Abstract
RNA editing can yield protein products that differ from those directly encoded by genomic DNA. This process is pervasive in the mitochondria of many eukaryotes, where it predominantly results in the restoration of ancestral protein sequences. Nuclear mRNAs in metazoans also undergo editing (adenosine-to-inosine or ‘A-to-I’ substitutions), and most of these edits appear to be nonadaptive ‘misfirings’ of adenosine deaminases. However, recent analysis of cephalopod transcriptomes found that many editing sites are shared by anciently divergent lineages within this group, suggesting they play some adaptive role. Recent discoveries have also revealed that some fungi have an independently evolved A-to-I editing mechanism, resulting in extensive recoding of their nuclear mRNAs. Here, phylogenetic comparisons were used to determine whether RNA editing generally restores ancestral protein sequences or creates derived variants. Unlike in mitochondrial systems, RNA editing in metazoan and fungal nuclear transcripts overwhelmingly leads to novel sequences not found in inferred ancestral proteins. Even for the subset of RNA editing sites shared by deeply divergent cephalopod lineages, the primary effect of nuclear editing is an increase—not a decrease—in protein divergence. These findings suggest fundamental differences in the forces responsible for the evolution of RNA editing in nuclear versus mitochondrial systems.
Keywords: A-to-I RNA editing, ADAR, amino acid substitutions, constructive neutral evolution, proteome diversity
1. Introduction
The ‘central dogma of molecular biology’ holds that genetic information stored in DNA is decoded into functional proteins, with messenger RNAs (mRNAs) acting as faithful intermediates. An important caveat is that RNA editing can result in amino acid sequences that differ from those encoded in the genome [1]. Editing can involve individual base substitutions as well as short indels. Such changes are often important for proper cellular and organismal function, as disruption of editing can have deleterious and even lethal phenotypic consequences [2,3]. It is less clear, however, whether editing can be considered adaptive in the sense that it provides some fitness benefit over direct encoding of corrected sequences in genomic DNA. Many adaptive hypotheses have been advanced (e.g. related to mutational buffering, gene regulation, proteome diversification and genomic GC content optimization), but non-adaptive mechanisms for the proliferation of editing sites have also been described [4].
Some insight into the origins and maintenance of mRNA editing may be gleaned by investigating its effects on protein conservation and diversity. Editing is well studied and often pervasive in the mitochondria of diverse eukaryotes, including land plants, trypanosomes, diplonemids, dinoflagellates, heteroloboseans, myxomycetes and some metazoans [1,5–7]. Editing in these mitochondrial systems is generally restorative, meaning that it tends to produce ancestral-like protein sequences that more closely resemble homologues in other eukaryotes [1]. In fact, a simple and effective method to predict mRNA editing sites in land plant organelle genomes (where cytidine-to-uridine or ‘C-to-U’ editing is common) is to scan genes for sites where C-to-T changes would increase protein sequence conservation with related species [8]. The restorative effects in systems with insertional editing are even more dramatic because they generally ‘correct’ shifted reading frames that would otherwise produce completely unrelated proteins. In many cases, mitochondrial mRNA editing is so extensive that the unedited gene sequences are essentially unrecognizable [6].
Metazoans have a nuclear RNA editing system, in which adenosine-to-inosine (A-to-I) substitutions are introduced by a specific class of adenosine deaminases known as ADARs [2]. During translation, inosine is read as a guanosine, so A-to-I editing can result in changes to amino acid sequences. The effects of nuclear A-to-I editing on protein conservation are less clear than those of the mitochondrial systems described above. A-to-I editing is often described as a mechanism that diversifies the proteome [9], but it has also been argued that it preferentially acts on sites that experienced a historical G-to-A change at the genomic level and thereby restores ancestral protein sequences [10,11]. Observed patterns of mRNA editing in humans have led to the conclusion that changes in protein-coding sequences are generally nonadaptive. Very few editing sites are shared with other mammals [10], and editing appears to be more common at sites that are less functionally important (e.g. synonymous sites) [12], suggesting that most edits are just tolerable by-products of promiscuous enzyme activity. This conclusion is thought to extend to other metazoan systems, but recent research has indicated that A-to-I mRNA editing is much more extensive and potentially adaptive in coleoid cephalopods [13]. Notably, some identified editing sites are even shared across representatives of divergent cephalopod groups that span hundreds of millions of years of evolution (i.e. octopus, squid and cuttlefish) [14].
A-to-I editing of nuclear mRNAs has also been discovered in the fungal genus Fusarium [15] and other filamentous ascomycetes [16], with large predicted effects on protein sequences during sexual development. Interestingly, this RNA editing system appears to be independently evolved because fungi lack ADARs, which are responsible for mRNA editing in metazoans.
Here, the consequences for protein diversity resulting from A-to-I editing of nuclear mRNA transcripts in metazoan and fungal lineages are compared to the well-documented restorative effects in mitochondrial systems. This analysis reveals that nuclear and mitochondrial editing systems have strikingly opposite effects on protein conservation. Rather than restoring ancestral protein sequences, the vast majority of A-to-I mRNA edits introduce evolutionarily derived amino acid changes.
2. Material and methods
Published datasets were obtained for A-to-I editing of nuclear transcripts in four focal species—Homo sapiens [12], Drosophila melanogaster [17], Octopus bimaculoides [14] and Fusarium graminearum [15]—and for C-to-U editing of mitochondrial transcripts in the angiosperm Arabidopsis thaliana [18]. For each species, edited protein sequences were mapped with NCBI BLAST v. 2.2.30+ to either protein (blastp) or genome/transcriptome (tblastn) databases from two successive outgroup species (table 1) to identify the amino acid states at orthologous positions. The chosen metazoan outgroups were previously shown to share a negligibly small fraction of editing sites with the focal species [10,14,17], and the proportion of shared editing sites also appears to be very low among filamentous fungi [16]. BLAST searches and extraction of sequence information were automated with custom BioPerl scripts. Analysis was restricted to nonsynonymous editing sites for which both outgroups share the same amino acid so that the ancestral state could be confidently inferred. Editing was defined as ‘restorative’ or ‘diversifying’ if the ancestral amino acid matched the edited and unedited state, respectively. Sites were excluded if the edited and unedited states were both different than the ancestral amino acid (electronic supplementary material, table S1). Statistical analysis was performed in R v. 3.3.3 (see electronic supplementary material, Methods).
Table 1.
Focal species and outgroups for analysis of RNA editing and reconstruction of ancestral states.
| focal species | editing type | editing data | Outgroup 1 | Outgroup 2 |
|---|---|---|---|---|
|
Drosophila melanogaster FlyBase r5.53 |
A-to-I (nuclear) | Ref [17] |
Glossina morsitans VectorBase GmorY1.6 |
Aedes
aegypti VectorBase AaegL3.4 |
|
Homo sapiens Ensembl GRCh37.p13 |
A-to-I (nuclear) | Ref [12] |
Mus musculus Ensembl GRCm38.p2 |
Canis lupus familiaris Ensembl CanFam3.1 |
|
Octopus bimaculoides Ref [14] |
A-to-I (nuclear) | Ref [14] |
Nautilus pompilius Ref [14] |
Aplysia californica Ref [14] |
|
Fusarium graminearum PH-1 Ensembl RR1 |
A-to-I (nuclear) | Ref [15] |
Claviceps purpurea Ensembl ASM34735v1 |
Neurospora crassa Ensembl NC12 |
|
Arabidopsis thaliana Ref [16] |
C-to-U (mito) | Ref [18] |
Roya obtusa GenBank NC_022863.1 |
Chara vulgaris GenBank NC_005255.1 |
3. Results
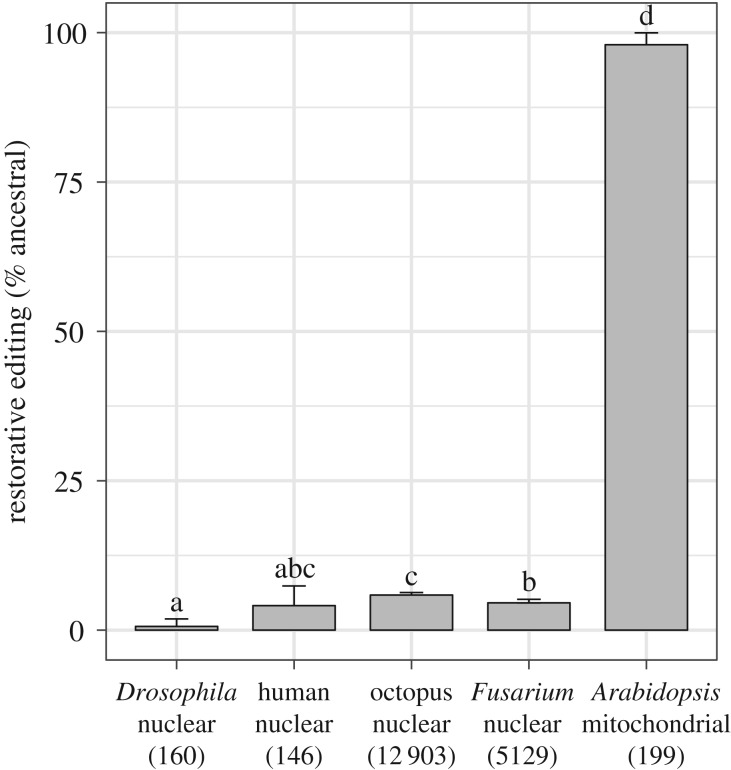
Analysis of C-to-U editing sites in A. thaliana confirmed that mRNA editing in plant mitochondria generally increases protein similarity across taxa. At 98.0% of the analysed sites, editing restores the amino acid found in two green algal outgroups (figure 1). Conversely, just 2.0% of these edits replace the ancestral state with a derived amino acid that differs from the two outgroups. The effects of A-to-I nuclear mRNA editing in metazoans are dramatically different. Only a small fraction of the analysed A-to-I sites lead to restoration of the ancestral state: 0.6%, 4.1% and 5.9% in D. melanogaster, H. sapiens and O. bimaculoides, respectively (figure 1).
Figure 1.
Differences in rates of restorative changes for nuclear A-to-I editing in four different species and mitochondrial C-to-U editing in the angiosperm Arabidopsis. Whereas mRNA editing generally restores ancestral-like protein sequences in most mitochondrial systems (including land plants as shown here), nuclear A-to-I editing is rarely restorative. Lettering is based on post hoc comparisons of each pairwise combination of species (electronic supplementary material, Methods). Species that do not share a letter in common are significantly different from each other. Error bars represent two standard errors of the proportion. The number of analysed sites is indicated in parentheses.
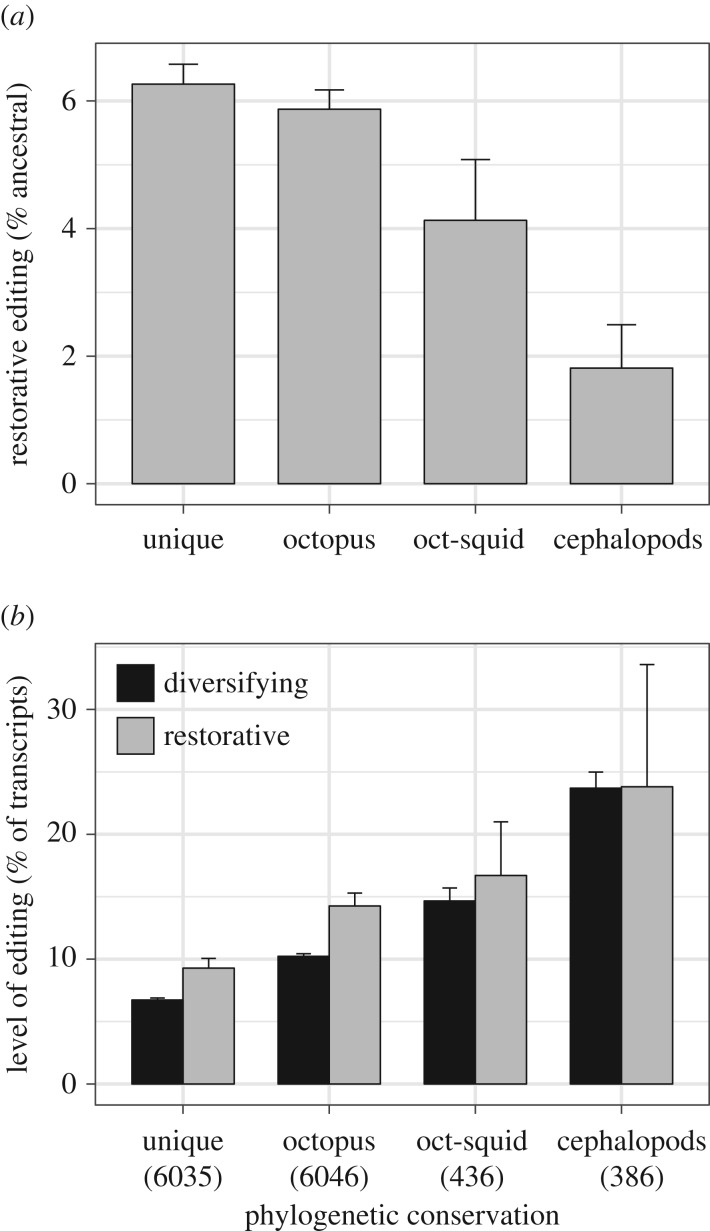
RNA editing is especially abundant in coleoid cephalopods, and previous analysis of O. bimaculoides has shown that most of its editing sites are either unique to that species or shared only with its closely related congener O. vulgaris [14]. However, a small proportion of the editing sites were identified as being shared with other anciently divergent lineages of coleoid cephalopods [14]. These shared sites, which represent the most likely candidates to play functionally important roles, also exhibit a low rate of restorative changes. In fact, in O. bimaculoides, the ratio of restorative to diversifying changes decreases to even lower values for the editing sites that are more widely shared with other cephalopods (figure 2a; table 2).
Figure 2.
Octopus bimaculoides editing sites were distinguished based on whether they are found only in O. bimaculoides (‘unique’), shared with the congener O. vulgaris but not with other cephalopods (‘octopus’), shared with the squid Doryteuthis pealeii but not with all sampled cephalopods (‘oct-squid’), or shared with all sampled cephalopods (‘cephalopods’) [14]. (a) The proportion of edits that restore the ancestral protein sequence decreases for classes of edit sites that are more widely shared across cephalopods. (b) More widely shared sites also have a higher editing frequency (i.e. a larger percentage of transcripts are edited at that site) [14]. Within each group, however, edits that restore the ancestral amino acid have a higher average editing frequency than those that result in a derived change. Errors bars represent one standard error of the proportion (a) or of the mean (b). The number of analysed sites is indicated in parentheses.
Table 2.
Logit model predicting the probability that an edit will be restorative based in the editing frequency at the site and extent of phylogenetic conservation of the editing site. Level of editing is expressed as a per cent (0–100), and the phylogenetic conservation parameters are expressed relative to the most widely shared ‘cephalopod’ category (figure 2).
| model parameter | estimate | z-score | p |
|---|---|---|---|
| intercept | −4.327 | −11.16 | <1 × 10−10 |
| level of editing | 0.012 | 5.80 | 7 × 10−9 |
| phylogenetic conservation | |||
| unique | 1.527 | 3.93 | 8 × 10−5 |
| octopus | 1.407 | 3.63 | 3 × 10−4 |
| oct-squid | 0.970 | 2.14 | 0.03 |
In addition to having low rates of restorative editing, the most widely conserved editing classes in O. bimaculoides were previously shown to exhibit the highest levels of editing (i.e. the fraction of transcripts that are edited at a given site) [14] (figure 2b). Overall, however, editing sites that restore the ancestral amino acid state are edited at a significantly higher level (11.9%) on average than those that produce a derived change (9.2%; table 2). This effect is driven by the higher level of editing at restorative sites within each of the phylogenetic-conservation classes—particularly for editing sites that are unique to O. bimaculoides or only shared within the Octopus genus (figure 2b)—which more than offsets the negative association that exists across classes between the average level of editing and rates of restorative editing (figure 2).
Although A-to-I editing in filamentous fungi appears to be evolutionarily independent from the ADAR system in metazoans, the fungus F. graminearum exhibits similarly low rates of restorative editing. Only 4.6% of the analysed A-to-I sites in F. graminearum nuclear mRNAs lead to restoration of the ancestral state (figure 1).
4. Discussion
Previous research on A-to-I mRNA editing in metazoans led to conclusions that ‘editing can mediate RNA memory on evolutionary time scales to maintain ancestral genetic information’ [11] and ‘editing serves as a mechanism to compensate for a loss of phenotype caused by G-to-A evolution’ [10]. These studies have shown that A-to-I editing sites are more likely to have experienced a previous G-to-A change in DNA sequence than a C-to-A or T-to-A change. Such patterns are important but may largely be explained by the fact that sites that historically accommodated a G will tend to be more permissive of A-to-I editing [12]. From this perspective, focusing on the role of A-to-I editing in reversing G-to-A mutations may arguably miss the bigger picture that it is far more common for A-to-I editing to introduce novel, derived amino acids than to restore ancestral protein sequences (figure 1). The present analysis has shown that this pattern applies not only to diverse metazoan lineages but also to an independent origin of A-to-I nuclear editing in fungi.
This feature of nuclear mRNA editing distinguishes it from mitochondrial editing systems, which generally have restorative effects on protein sequences. Such effects are exemplified here by the C-to-U edits in land plant mitochondrial genomes (figure 1), but they have been documented in numerous mitochondrial systems [1,5–7]. The contrasting effects of nuclear A-to-I editing may, in part, reflect mechanistic differences. In many mitochondrial systems, there is (relatively) precise determination of editing sites based on trans-acting factors or strict cis sequence motifs [19], whereas the adenosine deaminases responsible for A-to-I editing in metazoans appear to have limited specificity. The profile of A-to-I nuclear mRNA editing is also dominated by sites with low editing frequencies. Analysis of the O. bimaculoides transcriptome identified tens of thousands of editing sites in protein-coding sequences [14], but the median level of editing was only 3.4%. Thus, even in cephalopods, where there is evidence that A-to-I editing of protein-coding sequences plays a larger and more adaptive role than in other metazoans [13,14], it is likely that most identified editing sites are nonetheless the result of nonadaptive, off-target activity.
Even so, the differences between mitochondrial RNA editing systems and nuclear A-to-I editing cannot be attributed entirely to differences in enzyme promiscuity. Rates of restorative changes are also extremely low for the subset of A-to-I sites that are edited at high levels and shared among distant relatives (figure 2). One leading hypothesis to explain the proliferation of RNA editing is that the existence of editing activity facilitates the neutral spread by genetic drift of otherwise deleterious mutations. Upon reaching fixation, such mutations would make the formerly nonadaptive editing activity a functional necessity [4]. Now known as ‘constructive neutral evolution’ [20], this nonadaptive model provides a cogent explanation for the extensive restorative editing in mitochondrial genomes but is a seemingly poor fit for the editing patterns in nuclear genes. Instead, it is likely that the evolution of nuclear A-to-I editing and its effects at key functional sites in protein-coding sequences can be attributed to more conventional adaptive explanations associated with regulation and expansion of proteome diversity.
Supplementary Material
Supplementary Material
Acknowledgements
This manuscript was improved based on comments from two anonymous reviewers.
Data accessibility
Site-by-site data are provided as electronic supplementary material, File S1.
Competing interests
The author has no competing interests.
Funding
This work was supported by Colorado State University and the National Science Foundation (MCB-1412260).
References
- 1.Knoop V. 2011. When you can't trust the DNA: RNA editing changes transcript sequences. Cell. Mol. Life Sci. 68, 567–586. ( 10.1007/s00018-010-0538-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishikura K. 2010. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79, 321–349. ( 10.1146/annurev-biochem-060208-105251) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chateigner-Boutin AL, Small I. 2010. Plant RNA editing. RNA Biol. 7, 213–291. ( 10.4161/rna.7.2.11343) [DOI] [PubMed] [Google Scholar]
- 4.Covello PS, Gray MW. 1993. On the evolution of RNA editing. Trends Genet. 9, 265–268. ( 10.1016/0168-9525(93)90011-6) [DOI] [PubMed] [Google Scholar]
- 5.Lavrov DV, Adamski M, Chevaldonné P, Adamska M. 2016. Extensive mitochondrial mRNA editing and unusual mitochondrial genome organization in calcaronean sponges (Calcarea, Porifera). Curr. Biol. 26, 86–92. ( 10.1016/j.cub.2015.11.043) [DOI] [PubMed] [Google Scholar]
- 6.Burger G, Moreira S, Valach M. 2016. Genes in hiding. Trends Genet. 32, 553–565. ( 10.1016/j.tig.2016.06.005) [DOI] [PubMed] [Google Scholar]
- 7.Rüdinger M, Fritz-Laylin L, Polsakiewicz M, Knoop V. 2011. Plant-type mitochondrial RNA editing in the protist Naegleria gruberi. RNA 17, 2058–2062. ( 10.1261/rna.02962911) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mower JP. 2009. The PREP suite: predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 37, W253–W259. ( 10.1093/nar/gkp337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farajollahi S, Maas S. 2010. Molecular diversity through RNA editing: a balancing act. Trends Genet. 26, 221–230. ( 10.1016/j.tig.2010.02.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinto Y, Cohen HY, Levanon EY. 2014. Mammalian conserved ADAR targets comprise only a small fragment of the human editosome. Genome Biol. 15, R5 ( 10.1186/gb-2014-15-1-r5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L. 2013. Characterization and comparison of human nuclear and cytosolic editomes. Proc. Natl Acad. Sci. USA 110, E2741–E2747. ( 10.1073/pnas.1218884110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu G, Zhang J. 2014. Human coding RNA editing is generally nonadaptive. Proc. Natl Acad. Sci. USA 111, 3769–3774. ( 10.1073/pnas.1321745111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alon S, Garrett SC, Levanon EY, Olson S, Graveley BR, Rosenthal JJC, Eisenberg E. 2015. The majority of transcripts in the squid nervous system are extensively recoded by A-to-I RNA editing. Elife 4, e05198 ( 10.7554/eLife.05198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liscovitch-Brauer N, et al. 2017. Trade-off between transcriptome plasticity and genome evolution in cephalopods. Cell 169, 191–202. ( 10.1016/j.cell.2017.03.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu H, et al. 2016. Genome-wide A-to-I RNA editing in fungi independent of ADAR enzymes. Genome Res. 26, 499–509. ( 10.1101/gr.199877.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teichert I, Dahlmann TA, Kück U, Nowrousian M. 2017. RNA editing during sexual development occurs in distantly related filamentous ascomycetes. Genome Biol. Evol. 9, 855–868. ( 10.1093/gbe/evx052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu Y, Zhou H, Kong Y, Pan B, Chen L, Wang H, Hao P, Li X. 2016. The landscape of A-to-I RNA editome is shaped by both positive and purifying selection. PLoS Genet. 12, e1006191 ( 10.1371/journal.pgen.1006191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mower JP. 2008. Modeling sites of RNA editing as a fifth nucleotide state reveals progressive loss of edited sites from angiosperm mitochondria. Mol. Biol. Evol. 25, 52–61. ( 10.1093/molbev/msm226) [DOI] [PubMed] [Google Scholar]
- 19.Sloan DB, Wu Z. 2016. Molecular evolution: the perplexing diversity of mitochondrial RNA editing systems. Curr. Biol. 26, R22–R24. ( 10.1016/j.cub.2015.11.009) [DOI] [PubMed] [Google Scholar]
- 20.Lukeš J, Archibald JM, Keeling PJ, Doolittle WF, Gray MW. 2011. How a neutral evolutionary ratchet can build cellular complexity. IUBMB Life 63, 528–537. ( 10.1002/iub.489) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Site-by-site data are provided as electronic supplementary material, File S1.