Cytol., 162A, 1–65. [DOI] [PubMed] [Google Scholar]
- 15.Djeliova V., Russev,G. and Anachkova,B. (2001) Distribution of DNA replication origins between matrix-attached and loop DNA in mammalian cells. J. Cell. Biochem., 80, 353–359. [DOI] [PubMed] [Google Scholar]
- 16.Leu T.-H., Anachkova,B. and Hamlin,J.L. (1990) Repetitive sequence elements in an initiation locus of the amplified dihydrofolate reductase domain in CHO cells. Genomics, 7, 428–433. [DOI] [PubMed] [Google Scholar]
- 17.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1992) Short Protocols in Molecular Biology. John Wiley & Sons, New York, NY.
- 18.Kitsberg D., Selig,S., Keshet,J. and Cedar,H. (1993) Replication structure of the human β-globin gene domain. Nature, 368, 588–590. [DOI] [PubMed] [Google Scholar]
- 19.Jackson D.A., Dickinson,P. and Cook,P.R. (1990) Attachment of DNA to the nucleoskeleton of HeLa cells examined using physiological conditions. Nucleic Acids Res., 18, 4385–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi T., Rein,T. and DePamphilis,M.L. (1998) Identification of the primary initiation sites for DNA replication in the hamster dihydrofalate reductase gene initiation zone. Mol. Cell. Biol., 18, 3266–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altman A.L. and Fanning,E. (2001) The Chinese hamster difydrofolate reductase replication origin beta is active at multiple ectopic chromosomal locations and requires specific DNA sequence elements for activity. Mol. Cell. Biol. 21, 1098–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burhans W.C., Selegue,J.E. and Heintz,N.H. (1986) Isolation of the origin of replication associated with the amplified Chinese hamster dihydrofolate reductase domain. Proc. Natl Acad. Sci. USA, 83, 7790–7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anachkova B. and Hamlin,J.L. (1989) Replication of the amplified dihydropholate reductase domain in CHO cells may initiate at two distinct sites, one of which is a repetitive sequence element. Mol. Cell. Biol., 9, 532–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leu T.-H. and Hamlin,J.L. (1989) High-resolution mapping of replication fork movement through the amplified dihydrofolate reductase domain in CHO cells by in-gel renaturation analysis. Mol. Cell. Biol., 9, 523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burhans W.C., Vassilev,L.T., Caddle,M.S., Heintz,H. and DePamphilis,M.L. (1990) Identification of origin of bidirectional DNA replication in mammalian chromosomes. Cell, 62, 955–965. [DOI] [PubMed] [Google Scholar]
- 26.Vassilev L.T., Burhans,W.C. and DePamphilis,M.L. (1990) Mapping an origin of DNA replication at a single-copy locus in exponentially proliferating mammalian cells. Mol. Cell. Biol., 10, 4685–4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burhans W.C., Vassilev,L.T., Wu,J., Sogo,J.M., Nallaseth,F.S. and DePamphilis,M.L. (1991) Emetine allows identification of origins of mammalian DNA replication by imbalanced DNA synthesis, not through conservative nucleosome segregation. EMBO J., 10, 4351–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pelizon C., Diviacco,S., Falaschi,A. and Giacca,M. (1996) High-resolution mapping of the origin of DNA replication in the hamster dihydrofolate reductase gene domain by competitive PCR. Mol. Cell. Biol., 16, 5358–5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li C., Bogan,J.A., Natale,D.A. and DePamphilis,M.L. (2000) Selective activation of pre-replication complexes in vitro at specific sites in mammalian nuclei. J. Cell Sci., 113, 887–898 [DOI] [PubMed] [Google Scholar]
- 30.Aladjem M.I., Rodewald,L.W., Kolman,J.L. and Wahl,G.M. (1998) Genetic dissection of mammalian replicator in the human β-globin locus. Science, 281, 1005–1009. [DOI] [PubMed] [Google Scholar]
- 31.Berezney R. and Coffey,D.S. (1975) Nuclear protein matrix: association with newly synthesized DNA. Science, 189, 291–293. [DOI] [PubMed] [Google Scholar]
- 32.Jackson D.A. and Cook,P.S. (1986) Replication occurs at a nucleoskeleton. EMBO J., 5, 1403–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaughn J.P., Dijkwel,P.A., Mullenders,L.H.F. and Hamlin,J.L. (1990) Replication forks are associated with the nuclear matrix. Nucleic Acids Res., 18, 1965–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerdes M.G., Carter,K.C., Moen,P.T. and Lawrence,J.B. (1994) Dynamic changes in the higher-level chromatin organization of specific sequences revealed by in situ hybridization to nuclear halos. J. Cell Biol., 126, 289–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ortega M.J. and DePamphilis,M.L. (1998) Nucleoskeleton and initiation of DNA replication in metazoan cells. J. Cell Sci., 111, 3663–3673. [DOI] [PubMed] [Google Scholar]
- 36.Cook P.R. (1999) The organization of replication and transcription. Science, 284, 1790–1795. [DOI] [PubMed] [Google Scholar]
- 37.Heintz N.H. and Hamlin,J.L. (1982) An amplified chromosomal sequence that includes the gene for dihydrofolate reductase initiates replication within specific restriction fragments. Proc. Natl Acad. Sci. USA, 79, 4083–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taljanidisz J., Popowski,J. and Sarkar,N. (1989) Temporal order of gene replication in Chinese hamster ovary cells. Mol. Cell. Biol., 9, 2881–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pemov A., Bavykin,S. and Hamlin,J.L. (1998) Attachment to the nuclear matrix mediates specific alterations in chromatin structure. Proc. Natl Acad. Sci. USA, 95, 14757–14762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lagarkova M.A., Svetlova,E., Giacca,M., Falaschi,A. and Razin,S.V. (1998) DNA loop anchorage region colocalizes with the replication origin located downstream of the human gene encoding lamin B2. J. Cell. Biochem., 69, 13–18. [DOI] [PubMed] [Google Scholar]
- 41.Giacca M., Zentilin,L., Norio,P., Diviacco,S., Dimitrova,D., Contreas,G., Biamonti,G., Perini,G., Weighardt,F., Riva,S. and Falaschi,A. (1994) Fine mapping of a replication origin of human DNA. Proc. Natl Acad. Sci. USA, 91, 7119–7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newport J. and Yan,H. (1996) Organization of DNA into foci during replication. Curr. Opin. Cell Biol., 8, 365–369. [DOI] [PubMed] [Google Scholar]
- 43.Zink D. and Cremer,T. (1998) Cell nucleus: chromosome dynamics in nuclei of living cells. Curr. Biol., 8, R321–R324. [DOI] [PubMed] [Google Scholar]
- 44.Bridger J.M. and Bickmore,W.A. (1998) Putting the genome on the map. Trends Genet., 14, 403–409. [DOI] [PubMed] [Google Scholar]
- 45.Zink D., Cremer,T., Saffrich,R., Fisher,R., Trendelenburg,M.F., Ansorge,W. and Stelzer,E.H. (1998) Structure and dynamic of human interphase chromosome territories in vivo. Hum. Genet., 102, 214–251. [DOI] [PubMed] [Google Scholar]