Abstract
Herein we present a framework to characterize different sources of protein expression variability in Turing patterned tissues. In this context, we introduce the concept of granular noise to account for the unavoidable fluctuations due to finite cell-size effects and show that the nearest-neighbours autocorrelation function provides the means to measure it. To test our findings, we perform in silico experiments of growing tissues driven by a generic activator–inhibitor dynamics. Our results show that the relative importance of different sources of noise depends on the ratio between the characteristic size of cells and that of the pattern domains and on the ratio between the pattern amplitude and the effective intensity of the biochemical fluctuations. Importantly, our framework provides the tools to measure and distinguish different stochastic contributions during patterning: granularity versus biochemical noise. In addition, our analysis identifies the protein species that buffer the stochasticity the best and, consequently, it can help to determine key instructive signals in systems driven by a Turing instability. Altogether, we expect our study to be relevant in developmental processes leading to the formation of periodic patterns in tissues.
Keywords: patterning, noise, Turing, tissue, development
1. Introduction
Tissue patterning sets the developmental roadmap that provides positional information to cells and confers their unique identities [1–4]. Thus, as a response to distinct protein expression levels in a primordium, cells commit to different fates, may undergo apoptosis, increase/decrease the proliferation rate, or change their division mode to ultimately put into action the developmental plan that shapes the organism. As for the mechanisms of pattern formation, short-range signalling elicits local responses that may propagate in the tissue, e.g. Notch–Delta interactions leading to lateral inhibition/induction [5,6]. On the other hand, long-range signalling driven by diffusive molecules, morphogens, enables positional information to cells at a larger, tissue-level scale and induces pattern formation by following either the French flag model or other mechanisms [4,7]. In particular, Alan Turing proposed a long-range patterning mechanism where interacting species (activators and inhibitors) with different diffusive properties lead to periodic protein expression profiles [8,9]. Turing's proposal meant a breakthrough in mathematical biology; however, besides its success in explaining animal coating [9–11], its relevance in the field of development remained elusive for many years. Notably, during the last decade a number of developmental structures have been found to be shaped by this mechanism [12–14].
The characteristic size of the domains patterned by diffusive processes is controlled by the physical constraints set by the size of morphogen molecules, their production and degradation rates, and the time scales associated with exocytosis and endocytosis among other factors [15,16]. As a result, the reported ratio between the size of patterned domains and that of cells is typically of the order of 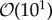 [12,14]. Moreover, cell growth leads to size doubling that leads to perturbations of this ratio by around 20%. These facts raise the intriguing question of how cells deal with aliasing-like effects. By ‘aliasing-like effects’, we mean the consequences in terms of protein variability derived from the sampling process since, as illustrated in figure 1, cells must perform an analogue-to-digital conversion of the, otherwise idealized, continuous periodic pattern into a discretized version due to finite cell-size effects. Here we coin the term granular noise to describe such variability. We point out that the granular effect relies on the hypothesis that proteins are well mixed at the single-cell level and, hence, cells cannot ‘sample’ protein numbers with a subcellular resolution. This hypothesis is sustained by the fact that patterning depends ultimately on morphogens that are driven by a homogenizing transport mechanism: diffusion.
[12,14]. Moreover, cell growth leads to size doubling that leads to perturbations of this ratio by around 20%. These facts raise the intriguing question of how cells deal with aliasing-like effects. By ‘aliasing-like effects’, we mean the consequences in terms of protein variability derived from the sampling process since, as illustrated in figure 1, cells must perform an analogue-to-digital conversion of the, otherwise idealized, continuous periodic pattern into a discretized version due to finite cell-size effects. Here we coin the term granular noise to describe such variability. We point out that the granular effect relies on the hypothesis that proteins are well mixed at the single-cell level and, hence, cells cannot ‘sample’ protein numbers with a subcellular resolution. This hypothesis is sustained by the fact that patterning depends ultimately on morphogens that are driven by a homogenizing transport mechanism: diffusion.
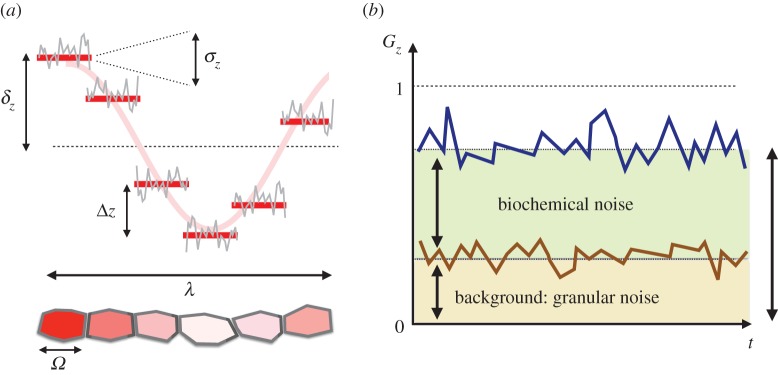
Figure 1.
(a) In a patterning situation, the finite cell size, Ω, sets a characteristic protein concentration variability for species Z between neighbouring cells, Δz, that accounts for the analogue-to-digital conversion of the continuous concentration levels: the so-called granular noise. The biochemical fluctuations at the single-cell level (grey line) additionally contribute to the concentration variability by an amount σz. Finally, patterning itself implies protein differences of order δz (pattern amplitude) at length scales of the order of the pattern wavelength, λ. (b) In a periodic pattern, the nearest-neighbours autocorrelation function, Gz, characterizes the variability at the single-cell level (blue line) by filtering out global patterning effects and allows us to differentiate between the background level due to granularity (brown line/shaded area) and the effective contribution of biochemical fluctuations (green shaded area).
During the last decade, much progress has been attained in the field of noise in gene expression at the single-cell level [17–26]. On the other hand, a number of studies have pointed out the ordering role of fluctuations in spatially extended systems [27–34]. In the context of developmental processes, the role played by cell-to-cell variability, modelled as biochemical noise, in morphogen patterned systems to produce robust and precise positional information has been explored [35]. Also, the effects of embryo size variability, the interactions that help to attenuate the associated noise and the scaling properties of positional information in growing tissues have been recently addressed [36,37].
However, the identification, the characterization and the function of different sources of variability at the collective tissue level within a biological context remain barely explored [38–41]. Notably, to the best of our knowledge, granularity effects have been neglected altogether. Moreover, so far there are no tools that allow us to distinguish among different stochastic contributions, i.e. to measure independently the variability arising from biochemical noise and that from the, unavoidable, granularity. Major difficulties in performing such analysis include the competing effects of various sources of variability at different spatio-temporal scales, the dynamical character of growing tissues and the role played by cell communication that effectively propagates noise in the local cellular environment. Here, we address these questions and present a framework to study gene expression variability, and differentiate among various contributions, in Turing-like patterned tissues (i.e. patterns showing a periodic motif). To that end, we introduce a formalism based on the autocorrelation function that can be easily implemented when processing experimental data. We illustrate our findings by means of a generic activator–inhibitor system and perform realistic numerical simulations of growing tissues using a vertex-model approach that includes mechanical interactions between cells, cell-cycle variability, binomial partition of molecules between daughter cells following division and biochemical noise. Our results confirm that the autocorrelation function is a robust method to compute the granular noise and to estimate also the levels of biochemical fluctuations. The applicability of our methodology to specific developmental processes patterned by the Turing instability is straightforward and we provide simple guidelines to assess and quantify the importance of granularity versus other protein variability sources.
2. Results
As for the various contributions to cell-to-cell variability, finite cell-size effects in protein variability are relevant at length scales of the order of Ω (cell's size) and set protein concentrations quanta: a characteristic protein concentration difference between neighbouring cells that results in a discontinuous protein concentration profile (figure 1). In addition, at the single-cell level, the effects of the biochemical noise further contribute to increasing the variability by a factor σz (the effective intensity of the biochemical fluctuations). Finally, patterning leads to protein concentration differences of order δz (pattern's amplitude) over length scales of order λ (pattern's wavelength).
To filter out the influence of the global patterning on the concentration variability from our analyses, we make use of the nearest-neighbours autocorrelation function
| 2.1 |
where zn stands for the protein concentration levels of species Z at cell n and the average, 〈•〉, is performed over cells (see below for considerations about time averaging). By estimating the concentration differences between neighbouring cells, Gz effectively measures the mutual information in the local cellular environment. Note that Gz also provides an (indirect) estimation of the value of the protein quanta, Δz = [〈(zn+1 − zn)2〉]1/2/〈zn〉.
Note that if there is no biochemical noise, either in the case of constant functions (no patterning) or in the case that there is patterning but it is a continuous solution, 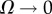 , then zn+1∼ zn and consequently Gz ∼ 0 (maximal similarity in the local cellular environment). On the other hand, when biochemical fluctuations are dominant, zn behaves as a spatial white noise, 〈znzm〉 ∼ δnm/Ω (δnm being the Kronecker delta) and Gz ∼ 1 (minimal similarity among neighbouring cells).
, then zn+1∼ zn and consequently Gz ∼ 0 (maximal similarity in the local cellular environment). On the other hand, when biochemical fluctuations are dominant, zn behaves as a spatial white noise, 〈znzm〉 ∼ δnm/Ω (δnm being the Kronecker delta) and Gz ∼ 1 (minimal similarity among neighbouring cells).
If further sampling over configurations, i.e. noise realizations, is needed (see below) then 〈•〉 accounts for a time average too. Ideally, the sampling frequency for time averages must be smaller than the typical protein turnover rate (to ensure that the pattern has reached stationary conditions) but larger than the characteristic frequency for pattern remodelling (to render enough statistics). Alternatively, if the focus of the study is to characterize the non-equilibrium fluctuations during the process of pattern formation, the analysis should be performed using a sampling frequency larger than the typical protein turnover rate. As for pattern remodelling, as the size of the tissue increases due to cellular growth more of the pattern's domains, i.e. wavelengths, can fit into the size-increasing primordium, thus leading to rearrangements (see the electronic supplementary material, movies S1–S4). Assuming exponential growth conditions, remodelling occurs at a rate ∼ (τlog(λ/Ω))−1 : τ being the average cell-cycle duration. Notably, pattern remodelling relies on the ability of cells to change their expression profile depending on their local environment as experimentally reported [14].
To separate the effects arising from different sources, we analyse first the case when the biochemical noise is negligible and show that we can provide an accurate theoretical estimation of the background granularity and its confidence bounds. If a periodic pattern develops, then zn can be approximated as zn = z0 + δzcos (q*nΩ) along, at least, one of the symmetry axes, where z0 is the average protein concentration and q* = 2π/λ is the most unstable Fourier mode. Under these conditions, Gz can be estimated (see Material and methods)
| 2.2 |
where Ω is the characteristic mean cell size and σΩ is its standard deviation. The error band of Gz sets the confidence bounds of the theoretical prediction and helps to (i) spot pattern misalignment with sampling boxes and (ii) assess if granularity is the dominant source of protein variability. Expression (2.2) determines the, unavoidable, background variability among cells in a local environment due to finite-size effects: the so-called granular noise. Note that this expression is independent of the expression levels of protein species and just depends on geometrical constraints: the ratio between the pattern wavelength and the cell size. Importantly, these quantities, together with σΩ, can be characterized and, consequently, one can compute the basal value of the autocorrelation in the absence of biochemical noise, Gz|σ2z = 0. As shown below, the relative difference of the estimated autocorrelation, Gz|σ2z = 0, from its actual, measured, value in the presence of noise, Gz|σ2z≠0, allows us to distinguish between stochastic sources of protein variability.
In regards of the protein variability due to the biochemical noise, in reaction–diffusion systems the effective magnitude of the fluctuations depends on the regulatory interactions between species, which can either enhance or damp out the noise, and on the diffusive process, which propagates the noise among cells in their local environment and also contributes to average out the fluctuations. We consider that the overall effect of this source of noise can be described as a random additive contribution, ηzn, to the protein concentration: zn = z0 + δzcos(q*nΩ) + ηzn. Thus, we expect the fluctuations to be more relevant at locations where cos(q*nΩ) ≃ 0 and the ratio between the noise and the pattern solution is maximal: the domain boundaries of the pattern. If there is enough statistics then either spatial or time averages, assuming ergodicity, sample the pattern configuration space (noise realizations): 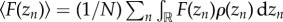 . In that case (see Material and methods)
. In that case (see Material and methods)
| 2.3 |
Consequently, the noise-to-signal ratio can be written as
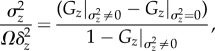 |
2.4 |
where Gz|σ2z = 0 is the background granular noise, which can be estimated by measuring λ and Ω by using equation (2.2), and Gz|σ2z≠0 is the actual nearest-neighbour autocorrelation measured in the presence of biochemical noise using equation (2.1). Table 1 summarizes the most relevant parameters defined in our analysis to estimate the granular noise and measure the effective value of the biochemical fluctuations.
Table 1.
Glossary of relevant symbols.
| symbol | meaning |
|---|---|
| z | concentration of species Z |
| n | cell index |
| σz | effective intensity of the biochemical fluctuations of species Z |
| z0 | average protein concentration of species Z |
| δz | average amplitude of the pattern of species Z |
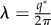 |
wavelength of the pattern |
| Ω | average cell diameter |
| σΩ | standard deviation of the average cell diameter |
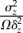 |
noise-to-signal ratio of protein species Z |
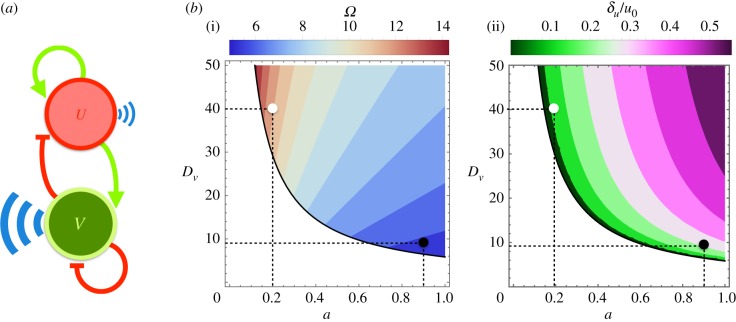
To test and illustrate our findings, we perform computer simulations of growing tissues driven by a generic activator–inhibitor protein dynamics (see Material and methods). Figure 2 shows the regulatory interactions between species and the region where a Turing instability develops and patterns the growing tissue. We stress that the formalism introduced herein does not depend on the specific details of the model (e.g. the number of species or the functional form of the equations) but simply on the existence of a patterning solution with a well-defined periodicity regardless of the mechanism. We simulate the regulatory interactions, the cell's biomechanics and growth/division effects (including a binomial redistribution of proteins between daughter cells following a division) by means of a vertex model approach (see Material and methods). We consider parameter sets with (i) distinct aspect ratios between the pattern domain size and the cell size and (ii) different pattern amplitudes (figure 2b).
Figure 2.
(a) In our simulations, we use a generic activator–inhibitor protein interaction scheme as a model system. In a tissue, a cell's morphogen species U (activator) and V (inhibitor), driven by opposing feedback loops and distinct diffusion rates, develop into a pattern solution at the tissue level. (b) The colour regions in the plots indicate the patterning region as a function of the dimensionless parameters a (activation versus inhibition strength) and Dv (inhibitor's diffusion coefficient in units of that of the activator). The scales stand for the size of pattern domains in units of the cell size (i) and for the ratio between the amplitude of the pattern of species U and its average concentration u0 (ii). We perform computer simulations of a developing tissue using a vertex model approach using the following parameters: (a, Dv) = (0.2, 40), a large ratio of the pattern domain size versus cell size, around 11, and a pattern amplitude with respect to the average levels of around 11% (white circle); and (a, Dv) = (0.9, 9), a small ratio for the pattern domain size versus cell size, around 6, and a global pattern variability of around 25% (black circle).
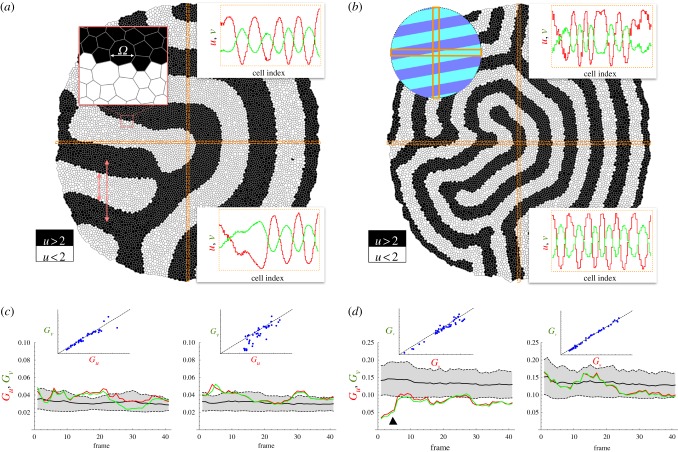
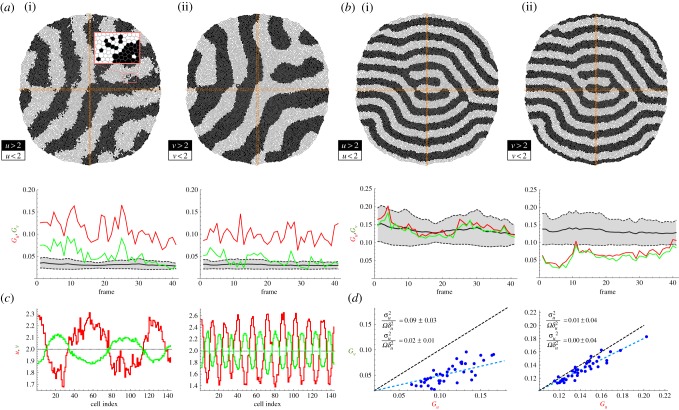
In order to assess whether our theoretical estimation of Gz|σ2z = 0 captures correctly the background granularity, we first performed numerical simulations of the growing tissue in the absence of biochemical fluctuations. Figure 3 shows snapshots of the growing, patterned tissue for different Ω/λ ratios (see the electronic supplementary material, movies S1 and S3). To quantify Gz using equation (2.1), we sampled the tissue at regular time intervals (approx. 40 frames per cell cycle). We show the importance of sampling the periodicity of the patterned solution correctly by sampling cells from the tissue along perpendicular, fixed stripes with a large aspect ratio: approximately one cell diameter width and whole-tissue length (figure 3b). Note that in the case of stripe-like patterns it is possible to find a large degree of alignment of the pattern with a sampling box (thus masking the pattern periodicity). In our analysis, the cell's characteristic diameter is estimated as the square root of the apical area of the sampled cells.
Figure 3.
(a,b) Tissue simulation snapshots (final frames). The concentration of species U is plotted using a binary representation (above/below the average concentration) to better visualize the pattern domains. (a/b) Simulation parameters as indicated by the black/white circles in figure 2b. The zoomed-in area in (a) helps to compare the typical cell size, Ω, with the pattern wavelength, λ, and the domain size. Insets in (a/b) show the concentration profiles of u and v as a function of the cell index (cell sizes have been normalized) of the cells within the horizontal/vertical sampling stripes (orange boxes). The top-left inset in (b) sketches a patterning condition where the horizontal sampling region would not capture the spatial periodicity adequately because of its large degree of alignment with the pattern (see (d)). (c,d) Quantification of Gz. The shaded region accounts for the theoretical estimation, equation (2.2): black solid/dashed lines stand for the mean/standard deviation. Granular effects are more pronounced as the ratio Ω/λ increases (notice the scale difference between (c) and (d)): both the background granular noise, Gz, and the protein quanta, Δz, increase approximately fourfold in (b) with respect to (a). Pattern/sampling-box alignment events can be spotted by the underestimation of Gz with respect to the theoretical background levels (black triangle in (d)) and help to discard data that might lead to artefacts in the analysis (wrong sampling of the pattern periodicity). (c,d) insets: Gv versus Gu. If no other noisy contribution is present then Gu ∼ Gv and the granular noise shows an extrinsic-like behaviour thus affecting all protein species equally.
As shown in figure 3, the quantification of the background granular noise is in agreement with the theoretical estimation, equation (2.2). As expected, finite cell-size effects are more pronounced as the ratio Ω/λ increases, that is, as the ratio of the pattern wavelength to the cell size decreases. Note also that the granular noise has an extrinsic character [19] since, as predicted, it affects all protein species equally and independently of their expression levels, i.e. Gu ≃ Gv. Thus, in a periodic patterning situation, if the pattern wavelength and the characteristic cell size are determined, then equation (2.2) accounts for the variability in protein concentration due to finite cell-size effects and allows us to distinguish among different noisy sources as shown below.
In order to explore the competing effects between granular noise and intrinsic/extrinsic fluctuations, we implement a stochastic dynamics for proteins U and V and study different noise-to-signal ratios (see Material and methods). Figure 4 shows the quantification of Gz|σ2z≠0 as measured by equation (2.1), the theoretical estimations of Gz|σ2z = 0, the noise-to-signal ratio, and the patterning domains of U and V (see the electronic supplementary material, movies S2 and S4). As the levels of Gz indicate, the protein concentration profiles of the activator (slow-diffusive) species, U, are noisier than those of V and these stochastic effects are specially relevant at the domain boundaries. In addition, the effects due to fluctuations are more significant as the background granular noise decreases. These results reveal that the noise-to-signal ratio is dominant in (i) slow versus fast diffusing protein species, because noise averages out more easily in the second case, (ii) low versus high background granular noise situations, because otherwise the latter prevails, and (iii) activator versus inhibitor species, because the positive auto-regulation of the former contributes to amplify the noise. Note also that Gz deviates from the extrinsic behaviour as predicted by equation (2.3).
Figure 4.
(a,b) Competing effects in protein variability between granular noise and intrinsic/extrinsic fluctuations in different patterning conditions. Parameter values of the simulations as indicated in figure 1: white/black circles correspond to (a/b). In the case that the background granular noise is weak and the pattern amplitude is of the order of the intrinsic/extrinsic fluctuations (a), then the latter are dominant (see (c)). The level of noise is effectively smaller in the inhibitor species, V , than in the activator species, U, due to the diffusive process, which is faster for V and thus helps fluctuations to average out, and to the regulatory interactions, since the autocatalytic feedback in U amplifies noisy effects. The zoom-in inset in (a) shows that noisy effects are specially relevant in the regions close to the boundary domains. (c) Profiles of U and V for (a(i)) and (b(ii)) along the sampling box that better capture the pattern periodicity (vertical box in both cases). Noise effects are more evident in species U as quantified by Gz. (d) Gv versus Gu for (a(i)) and (b(ii)) as registered by the vertical sampling boxes. If intrinsic/extrinsic fluctuations perturb the patterning, then Gz deviates from the extrinsic behaviour (cf. figure 3). In both cases, but especially in (a), Gu versus Gv deviates systematically below the diagonal, indicating that the fluctuations are larger at the level of U, as confirmed by the quantification of the noise-to-signal ratio (inset values).
3. Discussion
We have shown that by using the nearest-neighbours autocorrelation function the variability due to global patterning effects can be filtered out. Such variability of order δz can be estimated by Fourier analysis (amplitude of the most unstable mode). Aliasing effects and biochemical noisy contributions (intrinsic and/or extrinsic fluctuations) can be separated and determined by means of Gz when combining theoretical expressions and measurements. As for the applicability of our approach, experimental results have revealed that the ratio λ/Ω, pattern wavelength versus cell size, can be small and patterning domains may comprise around 8–10 cells [12–14]. Thus, we predict that the granular noise contributes significantly to protein variability. In the particular case of the limb bud, recent results have shown that this primordium is patterned by a Turing instability resulting from the interactions between two morphogens, Bmp and Wnt, and the skeletal marker Sox9 [14]. A quick assessment indicates that λ ∼ 10Ω and consequently the background granularity is of the order of around 20%. Evidence supporting the buffer effect of diffusion over the fluctuations can also be found in the limb bud. Both morphogens, Bmp and Wnt, lack a self-regulatory motif that in turn helps to damp the biochemical fluctuations, as shown in our simulations (see below for further considerations about the instructive role of inhibitors in Turing systems). However, the values of the diffusion coefficients satisfy that DBmp ≫ DWnt and one would expect, according to our analysis, that Wnt has a noiser profile than Bmp. The experimentally reported profiles of these species during limb bud patterning [14] seem to be in agreement with this prediction.
Importantly, we have presented our results using a methodology that is feasible in experimental situations. In particular, since the dimensionality of the pattern is not relevant for estimating these effects, by determining the pattern wavelength and by sampling the size and protein expression levels for (i) a few cells, 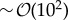 , along a direction that captures the best the pattern periodicity and (ii) a small number of time points in the tissue dynamics,
, along a direction that captures the best the pattern periodicity and (ii) a small number of time points in the tissue dynamics, 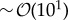 , then the background granularity and the biochemical noise can be specified. Further sampling can provide better, more accurate results but the fact that the method works with a limited statistics reveals its robustness. We acknowledge that the experimental quantification of protein concentrations at the single-cell level requires an accurate calibration of the fluorescence intensity [42]. However, we stress that the background granularity levels, as prescribed by the theoretical expression equation (2.2), are independent of the protein levels. Thus, as long as the fluorescence levels of protein species are normalized, our method provides a way to compute the relative noise-to-signal ratio and weight the importance of biochemical noise with respect to granularity.
, then the background granularity and the biochemical noise can be specified. Further sampling can provide better, more accurate results but the fact that the method works with a limited statistics reveals its robustness. We acknowledge that the experimental quantification of protein concentrations at the single-cell level requires an accurate calibration of the fluorescence intensity [42]. However, we stress that the background granularity levels, as prescribed by the theoretical expression equation (2.2), are independent of the protein levels. Thus, as long as the fluorescence levels of protein species are normalized, our method provides a way to compute the relative noise-to-signal ratio and weight the importance of biochemical noise with respect to granularity.
The proposed method relies on several assumptions that are worth commenting on. First, we have assumed that a pattern with a well-defined wavelength develops. In any particular patterning situation, a Fourier analysis on the protein profile, as obtained, for example, by fluorescence microscopy, will reveal the validity of this assumption. We point out that, even if several Fourier modes are relevant, the methodology remains valid. However, the analytical expressions to estimate the background granularity, equation (2.2), and the noise-to-signal ratio, equation (2.4), would need revision. In any case, if the pattern solution is a collection of waves then the calculations can be carried out easily (Material and methods). Second, we have assumed that, besides the pattern's rearrangement dynamics due to cell growth, stationary conditions apply. In this regard, we notice that if the amplitude of the pattern is modulated in time then our results still hold. However, we stress that time averaging must be done carefully to ensure that the pattern configuration space is properly sampled. Third, we have considered that the overall effect of the biochemical fluctuations renders a Gaussian-like, white behaviour that is independent of the value of the pattern amplitude. This approximation is motivated by the central limit theorem [43] and makes it possible to obtain analytical expressions for the noise-to-signal ratio. However, it may raise questions about its fidelity since other statistical properties for the noise are certainly possible [21,41]. Yet, the background granular noise is independent of the statistical properties of the noise since it merely depends on the geometrical constraints of the problem (cell size and pattern wavelength). Consequently, the difference between the nearest-neighbours autocorrelation and the estimated theoretical background granular noise still provides an estimation of the levels of biochemical fluctuations.
As for additional implications of our study, our results indicate that variability due to the biochemical noise is larger for cells at domain boundaries, especially for activator species. According to this, fate commitment, as a result of patterning, would be more difficult for those cells. In addition, boundary lines among cell populations would be wiggly, thus suggesting that the formation of developmental structures, e.g. the fingering pattern in the case of the limb bud [14], could be challenged. However, the robustness displayed by biological systems argues against these ideas. This raises the intriguing question about the mechanisms that are able to buffer noisy effects in tissue patterning. In this regard, we note that the level of variability is reduced in inhibitor species (figure 4). The latter suggests a possible instructive role for this species in fate decision-making (less noisy). In addition, it may explain the recurrent feedback found in developmental boundaries between patterning and cell mechanical properties to refine compartmentalization [44–49].
We expect our study to be relevant in developmental processes leading to the formation of periodic patterns in tissues when cell-to-cell variability needs to be characterized to, for example, better understand fate decision-making. The extension of the proposed method to other patterning situations, e.g. morphogen gradient profiles, or the implementation of feedback between signalling and cell mechanics to buffer the fluctuation effects are promising avenues of research that can shed further light on the role of noise in developmental patterning systems. Work in these directions is in progress.
4. Material and methods
4.1. Tissue simulations
The tissue dynamics is implemented in our simulations using a vertex model approach [50]. Each cell in the tissue is represented by a discrete set of points: the vertexes that define its shape. The energy associated with a vertex i reads
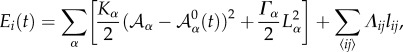 |
where the sums indexed by α and 〈ij〉 run, respectively, over the cells α and vertices j sharing vertex i. 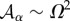 is the cell apical area, Kα is proportional to the Young modulus, Λij is a line tension that weights the cell adhesion effects (lij being the length of the edge connecting neighbouring vertices i and j), Γα is a term that accounts for the contraction effect of the actomyosin cortical ring and Lα is the cell perimeter (see [44] for details). The parameters used in our simulations are (dimensionless):
is the cell apical area, Kα is proportional to the Young modulus, Λij is a line tension that weights the cell adhesion effects (lij being the length of the edge connecting neighbouring vertices i and j), Γα is a term that accounts for the contraction effect of the actomyosin cortical ring and Lα is the cell perimeter (see [44] for details). The parameters used in our simulations are (dimensionless): 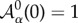 , K = 1, Γ = 2 × 10−2 and Λ = 10−2. In addition we consider that in the tissue periphery Λ = 5 × 10−2. The cell-cycle duration, τ, is stochastic in our simulations and follows the rule τ = ɛtdet + (1 − ɛ) tsto, where tdet is a deterministic time scale that accounts for a mean cell-cycle duration in the absence of mechanical stress and tsto is a random variable that accounts for the variability of cell-cycle duration,
, K = 1, Γ = 2 × 10−2 and Λ = 10−2. In addition we consider that in the tissue periphery Λ = 5 × 10−2. The cell-cycle duration, τ, is stochastic in our simulations and follows the rule τ = ɛtdet + (1 − ɛ) tsto, where tdet is a deterministic time scale that accounts for a mean cell-cycle duration in the absence of mechanical stress and tsto is a random variable that accounts for the variability of cell-cycle duration, 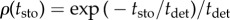 . The parameter ɛ ∈ [0, 1] weights the stochasticity of the cell-cycle duration. In our simulations, ɛ = 0.8.
. The parameter ɛ ∈ [0, 1] weights the stochasticity of the cell-cycle duration. In our simulations, ɛ = 0.8.
Cellular growth is implemented by prescribing the following dynamics for the preferred apical cell area 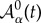 (dimensionless): cells start to grow their apical area linearly (towards doubling) at the moment they reach the middle of the cell-cycle progression [44]. With respect to the cleavage orientation, the code evaluates the inertia tensor of the cell with respect to its centre of mass assuming that a proper representation of the former is a polygonal set of rods, i.e. the cell edges. The principal inertia axes indicate the symmetry axes of the cell: the longest axis of the cell is orthogonal to the largest principal inertia axis. Cells divide following the Hertwig rule: the cleavage plane is perpendicular to the longest axis [51]. Cleavage is assumed to happen instantaneously. In our in silico experiments, tissues are typically grown from approximately 2.5 × 103 to approximately 1.2 × 104 cells (approx. two cell cycles). Transients effects in the tissue dynamics due to the initial conditions are eliminated from our analysis by discarding data from the first cell cycle.
(dimensionless): cells start to grow their apical area linearly (towards doubling) at the moment they reach the middle of the cell-cycle progression [44]. With respect to the cleavage orientation, the code evaluates the inertia tensor of the cell with respect to its centre of mass assuming that a proper representation of the former is a polygonal set of rods, i.e. the cell edges. The principal inertia axes indicate the symmetry axes of the cell: the longest axis of the cell is orthogonal to the largest principal inertia axis. Cells divide following the Hertwig rule: the cleavage plane is perpendicular to the longest axis [51]. Cleavage is assumed to happen instantaneously. In our in silico experiments, tissues are typically grown from approximately 2.5 × 103 to approximately 1.2 × 104 cells (approx. two cell cycles). Transients effects in the tissue dynamics due to the initial conditions are eliminated from our analysis by discarding data from the first cell cycle.
As for the protein dynamics, we assume each cell to be a well-stirred system, where spatial effects can be disregarded. Protein concentration values in each cell are obtained by integrating numerically the discretized model equations using an Euler algorithm. We take into account cell growth (dilution effects) to determine protein concentrations. Also, proteins are distributed binomially between daughter cells as a consequence of a division event. The morphogen diffusion process is mimicked by an out-of-lattice, discretized Laplacian operator that conserves the number of molecules [52],
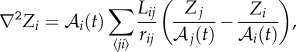 |
where Zi stands for the number of proteins at cell i, 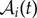 is its area, the sum runs over its nearest neighbours j, Lij is the length of the membrane shared among cells and rij is the distance between cell centres. This definition captures rigorously the discretization of the Laplacian operator as long as the well-mixed hypothesis holds (see Introduction).
is its area, the sum runs over its nearest neighbours j, Lij is the length of the membrane shared among cells and rij is the distance between cell centres. This definition captures rigorously the discretization of the Laplacian operator as long as the well-mixed hypothesis holds (see Introduction).
Biochemical noise effects in protein species Z at cell n and time t, ξzn(t), are implemented by means of additive uncorrelated Gaussian fluctuations: 〈ξzn(t)〉 = 0 and 〈ξzn(t)ξz′m(t′)〉 = (σ2ξ/Ω)δnmδzz′δ(t − t′), where σ2ξ is the intensity of the noise. Our simulations explore the conditions σξ < δz (black circle in figure 1b) and σξ ∼ δz (white circle in figure 1b). We do not consider the case σξ > δz since it leads to unrealistic fluctuation-controlled expression profiles in which patterning does not play a key role. In all cases σξ = 10−1. We point out that the value of σξ does not represent the effective intensity of the fluctuations σz since the latter depends on the regulatory interactions and on the morphogen diffusion process. That is, ξn are the input fluctuations with intensity σξ that we use to represent the biochemical noise at the single-cell level in the simulations, and ηn are the output fluctuations with intensity σz that, as a result of the interactions and diffusion, the proteins effectively experience and we register using the autocorrelation method. The code for carrying out the numerical simulations is provided as the electronic supplementary material.
4.2. Analytical calculations
Given the expression of the autocorrelation function, equation (2.2), and taking into account that 〈zn+1〉 = 〈zn〉, then Gz can be rewritten as
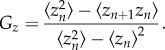 |
4.1 |
Thus, to characterize Gz the moments 〈zn〉, 〈z2n〉 and 〈zn+1zn〉 need to be estimated. Given the functional form of the Turing patterning solution, zn = z0 + δz cos(q*nΩ), and the following identities (valid if N = L/Ω ≫ 1, L being the size of the tissue), 〈cos(q*nΩ)〉 ≃ 0, 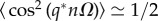 and 〈cos(q*nΩ)cos(q*(n + 1)Ω)〉 ≃ cos(q*Ω)/2, those moments read
and 〈cos(q*nΩ)cos(q*(n + 1)Ω)〉 ≃ cos(q*Ω)/2, those moments read
| 4.2 |
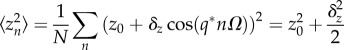 |
4.3 |
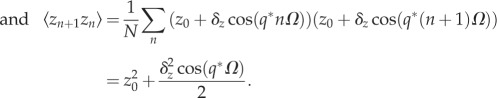 |
4.4 |
Substituting the above expressions into equation (4.1), we obtain Gz = 1 − cos(q*Ω). The confidence bounds for Gz are determined by the variation of the parameters q* and Ω. Yet, Turing patterning tissues show a precise wavelength and the main source of variability is the cell size, which may show variations up to 100% due to size doubling during cell-cycle progression. Thus, given the standard deviation of the cell size in the tissue, σΩ , the error propagates to Gz as ± |∂Gz/∂Ω|σΩ = ± σΩq*|sin(q*Ω)|.
In the case where we assume a random perturbation, η, of the patterning solution due to the biochemical noise, the functional form of the protein concentration profile now reads
| 4.5 |
By invoking the central limit theorem [43], here we implement a Gaussian approximation and assume that ηzn satisfies the distribution
where 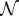 is a normalization constant and σ2z/Ω is the effective intensity of the biochemical noise of species Z. In addition, we consider that the fluctuations are uncorrelated in space and time: 〈ηnηm〉 = σ2z(δnm/2Ω)δ(t − t′), where δij stands for the Kronecker delta and δ(s) for the Dirac delta. Note that the cell size, Ω, is required in the definition of the noise autocorrelation such that in the continuous limit the autocorrelation satisfies a white noise character in space:
is a normalization constant and σ2z/Ω is the effective intensity of the biochemical noise of species Z. In addition, we consider that the fluctuations are uncorrelated in space and time: 〈ηnηm〉 = σ2z(δnm/2Ω)δ(t − t′), where δij stands for the Kronecker delta and δ(s) for the Dirac delta. Note that the cell size, Ω, is required in the definition of the noise autocorrelation such that in the continuous limit the autocorrelation satisfies a white noise character in space: 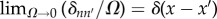 . Formally, one may choose a definition of the noise intensity leaving out the cell size Ω and integrating the latter in the correlation properties of the fluctuations (i.e. in the delta function). Yet, herein we preferred to explicitly include this dependence of the cell size effectively in the definition of the biochemical fluctuations. The Gaussian approximation is valid as long as the intensity of the noise does not overcome the patterning solution, that is, σz < z0 − δz. Otherwise the noise could lead to unphysical values (negative) of the protein concentration. Given ρ(ηzn) the probability distribution for zn reads
. Formally, one may choose a definition of the noise intensity leaving out the cell size Ω and integrating the latter in the correlation properties of the fluctuations (i.e. in the delta function). Yet, herein we preferred to explicitly include this dependence of the cell size effectively in the definition of the biochemical fluctuations. The Gaussian approximation is valid as long as the intensity of the noise does not overcome the patterning solution, that is, σz < z0 − δz. Otherwise the noise could lead to unphysical values (negative) of the protein concentration. Given ρ(ηzn) the probability distribution for zn reads
| 4.6 |
In this case, the moments 〈zn〉, 〈z2n〉 and 〈zn+1zn〉, by averaging over noise realizations, read
| 4.7 |
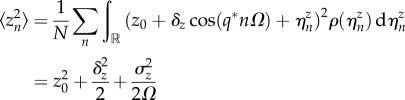 |
4.8 |
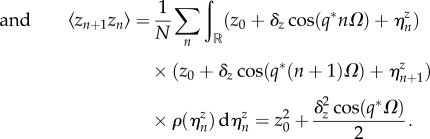 |
4.9 |
Consequently,
| 4.10 |
and
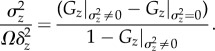 |
4.11 |
We notice that in the case where the assessment of the patterning solution reveals more than one unstable Fourier mode, e.g. zn = z0 + δ(1)zcos(q*(1)nΩ) + δ(2)zcos(q*(2)nΩ), the above calculations are straightforward. The generalization of our framework to other patterning situations, e.g. morphogen gradients, can also be easily implemented. By taking into account the morphogen decay length, λ, the protein profile reads [15], zn = z0 e−nΩ/λ. Thus, the moments 〈zn〉, 〈z2n〉, and 〈zn+1zn〉 can be estimated and the background granularity reads
As in the case of Turing patterning, the background granularity goes to zero as the ratio Ω/λ does.
4.3. Activator–inhibitor Turing patterning systems
For the sake of simplicity, we restrict our analysis to a system with two coupled reaction–diffusion equations in one spatial dimension. Yet, the analysis presented below can be easily generalized. Thus, the following dimensionless system of equations describes the reaction and diffusion terms of two protein species U and V:
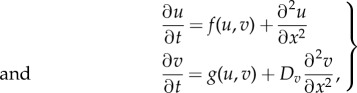 |
4.12 |
where u = u(x, t) and v = v(x, t) represent protein concentrations. The reaction terms f and g are supposed to have a single equilibrium point, 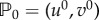 , such that f(u0, v0) = g(u0, v0) = 0 and the point
, such that f(u0, v0) = g(u0, v0) = 0 and the point 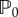 is assumed to be stable in the absence of diffusion. That is, by defining
is assumed to be stable in the absence of diffusion. That is, by defining 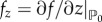 and
and 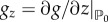 , where z stands for either the field u or the field v, the following conditions hold:
, where z stands for either the field u or the field v, the following conditions hold:
| 4.13 |
A Fourier analysis reveals that a mode, q*≠0, becomes destabilized and a pattern develops if 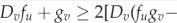
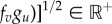 [9,33]. It is easy to prove that in terms of the signs of the entries of the Jacobian matrix
[9,33]. It is easy to prove that in terms of the signs of the entries of the Jacobian matrix
only four out of the possible eight options can lead to a Turing pattern instability:
Here we choose the first option where species U stands for an activator and V for an inhibitor: {fu, fv} > 0, {gu, gv} < 0. If fu < |gv|, and fu|gv| < fv|gu|, then conditions (4.11) are satisfied. In addition, if fu = fv = a, gu = − 2 and gv = − 1 a pattern develops if
The modelling equations require nonlinear terms to saturate to a finite value the amplitude of the pattern, which otherwise diverges. By choosing a cubic non-linearity and 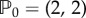 , we obtain the following modelling equations describing an activator–inhibitor system:
, we obtain the following modelling equations describing an activator–inhibitor system:
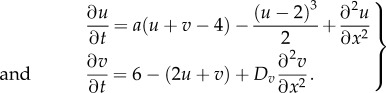 |
4.14 |
Supplementary Material
Data accessibility
The simulation code and compilation instructions for generating all data shown in the manuscript are available at https://doi.org/10.6084/m9.figshare.5146564.
Competing interests
I declare I have no competing interests.
Funding
No funding has been received for this article.
References
- 1.Gilbert SF. 2014. Developmental biology. Sunderland, MA: Sinauer Associates Inc. [Google Scholar]
- 2.Kicheva A, Cohen M, Briscoe J. 2012. Developmental pattern formation: insights from physics and biology. Science 338, 210–212. ( 10.1126/science.1225182) [DOI] [PubMed] [Google Scholar]
- 3.Tickle C. 2003. Patterning in vertebrate development. Oxford, UK: Oxford University Press. [Google Scholar]
- 4.Torii KU. 2012. Two-dimensional spatial patterning in developmental systems. Trends Cell Biol. 22, 438–446. ( 10.1016/j.tcb.2012.06.002) [DOI] [PubMed] [Google Scholar]
- 5.Bray SJ. 2016. Notch signalling in context. Nat. Rev. Mol. Cell. Biol. 17, 722–735. ( 10.1038/nrm.2016.94) [DOI] [PubMed] [Google Scholar]
- 6.Collier JR, Monk NA, Maini PK, Lewis JH. 1996. Pattern formation by lateral inhibition with feedback: a mathematical model of delta-notch intercellular signalling. J. Theor. Biol. 183, 429–446. ( 10.1006/jtbi.1996.0233) [DOI] [PubMed] [Google Scholar]
- 7.Hillenbrand P, Gerland U, Tkacik G. 2016. Beyond the French flag model: exploiting spatial and gene regulatory interactions for positional information. PLoS ONE 11, e0163628 ( 10.1371/journal.pone.0163628) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turing A. 1952. The chemical basis of morphogenesis. Phil. Trans. R. Soc. Lond. B 237, 37–72. ( 10.1098/rstb.1952.0012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray JD. 2011. Mathematical biology II. Spatial models and biomedical applications. New York, NY: Springer. [Google Scholar]
- 10.Kondo S, Iwashita M, Yamaguchi M. 2009. How animals get their skin patterns: fish pigment pattern as a live Turing wave. Int. J. Dev. Biol. 53, 851–856. ( 10.1387/ijdb.072502sk) [DOI] [PubMed] [Google Scholar]
- 11.Kondo S, Miura T. 2010. Reaction-diffusion model as a framework for understanding biological pattern formation. Science 329, 1616–1620. ( 10.1126/science.1179047) [DOI] [PubMed] [Google Scholar]
- 12.Economou AD, Ohazama A, Porntaveetus T, Sharpe PT, Kondo S, Basson MA, Gritli-Linde A, Cobourne MT, Green JBA. 2012. Periodic stripe formation by a Turing mechanism operating at growth zones in the mammalian palate. Nat. Genet. 44, 348–351. ( 10.1038/ng.1090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheth R, Marcon L, Bastida MF, Junco M, Quintana L, Dahn R, Kmita M, Sharpe J, Ros MA. 2012. Hox genes regulate digit patterning by controlling the wavelength of a Turing-type mechanism. Science 338, 1476–1480. ( 10.1126/science.1226804) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raspopovic J, Marcon L, Russo L, Sharpe J. 2014. Digit patterning is controlled by a Bmp-Sox9-Wnt Turing network modulated by morphogen gradients. Science 345, 566–570. ( 10.1126/science.1252960) [DOI] [PubMed] [Google Scholar]
- 15.Kicheva A, Pantazis P, Bollenbach T, Kalaidzidis Y, Bittig T, Jülicher F, González-Gaitán M. 2007. Kinetics of morphogen gradient formation. Science 315, 521–525. ( 10.1126/science.1135774) [DOI] [PubMed] [Google Scholar]
- 16.Truskey GA, Yuan F, Katz DF. 2009. Transport phenomena in biological systems. Englewood Cliffs, NJ: Prentice-Hall. [Google Scholar]
- 17.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. 2002. Regulation of noise in the expression of a single gene. Nat. Genet. 31, 69–73. ( 10.1038/ng869) [DOI] [PubMed] [Google Scholar]
- 18.Elowitz MB, Levine AJ, Siggia ED, Swain PS. 2002. Stochastic gene expression in a single cell. Science 297, 1183–1186. ( 10.1126/science.1070919) [DOI] [PubMed] [Google Scholar]
- 19.Swain PS, Elowitz MB, Siggia ED. 2002. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc. Natl Acad. Sci. USA 99, 12 795–12 800. ( 10.1073/pnas.162041399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao CV, Wolf DM, Arkin AP. 2002. Control, exploitation and tolerance of intracellular noise. Nature 420, 231–237. ( 10.1038/nature01258) [DOI] [PubMed] [Google Scholar]
- 21.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. 2005. Gene regulation at the single-cell level. Science 307, 1962–1965. ( 10.1126/science.1106914) [DOI] [PubMed] [Google Scholar]
- 22.Pedraza JM, van Oudenaarden A. 2005. Noise propagation in gene networks. Science 307, 1965–1969. ( 10.1126/science.1109090) [DOI] [PubMed] [Google Scholar]
- 23.Pedraza JM, Paulsson J. 2008. Effects of molecular memory and bursting on fluctuations in gene expression. Science 319, 339–343. ( 10.1126/science.1144331) [DOI] [PubMed] [Google Scholar]
- 24.Raser JM, O'Shea EK. 2005. Noise in gene expression: origins, consequences, and control. Science 309, 2010–2013. ( 10.1126/science.1105891) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eldar A, Elowitz MB. 2010. Functional roles for noise in genetic circuits. Nature 467, 167–173. ( 10.1038/nature09326) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munsky B, Neuert G, van Oudenaarden A. 2012. Using gene expression noise to understand gene regulation. Science 336, 183–187. ( 10.1126/science.1216379) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.García-Ojalvo J, Sancho JM. 2012. Noise in spatially extended systems. New York, NY: Springer. [DOI] [PubMed] [Google Scholar]
- 28.Sagués F, Sancho JM, García-Ojalvo J. 2007. Spatiotemporal order out of noise. Rev. Mod. Phys. 79, 829–882. ( 10.1103/RevModPhys.79.829) [DOI] [Google Scholar]
- 29.Parrondo JMR, van den Broeck C, Buceta J, de la Rubia FJ. 1996. Noise-induced spatial patterns. Physica A 224, 153–161. ( 10.1016/0378-4371(95)00350-9) [DOI] [Google Scholar]
- 30.Buceta J, Lindenberg K, Parrondo JMR. 2002. Stationary and oscillatory spatial patterns induced by global periodic switching. Phys. Rev. Lett. 88, 024103 ( 10.1103/PhysRevLett.88.024103) [DOI] [PubMed] [Google Scholar]
- 31.Buceta J, Lindenberg K. 2003. Spatial patterns induced purely by dichotomous disorder. Phys. Rev. E 68, 011103 ( 10.1103/PhysRevE.68.011103) [DOI] [PubMed] [Google Scholar]
- 32.Buceta J, Ibañes M, Sancho JM, Lindenberg K. 2003. Noise-driven mechanism for pattern formation. Phys. Rev. E 67, 021113 ( 10.1103/PhysRevE.67.021113) [DOI] [PubMed] [Google Scholar]
- 33.Buceta J, Lindenberg K. 2003. Patterns in reaction-diffusion systems generated by global alternation of dynamics. Physica A 325, 230–242. ( 10.1016/S0378-4371(03)00202-4) [DOI] [Google Scholar]
- 34.Buceta J, Lindenberg K. 2002. Switching-induced Turing instability. Phys. Rev. E 66, 046202 ( 10.1103/PhysRevE.66.046202) [DOI] [PubMed] [Google Scholar]
- 35.Bollenbach T, Pantazis P, Kicheva A, Bökel C, González-Gaitán M, Jülicher F. 2008. Precision of the Dpp gradient. Development 135, 1137–1146. ( 10.1242/dev.012062) [DOI] [PubMed] [Google Scholar]
- 36.Rahimi N, Averbukh I, Haskel-Ittah M, Degani N, Schejter ED, Barkai N, Shilo B-Z. 2016. A WntD-dependent integral feedback loop attenuates variability in drosophila toll signaling. Dev. Cell 36, 401–414. ( 10.1016/j.devcel.2016.01.023) [DOI] [PubMed] [Google Scholar]
- 37.Averbukh I, Ben-Zvi D, Mishra S, Barkai N. 2014. Scaling morphogen gradients during tissue growth by a cell division rule. Development 141, 2150–2156. ( 10.1242/dev.107011) [DOI] [PubMed] [Google Scholar]
- 38.England JL, Cardy J. 2005. Morphogen gradient from a noisy source. Phys. Rev. Lett. 94, 078101 ( 10.1103/PhysRevLett.94.078101) [DOI] [PubMed] [Google Scholar]
- 39.Gregor T, Tank DW, Wieschaus EF, Bialek W. 2007. Probing the limits to positional information. Cell 130, 153–164. ( 10.1016/j.cell.2007.05.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saunders TE, Howard M. 2009. Morphogen profiles can be optimized to buffer against noise. Phys. Rev. E. Stat. Nonlin. Soft. Matter. Phys. 80, 041902 ( 10.1103/PhysRevE.80.041902) [DOI] [PubMed] [Google Scholar]
- 41.Scott M, Poulin FJ, Tang H. 2011. Approximating intrinsic noise in continuous multispecies models. Proc. R. Soc. A 467, 718–737. ( 10.1098/rspa.2010.0275) [DOI] [Google Scholar]
- 42.Verdaasdonk JS, Lawrimore J, Bloom K. 2014. Determining absolute protein numbers by quantitative fluorescence microscopy. Methods Cell Biol. 123, 347–365. ( 10.1016/B978-0-12-420138-5.00019-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Kampen NG. 2011. Stochastic processes in physics and chemistry. Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- 44.Canela-Xandri O, Sagués F, Casademunt J, Buceta J. 2011. Dynamics and mechanical stability of the developing dorsoventral organizer of the wing imaginal disc. PLoS Comput. Biol. 7, e1002153 ( 10.1371/journal.pcbi.1002153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monier B, Pélissier-Monier A, Brand AH, Sanson B. 2010. An actomyosin-based barrier inhibits cell mixing at compartmental boundaries in Drosophila embryos. Nat. Cell Biol. 12, 60–65. ( 10.1038/ncb2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Umetsu D, Dahmann C. 2010. Compartment boundaries: sorting cells with tension. Fly 4, 241–245. ( 10.4161/fly.4.3.12173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dahmann C, Oates AC, Brand M. 2011. Boundary formation and maintenance in tissue development. Nat. Genet. 12, 43–55. ( 10.1038/nrg2902) [DOI] [PubMed] [Google Scholar]
- 48.Calzolari S, Terriente J, Pujades C. 2014. Cell segregation in the vertebrate hindbrain relies on actomyosin cables located at the interhombomeric boundaries. EMBO J. 33, 686–701. ( 10.1002/embj.201386003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guillot C, Lecuit T. 2013. Mechanics of epithelial tissue homeostasis and morphogenesis. Science 340, 1185–1189. ( 10.1126/science.1235249) [DOI] [PubMed] [Google Scholar]
- 50.Nagai T, Honda H. 2001. A dynamic cell model for the formation of epithelial tissues. Philos. Mag. B 81, 699–719. ( 10.1080/13642810108205772) [DOI] [Google Scholar]
- 51.Minc N, Burgess D, Chang F. 2011. Influence of cell geometry on division-plane positioning. Cell 144, 414–426. ( 10.1016/j.cell.2011.01.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sukumar N, Bolander JE. 2003. Numerical computation of discrete differential operators on non-uniform grids. Comput. Model. Eng. Sci. 4, 691–706. ( 10.3970/cmes.2003.004.691) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The simulation code and compilation instructions for generating all data shown in the manuscript are available at https://doi.org/10.6084/m9.figshare.5146564.