Summary
Human fibrillin-1, the major structural protein of extracellular matrix (ECM) 10–12 nm microfibrils, is dominated by 43 calcium binding epidermal growth factor-like (cbEGF) and 7 transforming growth factor β binding protein-like (TB) domains. Crystal structures reveal the integrin binding cbEGF22-TB4-cbEGF23 fragment of human fibrillin-1 to be a Ca2+-rigidified tetragonal pyramid. We suggest that other cbEGF-TB pairs within the fibrillins may adopt a similar orientation to cbEGF22-TB4. In addition, we have located a flexible RGD integrin binding loop within TB4. Modeling, cell attachment and spreading assays, immunocytochemistry, and surface plasmon resonance indicate that cbEGF22 bound to TB4 is a requirement for integrin activation and provide insight into the molecular basis of the fibrillin-1 interaction with αVβ3. In light of our data, we propose a novel model for the assembly of the fibrillin microfibril and a mechanism to explain its extensibility.
Introduction
Human fibrillin-1 is a 350 kDa modular glycoprotein which assembles to form 10–12 nm microfibrils in the ECM (Sakai et al., 1986). Mutations in the fibrillin-1 (FBN1) gene cause the connective tissue disease Marfan syndrome (MFS) (Dietz and Pyeritz, 1995). Although fibrillin-1 is the major structural component, other proteins such as microfibril-associated glycoprotein (MAGP) and latent transforming growth factor β binding proteins (LTBPs) colocalize with microfibrils (Gibson et al., 1986; Taipale et al., 1996; Dallas et al., 2000; Isogai et al., 2003). Fibrillin-1, like other modular glycoproteins within the ECM, interacts with a number of cell matrix components including αVβ3 integrin (Pfaff et al., 1996; Sakamoto et al., 1996). Integrin interactions are likely to be particularly important in tissues where microfibrils are in close proximity to cells, such as in the elastic lamellae, and may play a role in the assembly of the microfibril network. The loss of cell-matrix interactions, as well as disruption of microfibrils, is likely to underlie the pleiotropic manifestations of MFS.
The modular architecture of fibrillin-1 is dominated by EGF and TB domains. Forty-three of the EGF domains have a calcium binding (cb) motif and are arranged in contiguous arrays separated usually by a TB domain (Figure 1) (Pereira et al., 1993). Analysis of fibrillin monomers by rotary shadowing electron microscopy (RSEM) show them to have an extended structure of ∼150 nm in length, and 2 nm diameter (Sakai et al., 1991). Fibrillin microfibrils have a “beads on a string” appearance when viewed by RSEM, with an average periodicity of ∼56 nm (Sakai et al., 1986; Kielty et al., 1991). Both the fibrillin monomer and microfibril polymer have a calcium-dependent structure. Removal of Ca2+ from microfibrils by chelation leads to a reduction in the bead to bead length (∼56 to 41 nm) and an increased flexibility (Cardy and Handford, 1998; Wess et al., 1998). These changes are consistent with the observed NMR structure and dynamics of a fibrillin-1 cbEGF 32-33 pair (which show that Ca2+ restricts conformational flexibility in the inter-domain region), the behavior of recombinant fibrillin fragments in velocity sedimentation studies (Downing et al., 1996; Reinhardt et al., 1997a; Werner et al., 2000) and the enhanced resistance to proteolysis of recombinant fibrillin-1 fragments containing cbEGF domains in the presence of Ca2+ (Reinhardt et al., 1997b ; McGettrick et al., 2000).
Figure 1.
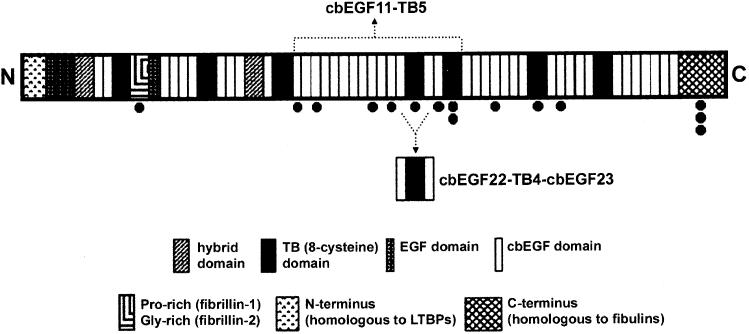
Domain Organization of Human Fibrillin-1 Potential glycosylation sites are indicated below the domain (●).
Structural analyses, epitope mapping, and transglutaminase cross-link identification support a parallel alignment of fibrillin monomers within the microfibril, but do not distinguish between the currently proposed models of either a staggered or unstaggered arrangement (Downing et al., 1996; Reinhardt et al., 1996; Qian and Glanville, 1997; Baldock et al., 2001). In the case of the latter, a folding back mechanism must be invoked to compress the 150 nm monomer into the 56 nm period commonly observed in normal untensioned microfibrils. Analysis of the mechanical properties of microfibrils suggests they are elastomeric/reversibly extensible and dependent upon Ca2+ for this property (Wess et al., 1998; Eriksen et al., 2001). The molecular basis for elasticity is unknown.
Since tandem repeats of fibrillin-1 cbEGFs are predicted to form rod-shaped structures in the presence of Ca2+, attention has focused on the TB domain as a potential source of flexibility via its pairwise interactions. The disulphide-rich, globular, TB module is found only in the fibrillin-LTBP family of proteins (Yuan et al., 1997) and high-resolution structures identifying intermodule connections mediated by this domain are currently lacking. A recent dynamics study of TB6-cbEGF32 identified a flexible linker, but the lack of sequence conservation within this region suggests that other TB-cbEGF domains may adopt different conformations (Yuan et al., 2002). There are no structural data available for the cbEGF-TB interaction.
In this study we have expressed and purified the cbEGF22-TB4-cbEGF23 fragment from fibrillin-1 which contains an RGD integrin binding site. Crystal structures of the apo, one-Ca2+ bound and holo (two-Ca2+ bound) forms identify novel pairwise interactions mediated by TB4, one of which may contribute to fibrillin elasticity. Further dissection of the integrin binding properties of cbEGF22-TB4-cbEGF23 implicates cbEGF22 as an accessory domain for fibrillin-mediated αVβ3 integrin activation. These data provide new information on the modular structure of fibrillin-1, Ca2+ coordination by cbEGF domains, fibrillin-1 integrin interactions, and microfibril organization.
Results and Discussion
Structure of cbEGF22-TB4-cbEGF23
Three different structures with varying Ca2+ occupancy (the two-Ca2+ bound holo, one-Ca2+ bound, and calcium-free apo forms) were determined (Table 1, Experimental Procedures). The highest resolution structure obtained (one-Ca2+ bound) has a crystallographic R factor of 20.2% for all data to 1.35 Å resolution and excellent stereochemistry (root-mean-square-deviation from the ideal bond lengths of 0.005 Å). The secondary and tertiary structures of this fragment are shown in Figures 2 and 3 respectively. The electron density map is of high quality (Figure 3A) with the exception of the flexible RGD loop region (residues 1539–1548). We have only attempted to interpret the electron density for this loop in the holo crystal form (see Experimental Procedures and Figure 3B). Overall cbEGF22-TB4-cbEGF23 has an elongated tetragonal pyramidal shape (Figure 3C), with dimensions 7 × 4 × 3 nm. In the holo form, a single Ca2+ ion is bound toward the N terminus of each of the cbEGF domains. The structures we have determined belong to two distinct crystal systems, giving views of the molecule in four markedly different crystal environments (Table 1). These structures are essentially indistinguishable (Figure 3D). The secondary structures of the two cbEGF domains are similar and consistent with that previously observed for this module (Downing et al., 1996). TB4 has a similar, but not identical, secondary structure arrangement to human fibrillin-1 TB6 (Yuan et al., 1997) which includes both a four-stranded and two-stranded antiparallel β sheet (Figure 2). Two α helices (A2 and A3) are arranged almost perpendicular to each other, with A3 positioned toward the cbEGF22-TB4 interface and A2 nearer to the TB4-cbEGF23 interface. The flexible RGD loop forms a rather long β hairpin (18 residues; Leu1536–Ile1553) between β strands B5 and B6 (Figures 2 and 3), as predicted by Yuan et al. (1997).
Table 1.
Data Collection, Phasing, and Refinement Statistics
| Data Set | Holo | One-Ca2+ Bound | Apo | SmAc |
|---|---|---|---|---|
| Space group | P21 | I222 | I222 | I222 |
| Unit cell dimensions (a, b, c) (Å) | 43.1, 105.6, 65.0 | 42.5, 70.8, 102.6 | 42.4, 70.8, 102.7 | 42.5, 65.1, 102.3 |
| β = 104.2° | ||||
| Molecules in the crystallographic asymmetric unit | 3 | 1 | 1 | 1 |
| Wavelength (Å) | 0.87 | 0.9787 | 1.5418 | 1.5418 |
| Resolution range (outer shell) (Å) | 30–2.25 (2.33–2.25) | 30–1.35 (1.40–1.35) | 20–2.40 (2.49–2.40) | 30–1.78 (1.84–1.78) |
| Measurements | 71,488 | 246,949 | 35,761 | 353,316 |
| Unique reflections | 25,753 | 33,911 | 6,275 | 13,997 |
| Number of images used | 180 | 337 | 163 | 971 |
| Completeness (%)a | 96.2 (92.6) | 98.3 (96.5) | 99.0 (98.5) | 99.5 (98.6) |
| I/σ(I)a | 9.7 (2.2) | 25.8 (2.7) | 19.4 (5.8) | 24.4 (5.3) |
| Rmerge (%)a,b | 9.9 (45.1) | 6.1 (42.4) | 8.7 (34.4) | 12.7 (47.9) |
| Phasing powerc | 4.34 | |||
| Figure of meritc | 0.6 | |||
| Rworking (%)d | 24.7 | 20.2 | 25.4 | 21 |
| Rfree (%)e | 31.8 | 26.1 | 33.0 | 25.4 |
| Solvent content (%) | 53 | 41 | 41 | 36 |
| Average B factors (Å2) | 40 | 27 | 36 | 36 |
| Average B factors for Ca2+ (Å2) | 30 | 19 | ||
| Rmsd from ideal values | ||||
| Bond lengths (Å) | 0.009 | 0.005 | 0.013 | 0.013 |
| Bond angles (°) | 1.6 | 1.7 | 1.8 | 1.8 |
| B rmsd (Å2) | ||||
| Main chain | 3.5 | 2.2 | 2.9 | 3.1 |
| Side chain | 4.8 | 3.4 | 4.3 | 4.7 |
| Total number of nonhydrogen atoms | ||||
| Protein | 3585 | 1123 | 1123 | 1123 |
| Water | 251 | 190 | 188 | |
| Other | 6 Ca2+ | 1 Ca2+, 1 SeMet | 1 SeMet | 3 Sm (III) |
Values in parentheses correspond to the highest resolution shell.
Rmerge = Σ|I − 〈I〉|/Σ〈I〉.
Phasing power represents rms heavy atom phasing signal to noise and the figure of merit is the cosine of the likely error in the phase angle. Values calculated for acentric reflections by SHARP (de La Fortelle and Bricogne, 1997).
Rworking = Σ||Fobs| − |Fcalc||/Σ|Fobs|.
Rfree is as for R factor but calculated using a 5% test set of randomly chosen reflections which were excluded from the refinement.
Figure 2.
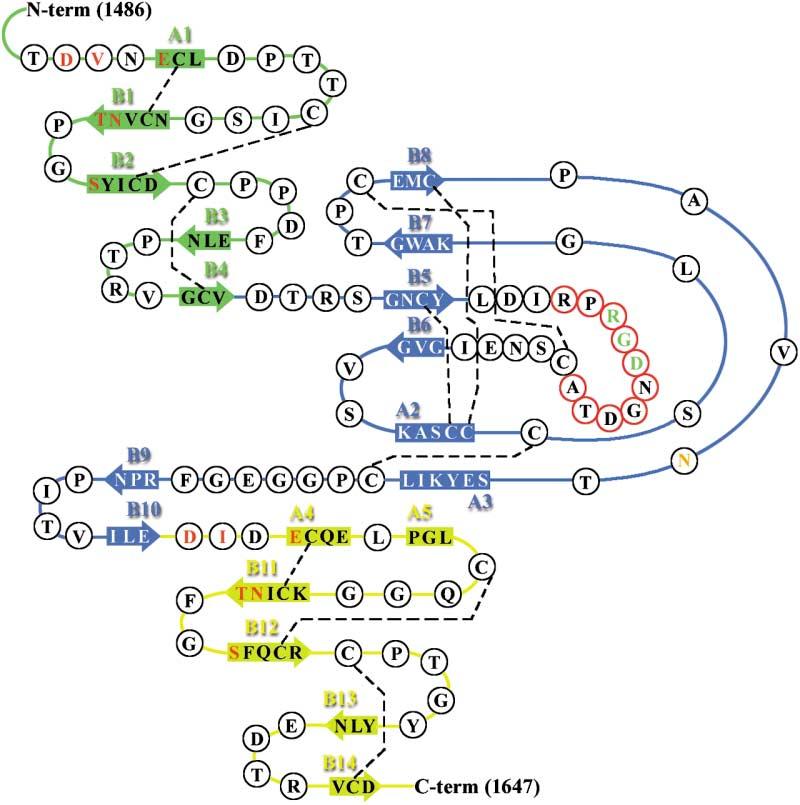
Secondary Structure of cbEGF22-TB4-cbEGF23 cbEGF22, TB4, and cbEGF23 are colored green, blue, and yellow, respectively. α helices and β strands are indicated by rectangular boxes and arrows and labeled A and B, respectively. Ca2+ ligands are colored red and the potential N-linked glycosylation site orange. Residues within the flexible RGD-loop region are encircled in red. The disulphide bonds are indicated by dashed lines.
Figure 3.
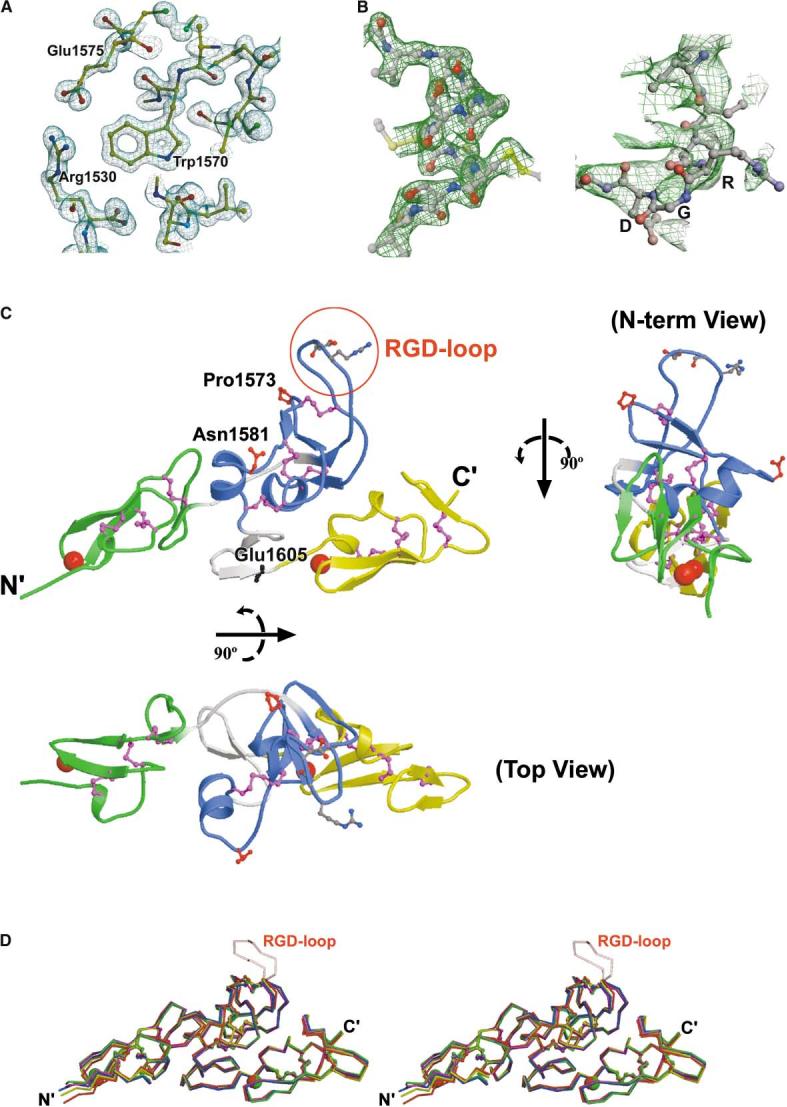
Crystal Structures of Human Fibrillin-1 Fragment cbEGF22-TB4-cbEGF23 (A) 1.35 Å electron density map (2Fo-Fc) of the one-Ca2+ bound form. (B) Electron density for the holo crystal form. Both panels show electron density for a 3-fold averaged 2.25 Å 2Fo-Fc SIGMAA-weighted electron density map (CNS) (Brunger et al., 1998), for which the RGD loop was omitted from the phasing model. The left-hand panel shows density around the Cys-Cys-Cys tripeptide within the TB4 domain. The right-hand panel shows the electron density in the region of the RGD loop. (C) Different views of the holo form (top view: rotated 90° about the horizontal axis, N term view: rotated 90° about the vertical axis). cbEGF22, TB4, and cbEGF23 are colored green, blue, and yellow, respectively, and the potential spring linking TB4 and cbEGF23 is colored gray (as is the linker region connecting cbEGF22 to TB4). The modeled RGD loop region is encircled in red; cis-proline 1573 and the potential N-linked glycosylation site (Asn1581) are also indicated. Location of a MFS-causing missense mutation Glu1605Lys in the spring is also labeled. (D) Stereo image showing superimposed Cα backbones of the three holo forms (red, yellow, blue), the one-Ca2+ bound form (magenta), and the apo form (green). Ca2+ ions are represented by spheres and colored accordingly. The average value of the rmsd between Cα atoms is ∼0.81 Å2 for all pairs of independent molecules.
Disulphide Bonds
In the absence of extensive secondary structure, conformational stability is maintained by disulphide bridges. In both cbEGF domains, cysteine residues pair up in a typical C1-3, 2-4, 5-6 arrangement (Downing et al., 1996). C1-3 and 2-4 provide structural integrity to the domain in the absence of a significant hydrophobic core. Interestingly, we observe isomerization of the C2-4 disulphide bond (Cys1497 and 1511) of cbEGF22 in the oneCa2+ bound form of the fragment. This has previously been proposed to explain the slow dynamics observed in the N-terminal domain of a cbEGF pair which has a low-affinity calcium binding site (Werner et al., 2000). In the TB4 domain, eight cysteine residues pair up in a C1-3, 2-6, 4-7, 5-8 arrangement.
Calcium Coordination by cbEGF22 and 23: A New Highly Conserved Ser Ligand
The cbEGF domain calcium binding consensus sequence comprises five residues defined as Asp/Asn-X-Asp/Asn-Glu/Gln-Xm-Asp*/Asn*-Xn-Tyr/Phe, where m and n are a variable number of residues and * denotes a potential β-hydroxylated residue (Handford et al., 1991). Each Ca2+ bound to cbEGF22 and cbEGF23 is coordinated by seven intradomain oxygen ligands (Asp1487C [OD1 and 2], Val1488 [O], Glu1490C [OE1], Asn1504C [OD1], Thr1505 [O], and Ser1508 [O] for cbEGF22, and Asp1606C [OD1 and 2], Ile1607 [O], Glu1609C [OE2], Asn1624C [OD1], Thr1625 [O], and Ser1628 [O] for cbEGF23; where “C” denotes a consensus residue), with approximate pentagonal bipyramidal geometry. Consensus residues from both cbEGF22 and 23 donate similar core oxygen ligands, while Asn1489 and Asp1608 (from cbEGF22 and 23) participate in “ASX” turns (Rao et al., 1995) and a serine residue on the central β hairpin (Ser1508 in cbEGF22 and Ser1628 in cbEGF23) donates a backbone carbonyl ligand to the Ca2+ (Figure 4). The hydroxyl groups of these serine residues stabilize their respective domains by a lattice work of hydrogen bonds with the side chains of Arg1523 and Arg1644 (projecting from the β hairpins B3/B4 and B13/B14, respectively) as well as nearby waters. The serine residues have unusual conformations allowing the correct presentation of the backbone carbonyl ligand and side chain hydroxyl group for hydrogen bonding. A corresponding serine residue is highly conserved in tandem cbEGF domains of the fibrillin-1 or class I type cbEGF repeats (Downing et al., 1996), and may enhance the affinity of cbEGF domains for Ca2+. It is remarkable that, despite the physiological importance of Ca2+, the atomic structure is barely affected by its removal (Figure 4); presumably the key difference is a loss of overall stability in the absence of Ca2+.
Figure 4.
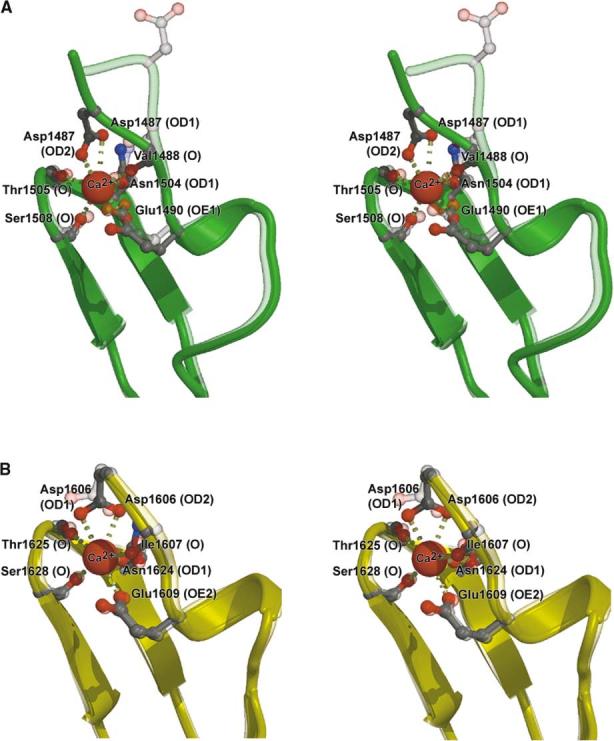
Stereoview of Ca2+ Bound and Ca2+-free Forms of cbEGF Domains Superimposed We use the highest resolution analysis available in each case. (A) cbEGF22 regions of the holo form (solid green) versus the one-Ca2+ bound form (transparent green) (B) cbEGF23 regions of the one-Ca2+ bound form (solid yellow) versus the apo form (transparent yellow).
TB4 Mediates Extensive Interdomain Contacts with Flanking cbEGF Domains
A striking feature of the cbEGF22-TB4-cbEGF23 structure is the extensive interface between adjacent domain surfaces (Figure 3C). This is very different from fibrillin-1 cbEGF domain pairs which have a comparatively small Ca2+ stabilized interface (Downing et al., 1996). For cbEGF22-TB4, the buried surface area is 550 Å2, averaged between the two surfaces, and the surface complementarity coefficient (SC) is 0.65 (0 = random and 1 = perfect fit [Lawrence and Colman, 1993]). For TB4-cbEGF23 the corresponding values are 670 Å2 and 0.69. In comparison, an antibody-antigen interface buries about 700 Å2 (Colman, 1988) with SC ≈ 0.67 (Garcia et al., 1998). Seven residue pairs involved in hydrophobic interactions stabilize the cbEGF22-TB4 interface, whereas the TB4-cbEGF23 interface comprises three hydrogen bonding and thirteen hydrophobic interaction residue pairs. Since these extensive interdomain contacts are found in a range of crystal packing environments, they are likely to be recapitulated in native fibrillin-1.
Pairwise Interaction of cbEGF22-TB4: Conservation in Fibrillins but Not LTBPs
TB domains are distributed exclusively among the fibrillin and LTBP families of proteins (Pereira et al., 1993; Zhang et al., 1994; Kanzaki et al., 1990; Moren et al., 1994; Yin et al., 1995; Giltay et al., 1997; Saharinen et al., 1998). Sequence alignment of cbEGF-TB linker regions identifies conservation of individual residues and linker length in the fibrillins only, suggesting that cbEGF22-TB4 can serve as a model for other cbEGF-TB interactions within the fibrillins, but not the LTBPs (Figure 5A). Within this linker region, Asp1528 and Arg1530 (Figure 5A, highlighted in green) form a salt bridge, as was observed previously for homologous residues in TB6 (Yuan et al., 1997). These residues also participate in both hydrophobic and polar interactions at the domain interface; for example, Arg1530 forms a hydrogen bond with Glu1575 (highlighted in green on Figure 5B) in TB4. Since all three residues are highly conserved, this structure is likely to be repeated at homologous sites within the fibrillins.
Figure 5.
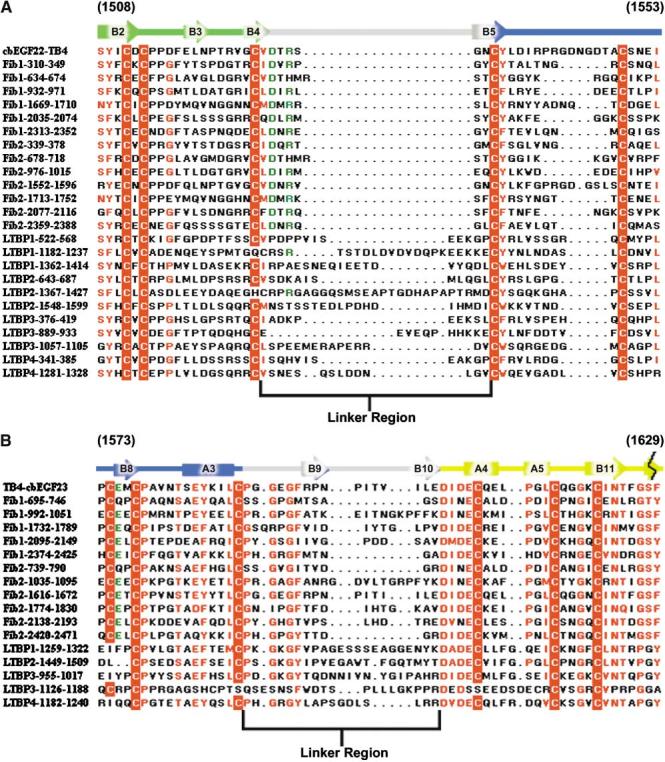
Sequence Alignment of cbEGF22-TB4 and TB4-cbEGF23 against Fibrillin-LTBP Family of Proteins Secondary structural elements for cbEGF22-TB4 (A) and TB4-cbEGF23 (B) are indicated above the sequence and color coded to represent different domains (cbEGF22 in green, TB4 in blue, cbEGF23 in yellow, and the linker regions in gray). Conserved residues are highlighted in red, and cysteine residues are color blocked in red. Highly conserved residues which, in cbEGF22-TB4 stabilize the domain interface, are colored green.
Pairwise Interaction of TB4-cbEGF23–An Elastic Linkage?
The extensive interdomain contacts observed for TB4-cbEGF23 contrast with those of fibrillin-1 TB6-cbEGF32 where calcium binding measurements, limited proteolysis, and dynamics analyses demonstrate a flexible interaction, despite the presence of a very similar length linker (Kettle et al., 1999; McGettrick et al., 2000; Yuan et al., 2002). Sequence alignment of the TB-cbEGF linker regions identifies much greater variation in linker length, even within the fibrillin family, than for cbEGF-TB (Figure 5B). Therefore, it is likely that other TB-cbEGF interactions will have to be determined empirically. Interestingly, the linker length for each TB-cbEGF pairing is conserved between fibrillin-1 and -2 (e.g., compare TB4-cbEGF23 with Fib-2 1616-1672, Figure 5B), suggesting that the linkers may have particular biological functions that are preserved between the two proteins.
The position of the B9/B10 β hairpin at the C terminus of TB4 (Figure 3C) raises the intriguing possibility that this linkage could act as a natural spring upon extension of fibrillin-1. Upon application of a tensional force the C-terminal β hairpin of TB4-cbEGF23 could unfold, allowing this region to extend at least 5 nm. Distortion of this, and other linker regions with similar secondary structures, may therefore contribute significantly to the observed variations in bead-to-bead length. Other un-characterized fibrillin-1 domains such as the pro-rich region may act similarly. Reversible unfolding of a linkage has not, to our knowledge, been previously reported; however, reversible unfolding of domains from the modular proteins fibronectin and titin has been postulated (Fowler et al., 2002; Oberhauser et al., 2002). A MFS-causing missense mutation Glu1605Lys (Krawczak and Cooper, 1997) is located within the C-terminal β hairpin of TB4 (Figure 3C). In the wild-type, the Glu side chain stabilizes the hairpin by hydrogen bonding to Asn1598 (ND2) and the change to a lysine is likely to electrostatically destabilize the structure and may thereby alter the biomechanical properties of the linker.
Homology Modeling of Fibrillin-1
The N- and C termini of cbEGF22-TB4-cbEGF23 lie at opposite ends of the structure, consistent with the observed extended structure of fibrillin-1, and the extensive pairwise interactions mediated by TB4 suggest that this fragment is rigid when untensioned. These data, together with the conservation of the cbEGF-TB interaction, have allowed us to model cbEGF11-TB5, a contiguous length of 16 domains within fibrillin-1 (Figure 6A). The model forms an extended structure with a kink introduced by the orientation of cbEGF22 relative to TB4. This kink, together with the gradual twist introduced by the cbEGF domains, may underlie the large-scale twist seen in 3D reconstructions of microfibrils (Baldock et al., 2001). Potential glycosylation sites and the RGD motif all map on one face of the structure, consistent with this forming the outside surface of the microfibril.
Figure 6.
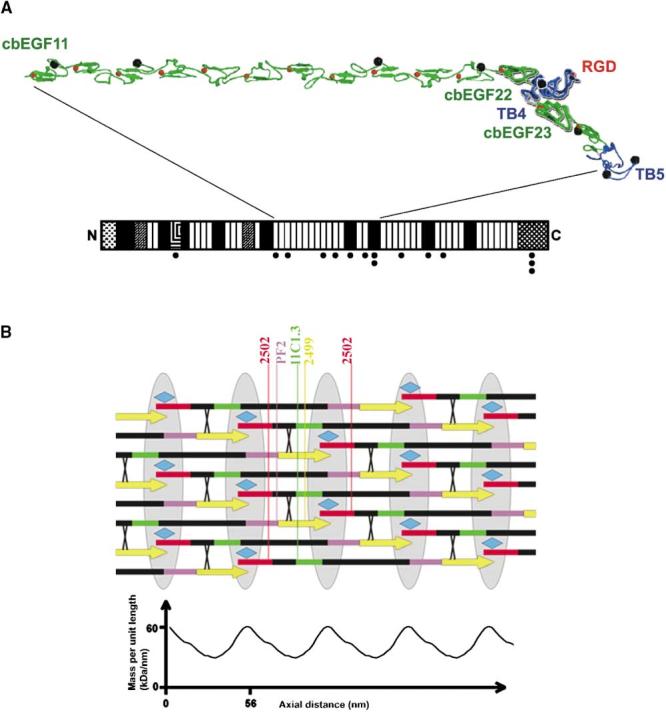
Models of Fibrillin-1 and Microfibril Organization (A) Homology model for cbEGF11-TB5. cbEGF and TB domains are colored green and blue, respectively. The crystal structure of cbEGF22-TB4-cbEGF23 fragment is highlighted in bold. Ca2+ and potential N-linked glycosylation sites are represented by red and black spheres, respectively. The RGD motif on the TB4 loop is indicated. (B) A simple model of the fibrillin microfibril. The arrows represent fibrillin-1 molecules with dimensions based on the knowledge of the component domains. The intermolecular transglutaminase cross-links are shown as X and the associated protein MAGP-1 as a turquoise diamond. A sketch of the STEM data of Baldock et al. (2001) is shown below. The colored regions mark the sites of known antibody epitopes (red, 2502; green, 11C1.3; purple, PF2; yellow, 2499). The bead regions are shown as gray ellipses.
Implications for Microfibril Organization
On the basis of a 3D reconstruction of the microfibril generated by automated electron tomography, Baldock et al. (2001) proposed that, in the untensioned state, folding back of three regions of the fibrillin monomer generates a 56 nm periodicity. This “jacknife” model is consistent with scanning transmission electron microscopy (STEM) data which demonstrate an asymmetric repeating bead-interbead organization (Sherratt et al., 1997, 2001). Our data do not reveal an obvious mechanism for folding back of the molecule, which would require interactions between closely associating module surfaces as well as a flexible linker. We therefore suggest that simpler models based on staggered arrangements be considered alongside the jacknife model. Our experimental and estimated dimensions for other fibrillin domains, predict that a Ca2+-stabilized fibrillin-1 molecule will have a length of ∼180 nm. Assuming fibrillin-1 molecules are arranged in a head-to-tail fashion, then an overlap of some 12 nm between the N- and C termini of successive molecules, together with a 1/3 staggered alignment, would give rise to the observed periodicity of 56 nm (Figure 6B). This very simple model agrees with the known data on transglutaminase cross-link sites (Qian and Glanville, 1997), antibody epitope sites (Baldock et al., 2001), and N- and C-terminal interactions of fibrillin-1 (Lin et al., 2002). The STEM data show a factor of ∼2 between the mass density of the bead and interbead regions, somewhat greater than predicted by our model. However, taking into account some additional mass from proteoglycans and MAGP-1 (molecular mass ∼21 kDa [Gibson et al., 1991]), which is suggested to be a component of the bead and binds to fibrillin-1 (Jensen et al., 2001; Trask et al., 2000), then our model has a reasonable fit with the observed mass ratio. It should be noted, however, that due to the stagger, the mass density in all beads would not be quite equal. Since the molecules are more extended, the staggered model would require a greater number of molecules to be involved in the microfibril at any point along its length than would the jacknife model. However, if the building block were assumed to be a dimer instead of a monomer, then the mass values fall roughly into line with the experimental determination from STEM. Overall, the staggered and jacknife models both account for the majority of the experimental data, and further evidence will be required to discriminate between them. Additional antibody labeling experiments may provide clues since the order of 11C1.3 and 2499 epitopes (defined in Baldock et al., 2001) produced by folding back of fibrillin-1 would be predicted to invert on stretching the mature microfibril (i.e., the two epitopes will lie on the opposite sides of the microfibril bead when the fibril is stretched). For now we suggest that domain linker extensibility be considered as a possible basis for the elastic properties of microfibrils.
Modeling of cbEGF22-TB4-cbEGF23 and Integrin αVβ3 Complex
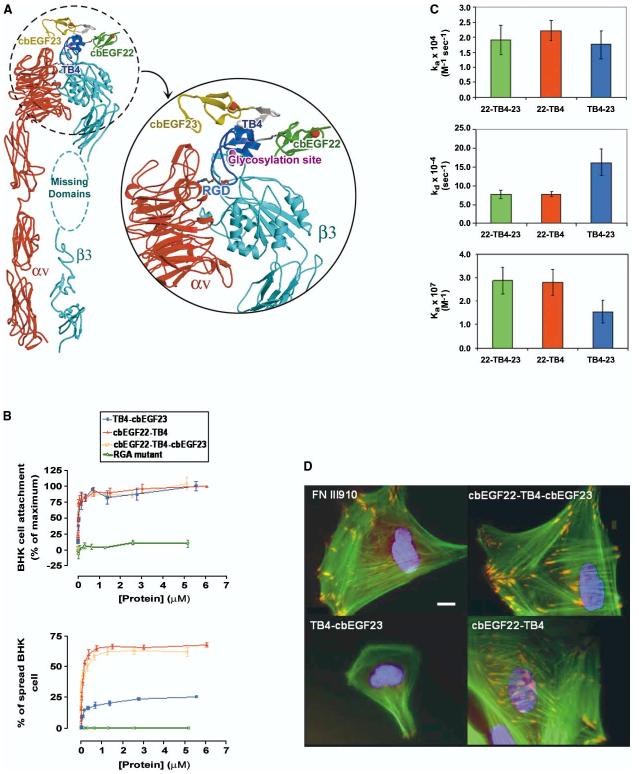
The RGD tripeptide located at the tip of a flexible loop in TB4 (Figure 3C) is likely to explain the previously observed fibrillin-1-integrin interactions (Pfaff et al., 1996; Sakamoto et al., 1996; Bax et al., 2003). We have demonstrated that cbEGF22-TB4-cbEGF23 contains the molecular determinants required to bind to αVβ3 and trigger downstream signaling events (see below). Cell adhesion experiments were performed using nonglycosylated protein and therefore demonstrate that a potential glycosylation site close to the RGD loop (Asn 1518) is not required for αVβ3 interaction. The recent determination of the structure of an integrin αVβ3+RGD ligand (Xiong et al., 2002) enables us to model the integrin/cbEGF22-TB4-cbEGF23 complex. The TB4 RGD lies in a 10 residue flexible loop; nevertheless, the electron density for the holo structure allows us to unambiguously place the RGD. Thus the right-hand panel for Figure 3B shows the electron density in the region of the RGD after averaging the three distinct crystal environments in the holo crystal. Although the density is diffuse, it demonstrates that this portion of structure does possess a consensus structure. The observed structure of the TB4 RGD allows us to fit the entire fibrillin fragment to the crystal structure of αVβ3 based upon the superposition of the TB4 RGD motif with the integrin-bound RGD cyclic pentapeptide. This places cbEGF22 in close proximity to the I-like domain of the β3 subunit, suggesting that contacts between these two domains, in addition to RGD contacts, may modulate integrin binding and signaling (Figure 7A).
Figure 7.
cbEGF22-TB4-cbEGF23 Interaction with αVβ3 Integrin Receptor (A) Crystal structure of cbEGF22-TB4-cbEGF23 docked onto the bound cyclic RGD-peptide of αVβ3 integrin receptor together with a closeup view of the modeled complex. cbEGF22, TB4, and cbEGF23 are colored green, blue, and yellow, respectively. Ca2+ ions are red spheres and the location of the potential N-linked glycosylation site in the TB4 is represented by a magenta sphere. The missing domains (I-EGF1, I-EGF2, part of I-EGF3, and PSI) of integrin β3 subunit are represented by the cyan ellipse. (B) Cell attachment and spreading assays. BHK cell attachment (upper panel) to fibrillin-1 constructs was estimated in a colorimetric assay. Note that cbEGF22-TB4 and TB4-cbEGF23 domain pairs have equal cell attachment ability which is dose dependent (and equal to that of the cbEGF22-TB4-cbEGF23). Results are expressed as percentages of maximum cell attachment.Values are mean ± SEM (n = 6). Baby hamster kidney (BHK) cell spreading (lower panel) in response to fibrillin-1 domain pairs demonstrates that the activity of TB4-cbEGF23 is substantially lower than that of equimolar cbEGF22TB4. Results are expressed as percentages of spread cells. Values are mean ± SEM (n = 6). Note that in both assays an RGA mutant showed minimal attachment and spreading. (C) Kinetics of αVβ3 interactions with fibrillin-1 constructs measured by surface plasmon resonance. The difference in Ka observed for the interaction of αVβ3 with fibrillin-1 fragments is mainly due to the faster dissociation of TB4-cbEGF23 from αVβ3 compared to cbEGF22-TB4. Error bars represent SD. (D) Immunofluorescence of human endometrial stromal fibroblasts (hESF) adhered to fibrillin-1 fragments and stained for actin (green), αVβ3 integrin (red), and nuclei (blue). In hESF cells attached to cbEGF22-TB4 and cbEGF22-TB4-cbEGF23, αVβ3 integrin is recruited to sites of focal adhesion, while in cells attached to TB4-cbEGF23, it shows a diffuse staining pattern. Scale bar, 10 μm.
Molecular Interactions between Fibrillin-1 and αVβ3
We investigated the possibility that additional sites in flanking cbEGF domains contribute to the integrin/fibrillin-1 interaction, as has been shown for other integrin binding molecules such as fibronectin (Obara et al., 1988). cbEGF22-TB4 and TB4-cbEGF23 domain pairs from fibrillin-1 and the cbEGF22-TB4-cbEGF23 triple domain protein were tested for cell adhesive capacity in BHK cell attachment and spreading assays. Quantification of cell attachment to protein-coated surfaces (Figure 7B) revealed that cbEGF22-TB4 and TB4-cbEGF23 have similar, dose-dependent capacity for mediating BHK cell attachment (and equal to that of the cbEGF22-TB4-cbEGF23). However, the two domain pairs exhibited significant differences in cell spreading, a secondary event in the process of cell adhesion. At a coating concentration of 1 μM, ∼70% of BHK cells adopted a spread morphology on cbEGF22-TB4 compared to ∼20% of spread cells on TB4-cbEGF23 (Figure 7B). Accessibility of the RGD loop in both proteins after their immobilization onto plastic surface was shown to be identical in an ELISA assay using polyclonal anti-fib1RGD antibody (data not shown).
The real-time kinetic parameters of fibrillin-αVβ3 integrin complex formation were monitored using surface plasmon resonance. A 2-fold difference in Ka for αVβ3 − cbEGF22-TB4 and αVβ3 − TB4-cbEGF23 interactions was observed (Figure 7C), due to the faster dissociation of TB4-cbEGF23 from αVβ3 ([1.6 ± 0.4] × 10−3 s−1) compared to cbEGF22-TB4 ([0.8 ± 0.05] × 10−3 s−1). No interaction with α5β1was observed (data not shown). The binding capacities of the chip, and accessibility of RGD loop after coupling to the chip, were the same for all fibrillin fragments used (data not shown). These results show that the cbEGF22 domain bound to TB4 is important for the stabilization of the integrin-ligand complex.
Immunofluorescent detection of αVβ3 in stromal fibroblasts cultured on fibrillin-1 constructs for 2 hr showed that, in cells adhering to cbEGF22-TB4 and cbEGF22-TB4-cbEGF23, integrin was sequestered in focal adhesion complexes at the tip of actin stress fibers. However, in cells attached to TB4-cbEGF23, there was diffuse staining for αVβ3 (Figure 7D). These data suggest that while molecular determinants required for the initial integrin binding event are contained within TB4, cbEGF22 contributes a synergistic activity to TB4 which is necessary for triggering downstream signaling events.
Our model of cbEGF22-TB4-cbEGF23 interaction with αVβ3 integrin based on the αVβ3-cyclic RGD structure (Xiong et al., 2002) provides one possible explanation for these functional data. It is well documented that integrin ligands contain accessory sites for receptor binding (Obara et al., 1988; Arnaout et al., 2002). Ligand binding specificity regions have been mapped for α I domain-lacking integrins to surface loops of the α subunit propeller (Irie et al., 1995; Zhang et al., 1999; Mould et al., 2000) and to the β I-like domain (Lin et al., 1997) which contains the so-called ligand specificity loop (Takagi et al., 1997). However, an alternative explanation could be that cbEGF22 acts indirectly, conferring conformational stability to TB4 and providing the correct topology for integrin binding, as shown for fibronectin (Altroff et al., 2001, 2003).
Experimental Procedures
Expression and Purification of cbEGF22-TB4-cbEGF23 and Related Constructs
Residues 1486–1647 of fibrillin-1 encompassing cbEGF22-TB4-cbEGF23 were expressed after PCR amplification of fibrillin-1 cDNA. Forward 5′-TAGTAGGCATGCATAGAAGGACGATCAGCAACAGAT GTGAATGAATGCCTGG-3′ and reverse 5′-TAGTAGGTCGACCTAT TAATCACACACTCGTGTATCTTC-3′ primers were used to clone the amplified DNA into pQE30 (Qiagen) which was transformed into E. coli NM554 [pREP4]. His6-tagged fusion protein was expressed, purified, and refolded in vitro using a previously described oxido-shuffling system (Knott et al., 1996). Protein was refolded at 0.2 mg/ml in 100 mM Tris-HCl (pH 8.3), 10 mM CaCl2 at 4°C for >24 hr. For crystallization trials, the final HPLC step was omitted and the purified FXa cleaved protein buffer exchanged into 10 mM MES, 10 mM CaCl2, pH 6.0 (holo form), and 0.01 M Tris-HCl, pH 7.5 (apo form). For the preparation of protein labeled with selenomethionine (SeMet), the expression construct was transformed into B834 [pREP4] and grown in Le Masters medium containing 50mg/l SeMet (Sigma) prior to purification by the above procedure. For cell binding studies cbEGF22-TB4 (residues 1486–1605) and TB4-cbEGF23 (residues 1527–1647) domain pairs were cloned using appropriate fibrillin-1 specific primers and purified as described above. The RGA mutant was produced by PCR mutagenesis according to standard procedures.
Cell Adhesion Assays and Immunocytochemistry
Human endometrial stromal fibroblasts (hESF) were isolated and cultured as described previously (Chobotova et al., 2002). Assays were performed with baby hamster kidney (BHK) fibroblasts as detailed elsewhere (Mardon and Grant, 1994). The data obtained are expressed as the means ± SEM of duplicate measurements in three independent experiments.
For immunocytochemistry, glass coverslips were coated with 100 μl of 100 μg/ml fibrillin-1 fragments overnight at 4°C. hESF were harvested with 0.5% trypsin-EDTA in PBS (Sigma), equilibrated in Dulbecco's minimum essential medium (DMEM), and replated on fibrillin-1 fragments-coated coverslips. Immunofluorescent detection of human integrin αVβ3 (clone 23C6; Serotec) and fluorescent labeling of actin were performed according to Hotchin et al. (1999). Mouse IgG (Coulter Immunotech), substituted for the primary antibody at 10 μg/ml, was used as a negative control (data not shown).
Surface Plasmon Resonance Studies
These were performed using a BIACORE 2000 instrument (Biacore, Uppsala, Sweden). Between 1000 and 2500 RU of fibrillin fragments were coupled to the surface of CM5 sensor chips (Biacore) via amine coupling according to the manufacturer's instructions (BIAapplications Handbook; Biacore). Both the association and the dissociation reactions were performed in Tris-buffered saline (TBS; 25 mM Tris, 150 mM NaCl, pH 7.4) containing MnCl2, MgCl2, and CaCl2 at 2 mM each. Either native αVβ3 or α5β1, purified from human placental extract (Altroff et al., 2001), was used as analyte. Integrin (120 to 8 nM) was injected for 5 min at a constant flow rate of 10 μl/min. A nonspecific fibrillin-1 fragment (TB6-cbEGF32) was used as reference. The reference run was subtracted from each sensorgram prior to data processing. Regeneration of surfaces was achieved using 100 μM EGTA in HEPES buffer saline. Data were analyzed by using the global fitting algorithm of the BIAevaluation 3.0 software package (Biacore), using a simple 1:1 kinetic model. At least three independent experiments were performed for each fibrillin-integrin couple.
Crystallization and Data Collection
Crystallization trials were performed at room temperature using microbridges (Harlos, 1992), in the presence and absence of a variety of divalent cations and in the presence of EDTA. Initially, crystals were grown in drops containing 3 μl of protein solution (5 mg/ml purified native protein fragment in 10 mM MES, 10 mM CaCl2, pH 6.0, buffer) which were simply dehydrated above a reservoir solution containing 15% PEG 8000, 10 mM MES, pH 6.0, and 10 mM CaCl2. These crystals grew as thin plates (<10 μm thick), were mechanically unstable, and tended to redissolve. It was only possible to collect a single native data set from these crystals. Crystals grown in the absence of Ca2+ were substantially thicker and led to the solution of the structure. In this case, the crystallization method was more conventional, the drop on the bridge containing 3 μl of protein solution (at 25 mg/ml in 0.01 M Tris-HCl, pH 7.5) plus 0.5 μl of 40% v/v polypropylene glycol P400 and 2.5 μl of reservoir solution (0.2 M lithium sulfate monohydrate, 0.1 M Tris-HCl, pH 8.5, 30% polyethylene glycol 4000, pH 7.5). Lastly, crystals of the apo form were obtained in a similar way, following concentration of the protein in 5 mM EDTA (5 mM EDTA was also added to the reservoir solution). X-ray diffraction data were collected at 100 K at the Synchrotron Radiation Source (SRS; Daresbury, UK) and in-house. Data were processed using the HKL program package (Otwinowski and Minor, 1997). Diffraction of the holo-form crystals (space group P21, Table 1) was highly anisotropic; nevertheless, a data set to 2.25 Å resolution was collected at station 9.6 (SRS), from a crystal cryoprotected in 20% glycerol. Crystals grown in the absence of Ca2+ were dehydated prior to data collection in 50% PEG4000, and frozen without further treatment. The best diffracting crystals (space group I222) were obtained from the SeMet derivatized protein, and data to 1.35 Å resolution were collected at station 14.2 (SRS) (one Ca2+ bound, Table 1). Native and SeMet derivatized crystals were isomorphous. Apo crystals were similar to these and a data set to 2.4 Å resolution was collected in-house (MAR 300 image plate detector, Rigaku rotating anode generator with Osmic mirrors) (Table 1).
Structure Determination and Refinement
A one-Ca2+ bound crystal of native protein soaked overnight in 10 mM samarium (III) acetate hydrate (SmAc) yielded the phase information required to solve the structure (1.78 Å resolution data were collected in house). This crystal was nonisomorphous with the one Ca2+ bound crystal (Table 1) and the structure was determined by single wavelength anomolous scattering (SAD) (some 1000° of data providing very high redundancy). As expected the two main binding sites for Sm were the Ca2+ binding pockets of cbEGF22 and cbEGF23. Heavy atom refinement using SHARP (de La Fortelle and Bricogne, 1997) and map modification with GAP (D.I.S. and J.M.G., unpublished data) led to an interpretable electron density map. The initial model was built manually and refined using CNS (Brunger et al., 1998). The structures of the one-Ca2+ bound and apo forms were solved by rigid-body refinement of this model, followed by standard refinement (SHELXL [Sheldrick and Schneider, 1997] and CNS respectively). The structure of the holo form was determined by molecular replacement with CNS using the cbEGF23 loaded form as a start point. Three molecules were found in the asymmetric unit and refined using CNS. No residues fell in disallowed regions of the Ramachandran plot (Laskowski et al., 1993). Final refinement statistics are given in Table 1.
Refinement of RGD Loop Density
The RGD loop region (residues 1539–1548) showed poor electron density in all of the structures obtained. The holo form was used for further analysis because it had a range of crystal packing interactions. A 3-fold averaged electron density map clearly showed the approximate position of the RGD tripeptide (Figure 3B). An atomic model was obtained, using an RGD loop from VP1 of foot-and-mouth disease virus (Logan et al., 1993) to guide the initial model building. During refinement the electron density improved, allowing RGD loop models to be built for all three molecules in the crystallographic asymmetric unit.
Homology and Interaction Modeling
The NMR structure of cbEGF32-33 (Downing et al., 1996) and the crystal structure of cbEGF22-TB4-cbEGF23 were used to homology model cbEGF11-TB5 (residues 1028–1758). The cbEGF32-33 structure was used as a template to pair up two cbEGF23 crystal structures to represent cbEGF11-12. This motif was the building block for further extensions to obtain models for cbEGF11-22 and cbEGF23-24 using the program IMPOSE (R. Esnouf, personal communication). Calpha (R. Esnouf, personal communication) was used to incorporate true amino acid sequences with canonical side chain orientations. The cbEGF22-TB4-cbEGF23 + integrin receptor complex was modeled using the crystal structure of a straightened form of liganded αVβ3 integrin receptor (Xiong et al., 2001, 2002). The fibrillin-1 fragment was docked by superimposing the RGD motif onto the cyclic RGD peptide bound on the integrin receptor (program SHP) (Stuart et al., 1979).
Structural Analyses and Visualization
Secondary structure assignment, structural superpositions, and sequence alignments used DSSP (Kabsch and Sander 1983), SHP (Stuart et al., 1979), and Clustal_W (Thompson et al., 1994) respectively. Interfaces were quantified using AREAIMOL, SC (CCP4, 1994). Figure 4 was created using ESPript (Gouet et al., 1999), other figures were drawn using Bobscript (Esnouf, 1999) and rendered with Raster3D (Merritt and Bacon, 1997).
Acknowledgments
We thank Janet Carver for excellent technical assistance, Harri Altroff, Sacha Jensen, Anton van der Merwe, and Pat Whiteman for helpful discussions, the MRC for financial support, and the staff of the SRS, Daresbury for help. We thank Cay Kielty for her kind gift of RGD antibody.
Footnotes
Accession Numbers
Coordinates and structure factors have been deposited in the Protein Data Bank, with accession codes as follows: holo, 1uzj; apo, 1uzq; one Ca2+, 1uzk; and Sm, 1uzp.
References
- Altroff H, van der Walle CF, Asselin J, Fairless R, Campbell ID, Mardon HJ. The eighth FIII domain of human fibronectin promotes integrin alpha5beta1 binding via stabilization of the ninth FIII domain. J. Biol. Chem. 2001;276:38885–38892. doi: 10.1074/jbc.M105868200. [DOI] [PubMed] [Google Scholar]
- Altroff H, Choulier L, Mardon HJ. Synergistic activity of the ninth and tenth FIII domains of human fibronectin depends upon structural stability. J. Biol. Chem. 2003;278:491–497. doi: 10.1074/jbc.M209992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaout MA, Goodman SL, Xiong JP. Coming to grips with integrin binding to ligands. Curr. Opin. Cell Biol. 2002;14:641–651. doi: 10.1016/s0955-0674(02)00371-x. [DOI] [PubMed] [Google Scholar]
- Baldock C, Koster AJ, Ziese U, Rock MJ, Sherratt MJ, Kadler KE, Shuttleworth CA, Kielty CM. The supra-molecular organization of fibrillin-rich microfibrils. J. Cell Biol. 2001;152:1045–1056. doi: 10.1083/jcb.152.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax DV, Bernard SE, Lomas A, Morgan A, Humphries J, Shuttleworth A, Humphries MJ, Kielty CM. Cell adhesion to fibrillin-1 molecules and microfibrils is mediated by alpha5 beta1 and alphaV beta3 integrins. J. Biol. Chem. 2003;278:34605–34016. doi: 10.1074/jbc.M303159200. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cardy CM, Handford PA. Metal ion dependency of microfibrils supports a rod-like conformation for fibrillin-1 cbEGF domains. J. Mol. Biol. 1998;276:855–860. doi: 10.1006/jmbi.1997.1593. [DOI] [PubMed] [Google Scholar]
- CCP4 (Collaborative Computational Project 4) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Chobotova K, Muchmore ME, Carver J, Yoo HJ, Manek S, Gullick WJ, Barlow DH, Mardon HJ. The mitogenic potential of heparin-binding EGF in the human endometrium is mediated by the EGF receptor and modulated by tumour necrosis factor-α. J. Clin. Endocrinol. Metab. 2002;87:5769–5777. doi: 10.1210/jc.2002-020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman PM. Structure of antibody-antigen complexes: implications for immune recognition. Adv. Immunol. 1988;43:99–132. doi: 10.1016/s0065-2776(08)60364-8. [DOI] [PubMed] [Google Scholar]
- Dallas SL, Keene DR, Bruder SP, Saharinen J, Sakai LY, Mundy GR, Bonewald LF. Role of LTBP1 in fibrillin-containing microfibrils in bone cells in vitro and in vivo. J. Bone Miner. Res. 2000;15:68–81. doi: 10.1359/jbmr.2000.15.1.68. [DOI] [PubMed] [Google Scholar]
- de La Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- Dietz HC, Pyeritz RE. Mutations in the human gene for fibrillin-1 (FBN1) in the Marfan syndrome and related disorders. Hum. Mol. Genet. 1995;4:1799–1809. doi: 10.1093/hmg/4.suppl_1.1799. [DOI] [PubMed] [Google Scholar]
- Downing AK, Knott V, Werner JM, Cardy CM, Campbell ID, Handford PA. Solution structure of a pair of cbEGF domains: implications for the Marfan syndrome and other genetic disorders. Cell. 1996;85:597–605. doi: 10.1016/s0092-8674(00)81259-3. [DOI] [PubMed] [Google Scholar]
- Eriksen TA, Wright DM, Purslow PP, Duance VC. Role of Ca2+ for the mechanical properties of fibrillin. Proteins. 2001;45:90–95. doi: 10.1002/prot.1127. [DOI] [PubMed] [Google Scholar]
- Esnouf RM. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D Biol. Crystallogr. 1999;55:938–940. doi: 10.1107/s0907444998017363. [DOI] [PubMed] [Google Scholar]
- Fowler SB, Best RB, Toca Herrera JL, Rutherford TJ, Steward A, Paci E, Karplus M, Clarke J. Mechanical unfolding of a titin Ig domain. J. Mol. Biol. 2002;322:841–849. doi: 10.1016/s0022-2836(02)00805-7. [DOI] [PubMed] [Google Scholar]
- Garcia KC, Degano M, Pease LR, Huang M, Peterson PA, Teyton L, Wilson IA. Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen. Science. 1998;279:1166–1172. doi: 10.1126/science.279.5354.1166. [DOI] [PubMed] [Google Scholar]
- Gibson MA, Hughes JL, Fanning JC, Cleary EG. The major antigen of elastin-associated microfibrils is a 31-kDa glycoprotein. J. Biol. Chem. 1986;261:11429–11436. [PubMed] [Google Scholar]
- Gibson MA, Sandberg LB, Grosso LE, Cleary EG. Complimentary DNA cloning establishes microfibril-associated glycoprotein (MAGP) to be a discrete component of the elastin-associated microfibrils. J. Biol. Chem. 1991;266:7596–7601. [PubMed] [Google Scholar]
- Giltay R, Kostka G, Timpl R. Sequence and expression of a novel member (LTBP-4) of the family of LTBPs. FEBS Lett. 1997;411:164–168. doi: 10.1016/s0014-5793(97)00685-6. [DOI] [PubMed] [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- Handford PA, Mayhew M, Baron M, Winship PR, Campbell ID, Brownlee GG. Key residues involved in calcium-binding motifs in EGF-like domains. Nature. 1991;351:164–167. doi: 10.1038/351164a0. [DOI] [PubMed] [Google Scholar]
- Harlos K. Micro-bridges for sitting-drop crystallizations. J. Appl. Crystallogr. 1992;25:536–538. [Google Scholar]
- Hotchin NA, Kidd AG, Altroff HA, Mardon HJ. Differential activation of focal adhesion kinase Rho and Rac by the ninth and tenth FIII domains of fibronectin. J. Cell Sci. 1999;112:2937–2946. doi: 10.1242/jcs.112.17.2937. [DOI] [PubMed] [Google Scholar]
- Irie A, Kamata T, Puzon-McLaughlin W, Takada Y. Critical amino acid residues for ligand binding are clustered in a predicted beta-turn of the third N-terminal repeat in the integrin alpha 4 and alpha 5 subunits. EMBO J. 1995;14:5550–5556. doi: 10.1002/j.1460-2075.1995.tb00242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai Z, Ono RN, Ushiro S, Keene DR, Chen Y, Mazzieri R, Charbonneau NL, Reinhardt DP, Rifkin DB, Sakai LY. LTBP1 interacts with fibrillin and is a microfibril associated protein. J. Biol. Chem. 2003;278:2750–2757. doi: 10.1074/jbc.M209256200. [DOI] [PubMed] [Google Scholar]
- Jensen S, Reinhardt DP, Gibson MA, Weiss AS. Protein interaction studies of MAGP-1 with tropoelastin and fibrillin-1. J. Biol. Chem. 2001;276:39661–39666. doi: 10.1074/jbc.M104533200. [DOI] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kanzaki T, Olofsson A, Moren A, Wernstedt C, Hellman U, Miyazono K, Claesson-Welsh L, Heldin CH. TGF-β1 binding protein: a component of the large latent complex of TGF-β1 with multiple repeat sequences. Cell. 1990;61:1051–1061. doi: 10.1016/0092-8674(90)90069-q. [DOI] [PubMed] [Google Scholar]
- Kettle S, Yuan X, Grundy G, Knott V, Downing AK, Handford PA. Defective calcium binding to fibrillin-1: consequence of an N2144S change for fibrillin-1 structure and function. J. Mol. Biol. 1999;285:1277–1287. doi: 10.1006/jmbi.1998.2368. [DOI] [PubMed] [Google Scholar]
- Kielty CM, Cummings C, Whittaker SP, Shuttleworth CA, Grant ME. Isolation and ultrastructural analysis of microfibrillar structures from foetal bovine elastic tissues. Relative abundance and supramolecular architecture of type VI collagen assemblies and fibrillin. J. Cell Sci. 1991;99:797–807. doi: 10.1242/jcs.99.4.797. [DOI] [PubMed] [Google Scholar]
- Knott V, Downing AK, Cardy CM, Handford P. Calcium binding properties of an EGF-like domain pair from human fibrillin-1. J. Mol. Biol. 1996;255:22–27. doi: 10.1006/jmbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- Krawczak M, Cooper DN. The human gene mutation database. Trends Genet. 1997;13:121–122. doi: 10.1016/s0168-9525(97)01068-8. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- Lin EC, Ratnikov BI, Tsai PM, Carron CP, Myers DM, Barbas CF, III, Smith JW. Identification of a region in the integrin β3 subunit that confers ligand binding specificity. J. Biol. Chem. 1997;272:23912–23920. doi: 10.1074/jbc.272.38.23912. [DOI] [PubMed] [Google Scholar]
- Lin G, Tiedemann K, Vollbrandt T, Peters H, Batge B, Brinckmann J, Reinhardt DP. Homo- and heterotypic fibrillin-1 and -2 interactions constitute the basis for assembly of microfibrils. J. Biol. Chem. 2002;277:50795–50804. doi: 10.1074/jbc.M210611200. [DOI] [PubMed] [Google Scholar]
- Logan D, Abu-Ghazaleh R, Blakemore W, Curry S, Jackson T, King A, Lea S, Lewis R, Newman J, Parry N, et al. Structure of a major immunogenic site on foot-and-mouth disease virus. Nature. 1993;362:566–568. doi: 10.1038/362566a0. [DOI] [PubMed] [Google Scholar]
- Mardon HJ, Grant KE. The role of the ninth and tenth type III domains of human fibronectin in cell adhesion. FEBS Lett. 1994;340:197–201. doi: 10.1016/0014-5793(94)80137-1. [DOI] [PubMed] [Google Scholar]
- McGettrick AJ, Knott V, Willis A, Handford PA. Molecular effects of calcium binding mutations in Marfan syndrome depend on domain context. Hum. Mol. Genet. 2000;9:1987–1994. doi: 10.1093/hmg/9.13.1987. [DOI] [PubMed] [Google Scholar]
- Merritt EA, Bacon DJ. Raster3D: photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- Moren A, Olofsson A, Stenman G, Sahlin P, Kanzaki T, Claesson-Welsh L, ten Dijke P, Miyazono K, Heldin CH. Identification and characterization of LTBP-2, a novel latent transforming growth factor-beta-binding protein. J. Biol. Chem. 1994;269:32469–32478. [PubMed] [Google Scholar]
- Mould AP, Askari JA, Humphries MJ. Molecular basis of ligand recognition by integrin alpha 5beta 1. I. Specificity of ligand binding is determined by amino acid sequences in the second and third NH2-terminal repeats of the alpha subunit. J. Biol. Chem. 2000;275:20324–20336. doi: 10.1074/jbc.M000572200. [DOI] [PubMed] [Google Scholar]
- Obara M, Kang MS, Yamada KM. Site-directed mutagenesis of the cell-binding domain of human fibronectin: separable, synergistic sites mediate adhesive function. Cell. 1988;53:649–657. doi: 10.1016/0092-8674(88)90580-6. [DOI] [PubMed] [Google Scholar]
- Oberhauser AF, Badilla-Fernandez C, Carrion-Vazquez M, Fernandez JM. The mechanical hierarchies of fibronectin observed with single-molecule AFM. J. Mol. Biol. 2002;319:433–447. doi: 10.1016/S0022-2836(02)00306-6. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pereira L, D'Alessio M, Ramirez F, Lynch JR, Sykes B, Pangilinan T, Bonadio J. Genomic organization of the sequence coding for fibrillin, the defective gene product in Marfan syndrome. Hum. Mol. Genet. 1993;2:961–968. doi: 10.1093/hmg/2.7.961. [DOI] [PubMed] [Google Scholar]
- Pfaff M, Reinhardt DP, Sakai LY, Timpl R. Cell adhesion and integrin binding to recombinant human fibrillin-1. FEBS Lett. 1996;384:247–250. doi: 10.1016/0014-5793(96)00325-0. [DOI] [PubMed] [Google Scholar]
- Qian RQ, Glanville RW. Alignment of fibrillin molecules in elastic microfibrils is defined by transglutaminase-derived cross-links. Biochemistry. 1997;36:15841–15847. doi: 10.1021/bi971036f. [DOI] [PubMed] [Google Scholar]
- Rao Z, Handford P, Mayhew M, Knott V, Brownlee GG, Stuart D. The structure of a cbEGF-like domain: its role in protein-protein interactions. Cell. 1995;82:131–141. doi: 10.1016/0092-8674(95)90059-4. [DOI] [PubMed] [Google Scholar]
- Reinhardt DP, Keene DR, Corson GM, Poschl E, Bachinger HP, Gambee JE, Sakai LY. Fibrillin-1: organization in microfibrils and structural properties. J. Mol. Biol. 1996;258:104–116. doi: 10.1006/jmbi.1996.0237. [DOI] [PubMed] [Google Scholar]
- Reinhardt DP, Mechling DE, Boswell BA, Keene DR, Sakai LY, Bachinger HP. Calcium determines the shape of fibrillin. J. Biol. Chem. 1997a;272:7368–7373. doi: 10.1074/jbc.272.11.7368. [DOI] [PubMed] [Google Scholar]
- Reinhardt DP, Ono RN, Sakai LY. Calcium stabilizes fibrillin-1 against proteolytic degradation. J. Biol. Chem. 1997b;272:1231–1236. doi: 10.1074/jbc.272.2.1231. [DOI] [PubMed] [Google Scholar]
- Saharinen J, Taipale J, Monni O, Keski-Oja J. Identification and characterization of a new LTBP, LTBP-4. J. Biol. Chem. 1998;273:18459–18469. doi: 10.1074/jbc.273.29.18459. [DOI] [PubMed] [Google Scholar]
- Sakai LY, Keene DR, Engvall E. Fibrillin, a new 350-kD glycoprotein, is a component of extracellular microfibrils. J. Cell Biol. 1986;103:2499–2509. doi: 10.1083/jcb.103.6.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai LY, Keene DR, Glanville RW, Bachinger HP. Purification and partial characterization of fibrillin, a cysteine-rich structural component of connective tissue microfibrils. J. Biol. Chem. 1991;266:14763–14770. [PubMed] [Google Scholar]
- Sakamoto H, Broekelmann T, Cheresh DA, Ramirez F, Rosen-bloom J, Mecham RP. Cell-type specific recognition of RGD and non-RGD-containing cell binding domains in fibrillin-1. J. Biol. Chem. 1996;271:4916–4922. [PubMed] [Google Scholar]
- Sheldrick GM, Schneider TR. SHELXL: high resolution refinement. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- Sherratt MJ, Holmes DF, Shuttleworth CA, Kielty CM. Scanning transmission electron microscopy mass analysis of fibrillin-containing microfibrils from foetal elastic tissues. Int. J. Biochem. Cell Biol. 1997;29:1063–1070. doi: 10.1016/s1357-2725(97)00028-9. [DOI] [PubMed] [Google Scholar]
- Sherratt MJ, Wess TJ, Baldock C, Ashworth J, Purslow PP, Shuttleworth CA, Kielty CM. Fibrillin-rich microfibrils of the extracellular matrix: ultrastructure and assembly. Micron. 2001;32:185–200. doi: 10.1016/s0968-4328(99)00082-7. [DOI] [PubMed] [Google Scholar]
- Stuart DI, Levine M, Muirhead H, Stammers DK. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 Å. J. Mol. Biol. 1979;134:109–142. doi: 10.1016/0022-2836(79)90416-9. [DOI] [PubMed] [Google Scholar]
- Taipale J, Saharinen J, Hedman K, Keski-Oja J. Latent TGF β1 and its binding protein are components of extracellular matrix microfibrils. J. Histochem. Cytochem. 1996;44:875–889. doi: 10.1177/44.8.8756760. [DOI] [PubMed] [Google Scholar]
- Takagi J, Kamata T, Meredith J, Puzon-McLaughlin W, Takada Y. Changing ligand specificities of alphaVbeta1 and alphaVbeta3 integrins by swapping a short diverse sequence of the beta subunit. J. Biol. Chem. 1997;272:19794–19800. doi: 10.1074/jbc.272.32.19794. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask BC, Trask TM, Broekelmann T, Mecham RP. The microfibrillar proteins MAGP-1 and fibrillin-1 form a terniary complex with the chondroitin sulphate proteoglycan decorin. Mol. Biol. Cell. 2000;11:1499–1507. doi: 10.1091/mbc.11.5.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner JM, Knott V, Handford PA, Campbell ID, Downing AK. Backbone dynamics of a cbEGF domain pair in the presence of calcium. J. Mol. Biol. 2000;296:1065–1078. doi: 10.1006/jmbi.1999.3513. [DOI] [PubMed] [Google Scholar]
- Wess TJ, Purslow PP, Sherratt MJ, Ashworth J, Shuttleworth CA, Kielty CM. Calcium determines the supramolecular organization of fibrillin-rich microfibrils. J. Cell Biol. 1998;141:829–837. doi: 10.1083/jcb.141.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Diefenbach B, Zhang R, Dunker R, Scott DL, Joachimiak A, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin alphaV beta3. Science. 2001;294:339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin alphaV beta3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- Yin W, Smiley E, Germiller J, Mecham RP, Florer JB, Wenstrup RJ, Bonadio J. Isolation of a novel latent transforming growth factor-beta binding protein gene (LTBP-3) J. Biol. Chem. 1995;270:10147–10160. doi: 10.1074/jbc.270.17.10147. [DOI] [PubMed] [Google Scholar]
- Yuan XM, Downing AK, Knott V, Handford PA. Solution structure of the TGF β-binding protein-like module, a domain associated with matrix fibrils. EMBO J. 1997;16:6659–6666. doi: 10.1093/emboj/16.22.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Werner JM, Lack J, Knott V, Handford PA, Campbell ID, Downing AK. Effects of the N2144S mutation on backbone dynamics of a TB-cbEGF domain pair from human fibrillin-1. J. Mol. Biol. 2002;316:113–125. doi: 10.1006/jmbi.2001.5329. [DOI] [PubMed] [Google Scholar]
- Zhang H, Apfelroth SD, Hu W, Davis EC, Sanguineti C, Bonadio J, Mecham RP, Ramirez F. Structure and expression of fibrillin-2, a novel microfibrillar component preferentially located in elastic matrices. J. Cell Biol. 1994;124:855–863. doi: 10.1083/jcb.124.5.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XP, Puzon-McLaughlin W, Irie A, Kovach N, Prokopishyn NL, Laferte S, Takeuchi K, Tsuji T, Takada Y. Alpha 3 β 1 adhesion to laminin-5 and invasin: critical and differential role of integrin residues clustered at the boundary between α 3 N-terminal repeats 2 and 3. Biochemistry. 1999;38:14424–14431. doi: 10.1021/bi990323b. [DOI] [PubMed] [Google Scholar]