Abstract
Schistosoma japonicum (S. japonicum) is a worldwide spread pathogen which penetrates host skin and then induces several diseases in infected host, such as fibrosis, formation of granulomas, hepatocirrhosis, and hepatomegaly. In present study, for the first time, transcriptomic profiles of mouse livers and skins infected by fork-tailed S. japonicum cercaria or non-fork-tailed S. japonicum cercaria were analyzed by using RNA-seq. The present findings demonstrated that transcriptomic landscapes of livers and skins infected by fork-tailed S. japonicum cercaria or non-fork-tailed S. japonicum cercaria were different. S. japonicum has great influence on hepatic metabolic processes. Fork-tailed S. japonicum cercaria upregulated hepatic metabolic processes, while non-fork-tailed S. japonicum cercaria downregulated hepatic metabolic processes. For the metabolism process or the metabolism enzyme expressional change, the pharmacokinetics of host could be changed during S. japonicum infection, regardless the biotypes of S. japonicum cercariae. The changes of infected skins focused on upregulation of immune response. During the S. japonicum skin infection period, fork-tailed S. japonicum cercaria infection induced stronger immune response comparing with that immune response triggered by non-fork-tailed S. japonicum cercaria. The transcription factor enrichment analysis showed that Irf7, Stat1 and Stat2 could play important roles in gene regulation during fork-tailed S. japonicum cercaria infection.
Keywords: Schistosoma japonicum, transcriptome profiles, liver, skin, morphology distinct cercariae
Introduction
Schistosoma japonicum (S. japonicum) is a worldwide spread blood fluke which prevalent among dozens of countries, including Cambodia, Brazil, Syria, Turkey, Japan, and China. The parasite has a very wide definitive host range, including human, ruminant, carnivore and a lot of domestic animals, causing serious harms to human being and animals. Over 200 million people were suffered from schistosomiasis annually (Chitsulo et al., 2000). The infected hosts could show several clinical symptoms, such as infertility, abortion, stomachache, diarrhea, fibrosis, formation of granulomas, hepatocirrhosis, hepatomegaly, and splenomegaly.
S. japonicum lives a complex life cycle which consist of several developmental stages, including adult worm, egg, daughter sporocyst, miracidia, cercaria, schistosomula (Gobert et al., 2009). The adult worm shed egg to outside world through definitive host defecating. After for several hours hatching, miracidia burst out from the egg shell and develop into cercaria in the intermediate host snail (Oncomelania). Mature cercaria escape from Oncomelania, infecting definitive host through skin contact and then develop into schistosomula and adult worm. Adult female S. japonicum can lay egg in the liver of infected host and then induces granulomas. Several reports have confirmed that S. japonicum infection can alter host gene expression and then jeopardize host. For example, IL-4 (Oswald et al., 2001) and IL-13 (Bartley et al., 2006) were reported to be upregulated during S. japonicum infection. The alterations of these cytokines are connected with host pathological change closely. For example, inhibiting the expression of IL-4 greatly reduces fibrosis in infected host (Cheever et al., 1995), while upregulation of IL-13 plays an important role in the formation of granuloma-associated fibrosis (Chiaramonte et al., 2003; Mentink-Kane et al., 2004). In these studies, the objects were focused on specific cytokines, such as IL-4 and IL-13. With the deepening of research, we realize that the final result of cytokine response is not only depend on cytokine expressional change but also on its receptor expression. For example, during pathogen infection, some cytokines could be upregulated, while their receptors could be downregulated (He et al., 2016). Studying expressions of a few genes that are altered during S. japonicum infection will lead to a false result. So it is urgent for us to know about the global gene expressional change during S. japonicum infection. Transcriptomic technology shows great advantage in studying global gene expressional change, researching the interactions between hosts and pathogens, revealing the pathogenic mechanism of pathogens. During long time evolution, morphology distinct cercariae have evolved. For example, non-fork-tailed cercaria and fork-tailed cercaria of S. japonicum are discovered in Yunnan province, China. According to unpublished data, non-fork-tailed cercaria and fork-tailed cercaria show different influences on infected host. Although the transcriptomes of S. japonicum have been widely researched (Gobert et al., 2009; Wang et al., 2015; Yang et al., 2015; Cai et al., 2016) and the transcriptomic changes of infected tissues or host cells have been reported (Chuah et al., 2013; Cai et al., 2017; Chen et al., 2017b; Luo et al., 2017), the comparative transcriptomic analysis between morphology distinct cercaria infecting tissues has not been researched. In order to discover the different features between fork-tailed cercaria infecting tissues and non-fork-tailed cercaria infecting tissues, revealing how the infected tissues response to morphology distinct cercariae, comparative analysis of host transcriptomic profiles during fork-tailed S. japonicum cercaria and non-fork-tailed S. japonicum cercaria was performed.
Materials and Methods
Ethics Statement
All animals were handled strictly according to the Animal Ethics Procedures and Guidelines of the People’s Republic of China. The study was reviewed and approved by the Animal Ethics Committee of Yunnan Agricultural University.
Schistosoma japonicum Infection and Sample Collection
Fork-tailed S. japonicum cercaria and non-fork-tailed S. japonicum cercaria were isolated and maintained by our laboratory. In present study, 12 SPF kunming mice were assigned to six groups, including non-fork-tailed S. japonicum cercaria infecting skin (S-nonFT), fork-tailed S. japonicum cercaria infecting skin (S-FT), non-fork-tailed S. japonicum cercaria infecting liver (L-nonFT), fork-tailed S. japonicum cercaria infecting liver (L-FT), non-infected skin control group (SC) and non-infected liver control group (LC). Two replicates were assigned to each group. In the infected groups, 100 cercariae were used for subcutaneous infection. 36 h post infection, mice skins (including infected groups S-nonFT, S-FT and control group SC) were individually harvested post-euthanasia for transcriptomic analysis. Ten days post infection, mice livers (including infected groups L-nonFT, L-FT, and control group LC) were harvested post-euthanasia for transcriptomic analysis. All collected tissues were cryopreserved at -80°C until used for RNA extractions and transcriptomic library constructions.
RNA Extraction, Qualification and Transcriptomic Library Construction
Total RNA was prepared individually from the mice skin or liver by using TRIzol Reagent according to the manufacturer’s protocol (Invitrogen Co. Ltd.). The RNA prepare protocol details are described as following: (1) 100 mg of cryopreserved tissue were grinded into powders by using liquid nitrogen; (2) 1 ml TRIzol Reagent was added to lysis of the powers followed by adding of 0.5 mL of isopropanol to the aqueous phase; (3) incubated for 10 min; (4) Centrifuge for 10 min at 12,000 ×g at 4°C and then discarded the supernatant; (5) resuspend the pellet in 1 mL of 75% ethanol followed by centrifuge for 5 min at 7500 ×g at 4°C; (6) discard the supernatant and air dry the RNA pellet for 10 min; (7) resuspend the pellet in 50 μL of RNase-free water and stored at -80°C until use. The quality and quantity of extracted RNAs were tested by using Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States) and Nanodrop 2000 (Thermo Scientific, Wilmington, DE, United States) individually. Before RNA library construction and RNA sequencing, the RNAs that RIN (RNA integrity number) >7.0 were treated with 20 units of RNase-Free DNase (Ambion, Shanghai, China) to remove residual genomic DNA. Illumina TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA, United States) was used for constructions of transcriptomic libraries according to manufacturer’s instructions. Briefly, (1) the mRNA was isolated by using magnetic beads with Oligo (dT); (2) the mRNA was fragmented and then cDNA was synthesized using the mRNA fragments as templates; (3) sequencing adaptor was added to the cDNA fragment and then used the fragment for agarose gel electrophoresis; (4) After agarose gel electrophoresis, the suitable fragments (200 bp) were selected for PCR amplification; (5) ABI StepOnePlus Real-Time PCR System was used in quantification and qualification of the sample library.
RNA Sequencing, Reads Mapping, Gene Identification and Bioinformatics Analyses of Differentially Expressed Genes (DEGs)
The constructed transcriptomic libraries were sequenced by using Illumina HiSeq 2000 sequencing machine. The raw sequencing data are available at SRA database of NCBI (access number: PRJNA397519). Q20 (99% sequencing accuracy) was used for raw data filtering which discarded the reads with low sequencing quality. After the raw data filtering, the adapters of reads with high sequencing quality were removed and then the clean reads were mapped to the mouse reference genome (mm10) by using BWA and mapped to reference gene by using Bowtie. The expression level of gene was analyzed with FPKM method. NOIseq R package was used for statistical analysis and identification of differentially expressed gene. Differentially expressed gene was defined as fold change ≥2, probability value calculated by NOIseq R package ≥0.8 (Tarazona et al., 2012; Seok et al., 2013; Tanaka et al., 2013; Yan et al., 2013). Gene Ontology (GO) was applied for gene functional annotation and enrichment. Kyoto Encyclopedia of Genes and Genomes database (KEGG) was applied for pathway annotation and enrichment. Significantly enriched GO term or KEGG pathway was defined using hypergeometric test followed by FDR correction (Benjamini and Hochberg, 1995). The FDR corrected P-value < 0.01 was used as cutoff of significantly enrichment. Comparative Toxicogenomics Database1 was applied for analysis of xenobiotics or drugs that are metabolized by differentially expressed enzymes (Davis et al., 2017). Transcription factor enrichment of differentially expressed gene was analyzed by Pscan (Zambelli et al., 2009).
Validation of Gene Expression Results of RNA Sequencing
Real-time quantitative PCR (Q-PCR) was applied for validating results of RNA sequencing. The RNAs that used for RNA-seq were reverse-transcribed to cDNA by using PrimeScriptTM RT reagent Kit (TaKaRa, Dalian, China). Nine genes were randomly selected for Q-PCR validation and Actb was chosen as endogenous reference gene. The primers used for Q-PCR are listed in Table 1. SYBR green (TaKaRa, Dalian, China) was used for fluorescence intensity detection. Melting curve analysis was performed to ensure the specificities of Q-PCR reactions. The Q-PCR cycle conditions were as follows: 95°C for 3 min followed by 40 cycles of 95°C for 30 s, 56°C for 30 s, 72°C for 2 min; melting curve analysis ranged from 72 to 95°C to ensure that specific product was amplified in each reaction. The 2-ΔΔCT relative expression calculating method was used to calculate the relative gene expression (Livak and Schmittgen, 2001).
Table 1.
Primers used of quantitative PCR (Q-PCR).
| Primer names | Sequence (5′->3′) | Product size |
|---|---|---|
| Actb-F | GAGACCTTCAACACCCCAGCC | 194 bp |
| Actb-R | GGCCATCTCTTGCTCGAAGTC | |
| Ces2c-F | CCAGTATCGTCTGGGTGTCC | 122 bp |
| Ces2c-R | CAAAGTGAGCGATGTTCTGC | |
| CD74-F | GTCTCTGTCCTGGTGGCTCT | 107 bp |
| CD74-R | AGGTTCTGGGAGGTGATGG | |
| Cmbl-F | TCAGGTTGAGCACATCAAGG | 119 bp |
| Cmbl-R | CCATTTCTGGCAATCATGTC | |
| Eci 2-F | GCCTCTGGTTGCGGTAGTAA | 113 bp |
| Eci 2-R | GGCTGAATGGAGTATGAAACG | |
| Ftl1-F | CATCAAGAAGATGGGCAACC | 110 bp |
| Ftl1-R | GGCGCTCAAAGAGATACTCG | |
| HBB-BS-F | GTGAACGCCGATGAAGTTG | 108 bp |
| HBB-BS-R | AGCAGAGGCAGAGGATAGGTC | |
| Irgm1-F | AGTTCAGCAGGTAGCCCAGA | 110 bp |
| Irgm1-R | TCAGCCTCAGTTTCCAGTCC | |
| Psme1-F | AGCCAAGGTGGATGTGTTCC | 172 bp |
| Psme1-R | TGGATCGGGTACTGGGATGT | |
| Mup17-F | CTCTATGGCCGAGAACCAGA | 113 bp |
| Mup17-R | CGATTGGCATTGGATAGGTC |
Results
RNA Extraction, RNA-seq and Differentially Expressed Gene
According to Agilent2100 Bioanalyzer analysis, the RINs of all RNA samples were greater than 7.0. In each group, Q20 percentage over 95%. About 50 million clean reads were obtained in each sample. More than fifteen thousand genes were detected in present study. 158 upregulated genes and 242 downregulated genes were found in non-fork-tailed S. japonicum cercaria infecting liver (L-nonFT). However, 157 upregulated genes and 118 downregulated genes were found in fork-tailed S. japonicum cercaria infecting liver (L-FT). 138 upregulated genes and 69 downregulated genes differentially expressed between L-FT and L-nonFT (Figure 1). In the infected skins, 225 upregulated genes and 124 downregulated genes were found in non-fork-tailed cercaria infecting skin (S-nonFT). 295 upregulated genes and 54 downregulated genes were found in fork-tailed cercaria infecting skin (S-FT). 182 upregulated genes and 29 downregulated genes differentially expressed between S-FT and S-nonFT. The pearson correlation coefficient of DEGs between L-FT and L-nonFT was 0.67, while the pearson correlation coefficient of DEGs between S-FT and S-nonFT was 0.74. The global gene expression pearson correlation coefficient between the two replicates were range from 0.90 to 0.98 (Figure 2). Most Q-PCR results shared the same expressional trends with RNA-seq (Supplementary Figure S1).
FIGURE 1.

Summary of differentially expressed genes (DEGs). (A) Pearson correlation between DEGs of L-nonFT and DEGs of L-FT. (B) Pearson correlation between DEGs of S-nonFT and DEGs of S-FT. (C) DEGs number in each group. L-nonFT represents liver infected by non-fork-tailed S. japonicum cercaria. L-FT represents liver infected by fork-tailed S. japonicum cercaria. S-nonFT represents skin infected by non-fork-tailed S. japonicum cercaria. S-FT represents skin infected by fork-tailed S. japonicum cercaria.
FIGURE 2.
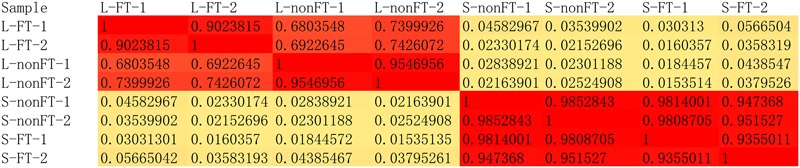
The global pearson correlation coefficient between expressional changes of the two replicates.
GO and KEGG Enrichment Analyses of Infected Livers or Skins
Gene Ontology and KEGG databases were applied for functional annotation and enrichment analysis. In infected livers, 52 GO terms were significantly enriched in both of L-nonFT and L-FT. Most of them are metabolism related GO terms (Supplementary Data Sheets 1, 2). Additionally, most enriched metabolism processes were downregulated in L-nonFT group, while upregulated metabolism processes were found in L-FT group. 46 significantly enriched GO terms were found between L-FT and L-nonFT. Most of the 46 GO terms were metabolism related GO terms and the metabolism processes were stronger in L-FT (Supplementary Data Sheet 3). Eleven pathways were significantly enriched in L-nonFT, while 21 pathways were significantly enriched in L-FT (Supplementary Data Sheet 2). Eighteen significantly enriched pathways were found between L-FT and L-nonFT (Supplementary Data Sheet 3) and most of these enriched pathways are involved in metabolism processes. Pathogenic Escherichia coli infection pathway was the only significantly enriched pathway which related to infection or immune response in L-nonFT. Caffeine metabolism, sulfur metabolism, porphyrin and chlorophyll metabolism and complement and coagulation cascade were significantly enriched in L-FT only. Toll-like receptor signaling pathway was not significantly enriched in infected liver, however, one key component (Irf7) of toll-like receptor signaling pathway was downregulated in both of L-nonFT and L-FT (Table 2).
Table 2.
Differentially expressed transcription factors in infected livers or skins.
| Transcription factor family | Transcription factor | L-nonFT(Log2 fold change) | L-FT(Log2 fold change) | S-nonFT(Log2 fold change) | S-FT(Log2 fold change) |
|---|---|---|---|---|---|
| Family:ARID | Arid5b | 1.744377 | 0.255159 | -0.10437 | -0.22878 |
| Family:bHLH | Arntl | -5.16863 | -2.37464 | -0.72068 | -1.3467 |
| Family:bHLH | Bhlhe41 | 6.531381 | 6.228819 | 1.766867 | 0.023723 |
| Family:bHLH | Hes6 | -0.26462 | 0.760736 | -0.5198 | -0.47499 |
| Family:bHLH | Myc | 1.401067 | -0.51821 | 0.237499 | 0.992387 |
| Family:bHLH | Srebf1 | -2.0337 | -0.69429 | 1.009808 | 0.468553 |
| Family:bHLH | Npas2 | -5.61738 | -4.21883 | -0.24101 | -3.19143 |
| Family:C/EBP | Cebpb | 1.609939 | 1.018074 | -0.12923 | 0.906537 |
| Family:C/EBP | Cebpa | 0.54264 | 0.504914 | 0.695452 | 2.107102 |
| Family:CUT | Onecut1 | 3.53318 | 1.118757 | No data | No data |
| Family:ESR-like | Ar | 4.459432 | 6.179909 | 1.991742 | 0.387604 |
| Family:ETS | Spi1 | -0.75176 | -0.80993 | 1.242256 | 1.996953 |
| Family:Forkhead | Foxa3 | 0.459164 | -0.59706 | -3.45943 | -3.45943 |
| Family:Forkhead | Foxq1 | 0.650976 | -1.20901 | 1.247928 | -0.41504 |
| Family:HMG | Sox18 | -1.22553 | -1.28279 | -1.09544 | -0.91334 |
| Family:HMGI/HMGY | Hmga1 | -0.04477 | -2.65896 | 0.143096 | -0.37148 |
| Family:Homeobox | Hopx | 1.65774 | 1.232161 | 0.184947 | -0.60868 |
| Family:Homeobox | Hhex | 0.437507 | -0.6039 | -0.51143 | -0.74959 |
| Family:IRF | Irf7 | -1.59553 | -1.654 | -0.11459 | 3.04564 |
| Family:IRF | Irf1 | -0.74215 | -0.48373 | 0.097016 | 1.704004 |
| Family:IRF | Irf9 | -0.99419 | -0.99613 | -0.24284 | 1.311586 |
| Family:Miscellaneous | Nr0b2 | -0.86414 | 0.318146 | -3.32193 | -3.32193 |
| Family:SAND | Sp110 | -0.46138 | -0.75995 | 1.119612 | 2.101329 |
| Family:STAT | Stat1 | -0.5378 | -0.69545 | 0.180428 | 2.79527 |
| Family:STAT | Stat2 | -0.72042 | -0.67314 | 0.423419 | 2.558745 |
| Family:TF-bZIP | Jun | 2.362894 | 0.61728 | -0.2881 | 0.256207 |
| Family:TF-bZIP | Tef | 2.225533 | 2.157897 | 0.38319 | 0.872954 |
| Family:TF-bZIP | Nfe2l2 | -0.67238 | 0.35344 | 0.185609 | 0.474892 |
| Family:TF-bZIP | Atf5 | -1.43063 | -0.83637 | 1.015755 | 0.475647 |
| Family:TF-bZIP | Maff | 2.814493 | 1.703519 | 0.589408 | 2.017427 |
| Family:TF-bZIP | Nfil3 | -2.13815 | -1.25659 | 0.182736 | 0.953936 |
| Family:TF-bZIP | Mafb | 2.857215 | 1.758992 | 0.837026 | 1.650973 |
| Family:TF-bZIP | Batf2 | -1.52725 | -1.97085 | 0.826548 | 3.892543 |
| Family:TF-bZIP | Junb | 1.562835 | 0.508301 | 0.16741 | 1.051094 |
| Family:TF-bZIP | Fos | 1.061164 | -0.71256 | -1.83582 | -2.1009 |
| Family:TF-bZIP | Dbp | 6.626142 | 5.875997 | 0.838356 | 2.344737 |
| Family:THR-like | Rorc | -0.59358 | -0.1619 | 1.186474 | 0.442295 |
| Family:THR-like | Nr1d1 | 2.450471 | 2.646729 | 0.04676 | -0.30544 |
| Family:THR-like | Ppard | -3.00331 | -2.80956 | -0.2797 | 0.048875 |
| Family:THR-like | Nr1d2 | 2.293341 | 2.09211 | 0.082757 | 0.45807 |
| Family:TSC22 | Tsc22d1 | -1.88268 | -1.7696 | -0.14401 | -0.40651 |
| Family:ZBTB | Bcl6 | -2.00565 | -0.12776 | 0.105905 | 0.314873 |
| Family:zf-C2H2 | Zfp445 | 0.710457 | 1.322984 | 0.535262 | 0.274248 |
| Family:zf-C2H2 | Gm14420 | 1.70156 | 3.722319 | 0.291651 | 0.635046 |
| Family:zf-C2H2 | Egr1 | -0.57262 | -3.54102 | -0.30641 | -1.3635 |
| Family:zf-C2H2 | Peg3 | -1.91136 | -2.84577 | 0.3109 | -0.42065 |
| Family:zf-C2H2 | Klf10 | 0.364352 | -0.22098 | 1.17353 | 0.182848 |
| Family:zf-C2H2 | Klf13 | 1.4753 | 1.02966 | 0.020408 | 0.329335 |
| Family:zf-C2H2 | Gm14308 | -0.0977 | 0.058894 | 8.301496 | 6.409391 |
| Family:zf-C2H2 | Zkscan7 | 0.547488 | 3.083017 | 0.139474 | No data |
| Family:zf-C2H2 | Etohi1 | -0.06447 | 1.513672 | 0.747907 | 0.212442 |
| Transcription co-factors | Ifi204 | -1.06675 | -0.99235 | 1.208517 | 2.746205 |
| Transcription co-factors | Ankrd1 | No data | No data | 5.484695 | 4.363299 |
| Transcription co-factors | Bcl3 | -0.3721 | -0.41914 | 0.950847 | 2.344383 |
| Transcription co-factors | Calr | -0.96153 | -1.2745 | 0.356958 | 1.017746 |
| Transcription co-factors | Pnrc1 | -1.29687 | -0.9161 | -0.93507 | -0.13759 |
| Transcription co-factors | Ccne1 | 2.492396 | 0.892144 | 0.121991 | 1.855052 |
| Transcription co-factors | Lpin2 | 2.163839 | 2.307111 | 0.852774 | 1.169925 |
| Transcription co-factors | Mnda | -1.1882 | -1.39102 | 0.346998 | 1.760863 |
| Transcription co-factors | Btg1 | -1.68806 | -0.80647 | -0.30055 | 0.079588 |
| Transcription co-factors | Cry1 | -2.1775 | -1.80425 | 0.364456 | 0.037475 |
| Transcription co-factors | Lmcd1 | -1.03171 | -1.16601 | 1.201395 | 0.36066 |
| Transcription co-factors | Tob1 | 1.20179 | 1.301431 | -0.14437 | -0.23932 |
| Transcription co-factors | Txnip | -0.0122 | -0.62252 | 0.687301 | -0.4335 |
| Chromatin remodeling factors | Hdac5 | -1.20928 | -0.53497 | -0.13199 | 0.09322 |
In the infected skins, 51 GO terms and 96 GO terms were significantly enriched in S-nonFT and S-FT, respectively (Supplementary Data Sheet 1). Sixty-three significantly enriched GO terms were found between S-FT and S-nonFT (Supplementary Data Sheet 4). Twenty pathways were significantly enriched in S-nonFT while 36 pathways were significantly enriched in S-FT (Supplementary Data Sheet 2). Twenty-six pathways were significantly enriched between S-FT and S-nonFT (Supplementary Data Sheet 4). Six pathways (including asthma, phagosome, staphylococcus aureus infection, natural killer cell mediated cytotoxicity, osteoclast differentiation, cell adhesion molecules) were significantly enriched in the three groups (S-nonFT, S-FT and S-FT/S-nonFT).
Comparative Toxicogenomic Analysis
As shown in Figure 3, 61 differentially expressed enzymes that involved in detoxification of drugs or xenobiotics were found in infected livers or skins, including 46 enzymes of metabolic pahse I and 15 enzymes of metabolic phase II. Thirteen enzymes significantly expressed higher in the L-FT group comparing with L-nonFT group. Only 1 enzyme (Inmt) expression was significantly higher in S-FT comparing with S-nonFT. No differentially expressed enzyme of metabolic phase II was found in the S. japonicum infecting skins. According to Comparative Toxicogenomics Database, thousands drugs or xenobiotics metabolisms will be altered by the differentially expressed enzymes that found in present study, including metabolisms of anti-S. japonicum drugs (such as artemisinin and oxadiazoles). Details of relationship between metabolism enzymes and substrate are listed in Supplementary Data Sheet 5.
FIGURE 3.
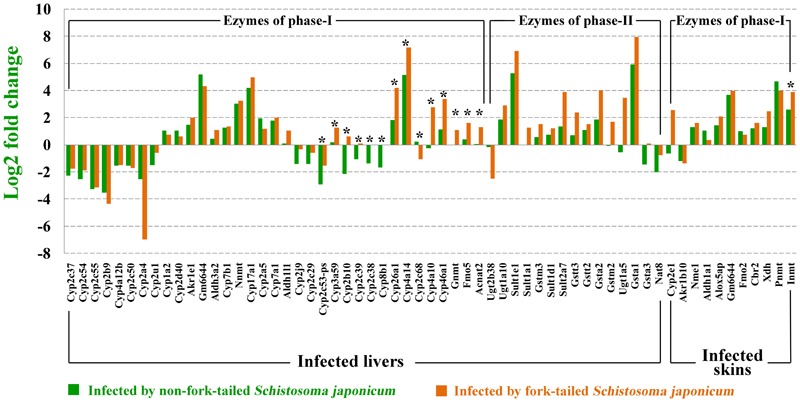
Differentially expressed metabolic enzymes in infected livers or skins. ∗Represents differentially expressed genes between non-fork-tailed cercaria infecting tissue and fork-tailed cercaria infecting tissue.
Cytokine Analysis
As shown in Figure 4, 31 cytokines or cytokine receptors were differentially expressed in infected livers or skins. Ccr3, Oit3, Pxdn, Prlr, Ifngr2, and Inhba were downregulated in infected livers or skins, especially in the infected livers. Cxcl10 and Cxcl9 were downregulated in L-nonFT and S-nonFT, however, upregulations of Cxcl10 and Cxcl9 were found in the S-FT. Il6ra, Il1r1, Il1r2, Ccl8, and Gm13305 were shown upregulated trends in present study. The significant upregulations of Ccl19, Tnfrsf12a, Cxcl13, Ccl12, Ccl4, Lep, Ccl9, Il1b, Ccl6, Csf1r, Ccl7, Ccr5, Ccl5, Ccl2, and Il4ra were only found in the infected skins. Expressions of Ccl2, Ccl5, Ccl6, Ccl7, Ccl8, Ccl9, Ccl12, Cxcl10, Cxcl9, Lep, Ccr5, and Il4ra were significantly higher expressed in S-FT comparing with S-nonFT. Il6ra and Il1r1 were significantly higher expressed in L-nonFT comparing with L-FT.
FIGURE 4.

Differentially expressed cytokines and cytokine receptors in infected livers or skins. L-nonFT represents liver infected by non-fork-tailed S. japonicum cercaria. L-FT represents liver infected by fork-tailed S. japonicum cercaria. S-nonFT represents skin infected by non-fork-tailed S. japonicum cercaria. S-FT represents skin infected by fork-tailed S. japonicum cercaria. ∗Represents differentially expressed genes between non-fork-tailed cercaria infecting tissue and fork-tailed cercaria infecting tissue.
Transcription Factor Analysis
In present study, as shown in Table 2, 65 differentially expressed transcription factors were found in infected livers or skins. These differentially expressed transcription factors can be clustered into 21 transcription factor families, including ARID, bHLH, C/EBP, CUT, ESR-like, ETS, Forkhead, HMG, HMGI/HMGY, Homeobox, IRF, Miscellaneous, SAND, STAT, TF-bZIP, THR-like, TSC22, ZBTB, zf-C2H2, transcription co-factors and chromatin remodeling factors. According to Z-score of Pscan analysis, top 10 transcription factors used by upregulated genes or downregulated genes of each group are shown in Figure 5. Some of these transcription factors were differentially expressed in present study, including five upregulated transcription factors (Cebpb of L-nonFT group, Stat1, Stat2, Irf1, and Irf7 of S-FT group) and 1 downregulated transcription factor (Ifr7 of L-FT group). Figure 6 shows the differentially expressed target genes of the differentially expressed transcription factors.
FIGURE 5.

Interactions between top10 TFs used by upregulated genes or doweregulated genes. L-nonFT represents liver infected by non-fork-tailed S. japonicum cercaria. L-FT represents liver infected by fork-tailed S. japonicum cercaria. S-nonFT represents skin infected by non-fork-tailed S. japonicum cercaria. S-FT represents skin infected by fork-tailed S. japonicum cercaria. Red presents upregulated gene.
FIGURE 6.
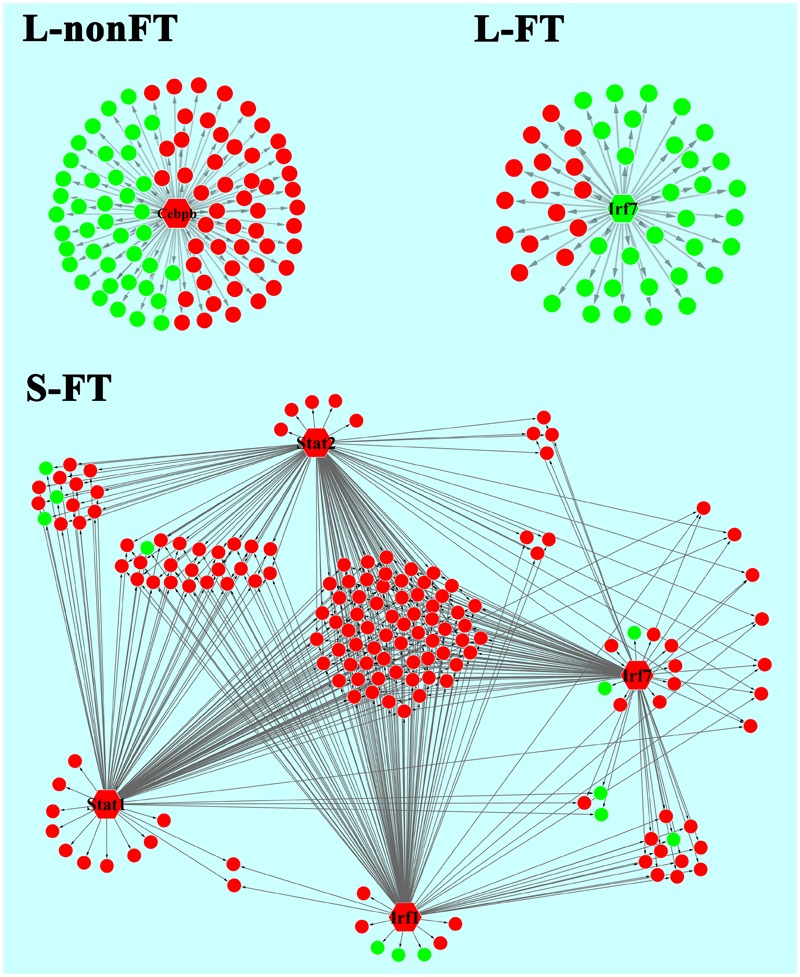
Relationship between differentially expressed TFs and the regulating genes. L-nonFT represents liver infected by non-fork-tailed S. japonicum cercaria. L-FT represents liver infected by fork-tailed S. japonicum cercaria. S-FT represents skin infected by fork-tailed S. japonicum cercaria. TF is presented as hexagon and pie presents the TF regulating gene that differentially expressed in present study. Red presents upregulated gene and green presents downregulated gene.
Discussion
In present study, all RNA integrity numbers (RIN) were >7.0. Previous report showed that RIN >7.0 is the precondition that ensures the quality of RNA-seq (Romero et al., 2014). Q20 percentage over 95% indicated that our sequencing quality was good enough for ensuring the transcriptomic analysis. The pearson correlation coefficient between the replicates were range from 0.90 to 0.98 (Figure 2). These analyses suggested that variability between the replicates was relatively low and the sequencing data was reliable. As shown in Figure 1C, most of the differentially expressed genes were upregulated in L-FT, S-nonFT, and S-FT, excepting the mouse liver infected by non-fork-tailed S. japonicum cercaria (L-nonFT). Comparison analysis between fork-tailed cercaria infection and non-fork-tailed cercaria infection showed that 138 upregulated genes and 69 downregulated genes were found between L-FT and L-nonFT, 182 upregulated genes and 29 downregulated genes were found between the S-FT and S-nonFT. The correlation between differentially expressed genes of L-nonFT and differentially expressed genes of L-FT was 0.67, while the correlation between differentially expressed genes of S-nonFT and differentially expressed genes of S-FT was 0.74 (Figures 1A,B). These results showed that morphology distinct cercariae of S. japonicum can induce some different host responses.
Several scientists have reported that S. japonicum infection result in metabolic disturbance in the infected liver (Olds et al., 1986; Chen et al., 2002; Wang et al., 2006), however, no comparative analysis has been performed to analyze the differences between fork-tailed S. japonicum cercaria infecting tissues and non-fork-tailed S. japonicum cercaria infecting tissues. According to GO enrichment analyses, most significantly enriched GO terms of L-nonFT or L-FT were metabolism related, such as lipid metabolic process, small molecule metabolic process, secondary metabolic process and arachidonic acid metabolism. The details of GO enrichment analysis are listed in Supplementary Data Sheet 1 which shows that most DEGs of each GO term of L-nonFT were downregulated, however, most DEGs of each GO term of L-FT were upregulated. The results of KEGG enrichment analysis consistent with GO enrichment analysis (Supplementary Data Sheet 2). These suggested that both of the two types S. japonicum cercariae induced hepatic metabolic changes, however, the final results of infections were different. Metabolic processes of non-fork-tailed cercaria infecting liver will be downregulated while metabolic processes of fork-tailed cercaria infecting liver will be upregulated.
Liver is a central organ that in charge of the detoxification of drugs or xenobiotics in mammal. Generally, the detoxification process in liver occurs in three alexipharmic phases including phase-I, phase-II, and phase-III. Phase-I is mainly mediated by cytochrome P450s, aldo-keto reductases, aldehyde dehydrogenases, flavin monooxygenases and other metabolic enzymes. After the drugs or xenobiotics are degraded by metabolic enzymes of phase-I, the products of phase-I will be further processed by enzymes of phase-II, such as glutathione S-transferases, UDP-glucuronosyltransferases, sulfotransferases, methyl transferases, and other enzymes. In the phase-III, all detoxified products of phase-II will be exported to intestinal tract through bile secretion and then excreted by excretory organs. In present study, 61 differentially expressed genes that involved in detoxification of drugs or xenobiotics were found in infected livers or skins. As shown in Figure 3, fork-tailed S. japonicum cercaria infection generally induced higher expressions of detoxifying enzymes, especially the enzymes of phase-I. According to Comparative Toxicogenomics Database, some medicines used for anti-S. japonicum were also targeted by the differentially expressed enzymes that were found in present study. For example, artemisinin and oxadiazoles can be used for treatment of S. japonicum infection (Fenwick and Webster, 2006; Sayed et al., 2008). In present study, Cyp2a5, which in charges of artemisinin metabolism, was upregulated in infected livers. The expressions of Gsta3 and Gsta1 that involved in oxadiazoles metabolism were also showed altered expressions in infected livers. The present results suggested that S. japonicum infection can alter host pharmacokinetics and then influences the effects of medical care. We don’t know whether there are substances listed in Supplementary Data Sheet 5 will contribute to anti-S. japonicum. As the developing of the medicinal development, more and more new anti-S. japonicum drugs will be found, our comparative toxicogenomics analysis results (Supplementary Data Sheet 5) could provide valuable data for studying pharmacokinetics of rising anti-S. japonicum drugs and then contributes to treatment of schistosomiasis.
In contrast with infected livers, most significantly enriched GO terms or KEGG pathways of infected skins were immunity related, such as response to cytokine stimulus, antigen processing and presentation, extracellular region and MHC protein complex (Supplementary Data Sheets 1, 2). MHC complex plays an essential role in innate immune response against S. japonicum (Waine et al., 1998; Tang et al., 2012; Chen et al., 2017a). In present study, MHC complex was significantly upregulated in the infected skins, however, MHC complex related GO terms was not significantly enriched in infected livers. Additionally, as shown in Supplementary Data Sheets 1, 2, only a few immunity related GO terms were enriched in infected livers. So S. japonicum cercaria infection showed different impacts on host liver and skin. S. japonicum cercaria infection in the liver influences host metabolism mainly and non-fork-tailed cercaria infection showed much lower metabolic ability comparing with fork-tailed cercaria infection, however, the S. japonicum cercaria infection in the skin influences host immune response mainly and the host skin showed much stronger immune response during fork-tailed S. japonicum cercaria infection (Figure 4).
Host immune response is connected with cytokine pathway closely. So analysis of cytokine pathway change during S. japonicum infection is necessary for us to understand the battle between host and S. japonicum. As shown in Figure 4, only nine cytokines or cytokine receptors were differentially expressed in infected livers. The cytokine expressional changes of L-nonFT and L-FT were similar, however, Lifr was downregulated in L-FT. Lifr is involved in suppression of granuloma formation (Okamura et al., 2010). The downregulation of Lifr in L-FT suggested that granuloma could be much more serious in the liver during for-tailed cercaria infection. Cxcl10 and Cxcl9 were downregulated in both L-nonFT, L-FT, and S-nonFT. According to published report, Cxcl9 and Cxcl10 are involved in recruitments of neutrophil, Th1, B cell, dendritic cell and eosinophil (Griffith et al., 2014). It suggested that chemotaxis of neutrophil, Th1, B cell, dendritic cell and eosinophil will be downregulated in the infected livers, regardless of biotype of S. japonicum cercaria. Although most differentially expressed cytokines or cytokine receptors were downregulated in infected livers, the infected skins showed an enhanced inflammation response. In present study, 21 cytokines or cytokine receptors were differentially expressed in infected skins. Most of the differentially expressed cytokines or cytokine receptors of infected skins were upregulated during S. japonicum infection, including Ccl2, Ccl4, Ccl5, Ccl6, Ccl7, Ccl8, Ccl9, Ccl12, Ccl19, Cxcl13, Il1b, Ccr5, Gm13305, Il1r2, and Tnfrsf12a. Additionally, most of these cytokines or receptors showed higher expressions in S-FT comparing with S-nonFT (Figure 4). According to published report (Griffith et al., 2014), Ccl19, Cxcl13, Ccl12, Ccl4, Ccl9, Ccl6, Ccl7, and Ccl2 involved in recruitments of T cell, Th1, Th2, B cell, DC, immature DC, NK, astrocyte, basophil, eosinophil, monocyte, macrophage, mast cell and neutrophil. So the present RNA-seq results indicated that the inflammatory responses in the infected skins were stronger than that of infected livers, especially in S-FT group. Published reports showed that IL-4 and IL-13 were upregulated during S. japonicum infection (Oswald et al., 2001; Bartley et al., 2006), however, both of these cytokine showed no differential expression in present study. The inconsistent could be the results of different host, sample collection time, detection technologies, criterion of differentially expressed gene or differential infection progresses. Although IL-4 showed no differentially expressed in present study, its receptor Il4ra was upregulated in the infected mouse skins, especially in the fork-tailed S. japonicum cercaria infecting skin (Figure 4). It is clear that Il4ra is one of the key components of IL-4 signaling pathway which involved in recruitment of mediators of cell growth, cellular resistance to apoptosis, gene activation and immune cell differentiation (Nelms et al., 1999). So, in present study, it is very likely that upregulation of Il4ra will enhance the effects of IL-4 signaling pathway under the condition that IL-4 shows no differentially expressed.
Expression of cytokine is regulated by various transcription factors, such as signal transducer and activator of transcription (Stat) and interferon regulatory factor (Irf). These transcription factors play important roles in host inflammation induction (Taniguchi et al., 2001; Hanada and Yoshimura, 2002). STAT1-/- mice shows decreased cytotoxicity of NK cell (Chen et al., 2016) which negatively regulates S. japonicum egg-induced liver fibrosis (Hou et al., 2012). Stat2 is one of the TFs in STAT signaling pathway which regulates host immune response (Decker et al., 2002), however, its role in S. japonicum infection still unclear. Irf7 has been confirmed as important regulator of innate immune response (Lu et al., 2002), however, its role in S. japonicum infection is needed to be illustrated too. In present study, 65 differentially expressed TF were found in infected livers or skins, including Stat1, Stat2, and Irf7 (Table 2). These 65 differentially expressed TFs can be clustered into 21 transcription factor families. As shown in Table 2, the members of transcription co-factors family and TF-bZIP family were affected most by S. japonicum infection and there were 13 transcription co-factors and 11 TF-bZIPs differentially expressed.
Pscan was applied to analyze TF regulating genes and String database was applied to analyze the interactions between the TFs. According to Z-score of Pscan analysis, top 10 TFs used by upregulated genes or downregulated genes are shown in Figure 6. As shown in Figure 6, there were five differentially expressed TF, including Cebpb, Stat1, Stat2, Irf1, and Irf7 (Figure 6). Cebpb, which was upregulated in present study, is a key hepatocyte TF which is known to regulate the function or regeneration of animal liver (Jakobsen et al., 2013). Irf7 is involved in regulation of type I interferon production, oncogenesis and apoptosis (Ning et al., 2011). In present study, Irf7 was downregulated in L-FT group. The downregulation of Irf7 consistent with that most of the DEGS that are regulated by Irf7 were downregulated in L-FT group (Figure 6). Irf7 was upregulated in S-FT group, it also agreed with that most DEGs that are regulated by Irf7 were upregulated in fork-tailed cercaria infecting skin (Figure 6). So, during the fork-tailed S. japonicum cercaria infection, it is very likely that the differential expression of Irf7 plays an important regulating role in host gene expressional regulation and then contributes to host immune responses against S. japonicum infection. Beside of Irf7, three upregulated TFs (Stat1, Stat2, and Irf1) were found in S-FT group. It also consistents with that most DEGs that are regulated by Stat1, Stat2, and Irf1 were upregulated in S-FT group, especially those genes regulated by Stat1, Stat2, Irf1, and Irf7 at the same time. These results indicated that S. japonicum can alter host TF expression and then regulates host biological processes indirectly. The differentially expressed TFs, such as Stat1, Stat2, Irf1, and Irf7, could play important regulatory roles during S. japonicum infection.
Conclusion
In present study, for the first time, we analyzed the transcriptomic changes of mouse livers and skins infected by non-fork-tailed S. japonicum cercaria or fork-tailed S. japonicum cercaria, analyzed the transcriptomic differences between the two cercariae infecting tissues. The present results will give us valuable data about the gene expressional differences between non-fork-tailed S. japonicum cercaria infecting tissues and fork-tailed S. japonicum cercaria infecting tissues. Our results indicate that, during the S. japonicum infection, host liver and skin show totally different transcriptomic landscapes. The functional changes of infected livers focus on alterations of metabolic processes. Additionally, non-fork-tailed S. japonicum cercaria infecting liver shows lower metabolic ability comparing with fork-tailed S. japonicum cercaria infecting liver. The gene expressional changes of infected skins focus on upregulation of inflammation response. Furthermore, fork-tailed cercaria infecting skin shows much stronger inflammation response than that of non-fork-tailed cercariae infecting skin. Comparative Toxicogenomic analysis shows that S. japonicum infection can alter the expressions of genes that involved in regulating pharmacokinetics of host (including pharmacokinetics of some anti-S. japonicum drugs). Transcription factor analysis reveals that some TF expressions can be altered during S. japonicum infection and the TF usage frequency depend on biotypes of S. japonicum cercariae and infected tissue attributes. Irf7, Stat1, and Stat2 could play important regulation roles in host inflammatory response during fork-tailed S. japonicum cercaria infection.
Author Contributions
F-CZ and J-JH conceived and designed the study, and critically revised the manuscript. YY, SH, and XX performed the experiments, analyzed the data and drafted the manuscript, J-FY and HC helped in study design, study implementation and manuscript revision. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Key project of applied basic research plan of Yunnan Province (Grant No. 2013FA037), and the National Natural Science Foundation of China (Grant No. 31360607). We thank also BGI-Shenzhen for technical assistance.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01648/full#supplementary-material
Quantitative PCR (Q-PCR) validation of the results of RNA-seq.
Significantly enriched GO terms.
Significantly enriched pathways.
Results of DEG comparative enrichment analysis between L-nonFT and L-FT.
Results of DEG comparative enrichment analysis between S-nonFT and S-FT.
Relationship between differentially expressed metabolism enzyme and substrate.
References
- Bartley P. B., Ramm G. A., Jones M. K., Ruddell R. G., Li Y., McManus D. P. (2006). A contributory role for activated hepatic stellate cells in the dynamics of Schistosoma japonicum egg-induced fibrosis. Int. J. Parasitol. 36 993–1001. 10.1016/j.ijpara.2006.04.015 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 57 289–300. [Google Scholar]
- Cai P., Liu S., Piao X., Hou N., Gobert G. N., McManus D. P., et al. (2016). Comprehensive transcriptome analysis of sex-biased expressed genes reveals discrete biological and physiological features of male and female Schistosoma japonicum. PLoS Negl. Trop. Dis. 10:e0004684 10.1371/journal.pntd.0004684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai P., Liu S., Piao X., Hou N., You H., McManus D. P., et al. (2017). A next-generation microarray further reveals stage-enriched gene expression pattern in the blood fluke Schistosoma japonicum. Parasit. Vectors 10:19 10.1186/s13071-016-1947-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheever A. W., Finkelman F. D., Cox T. M., Cox T. M. (1995). Anti-interleukin-4 treatment diminishes secretion of Th2 cytokines and inhibits hepatic fibrosis in murine schistosomiasis japonica. Parasite Immunol. 7 103–109. [DOI] [PubMed] [Google Scholar]
- Chen F., Cai W. M., Chen Z., Chen X. M., Liu R. H. (2002). Dynamic changes in the collagen metabolism of liver fibrosis at the transcription level in rabbits with schistosomiasis japonica. Chin. Med. J. 115 1637–1640. [PubMed] [Google Scholar]
- Chen J. L., Xu T. H., Zhu D. D., Wang J. X., Huang C. Q., Lyu L., et al. (2016). Egg antigen p40 of Schistosoma japonicum promotes senescence in activated hepatic stellate cells by activation of the STAT3/p53/p21 pathway. Cell Death Dis. 7:e2315 10.1038/Cddis.2016.228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Chen Q., Hou W., He L. (2017a). The dynamic changes of CD3e-CD11c+ dendritic cells in spleens and bone marrow of mice infected with Schistosoma japonicum. Parasitol. Res. 116 1007–1011. 10.1007/s00436-017-5381-6 [DOI] [PubMed] [Google Scholar]
- Chen L., Chen Q., Hou W., He L. (2017b). High-throughput dynamic analysis of differentially expressed genes in splenic dendritic cells from mice infected with Schistosoma japonicum. Immunol. Lett. 184 15–22. 10.1016/j.imlet.2017.02.001 [DOI] [PubMed] [Google Scholar]
- Chiaramonte M. G., Mentink-Kane M., Jacobson B. A., Cheever A. W., Whitters M. J., Goad M. E. P., et al. (2003). Regulation and function of the interleukin 13 receptor alpha 2 during a T helper cell type 2-dominant immune response. J. Exp. Med. 197 687–701. 10.1084/jem.20020903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitsulo L., Engels D., Montresor A., Savioli L. (2000). The global status of schistosomiasis and its control. Acta Trop. 77 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuah C., Jones M. K., Burke M. L., Owen H. C., Anthony B. J., McManus D. P., et al. (2013). Spatial and temporal transcriptomics of Schistosoma japonicum-induced hepatic granuloma formation reveals novel roles for neutrophils. J. Leukoc. Biol. 94 353–365. 10.1189/jlb.1212653 [DOI] [PubMed] [Google Scholar]
- Davis A. P., Grondin C. J., Johnson R. J., Sciaky D., King B. L., McMorran R., et al. (2017). The comparative toxicogenomics database: update 2017. Nucleic Acids Res. 45 D972–D978. 10.1093/nar/gkw838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker T., Stockinger S., Karaghiosoff M., Muller M., Kovarik P. (2002). IFNs and STATs in innate immunity to microorganisms. J. Clin. Invest. 109 1271–1277. 10.1172/Jci200215770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenwick A., Webster J. P. (2006). Schistosomiasis: challenges for control, treatment and drug resistance. Curr. Opin. Infect. Dis. 19 577–582. 10.1097/01.qco.0000247591.13671.6a [DOI] [PubMed] [Google Scholar]
- Gobert G. N., Moertel L., Brindley P. J., McManus D. P. (2009). Developmental gene expression profiles of the human pathogen Schistosoma japonicum. BMC Genomics 10:128 10.1186/1471-2164-10-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J. W., Sokol C. L., Luster A. D. (2014). Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu. Rev. Immunol. 32 659–702. 10.1146/annurev-immunol-032713-120145 [DOI] [PubMed] [Google Scholar]
- Hanada T., Yoshimura A. (2002). Regulation of cytokine signaling and inflammation. Cytokine Growth Factor Rev. 13 413–421. [DOI] [PubMed] [Google Scholar]
- He J. J., Ma J., Song H. Q., Zhou D. H., Wang J. L., Huang S. Y., et al. (2016). Transcriptomic analysis of global changes in cytokine expression in mouse spleens following acute Toxoplasma gondii infection. Parasitol. Res. 115 703–712. 10.1007/s00436-015-4792-5 [DOI] [PubMed] [Google Scholar]
- Hou X., Yu F. Z., Man S. Q., Huang D. K., Zhang Y. X., Liu M., et al. (2012). Negative regulation of Schistosoma japonicum egg-induced liver fibrosis by natural killer cells. PLoS Negl. Trop. Dis. 6:e1456 10.1371/journal.pntd.0001456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen J. S., Waage J., Rapin N., Bisgaard H. C., Larsen F. S., Porse B. T. (2013). Temporal mapping of CEBPA and CEBPB binding during liver regeneration reveals dynamic occupancy and specific regulatory codes for homeostatic and cell cycle gene batteries. Genome Res. 23 592–603. 10.1101/gr.146399.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu R. Q., Moore P. A., Pitha P. M. (2002). Stimulation of IRF-7 gene expression by tumor necrosis factor alpha - Requirement for NF kappa B transcription factor and gene accessibility. J. Biol. Chem. 277 16592–16598. 10.1073/jbc.M111440200 [DOI] [PubMed] [Google Scholar]
- Luo J., Liang Y., Kong F., Qiu J., Liu X., Chen A., et al. (2017). Vascular endothelial growth factor promotes the activation of hepatic stellate cells in chronic schistosomiasis. Immunol. Cell Biol. 95 399–407. 10.1038/icb.2016.109 [DOI] [PubMed] [Google Scholar]
- Mentink-Kane M. M., Cheever A. W., Thompson R. W., Hari D. M., Kabatereine N. B., Vennervald B. J., et al. (2004). IL-13 receptor alpha 2 down-modulates granulomatous inflammation and prolongs host survival in schistosomiasis. Proc. Natl. Acad. Sci. U.S.A. 101 586–590. 10.1073/pnas.0305064101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelms K., Keegan A. D., Zamorano J., Ryan J. J., Paul W. E. (1999). The IL-4 receptor: signaling mechanisms and biologic functions. Annu. Rev. Immunol. 17 701–738. 10.1146/annurev.immunol.17.1.701 [DOI] [PubMed] [Google Scholar]
- Ning S., Pagano J. S., Barber G. N. (2011). IRF7: activation, regulation, modification and function. Genes Immun. 12 399–414. 10.1038/gene.2011.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura Y., Nomoto S., Kanda M., Li Q., Nishikawa Y., Sugimoto H., et al. (2010). Leukemia inhibitory factor receptor (LIFR) is detected as a novel suppressor gene of hepatocellular carcinoma using double-combination array. Cancer Lett. 289 170–177. 10.1016/j.canlet.2009.08.013 [DOI] [PubMed] [Google Scholar]
- Olds G. R., Finegan C., Kresina T. F. (1986). Dynamics of hepatic glycosaminoglycan accumulation in murine Schistosoma japonicum infection. Gastroenterology 91 1335–1342. [DOI] [PubMed] [Google Scholar]
- Oswald I. P., Dozois C. M., Barlagne R., Fournout S., Johansen M. V., Bogh H. O. (2001). Cytokine mRNA expression in pigs infected with Schistosoma japonicum. Parasitology 122 299–307. [DOI] [PubMed] [Google Scholar]
- Romero I. G., Pai A. A., Tung J., Gilad Y. (2014). RNA-seq: impact of RNA degradation on transcript quantification. BMC Biol. 12:42 10.1186/1741-7007-12-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed A. A., Simeonov A., Thomas C. J., Inglese J., Austin C. P., Williams D. L. (2008). Identification of oxadiazoles as new drug leads for the control of schistosomiasis. Nat. Med. 14 407–412. 10.1038/nm1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok J., Warren H. S., Cuenca A. G., Mindrinos M. N., Baker H. V., Xu W., et al. (2013). Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. U.S.A. 110 3507–3512. 10.1073/pnas.1222878110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Nishimura M., Ihara F., Yamagishi J., Suzuki Y., Nishikawa Y. (2013). Transcriptome analysis of mouse brain infected with Toxoplasma gondii. Infect. Immun. 81 3609–3619. 10.1128/IAI.00439-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G. X., Zhou H. J., Xu J. W., Xu J. M., Ji M. J., Wu H. W., et al. (2012). Schistosoma japonicum soluble egg antigens attenuate IFN-gamma-induced MHC class II expression in RAW 264.7 macrophages. PLoS ONE 7:e49234 10.1371/journal.pone.0049234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T., Ogasawara K., Takaoka A., Tanaka N. (2001). IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19 623–655. 10.1146/annurev.immunol.19.1.623 [DOI] [PubMed] [Google Scholar]
- Tarazona S., García F., Ferrer A., Dopazo J., Conesa A. (2012). NOIseq: a RNA-seq differential expression method robust for sequencing depth biases. EMBnet. J. 17 18–19. 10.14806/ej.17.B.265 [DOI] [Google Scholar]
- Waine G. J., Ross A. G., Williams G. M., Sleigh A. C., McManus D. P. (1998). HLA class II antigens are associated with resistance or susceptibility to hepatosplenic disease in a Chinese population infected with Schistosoma japonicum. Int. J. Parasitol. 28 537–542. [DOI] [PubMed] [Google Scholar]
- Wang X., Xu X., Lu X., Zhang Y., Pan W. (2015). Transcriptome bioinformatical analysis of vertebrate stages of Schistosoma japonicum reveals alternative splicing events. PLoS ONE 10:e0138470 10.1371/journal.pone.0138470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Utzinger J., Xiao S. H., Xue J., Nicholson J. K., Tanner M., et al. (2006). System level metabolic effects of a Schistosoma japonicum infection in the Syrian hamster. Mol. Biochem. Parasitol. 146 1–9. 10.1016/j.molbiopara.2005.10.010 [DOI] [PubMed] [Google Scholar]
- Yan L., Yang M., Guo H., Yang L., Wu J., Li R., et al. (2013). Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 20 1131–1139. 10.1038/nsmb.2660 [DOI] [PubMed] [Google Scholar]
- Yang J., Fu Z., Hong Y., Wu H., Jin Y., Zhu C., et al. (2015). The differential expression of immune genes between water buffalo and yellow cattle determines species-specific susceptibility to Schistosoma japonicum infection. PLoS ONE 10:e0130344 10.1371/journal.pone.0130344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambelli F., Pesole G., Pavesi G. (2009). Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 37 W247–W252. 10.1093/nar/gkp464 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantitative PCR (Q-PCR) validation of the results of RNA-seq.
Significantly enriched GO terms.
Significantly enriched pathways.
Results of DEG comparative enrichment analysis between L-nonFT and L-FT.
Results of DEG comparative enrichment analysis between S-nonFT and S-FT.
Relationship between differentially expressed metabolism enzyme and substrate.


