Abstract
We report three signatures produced from SHARPIN gene copy number increase (GCN-Increase) and their effects on patients with breast cancer (BC). In the Metabric dataset (n = 2059, cBioPortal), SHARPIN GCN-Increase occurs preferentially or mutual exclusively with mutations in TP53, PIK3CA, and CDH1. These genomic alterations constitute a signature (SigMut) that significantly correlates with reductions in overall survival (OS) in BC patients (n = 1980; p = 1.081e − 6). Additionally, SHARPIN GCN-Increase is associated with 4220 differentially expressed genes (DEGs). These DEGs are enriched in activation of the pathways regulating cell cycle progression, RNA transport, ribosome biosynthesis, DNA replication, and in downregulation of the pathways related to extracellular matrix. These DEGs are thus likely to facilitate the proliferation and metastasis of BC cells. Additionally, through forward (FWD) and backward (BWD) stepwise variate selections among the top 160 downregulated and top 200 upregulated DEGs using the Cox regression model, a 6-gene (SigFWD) and a 50-gene (SigBWD) signature were derived. Both signatures robustly associate with decreases in OS in BC patients within the Curtis (n = 1980; p = 6.16e − 11 for SigFWD; p = 1.06e − 10, for SigBWD) and TCGA cohort (n = 817; p = 4.53e − 4 for SigFWD and p = 0.00525 for SigBWD). After adjusting for known clinical factors, SigMut (HR 1.21, p = 0.0297), SigBWD (HR 1.25, p = 0.0263), and likely SigFWD (HR 1.17, p = 0.062) remain independent risk factors of BC deaths. Furthermore, the proportion of patients positive for these signatures is significantly increased in ER −, Her2-enriched, basal-like, and claudin-low BCs compared to ER + and luminal BCs. Collectively, these SHARPIN GCN-Increase-derived signatures may have clinical applications in management of patients with BC.
Keywords: SIPL1/SHARPIN, Cancer, Breast cancer, Overall survival
Highlights
-
•
SHARPIN genomic increase correlates with poor prognosis in breast cancer patients
-
•
SHARPIN genomic increase associates with enrichment of mutations in TP53 and others
-
•
SHARPIN genomic increases occur along with many differentially expressed genes (DEGs)
-
•
These DEGs enhance breast cancer cell proliferation and reduces extracellular matrix
-
•
Enriched mutations and DEGs strongly associate with reductions in overall survival
1. Introduction
Breast cancer (BC) is a leading cause of cancer death in women with approximately 1.7 million new cases and 500,000 fatalities annually [1]. Clinically, BC is classified based on the expression of estrogen receptor (ER) and Her2 and is categorized into ER +, Her2 +, or triple negative (TN; negative for ER, progesterone receptor (PR), and Her2) subclasses. Additionally, it is also classified according to gene expression profiles. Six intrinsic subtypes have been identified: luminal A and B (ER +), normal-like, Her2-enriched, basal-like, and claudin-low (the latter two being TN) BCs [2], [3], [4], [5]. Furthermore, combination of gene copy number (GCN) variation and gene expression profile has divided BC into 10 integrative clustering sub-groups [6]. In addition to GCN variation, BC contains several common mutations including TP53, PIK3CA, GATA3, and CDH1 [7], [8], [9]. Despite this rich knowledge of BC tumorigenesis, our understanding on the etiology leading to BC tumorigenesis and progression remains limited.
SHARPIN (Shank-associated RH domain interacting protein) or SIPL1 (Shank-Interacting Protein-Like 1) is a Shank-binding protein in the postsynaptic density [10]. SHARPIN is also a major component of the linear ubiquitin chain assembly complex (LUBAC), an E3 ubiquitin-protein ligase complex that activates NF-κB signalling [11], [12], [13], [14]. Additionally, SHARPIN/SIPL1 is physically associated with PTEN, resulting in PTEN inactivation [15], [16]. In line with the important contributions of NF-κB signalling and PTEN inactivation in tumorigenesis [15], [17], SHARPIN possesses multiple oncogenic activities in vitro, including suppression of apoptosis [18], [19], enhancement of cell detachment and migration [20], [21], and AKT activation [15]. In vivo, SHARPIN promotes the tumorigenesis of cervical cancer [15] and hepatocellular carcinoma [22]. In patients, upregulation of SHARPIN occurs in ovarian cancer, cervical cancer, liver cancer, and prostate cancer [15], [20], [22], [23].
NF-κB signalling, PI3K activation, and PTEN inactivation are well demonstrated oncogenic events that occur during BC tumorigenesis [24]. Of note, upregulation of SHARPIN expression was detected in BC [16], [25], [26]. SHARPIN plays a role in BC metastasis [25] and inhibition of p53-mediated tumor suppression [26]. Elevations in SHARPIN expression modestly associate with reductions in overall survival (OS) in patients with breast cancer [16], [25], [26]. In supporting these observations, an increase in SHARPIN gene copy number (GCN) was also observed and this genomic alteration modestly correlates with shortening of OS in patients with BC [16].
To further examine the association of SHARPIN GCN increase with BC prognosis, we have taken a thorough in silico investigation of SHARPIN GCN increase, its-associated enrichment in mutations and gene expression, and the impact of these events on OS in patients with breast cancer. The two most comprehensive datasets, the Metabric (n = 2509) and TCGA-Cell (n = 817) cohorts within the cBioPortal database were used. We report here the identification of SHARPIN GCN increase-associated mutations of TP53, PI3KCA, GATA3, CDH1, AKT, and ASXL1 and > 4000 differentially expressed genes (DEGs). These DEGs function in multiple aspects of cell proliferation and extracellular matrix processes. Furthermore, these enrichments in mutations and DEGs form three signatures that robustly associate with shortening of OS in patients with breast cancer.
2. Materials and methods
2.1. cBioPortal
The Metabric and TCGA-Cell 2015 (TCGA) datasets within cBioPortal [27], [28] (http://www.cbioportal.org/index.do) contain 2509 and 815 patients with breast cancer, respectively. The Metabric dataset constitutes two sub-datasets with one being the Curtis [6] containing 1980 patients with a follow-up period up to 350 months (http://www.cbioportal.org/index.do). The TCGA dataset has 816 tumors (from 815 patients) with RNA sequencing and copy number variation data and a follow-up period of 300 months [29] (http://www.cbioportal.org/index.do).
2.2. Establishment of three SHARPING GCN increase-derived signatures: SigMut, SigFWD, and SigBWD
Enrichment data of mutations and gene expression (differentially expressed genes/DEGs) with respect to SHARPIN GCN increases were extracted from the Metabric dataset. The six enriched mutations (Table 1) were analyzed for contributions to hazard ratio (HR) using the Cox regression model (SPSS Statistics version 23). The top 160 downregulated DEGs and top 200 upregulated DEGs were selected for their effects on HR by either forward (FWD) stepwise or backward (BWD) stepwise method, using the multivariate Cox regression model (SPSS Statistics version 23). These analyses resulted in three signatures: SigMut, SigWFD, and SigBWD.
Table 1.
Co-alteration of mutations with SHARPIN GCN amplification.
| Gene | locus | AMP +a | AMP −a | Log Rc | p-Value | q-Value |
|---|---|---|---|---|---|---|
| TP53b | 17q13.1 | 232 (53.33%)d | 509 (31.50%)e | 0.76 | 9.27e − 17 | 1.60e − 14 |
| PIK3CAb | 3q26.3 | 136 (31.26%) | 719 (44.49%) | − 0.51 | 3.36e − 7 | 2.91e − 15 |
| GATA3b | 10p15 | 24 (5.52%) | 217 (13.43%) | − 1.28 | 8.45e − 7 | 4.87e − 5 |
| CDH1b | 16q22.1 | 23 (5.29%) | 175 (10.83%) | − 1.03 | 1.75e − 4 | 7.56e − 3 |
| AKT1b | 14q32.32 | 6 (1.38%) | 80 (4.95%) | − 1.84 | 2.40e − 4 | 8.31e − 3 |
| ASXL1b | 20q11 | 2 (0.46%) | 44 (2.72%) | − 2.57 | 1.40e − 3 | 0.0403 |
SHARPIN genomic amplification positive and negative.
These mutations were co-altered with SHARPIN genomic amplification.
Log2-based ratio of percentage in altered group/percentage in unchanged group; positive and negative ratios are for co-occurrence and mutual exclusiveness, respectively.
Number of mutation cases/number of cases with SHARPIN amplification × 100.
Number of mutation cases/number of cases without SHARPIN amplification × 100.
2.3. Gene set and pathway enrichment analyses
The GAGE package in R was used to analyze DEGs for gene set enrichment within the KEGG (Kyoto Encyclopedia of Genes and Genomes) and GO (gene ontology) databases [30]. Pathway enrichment was performed using the Reactome package in R [31].
2.4. Statistical analysis
Fisher's exact test was performed using the GraphPad Prism 5 software. Kaplan-Meier surviving curves and log-rank test were carried out using the R survival package, SPSS Statistics version 23, and tools provided by cBioPortal. Multivariate Cox regression analysis was performed using the R survival package and SPSS Statistics version 23. A value of p < 0.05 is considered statistically significant.
3. Results
3.1. Increase in SHARPIN GCN associates with mutations in TP53, PIK3CA, CDH1, and others
The SHARPIN gene resides at 8q24.3 which is frequently amplified in breast cancers (BC) [32], [33], [34], [35], [36], [37]. SHARPIN GCN increase was reported to correlate with a reduction in OS in patients with BC [16]. In accordance with these observations, we observed SHARPIN GCN amplification in 403 breast tumors among 1980 BCs in the Curtis sub-dataset [6] of the Metabric dataset within the cBioPortal database; the amplification modestly associates with a decrease in OS in BC patients [38] (see Fig. 1 in Ref [38]). This modest association is consistent with the consensus that individual factors have limited biomarker values and that combination of multiple factors is needed to yield a robust signature. To improve SHARPIN GCN amplification-derived association with the OS reduction, we have extracted the genomic events (mutations and gene copy number variation) from the Metabric dataset, which are enriched with SHARPIN GCN amplification. Since the 8q24 region is commonly amplified in BC, a large number of loci are co-amplified with SHARPIN, which is attributable to “the neighboring effect”. As such, their co-amplification may not contribute to the biomarker value of SHARPIN GCN increase. We thus focused on enrichment in gene mutations. With enrichment being defined at q value < 0.05, we have extracted a set of enriched mutations with either co-occurrence or mutual exclusiveness with SHARPIN GCN increase (Table 1). Interestingly, these enriched mutations include TP53, PIK3CA, GATA3, and CDH1 (Table 1). These genes are frequently mutated in BC and play important roles in BC tumorigenesis [6], [7], [29], [39]. Nonetheless, the potential role of ASXL1 (additional sex comb-like 1) in BC remains unclear.
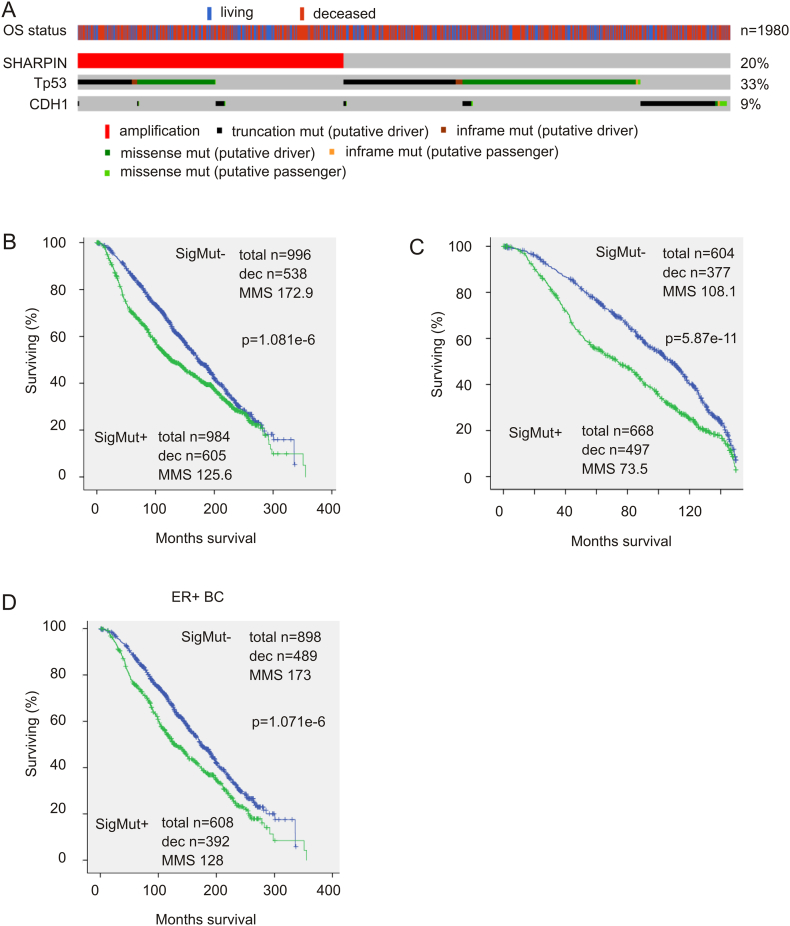
We subsequently determined the contributions of these mutations to SHARPIN genomic amplification-associated reductions in OS in BC patients. Through multivariate Cox proportional hazards regression analyses of SHARPIN GCN increase, together with those altered mutations (Table 1), we were able to remove variates with either negative co-efficient or non-significant HR. After three rounds of selection, a mutation-related signature (SigMut) consisting of SHARPIN GCN increase (HR 1.155, 95% CI: 1.00–1.334, p = 0.05), TP53 mutation (HR 1.269, 95% CI: 1.20–1.438, p < 0.001) and CDH1 mutation (HR 1.263, 95% CI: 1.039–1.537, p = 0.019) was formulated (Fig. 1A). Removal of either TP53 mutation or CDH1 mutation decreased the signature's association with reductions in OS from p = 1.081e − 6 (Fig. 1A) to 2.93e − 3 or 1.205e − 5. In the total recurrent cases (605 + 538 = 1143, Fig. 1A), 52.93% were SigMut-positive, which was reduced to 43.57% after removal of “SHARPIN GCN increase” from the signature; the difference was statistically significant (p < 0.0001 by chi-squared test). While SigMut correlates with OS reductions at p = 1.081e − 6, the correlation is at p = 2.358e − 6 after removal of “SHARPIN GCN increase” from it. In this regard, individual components make unique contributions to SigMut's biomarker value.
Fig. 1.
SHARPIN GCN increase-derived mutation-related signature (SigMut) is associated with reductions of OS in patients with breast cancer. (A) Genomic alterations of SigMut. Data was extracted from the Curtis sub-dataset (n = 1980) [6] of the Metabric dataset within the cBioPortal database. SHARPIN gene copy number increases, TP53 mutations, CDH1 mutations, and overall survival (OS) status of patients are indicated. Only the proportion of patients with SigMut alterations is included. (B–D) The OS in patients with SigMut-positive or -negative breast cancer in the entire cohort with 350-month follow-up period (B) or 150-month follow up period (C) as well as in patients with ER + breast cancer (D) was examined using the Kaplan-Meier surviving curve. Statistical analysis was performed using Log-rank test. Dec: deceased cases; MMS: median months survival.
In comparison to SHARPIN GCN increase alone (see Fig. 1 in Ref [38]), SigMut is much more robust in association with shortening of OS in patients with breast cancer (Fig. 1B). The association is also stronger during the first 150 month follow-up (Fig. 1C) when compared to the entire follow-up period of 350 months (Fig. 1B). Furthermore, in the ER expression-based BC classification system, SigMut correlates with decreases of OS only in patients with ER + BC (Fig. 1D). However, in comparison to 40% of ER + tumors being SigMut-positive, a significantly higher proportion of ER − BCs (79.3%) are SigMut-positive (Table 2). Collectively, the above observations support the association of SigMut with poor prognosis in patients with BC.
Table 2.
Distribution of SigMut, SigFWD, and SigBWD in breast cancer subtypes.
| ER + | ER − | Luminal | Normal | Her2 | Basal | Claudin-low | |
|---|---|---|---|---|---|---|---|
| Total (n) | 1506 | 474 | 1175 | 148 | 224 | 209 | 218 |
| SigMut − | 898 (59.6%) | 98 (20.7%) | 727 (61.9%) | 89 (60.1%) | 50 (22.3%) | 24 (11.5%) | 101 (46.3%) |
| SigMut + | 608 (40.4%) | 376 (79.3%) | 448 (38.1%) | 59 (39.9%) | 174 (77.8%) | 185 (88.5%) | 117 (53.7%) |
| p | ⁎ | ⁎, # | ⁎, #, $ | ⁎, #, $, & | |||
| SigFWD − | 952 (63.1%) | 175 (36.9%) | 765 (65.1%) | 94 (63.5%) | 46 (20.5%) | 67 (32.1%) | 152 (69.7%) |
| SigFWD + | 554 (36.8%) | 299 (63.1%) | 410 (34.9) | 54 (36.5%) | 178 (79.5%) | 142 (67.9%) | 66 (30.3%) |
| p | ⁎ | ⁎, # | ⁎, #, $ | ⁎, $, & | |||
| SigBWD − | 532 (35.3%) | 39 (8.2%) | 419 (35.6%) | 71 (48%) | 12 (5.4%) | 11 (5.3%) | 56 (25.7%) |
| SigBWD + | 974 (64.7%) | 435 (91.8%) | 756 (64.3%) | 77 (52%) | 212 (94.6%) | 198 (94.7%) | 162 (74.3%) |
| p | ⁎ | ⁎ | ⁎, # | ⁎, #, $ | ⁎, #, & |
Statistical analysis was performed using Fisher's exact test.
P < 0.01 in comparison to ER + and Luminal BC, respectively.
P < 0.01 in comparison to normal-like BC.
P < 0.01 in comparison to Her2-enriched BC.
P < 0.001 in comparison to basal-like BC.
3.2. Identification and characterization of differentially expressed genes (DEGs) that are associated with SHARPIN GCN increase
To further examine the impact of SHARPIN GCN increase, we determined DEGs in breast tumors with and without SHARPIN GCN increase. From the Metabric dataset (n = 2509) within the cBioPortal database, we retrieved 4220 DEGs that were defined by q < 0.001 (see Table 1 in Ref [38]). SHARPIN is among the upregulated DEGs (item #4157, see Table 1 in Ref [38]), an observation that provides additional support for the relevance of SHARPIN GCN increase in reducing OS in patients with breast cancer.
To characterize these DEGs, we performed enrichment analysis for gene sets using the GAGE package in R [30]. Enrichment was defined by q < 0.05. Using the KEGG gene set database, 58, 44, 32, and 21 genes among DEGs are respectively upregulated in the gene sets of cell cycle regulation, RNA transport, ribosome biosynthesis, and DNA replication (Table 3). Similar enrichment in gene set was also obtained using the Gene Ontology (GO) go.gs and go.sets.hs database (Table 3). Nonetheless, additional gene sets are upregulated in the go.gs and go.sets.hs datasets; these gene sets function in RNA processing, DNA metabolism, translation, protein metabolism, and cytoskeleton organization (see Tables 2 and 3 in Ref [38]). Interestingly, downregulation of gene sets involved in extracellular matrix was also observed within the go.set.hs database (Table 3). These observations agree well with the reported roles of SHARPIN in cytoskeleton organization [20], [21], particularly in regulating collagen and extracellular matrix structure [40].
Table 3.
Alteration of gene expression within specific gene sets in SHARPIN genomic amplification-associated DEGs.a
| Gene sets | Set sizeb | p-Value | q-Value |
|---|---|---|---|
| hsa04110 Cell cyclec | 58 | 2.81e − 8 | 2.86e − 6 |
| hsa03013 RNA transportc | 44 | 5.92e − 5 | 0.003018 |
| hsa03008 ribosome biogenesis in eukaryotesc | 32 | 0.001106 | 0.032013 |
| hsa03030 DNA replicationc | 21 | 0.001255 | 0.032013 |
| GO:0000279 M phased | 128 | 4.19e − 14 | 6.76e − 12 |
| GO:0000278 mitotic cell cycled | 154 | 4.06e − 13 | 2.86e − 11 |
| GO:0000087 M phase of mitotic cell cycled | 102 | 1.88e − 11 | 6.64e − 10 |
| GO:0000280 nuclear divisiond | 102 | 1.88e − 11 | 6.64e − 10 |
| GO:0006259 DNA metabolic processd | 178 | 6.48e − 7 | 1.59e − 5 |
| GO:0006260 DNA replicationd | 89 | 6.76e − 7 | 1.59e − 5 |
| GO:0022403 cell cycle phasee | 311 | 6.06e − 19 | 1.76e − 15 |
| GO:0000278 mitotic cell cyclee | 297 | 4.16e − 17 | 6.05e − 14 |
| GO:0000279 M phasee | 196 | 1.42e − 15 | 1.37e − 12 |
| GO:0022402 cell cycle processe | 374 | 3.49e − 15 | 2.54e − 12 |
| GO:0000087 M phase of mitotic cell cyclee | 153 | 8.38e − 14 | 4.88e − 11 |
| GO:0048285 organelle fissione | 157 | 2.69e − 13 | 1.12e − 10 |
| GO:0005576 extracellular regionf | 402 | 8.14e − 9 | 2.37e − 5 |
| GO:0031012 extracellular matrixf | 103 | 1.49e − 7 | 2.17e − 4 |
| GO:0005578 proteinaceous extracellular matrixf | 90 | 1.30e − 6 | 0.001257 |
| GO:0044421 extracellular region partf | 242 | 6.32e − 6 | 0.004593 |
| GO:0044459 plasma membrane partf | 383 | 2.08e − 5 | 0.012111 |
| GO:0031226 intrinsic to plasma membranef | 229 | 4.54e − 5 | 0.020163 |
| GO:0005887 integral to plasma membranef | 223 | 5.65e − 5 | 0.020163 |
| GO:0038023 signalling receptor activityf | 140 | 5.85e − 5 | 0.020163 |
| GO:0004872 receptor activityf | 189 | 6.24e − 5 | 0.020163 |
| GO:0032989 cellular component morphogenesisf | 240 | 9.44e − 5 | 0.02747 |
| GO:0000902 cell morphogenesisf | 299 | 1.18e − 4 | 0.031194 |
Enrichment analysis for gene sets was performed using the GAGE package in R.
Number of genes in DEGs that are enriched in the individual gene sets.
Gene sets upregulated in KEGG gene sets.
Upregulated gene sets in the Gene Ontology (GO) gene set database.
Enhanced gene sets in the go.sets.hs database.
Downregulated gene sets in the go.sets.hs database.
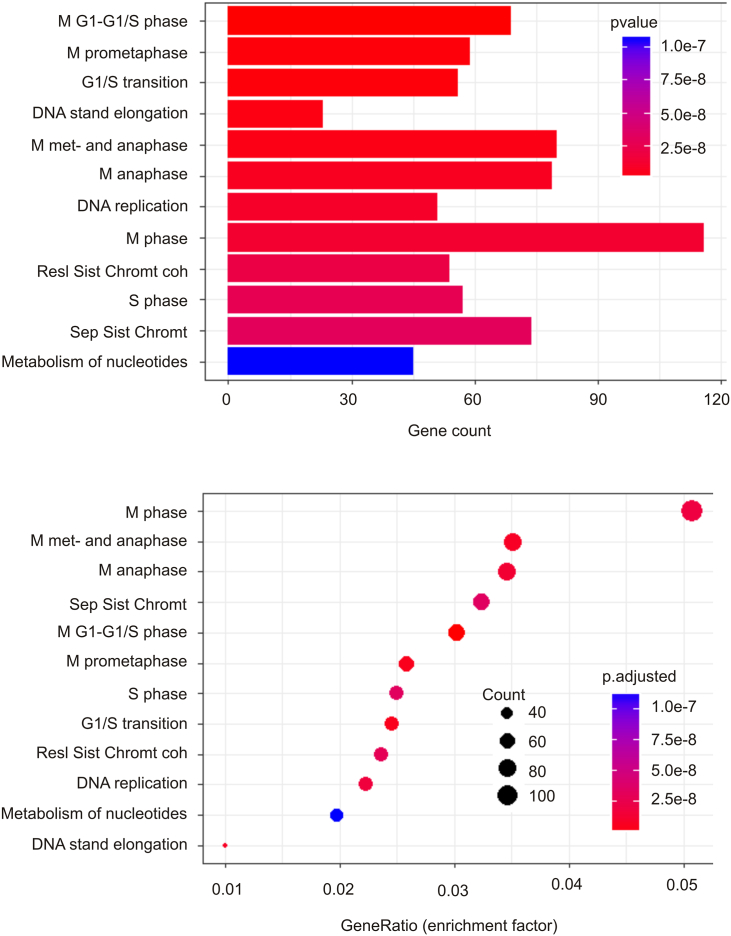
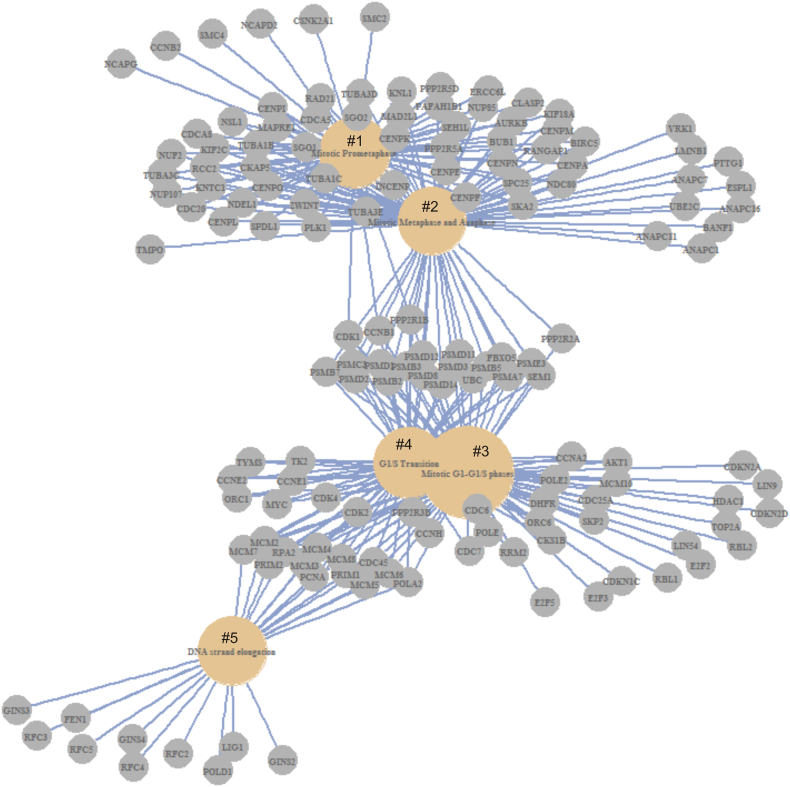
We subsequently carried out pathway enrichment analysis using the Reactome package in R [31]. A set of 87 pathways are upregulated among SHARPIN GCN increase-associated DEGs. These pathways function in cell cycle regulation, RNA processing, DNA metabolism, amino acid metabolism, purine metabolism, and p53-regulated transcription of cell cycle genes (see Table 4 in Ref [38]). The enrichment in p53-regulated cell cycle events is supported by the enrichment of TP53 mutations in tumors with increases in SHARPIN GCN (Table 1; Fig. 1A). The top 12 enriched pathways include those functioning in cell cycle regulation, chromatid separation, and DNA replication (Fig. 2). The network nature of DEGs contributing to the pathway enrichment can be documented (Fig. 3). Taken together, evidence supports that SHARPIN GCN increase-associated DEGs promote cell proliferation, DNA metabolism, and other essential cellular processes that are involved in cell proliferation.
Fig. 2.
SHARPIN GCN increase-associated DEGs are enriched in upregulation of a set of the pathways promoting cell proliferation. The DEG data was extracted from the Metabric dataset (n = 2509) within the cBioPortal database and analyzed using the Reactome package in R [31]. The number of DEGs upregulated in the top 12 pathways (top panel) and the enrichment factor (Gene ratio) for each enriched pathway (bottom panel) are shown.
Fig. 3.
Five enriched pathways and their contributing DEGs are shown. The enriched pathways are for Mitotic Prometaphase (#1), Mitotic Metaphase and Anaphase (#2), Mitotic G1–G1/S phase (#3), G1/S phase Transition (#4), and DNA strand elongation (#5). Analysis for pathway enrichment was carried out using the Reactome package in R [31].
3.3. Building two signatures with dynamic association with OS in patients with breast cancer
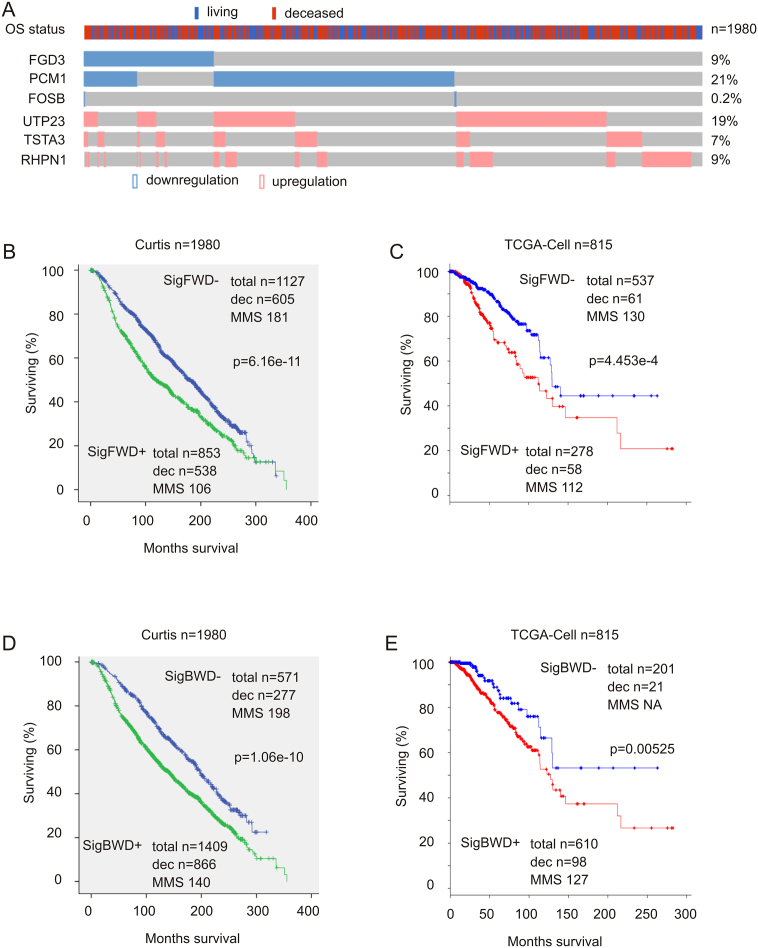
Elevation in cell proliferation is a major contributor to tumor progression; the observed upregulation above in gene sets and pathways promoting cell cycle progression and other aspects of cell proliferation among DEGs related to SHARPIN GCN increase strongly indicates an association of the DEGs with clinical outcome, more specifically, with OS in BC patients. To address this possibility, we selected 360 DEGs for signature building. These DEGs consist of the top 160 downregulated and top 200 upregulated DEGs, based on the level of changes in tumors with SHARPIN GCN increase vs those without the increase (see Table 1 in Ref [38]). For the 160 downregulated DEGs, tumors (patients) were defined to express reduced DEG genes if the genes are expressed at levels 1.5 standard derivation lower than (− 1.5SD) the reference population mean expression; for the upregulated 200 DEGs, patients were stratified into two groups: one with upregulation of specific DEGs if the DEGs are expressed at levels > 2SD above the reference population mean expression and one without specific DEG upregulation when the DEG genes are expressed at levels not exceeding 2SD above the population mean. By using multiple Cox regression analysis, we first removed those variates with negative co-efficient ≤ − 0.1 and then selected 57 DEGs. These DEGs were then imputed into the Cox regression model (SPSS Statistics version 23) for both forward and backward stepwise method of co-variate selection. Each method of selection respectively resulted in a 6-gene forward (FWD) signature (SigFWD) (Fig. 4A) and a 50-gene backward signature (SigBWD) (see Table 5 in Ref [38]).
Fig. 4.
SigFWD and SigBWD robustly correlate with reductions in OS in patients with breast cancer. (A) Expression profiles of SigFWD. Data were extracted from the Curtis dataset (n = 1980) [6]. Downregulation and upregulation for the indicated genes and the OS status for the patients involved are shown. Only the proportion of patients with SigBWD alterations is included. (B, C) The impacts of SigFWD on OS in patients with breast cancer in the Curtis and TCGA-Cell cohorts. (D, E) The impact of SigBWD on OS in patients with breast cancer in the Curtis and TCGA-Cell cohorts. Statistical analysis was performed using Log-rank test. Dec: deceased cases; MMS: median months survival.
SigFWD consists of downregulation in FGD3, PCM1, and FOSB as well as upregulation of UTP23, TSTA3, and RHPN1 (Fig. 4A). FGD3 and SUSD3 form a metagene with a strong prediction of OS in patients with BC from the Metabric dataset [41]. PCM1 is a candidate of tumor suppressor in breast cancer [42]. A very recent report revealed that FOSB possesses tumor suppression activities toward triple negative breast cancer [43]. UTP23 functions in ribosome RNA processing [44], which is likely a risk factor for colorectal cancer [45]. TSTA3 is a potential oncogenic factor in breast cancer [46]. RHPN1 regulates cytoskeleton in podocytes [47], but its potential role in tumorigenesis remains unclear. Collectively, the general function of the individual components of SigFWD in tumorigenesis supports an association of SigFWD with poor prognosis of patients with BC. In this regard, SigFWD robustly associates with decreases in OS in two independent cohorts of patients with BC, the Metabric (n = 1980; Fig. 4B) and the TCGA-Cell (n = 815; Fig. 4C).
SigBWD constitutes 50 DEGs, including SHARPIN (item #38, see Table 5 in Ref [38]). SigBWD dramatically correlates with reductions in OS in BC patients from the Metabric and TCGA datasets (Fig. 4D, E).
3.4. Study of the association of SigMut, SigFWD, and SigBWD with BC-derived poor prognosis
Gene copy number increase and elevation of SHARPIN (SIPL1) gene expression displayed a modest association with reductions in OS largely in patients with ER + breast cancer [16]. Of note, SigMut correlates with OS reductions in patients with ER + BC (Fig. 1D) but not those with ER − BC (p = 0.438). Both SigFWD and SigBWD significantly associate with OS reduction in patients with either ER + or ER − BC (see Fig. 2 in Ref [38]). For PAM50 classified intrinsic BC subtypes, SigMut, SigFWD, and SigBWD all correlate with shortening of OS in patients with luminal or normal-like BCs, but not in those with basal-like or Her2-enriched breast tumors (Table 4). SigFWD and SigBWD display a relationship with OS reduction in clauding-low BC subclass at a statistically significant level and close to a significant level, respectively (Table 4). Normal-like BCs are more aggressive than luminal BC and less aggressive than those of basal-like, Her2-enriched, and Claudin-low breast tumors [48]. The discussions above thus suggest a tendency of all three signatures in association with OS decreases in patients with less aggressive breast tumors. Nonetheless, SigMut, SigFWD, and SigBWD are expressed at a significant higher rate in aggressive BC sub-types compared to those of a less aggressive nature. For example, they are expressed at a higher rate in ER − vs ER + subgroups, basal-like and Her2-enriched breast tumors vs luminal breast cancers, and basal-like and Her2-enriched vs normal-like subclasses (Table 2). Furthermore, after adjusting for the age at diagnosis, cellularity, Integrative Cluster, Neoplasm Histologic Grade, Nottingham prognostic index, tumor size and tumor stage, SigMut and SigBWD as well as likely SigFWD (Table 5) remain risk factors for breast cancer fatality. Taken together, the above evidence supports an association of SigMut, SigFWD, and SigBWD with poor prognosis in patients with breast cancer.
Table 4.
Signature-associated overall survival in patients with intrinsic subtypes of breast cancer.
| Luminal | Basal-like | Claudin-low | Her2 | Normal-like | |
|---|---|---|---|---|---|
| SigMut − | 720/410 169.8 (156.3–184.3) |
24/11 | 101/40 | 50/34 | 89/39 191.9 (145–239) |
| SigMut + | 446/283 129.3 (112.4–146.2) |
185/104 | 117/54 | 174/123 | 59/40 104.7 (43–167) |
| p-Value | 0.00051⁎ | 0.267 | 0.504 | 0.530 | 0.001⁎ |
| SigFWD − | 765/434 170 (157–187) |
67/35 183 (54.8–NA) |
152/60 228 (205.6–NA) |
46/32 154 (99–245) |
94/41 198 (159–NA) |
| SigFWD + | 410/259 126 (111–151) |
142/80 112 (83.4–207) |
66/34 163 (92.8–NA) |
178/125 99 (75–142) |
54/38 100 (78–159) |
| p-Value | 3.9e − 5⁎ | 0.524 | 0.0104⁎ | 0.175 | 4.03e − 4⁎ |
| SigBWD − | 419/216 189 (169–204) |
11/4 NA (45.6–NA) |
56/20 229 (209–NA) |
12/7 199 (192–NA) |
71/28 216 (172–NA) |
| SigBWD + | 756/477 144 (130–159) |
198/111 129 (83.4-NA) |
162/74 195 (151-NA) |
212/150 101 (87.2–142) |
77/51 111 (86–159) |
| p-Value | 5.25e − 6⁎ | 0.316 | 0.0651 | 0.152 | 1.78e − 4⁎ |
Note: summary of OS data determined by Kaplan-Meier survival curve with statistical significance examined by Log-rank test. Data are presented in the form of total cases/deceased cases and median months survival (95% confidence interval); NA: not available, i.e. the up level of 95% CI could not be determined.
Shows the individual signatures are associated with a decrease in OS in the indicated BC subtypes.
Table 5.
Multivariate Cox analysis of SigMut, SigFWD, and SigBWD.
| Clin var and siga | HRb | 95% CIc | p-Value |
|---|---|---|---|
| SigMut | 1.21 | 1.02–1.43 | 0.0297⁎ |
| SigFWD | 1.17 | 0.99–1.38 | 0.062 |
| SigBWD | 1.25 | 1.03–1.52 | 0.0263⁎ |
| Age at diagnosis | 1.04d | 1.03–1.04 | 2e − 16⁎ |
| 1.04e | 1.03–1.04 | 2e − 16⁎ | |
| 1.03f | 1.03–1.04 | 2e − 16⁎ | |
| Cellularity | 1.01 | 0.9–1.12 | 0.918 |
| 1.01 | 0.9–1.13 | 0.891 | |
| 0.99 | 0.89–1.11 | 0.904 | |
| Integrat clusterg Cluster 5 |
1.63 | 1.16–2.29 | 0.0052⁎ |
| 1.69 | 1.23–2.37 | 0.0023⁎ | |
| 1.68 | 1.2–2.36 | 0.0026⁎ | |
| Neo His Gh | 0.76 | 0.63–0.93 | 0.0065⁎ |
| 0.77 | 0.64–0.94 | 0.0088⁎ | |
| 0.75 | 0.62–0.92 | 0.0049⁎ | |
| N Prog indexi | 1.40 | 1.24–1.59 | 1.30e − 7⁎ |
| 1.41 | 1.23–1.58 | 1.70e − 7⁎ | |
| 1.41 | 1.25–1.9 | 7.71e − 8⁎ | |
| Tumor size | 1.01 | 1.01–1.01 | 2.15e − 5⁎ |
| 1.01 | 1.01–1.01 | 2.23e − 5⁎ | |
| 1.01 | 1.01–1.01 | 1.24e − 5⁎ | |
| Tumor stage | 1.13 | 0.96–1.33 | 0.152 |
| 1.12 | 0.95–1.32 | 0.180 | |
| 1.11 | 0.94–1.32 | 0.208 |
a: Clinical variables and Signatures.
b: Hazard ratio.
c: Confidence interval.
d–f: In analysis with SigMut, SigFWD, and SigBWD, respectively.
g: Integrative cluster; h: neoplasm histologic grade.
i: Nottingham prognostic index.
Shows that the indicated signatures and pathological events are significantly associated with BC fatality.
3.5. SigMut, SigFWD, and SigBWD associate with poor prognosis in overlapped and different populations of patients with BC
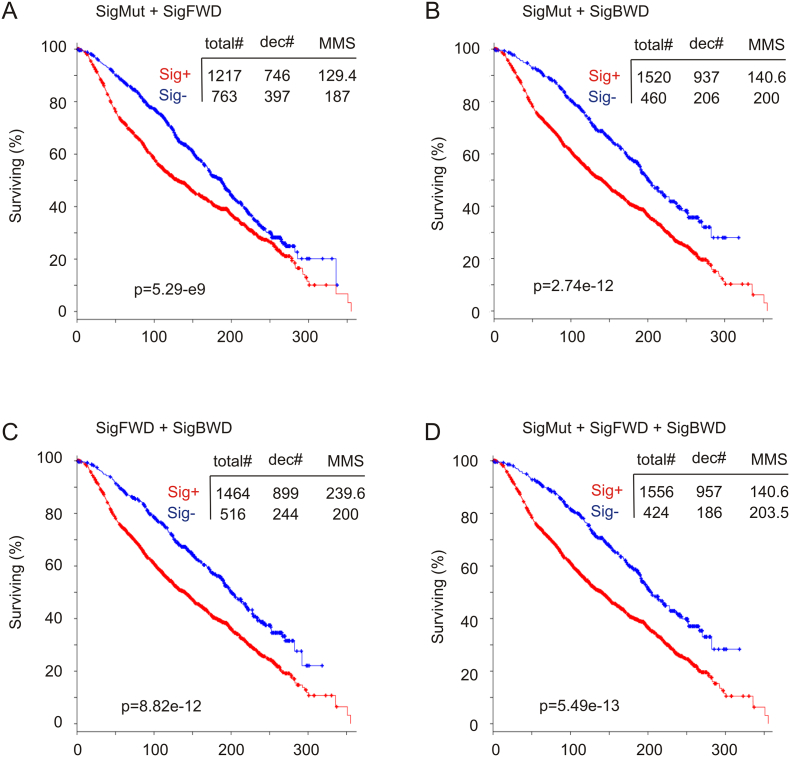
The fact that all the three signatures were derived from SHARPIN GCN increase indicates a relationship among these signatures in predicting a reduction in OS in patients with breast cancer. This concept is supported by their similarities in association with OS reductions in patients with ER + or normal-like BC but not with basal-like or Her2-enriched subtypes (Table 4). Additionally, alterations in the three signatures can be demonstrated in a core patient group (see Fig. 3 in Ref [38]). A combination of SigMut with SigFWD significantly associates with a decrease in OS in the Curtis cohort [risk cases 1217/fatality cases 746 and median months survival/MMS 129.4 months compared to non-risk cases (signature negative) 763/397, MMS 187; p = 5.29e − 9] (Fig. 5A). However, in comparison to SigFWD alone (853/538, MMS 106 vs 1127/605, MMS 181; p = 6.16e − 11) (Fig. 4B), SigMut plus SigFWD is clearly not more potent in association with the OS reductions (Fig. 5A). This suggests that both SigMut and SigFWD associate largely with an overlapped patient cohort. On the other hand, a combination of SigMut and SigBWD (Fig. 5B) is more potent than either of them alone (Figs. 1A and 4D), indicating the existence of a non-overlapped predictive value for this pair of signatures. This relationship is further supported by the combination of SigFWD and SigBWD, which results in enhancement of the association (Fig. 5C) compared to either (Fig. 4B, D). Finally, a combination of all three produced the most powerful signature examined in this manuscript (Fig. 5D). Collectively, we provide evidence supporting overlapped and independent features among SigMut, SigFWD, and SigBWD in their association with poor outcome in patients with BC.
Fig. 5.
Characterization of SigMut, SigFWD, and SigBWD-associated OS shortening in patients with breast cancer. The Curtis dataset (n = 1980) [6] was used to determine the effects of the indicated combination of SigMut, SigFWD, and SigBWD on OS in patients with breast cancer. Statistical analysis was performed using Log-rank test. Total#: total number of cases; dec#: number of deceased cases; MMS: median months survival.
4. Discussion
Since its initial identification as a Shank-binding protein in the postsynaptic density in 2001 [10], SHARPIN/SIPL1 has been emerging as a protein with multiple functions in the regulation of cell cytoskeleton [20], [21], [49], extracellular matrix structure [40], activation of NF-κB signalling [11], [12], [13], [14], as well as inhibition of PTEN- and p53-derived tumor suppression [15], [26]. Consistent with these oncogenic processes, accumulative evidence demonstrates a general tumor-promoting function of SHARPIN in multiple cancer types, including cervical cancer, hepatocellular carcinoma, ovarian cancer, prostate cancer, and breast cancer [15], [16], [20], [22], [23], [25], [26], [50]. However, the impact of SHARPIN on the outcome of patients with BC has not been thoroughly investigated, neither has the potential pathways underlying SHARPIN's influence on breast cancer prognosis.
By in silico analysis of the rich genomic alteration and gene expression data of breast cancer available in the cBioPortal database [27], [28] (http://www.cbioportal.org/index.do), we have formulated three signatures using the enriched mutation and gene expression data that are associated with SHARPIN GCN increase. While SigFWD and SigBWD predict OS reductions in patients with ER − breast cancer, all three signatures robustly correlate with OS decreases in patients with ER + BCs. The biomarker values of all three signatures in predicting OS shortening in patients with either basal-like or Her2-enriched subclasses are limited. It is thus suggested that all three signatures that were derived from SHARPIN GCN amplification have diagnostic and prognostic values in management of patients with ER + breast cancer. Among the three signatures, SigFWD is the most robust in association with OS reductions in two large and independent cohorts of patient with breast cancer (n = 1980, n = 815); its composition is also simple. SigFWD is thus appealing. However, its biomarker value needs to be further studied in comparison to other clinical biomarker sets in order to develop it for use in the clinical field. With the exception of FGD3 in SigFWD, the rest of the components have yet to be thoroughly investigated with respect to their contributions to BC tumorigenesis and progression; this will be an interesting area for future investigations.
Based on the apparent overlap between SigMut and SigFWD, as well as the observed compensation (or addictive values) between SigBWD and SigMut or SigFWD in association with OS reduction in patients with breast cancer, specific combinations among the three signatures can be envisaged, particularly the combination of all three signatures put together (Fig. 5D).
It should be stressed that although the three signatures are not able to predict OS decrease in patients with either basal-like or Her2-enriched breast tumors, the signatures are expressed in these BC subclasses at significantly higher rates compared to the luminal subtype. This implies that the imbalance in distribution between signature positive and signature negative tumors is likely a major cause for the insignificant association in these BC subtypes. On the other hand, it will be interesting to determine whether these signatures have applications in the differentiation of intrinsic subtypes of breast cancer.
The suggested mechanisms by which SHARPIN GCN increase-derived signatures associate with poor prognosis in patients with breast cancer include upregulation of multiple pathways promoting cell proliferation, such as the pathways of cell cycle progression, DNA replication and repair, as well as other processes. Intriguingly, elevations in SHARPIN GCN associate with downregulation of pathways regulating extracellular matrix. This is consistent with a recent report revealing SHARPIN's ability to facilitate breast epithelial cell-mediated invasion during mammary development by regulating extracellular matrix structure [40]. Finally, how SHARPIN regulates the above pathways in the context of breast cancer tumorigenesis and progression should be investigated in future.
Transparency document
Transparency document.
Acknowledgments
Acknowledgments
The results shown here are based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov/). D.O. is supported by a Graduate Studentship the Research Institute of St. Joe's Hamilton. This work was supported in part by a GAP funding from McMaster University and St. Joseph's Hospital in Hamilton to D.T., an award from Teresa Cascioli Charitable Foundation Research Award in Women's Health to D.T., and a grant from the Canadian Breast Cancer Foundation/the Canadian Cancer Society (grant #: 319412) to D.T.
Footnotes
The Transparency document associated with this article can be found, in online version.
References
- 1.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Perou C.M., Sorlie T., Eisen M.B., van de Rijn M., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Akslen L.A., Fluge O., Pergamenschikov A., Williams C., Zhu S.X., Lonning P.E., Borresen-Dale A.L., Brown P.O., Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Sorlie T., Perou C.M., Tibshirani R., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M.B., van de Rijn M., Jeffrey S.S., Thorsen T., Quist H., Matese J.C., Brown P.O., Botstein D., Lonning P.E., Borresen-Dale A.L. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haakensen V.D., Lingjaerde O.C., Luders T., Riis M., Prat A., Troester M.A., Holmen M.M., Frantzen J.O., Romundstad L., Navjord D., Bukholm I.K., Johannesen T.B., Perou C.M., Ursin G., Kristensen V.N., Borresen-Dale A.L., Helland A. Gene expression profiles of breast biopsies from healthy women identify a group with claudin-low features. BMC Med. Genet. 2011;4:77. doi: 10.1186/1755-8794-4-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X., Oprea-Ilies G.M., Krishnamurti U. New developments in breast cancer and their impact on daily practice in pathology. Arch. Pathol. Lab. Med. 2017;141:490–498. doi: 10.5858/arpa.2016-0288-SA. [DOI] [PubMed] [Google Scholar]
- 6.Curtis C., Shah S.P., Chin S.F., Turashvili G., Rueda O.M., Dunning M.J., Speed D., Lynch A.G., Samarajiwa S., Yuan Y., Graf S., Ha G., Haffari G., Bashashati A., Russell R., McKinney S., M. Group, Langerod A., Green A., Provenzano E., Wishart G., Pinder S., Watson P., Markowetz F., Murphy L., Ellis I., Purushotham A., Borresen-Dale A.L., Brenton J.D., Tavare S., Caldas C., Aparicio S. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.N. Cancer Genome Atlas Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis D.G., Siddiqui M.T., Oprea-Ilies G., Stevens K., Osunkoya A.O., Cohen C., Li X.B. GATA-3 and FOXA1 expression is useful to differentiate breast carcinoma from other carcinomas. Hum. Pathol. 2016;47:26–31. doi: 10.1016/j.humpath.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Dey N., Williams C., Leyland-Jones B., De P. Mutation matters in precision medicine: a future to believe in. Cancer Treat. Rev. 2017;55:136–149. doi: 10.1016/j.ctrv.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Lim S., Sala C., Yoon J., Park S., Kuroda S., Sheng M., Kim E. Sharpin, a novel postsynaptic density protein that directly interacts with the shank family of proteins. Mol. Cell. Neurosci. 2001;17:385–397. doi: 10.1006/mcne.2000.0940. [DOI] [PubMed] [Google Scholar]
- 11.Gerlach B., Cordier S.M., Schmukle A.C., Emmerich C.H., Rieser E., Haas T.L., Webb A.I., Rickard J.A., Anderton H., Wong W.W., Nachbur U., Gangoda L., Warnken U., Purcell A.W., Silke J., Walczak H. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471:591–596. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- 12.Emmerich C.H., Schmukle A.C., Walczak H. The emerging role of linear ubiquitination in cell signaling. Sci. Signal. 2011;4 doi: 10.1126/scisignal.2002187. [DOI] [PubMed] [Google Scholar]
- 13.Ikeda F., Deribe Y.L., Skanland S.S., Stieglitz B., Grabbe C., Franz-Wachtel M., van Wijk S.J., Goswami P., Nagy V., Terzic J., Tokunaga F., Androulidaki A., Nakagawa T., Pasparakis M., Iwai K., Sundberg J.P., Schaefer L., Rittinger K., Macek B., Dikic I. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature. 2011;471:637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tokunaga F., Nakagawa T., Nakahara M., Saeki Y., Taniguchi M., Sakata S., Tanaka K., Nakano H., Iwai K. SHARPIN is a component of the NF-kappaB-activating linear ubiquitin chain assembly complex. Nature. 2011;471:633–636. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- 15.He L., Ingram A., Rybak A.P., Tang D. Shank-interacting protein-like 1 promotes tumorigenesis via PTEN inhibition in human tumor cells. J. Clin. Invest. 2010;120:2094–2108. doi: 10.1172/JCI40778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Melo J., Tang D. Elevation of SIPL1 (SHARPIN) increases breast cancer risk. PLoS One. 2015;10 doi: 10.1371/journal.pone.0127546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoesel B., Schmid J.A. The complexity of NF-kappaB signaling in inflammation and cancer. Mol. Cancer. 2013;12:86. doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang Y., Sundberg J.P. SHARPIN regulates mitochondria-dependent apoptosis in keratinocytes. J. Dermatol. Sci. 2011;63:148–153. doi: 10.1016/j.jdermsci.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sieber S., Lange N., Kollmorgen G., Erhardt A., Quaas A., Gontarewicz A., Sass G., Tiegs G., Kreienkamp H.J. Sharpin contributes to TNFalpha dependent NFkappaB activation and anti-apoptotic signalling in hepatocytes. PLoS One. 2012;7 doi: 10.1371/journal.pone.0029993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung J., Kim J.M., Park B., Cheon Y., Lee B., Choo S.H., Koh S.S., Lee S. Newly identified tumor-associated role of human Sharpin. Mol. Cell. Biochem. 2010;340:161–167. doi: 10.1007/s11010-010-0413-x. [DOI] [PubMed] [Google Scholar]
- 21.Pouwels J., De Franceschi N., Rantakari P., Auvinen K., Karikoski M., Mattila E., Potter C., Sundberg J.P., Hogg N., Gahmberg C.G., Salmi M., Ivaska J. SHARPIN regulates uropod detachment in migrating lymphocytes. Cell Rep. 2013;5:619–628. doi: 10.1016/j.celrep.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka Y., Tateishi K., Nakatsuka T., Kudo Y., Takahashi R., Miyabayashi K., Yamamoto K., Asaoka Y., Ijichi H., Tateishi R., Shibahara J., Fukayama M., Ishizawa T., Hasegawa K., Kokudo N., Koike K. Sharpin promotes hepatocellular carcinoma progression via transactivation of Versican expression. Oncogene. 2016;5 doi: 10.1038/oncsis.2016.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang H., Du T., Zhang Y., Lai Y., Li K., Fan X., Zhu D., Lin T., Xu K., Huang J., Liu L., Guo Z. Elevation of SHARPIN protein levels in prostate adenocarcinomas promotes metastasis and impairs patient survivals. Prostate. 2017;77:718–728. doi: 10.1002/pros.23302. [DOI] [PubMed] [Google Scholar]
- 24.Ojo D., Wei F., Liu Y., Wang E., Zhang H., Lin X., Wong N., Bane A., Tang D. Factors promoting tamoxifen resistance in breast cancer via stimulating breast cancer stem cell expansion. Curr. Med. Chem. 2015;22:2360–2374. doi: 10.2174/0929867322666150416095744. [DOI] [PubMed] [Google Scholar]
- 25.Bii V.M., Rae D.T., Trobridge G.D. A novel gammaretroviral shuttle vector insertional mutagenesis screen identifies SHARPIN as a breast cancer metastasis gene and prognostic biomarker. Oncotarget. 2015;6:39507–39520. doi: 10.18632/oncotarget.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang H., Yu S., Wang W., Li X., Hou Y., Liu Z., Shi Y., Mu K., Niu G., Xu J., Wang H., Zhu J., Zhuang T. SHARPIN facilitates p53 degradation in breast cancer cells. Neoplasia. 2017;19:84–92. doi: 10.1016/j.neo.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., Antipin Y., Reva B., Goldberg A.P., Sander C., Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6 doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciriello G., Gatza M.L., Beck A.H., Wilkerson M.D., Rhie S.K., Pastore A., Zhang H., McLellan M., Yau C., Kandoth C., Bowlby R., Shen H., Hayat S., Fieldhouse R., Lester S.C., Tse G.M., Factor R.E., Collins L.C., Allison K.H., Chen Y.Y., Jensen K., Johnson N.B., Oesterreich S., Mills G.B., Cherniack A.D., Robertson G., Benz C., Sander C., Laird P.W., Hoadley K.A., King T.A., Network T.R., Perou C.M. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo W., Friedman M.S., Shedden K., Hankenson K.D., Woolf P.J. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu G., He Q.Y. ReactomePA: an R/bioconductor package for reactome pathway analysis and visualization. Mol. BioSyst. 2016;12:477–479. doi: 10.1039/c5mb00663e. [DOI] [PubMed] [Google Scholar]
- 32.Kallioniemi A., Kallioniemi O.P., Piper J., Tanner M., Stokke T., Chen L., Smith H.S., Pinkel D., Gray J.W., Waldman F.M. Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc. Natl. Acad. Sci. U. S. A. 1994;91:2156–2160. doi: 10.1073/pnas.91.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollack J.R., Perou C.M., Alizadeh A.A., Eisen M.B., Pergamenschikov A., Williams C.F., Jeffrey S.S., Botstein D., Brown P.O. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat. Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 34.Pollack J.R., Sorlie T., Perou C.M., Rees C.A., Jeffrey S.S., Lonning P.E., Tibshirani R., Botstein D., Borresen-Dale A.L., Brown P.O. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rennstam K., Ahlstedt-Soini M., Baldetorp B., Bendahl P.O., Borg A., Karhu R., Tanner M., Tirkkonen M., Isola J. Patterns of chromosomal imbalances defines subgroups of breast cancer with distinct clinical features and prognosis. A study of 305 tumors by comparative genomic hybridization. Cancer Res. 2003;63:8861–8868. [PubMed] [Google Scholar]
- 36.Seute A., Sinn H.P., Schlenk R.F., Emig R., Wallwiener D., Grischke E.M., Hohaus S., Dohner H., Haas R., Bentz M. Clinical relevance of genomic aberrations in homogeneously treated high-risk stage II/III breast cancer patients. Int. J. Cancer. 2001;93:80–84. doi: 10.1002/ijc.1296. [DOI] [PubMed] [Google Scholar]
- 37.Tirkkonen M., Tanner M., Karhu R., Kallioniemi A., Isola J., Kallioniemi O.P. Molecular cytogenetics of primary breast cancer by CGH. Genes Chromosomes Cancer. 1998;21:177–184. [PubMed] [Google Scholar]
- 38.Ojo D., Seliman M., Tang D. 2017. Detail of the signatures (SigMut, SigFWD, and SigBWD) derived from increase in SHARPIN gene copy number in breast cancer. Data in Brief, Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pereira B., Chin S.F., Rueda O.M., Vollan H.K., Provenzano E., Bardwell H.A., Pugh M., Jones L., Russell R., Sammut S.J., Tsui D.W., Liu B., Dawson S.J., Abraham J., Northen H., Peden J.F., Mukherjee A., Turashvili G., Green A.R., McKinney S., Oloumi A., Shah S., Rosenfeld N., Murphy L., Bentley D.R., Ellis I.O., Purushotham A., Pinder S.E., Borresen-Dale A.L., Earl H.M., Pharoah P.D., Ross M.T., Aparicio S., Caldas C. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peuhu E., Kaukonen R., Lerche M., Saari M., Guzman C., Rantakari P., De Franceschi N., Warri A., Georgiadou M., Jacquemet G., Mattila E., Virtakoivu R., Liu Y., Attieh Y., Silva K.A., Betz T., Sundberg J.P., Salmi M., Deugnier M.A., Eliceiri K.W., Ivaska J. SHARPIN regulates collagen architecture and ductal outgrowth in the developing mouse mammary gland. EMBO J. 2017;36:165–182. doi: 10.15252/embj.201694387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang T.H. Ou, Cheng W.Y., Zheng T., Maurer M.A., Anastassiou D. Breast cancer prognostic biomarker using attractor metagenes and the FGD3-SUSD3 metagene. Cancer Epidemiol. Biomark. Prev. 2014;23:2850–2856. doi: 10.1158/1055-9965.EPI-14-0399. [DOI] [PubMed] [Google Scholar]
- 42.Armes J.E., Hammet F., de Silva M., Ciciulla J., Ramus S.J., Soo W.K., Mahoney A., Yarovaya N., Henderson M.A., Gish K., Hutchins A.M., Price G.R., Venter D.J. Candidate tumor-suppressor genes on chromosome arm 8p in early-onset and high-grade breast cancers. Oncogene. 2004;23:5697–5702. doi: 10.1038/sj.onc.1207740. [DOI] [PubMed] [Google Scholar]
- 43.Ting C.H., Chen Y.C., Wu C.J., Chen J.Y. Targeting FOSB with a cationic antimicrobial peptide, TP4, for treatment of triple-negative breast cancer. Oncotarget. 2016;7:40329–40347. doi: 10.18632/oncotarget.9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoareau-Aveilla C., Fayet-Lebaron E., Jady B.E., Henras A.K., Kiss T. Utp23p is required for dissociation of snR30 small nucleolar RNP from preribosomal particles. Nucleic Acids Res. 2012;40:3641–3652. doi: 10.1093/nar/gkr1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hutter C.M., Chang-Claude J., Slattery M.L., Pflugeisen B.M., Lin Y., Duggan D., Nan H., Lemire M., Rangrej J., Figueiredo J.C., Jiao S., Harrison T.A., Liu Y., Chen L.S., Stelling D.L., Warnick G.S., Hoffmeister M., Kury S., Fuchs C.S., Giovannucci E., Hazra A., Kraft P., Hunter D.J., Gallinger S., Zanke B.W., Brenner H., Frank B., Ma J., Ulrich C.M., White E., Newcomb P.A., Kooperberg C., LaCroix A.Z., Prentice R.L., Jackson R.D., Schoen R.E., Chanock S.J., Berndt S.I., Hayes R.B., Caan B.J., Potter J.D., Hsu L., Bezieau S., Chan A.T., Hudson T.J., Peters U. Characterization of gene-environment interactions for colorectal cancer susceptibility loci. Cancer Res. 2012;72:2036–2044. doi: 10.1158/0008-5472.CAN-11-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun Y., Liu X., Zhang Q., Mao X., Feng L., Su P., Chen H., Guo Y., Jin F. Oncogenic potential of TSTA3 in breast cancer and its regulation by the tumor suppressors miR-125a-5p and miR-125b. Tumour Biol. 2016;37:4963–4972. doi: 10.1007/s13277-015-4178-4. [DOI] [PubMed] [Google Scholar]
- 47.Lal M.A., Andersson A.C., Katayama K., Xiao Z., Nukui M., Hultenby K., Wernerson A., Tryggvason K. Rhophilin-1 is a key regulator of the podocyte cytoskeleton and is essential for glomerular filtration. J. Am. Soc. Nephrol. 2015;26:647–662. doi: 10.1681/ASN.2013111195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yersal O., Barutca S. Biological subtypes of breast cancer: prognostic and therapeutic implications. World J. Clin. Oncol. 2014;5:412–424. doi: 10.5306/wjco.v5.i3.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rantala J.K., Pouwels J., Pellinen T., Veltel S., Laasola P., Mattila E., Potter C.S., Duffy T., Sundberg J.P., Kallioniemi O., Askari J.A., Humphries M.J., Parsons M., Salmi M., Ivaska J. SHARPIN is an endogenous inhibitor of beta1-integrin activation. Nat. Cell Biol. 2011;13:1315–1324. doi: 10.1038/ncb2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li J., Lai Y., Cao Y., Du T., Zeng L., Wang G., Chen X., Chen J., Yu Y., Zhang S., Zhang Y., Huang H., Guo Z. SHARPIN overexpression induces tumorigenesis in human prostate cancer LNCaP, DU145 and PC-3 cells via NF-kappaB/ERK/Akt signaling pathway. Med. Oncol. 2015;32:444. doi: 10.1007/s12032-014-0444-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document.