Abstract
Genetic variation within the transcription factor TCF4 locus can cause the intellectual disability and developmental disorder Pitt-Hopkins syndrome (PTHS), whereas single-nucleotide polymorphisms within noncoding regions are associated with schizophrenia. These genetic findings position TCF4 as a link between transcription and cognition; however, the neurobiology of TCF4 remains poorly understood. Here, we quantitated multiple distinct TCF4 transcript levels in human induced pluripotent stem cell-derived neural progenitors and differentiated neurons, and PTHS patient fibroblasts. We identify two classes of pharmacological treatments that regulate TCF4 expression: WNT pathway activation and inhibition of class I histone deacetylases. In PTHS fibroblasts, both of these perturbations upregulate a subset of TCF4 transcripts. Finally, using chromatin immunoprecipitation sequencing in conjunction with genome-wide transcriptome analysis, we identified TCF4 target genes that may mediate the effect of TCF4 loss on neuroplasticity. Our studies identify new pharmacological assays, tools, and targets for the development of therapeutics for cognitive disorders.
Keywords: Pitt-Hopkins syndrome, Schizophrenia, TCF4, WNT, Epigenetics, Histone deacetylase, Drug discovery
Introduction
The TCF4 gene (Entrez GeneID: 6925) at 18q21.2 is a complex locus with multiple promoters encoding multiple alternatively spliced isoforms of a member of the basic-helix loop helix (bHLH) transcription factor family involved in the development of multiple cell types throughout the human body, including neurons and glia in the brain [1]. In the brain, TCF4 has the potential to regulate transcription of target genes as a homodimer [2], or as a heterodimer with key bHLH proneural transcription factors, such as ATOH [3], ASCL1 [4], and NEUROD2 [5]. The critical roles played by TCF4 in the brain are underscored by the fact that exonic mutations of TCF4 cause the severe neurodevelopmental intellectual disability disorder Pitt-Hopkins syndrome (PTHS) [6, 7, 8], while common genetic variants and chromosomal abnormalities at the TCF4 locus confer risk for schizophrenia [9] and autism spectrum disorder [10, 11].
In animal models, knockdown or genetic ablation of TCF4 in vivo has been reported to result in specific defects in zebrafish embryo brain and eye differentiation [12], defects in the formation of synapses and proper neurite formation in Drosophila[13], altered mouse hindbrain development [3] and gastrointestinal function [14], and derepression of ion channel genes (e.g., KCNQ1 and SCN10A) that are critical for regulating intrinsic neuronal excitability in rat prefrontal neurons [15]. Conversely, the overexpression of TCF4 in mice causes cognitive and sensorimotor gating impairments [16]. Clearly, the expression and activity of TCF4 is critical for important aspects of development and function of the nervous system and other tissues.
Despite its importance to multiple aspects of nervous system function, significant gaps remain in our knowledge of the pathological consequences of genetic variation or mutations in TCF4 underlying neuropsychiatric disorders. While TCF4 haploinsufficiency appears to be the primary cause of PTHS [6, 7, 8], hypomorphic or dominant negative effects may occur in some patients [17]. Recent findings have further emphasized the complexity of the TCF4 locus with the identification of certain mutations that disrupt only a subset of the TCF4 isoforms, and are linked to intellectual disability rather than classical PTHS [18, 19]. In schizophrenia, risk-associated genetic variation at the TCF4 locus is intronic [9], presumably occurring in elements that control TCF4 gene expression, although specific expression quantitative trail loci or splicing or stability effects by a disease- associated variant have not yet been described. On the basis of these observations, translational efforts for TCF4-associated neuropsychiatric disorders will require precise delineation of brain cell-type specific TCF4 gene expression patterns, identification of genetic elements controlling these expression patterns, and of particular importance, determination of biological signaling pathways that can be manipulated pharmacologically to alter TCF4 gene expression in a therapeutically relevant manner in the central nervous system (CNS) and other tissues.
Evidence from studies in nonneuronal cancer cells suggests that TCF4 expression can be regulated by a major signaling pathway involved in brain development - the WNT (Wingless-related integration site)/β-catenin pathway [20]. In cancer cells, overexpression of the WNT signaling effector β-catenin has been shown to upregulate TCF4 [21]. Furthermore, chromatin remodeling complexes known to interact with β-catenin can bind to a TCF4 promoter in cancer cells [22]. Importantly, WNT pathway dysregulation may be a component of neuropsychiatric disease pathology, as genetic variation at WNT signaling gene loci has been associated with both schizophrenia and neurodevelopmental disorders [9, 23]. However, WNT pathway regulation of TCF4 expression in human neural progenitor cells (NPCs) and postmitotic neurons has not yet been studied and thus represents a critical question to address in order to delineate mechanisms of TCF4 regulation in the CNS.
Here, we present a detailed study of the expression of TCF4 mRNA and protein in human induced pluripotent stem cell (iPSC)-derived NPCs and differentiated, postmitotic neurons. We report on regulation of TCF4 mRNA and protein expression in NPC by pharmacological agents that alter the activity of the WNT signaling pathway. We also find that epigenetic perturbation caused by histone deacetylase (HDAC) inhibitors can enhance TCF4 gene expression. Lastly, we show that both WNT signaling activators and HDAC inhibition can produce potentially therapeutically relevant upregulation of TCF4 gene expression in fibroblasts derived from PTHS patients.
Materials and Methods
Human Cell Culture and Treatments
Human iPSC-derived Nestin and SOX1-expressing NPCs from a de-identified, healthy control individual 8330-8 [24] were cultured in neural stem (NS) media: 70% DMEM (Dulbecco's modified Eagle's Medium, High Glucose 1×, Gibco 11995), 30% Ham's F12 with L-glutamine (Modified, Cellgro/Mediatech #10-080-CV), 1× penicillin/streptomycin, 1× B27 Supplement (50×, Gibco #17504-044) supplemented with 20 ng/mL EGF (Epidermal Growth Factor, Sigma, #E9644) prepared as 20 μg/mL stock in DMEM, 20 ng/mL bFGF (basic Fibroblast Growth Factor, Stemgent #03-0002, prepared as 20 μg/mL stock in PBS) and 5 μg/mL heparin (Sigma, #H3149) prepared as 5 mg/mL stock in Ham's F12 media just before use as described [25]. NPCs were grown for experimental treatments on poly-ornithine (Sigma #P3655) and laminin (Sigma #L-2020)-coated 6-well plates to ∼80% confluency. Differentiation of human NPCs was induced by removal of growth factors (EGF, FGF, heparin) from the culture media. Cells were rinsed with 2 mL media, and then 2 mL media without growth factors was added. Media was changed every ∼3 days, and cells were allowed to differentiate for 30 days. Cell cultures were treated for 24 h with compounds at the indicated concentrations, or Wnt3a-conditioned media at the 10% concentration (Wnt3a-CM). Wnt3a-CM was harvested from a mouse L-cell line that secretes mouse Wnt3a protein as described [25]. Human fibroblasts were cultured in 6-well plates (Falcon #353046) in DMEM (Gibco #11965-092) supplemented with 10% FBS (BenchMark #100–106), 2 mML-glutamine (Gibco #25030-081), and 1× penicillin/streptomycin (Gibco #15140-122). Medium was changed every 3 days until fibroblasts reached ∼80% confluency. Fibroblasts were then treated with compound for 24 h prior to harvesting. Human fibroblasts from healthy controls and PTHS participants were collected for experimentation with informed consent covered under IRB-approved protocols at University of Texas, Health Sciences Center (HSC20000318H) and Massachusetts General Hospital/Partners HealthCare (2009-P-001142).
TaqMan Real-Time PCR (qPCR) Assays
Online supplementary Table 1 (for all online suppl. material, see www.karger.com/doi/10.1159/000475666) lists TCF4 TaqMan primer/probe sequences used in qPCR assays. cDNA from human 8330-8 fibroblasts or NPCs differentiated for 0, 3, and 6 weeks upon growth factor withdrawal was used to first validate that each of qPCR primer/probe set generated PCR products at the expected sizes based on the reference genome build (online suppl. Fig. 1). For human NPC and fibroblast assays, total RNA was harvested using the RNeasy kit (Qiagen), and first strand cDNA was synthesized with the High Capacity cDNA Reverse Transcriptase kit (Applied Biosystems #4374966). TaqMan qPCR assays were run according to the manufacturer's protocols (Life Technologies) on a Roche LightCycler 480 II (software version 1.5.1).
RNAi Knockdown of TCF4 and Microarray Gene Expression Analysis
Short-hairpin RNAs targeting several sequences (TCF4 RNAi #1-ATTCATAACTACTCAGACTTG; TCF4 RNAi #3-CGACAAGAAGGATATCAAA; TCF4 RNAi #4-GAGACTGAACGGCAATCTTTC; TCF4 RNAi#7-GAAAGGAATCTGAATCCGAAA) within the 3′ UTR or C-terminal exons common to all TCF4 transcripts, or control sequences (RFP RNAi #5-CTCAGTTCCAGTACGGCTCCA; LacZ RNAi #6-TCGTATTACAACGTCGTGACT; LUC RNAi #8-ACGCTGAGTACTTCGAAATGT; GFP RNAi #9-ACAACAGCCACAACGTCTATA), were obtained from the Broad Institute RNAi Consortium and/or cloned into the lentiviral vector pLK0.1 (http://www.broadinstitute.org/rnai/public/vector/details?vector=pLKO.1) and packaged into lentivirus particles by cotransfection with packaging plasmids pCMV-dR8.2 dvpr (Addgene plasmid #8455; gift from Dr. Bob Weinberg) and pMD2.G (Addgene plasmid #12259; gift from Dr. Didier Trono) into HEK293 cells exactly as described in Wang et al. [26]. Lentivirus particles were concentrated by ultracentrifugation (19,500 rpm for 2 h at 4°C in a Beckman SW32Ti rotor) through an underlay of 3 mL 20% sucrose (in PBS), and resuspended in PBS. Human NPCs were infected with lenti-shRNAs (M.O.I. = 10) with puromycin (Gibco #A11138-03) at a final concentration of 0.8 µg/mL added to cultures 24 h after infection. Following two passages after puromycin selection, cells were harvested for TCF4 mRNA analysis or Western blot analysis with anti-TCF4 antibody (Santa Cruz SC-101095).
Immunofluorescence
Cells were grown and differentiated in 6-well plates, fixed for 20 min in 4% formaldehyde (Tousimis #1008A), blocked/permeabilized 30 min, incubated in primary antibodies (diluted in blocking/permeabilization buffer: 1% BSA [Sigma #A7906], 0.05% Tween 20 [Fisher Scientific #BP337], 2% goat serum [Invitrogen #16210-072], 0.1% gelatin [Sigma #G1890], 0.1% Triton X-100 [Fisher Scientific #BP150], in PBS [MediaTech #21-040-CV] with pH 7.4) overnight at 4°C, washed 2× with 4 mL PBS, incubated in secondary antibodies (Alexa Fluor-488 goat anti-mouse [ThermoFisher #A-11001], Alexa Fluor-594 goat anti-chicken, [ThermoFisher #A-11042] and Hoechst 33342 [Life Technologies #H3570]) for 2.5 h at room temperature, washed 2× with 4 mL PBS, and imaged on a Zeiss Observer.Z1 fluorescence microscope with a ×10 objective. Primary antibodies were: MAP2 (EnCor Biotechnology #CPCA-MAP2, 1:5,000) and SMI312R (Covance #39422, 1:1,000).
Western Blotting
SDS-PAGE was performed using 7% Tris-Acetate gels (BioRad #345-0137). Blots were blocked for 1 h in 5% milk (LabScientific #732-291-1940) in TBS-Tween 20 at room temperature. Primary antibodies were diluted in 5% BSA in TBS-Tween 20, and incubated overnight at 4°C. After washing 4× with TBS-Tween 20, blots were incubated with horseradish peroxidase-conjugated secondary antibodies from Cell Signaling Technology (mouse #7076, rabbit #7074) both at 1:2,000 in 5% milk in TBS-Tween 20 for 1 h. After washing 4× with TBS-Tween 20, enhanced chemiluminescence kits (Pierce ECL Western Blot Substrate, catalog No. 32106; or Pierce Super Signal West Dura Extended Duration Substrate, catalog No. 34076, as needed) were used to detect and capture Western blot signals on photographic film (Kodak Biomax MR) processed on a Kodak X-OMAT 2000A developer. A Bio-Rad GS800 Calibrated Densitometer was used to digitize 16-bit, 400-dpi blot images. Image J (version 1.49v) was used to quantify band mean pixel intensity following background subtraction; background mean pixel intensity was the mean of three measurements from appropriate areas of the blot that had only background signal. Primary antibodies were: PSD-95 (NeuroMab clone K28/43 #75-028, 1:1,000), Synapsin1 (Synaptic Systems #106-001, 1:2,000), synaptophysin (Sigma #S5768, 1:500), actin (Sigma #A1978, 1:10,000), tubulin (Sigma #T6199, 1:10,000), and TCF4 (Santa Cruz Biotechnology #sc-101095, 1:500).
WNT Pathway Reporter Gene Assay
Human NPC lines engineered to express a WNT pathway reporter gene were described in detail in a previous publication [25]. Preparation of culture plasticware and media, expansion of NPCs, and execution of assays followed procedures exactly as previously described [25]. For assays, cells of the reporter NPCs were plated on precoated 384-well plates (Corning #3707) at 6,000 cells per well in 30 μL culture media; 24 h later, 10 μL of media or Wnt3a-conditioned media (Wnt3a-CM) was dispensed into each well for media-only or Wnt3a-CM (5% final concentration) conditions, which were followed by compound treatment. After 24 h of treatment, cells were lysed by dispensing 15 μL of SteadyGlo reagent (Promega E2550) and read for luminescence on an EnVision multilabel plate reader (PerkinElmer). Activities of WNT signaling reporter gene stimulation are expressed as fold change over DMSO-treated samples. Statistical comparisons across different stimuli effects on WNT reporter gene activity were performed by one-way ANOVA followed by Dunnett's multiple comparisons test comparing all stimuli with DMSO alone controls.
Transcriptome Analysis
For analysis of genome-wide gene expression changes induced by TCF4 RNAi, two stable human NPC lines expressing shRNAs (control RFP#5 and TCF4 RNAi#3) were differentiated by growing NPCs to confluency (∼1.2 million cells/well) in 6-well plates, and then replacing media with fresh NS media lacking EGF/FGF, and maintaining the cells in this media for 6 weeks, with media changes every 3–4 days. Following differentiation, total RNA was harvested using the RNeasy kit (Qiagen 74106) and the levels of TCF4 knockdown were first validated using a TaqMan assay for total TCF4 transcripts. RNA quality was assessed on an Agilent 2,100 Bioanalyzer with all samples having RIN scores >7.0. Total RNA was then analyzed using the HumanHT-12 v4 Expression BeadChip containing more than 47,000 probes derived from NCBI RefSeq Release 38 and other sources at the IDDRC Microarray Facility at Boston Children's Hospital using standard operating procedures from the manufacturer. A total of n = 3 biological replicates were analyzed for each of the control and TCF4 RNAi treatments. Raw data were annotated with Genome Studio (Illumina), and then quantile normalized and baseline transformed to the median of the control samples using GeneSpring GX software version 12 (Agilent). Probes failing to score as “present” in 100% of samples from at least one condition (control or TCF4 knockdown) were removed. A statistical threshold (p < 0.05, paired t test with Benjamini-Hochberg multiple comparisons correction) and a fold change criterion (≥1.3) were then applied to generate lists of genes up- and downregulated by TCF4 knockdown (online suppl. Table 4).
TCF4 Isoform 10 Chromatin Immunoprecipitation Sequencing
TCF4 transcript 10 (NCBI RefSeq NM_001243234.1), including an N-terminal 3X-Flag tag (DYKDHDGDYKDHDIDYKDDDDK), was cloned into the NheI-NotI sites of the lentivirus expression vector pCDH1 (System Biosciences catalog No. CD532A-2). pCDH1-3X-Flag-TCF4-transcript 10 was packaged into lentivirus particles by co-transfection into HEK293 cells with the packaging plasmids pCMV-dR8.2 dvpr (Addgene plasmid #8455; gift from Dr. Bob Weinberg) and pMD2.G (Addgene plasmid #12259; gift from Dr. Didier Trono). Lentivirus particles were concentrated by ultracentrifugation (19,500 rpm for 2 h at 4°C in a Beckman SW32Ti rotor) through an underlay of 3 mL 20% sucrose (in PBS), and resuspended in PBS. Concentrated lentivirus was used to infect human iPSC-derived 8330-8 NPCs cultured as described in Sheridan et al. [24]. In brief, NPC were plated at a density of 160,000 cells/well on poly-ornithine + laminin-coated 12-well plates in NS media (30% Ham's F12 media, 70% DMEM, 2% B27 supplement, 20 ng/ml EGF, 20 ng/ml bFGF, 5 mg/ml heparin, and 1× penicillin/streptomycin). Cells were infected with 20 µL of concentrated lentivirus preparation, and the next day, media were replaced with fresh media containing puromycin (Gibco #A11138-03) (0.8 µg/mL). Puromycin-selected cells were expanded, and Flag-TCF4-isoform 10 expression was verified by Western blotting with anti-Flag M2 antibody (Sigma #F1804). Neuronal differentiation was initiated in NPCs grown to confluency (∼20 million cells/dish) in 15-cm dishes (Falcon #353025) by replacing media with fresh NS media lacking EGF and FGF, and maintaining the cells in this media for 30 days, with media changes every 3–4 days. Chromatin immunoprecipitation (ChIP) using Flag antibody was performed as described by Bernstein's group in 2005 with the following exceptions: 40 million neurons (from two 15-cm plates combined) were used per each IP. Formaldehyde (Tousimis #1008A) cross-linking was quenched by adding 1.1 mL 2.5 M glycine (Sigma #G8898); incubating 5 min at RT with rotation, and washing with ice-cold PBS. Based on a method successfully employed for ChIP sequencing (ChIP-seq) for the related bHLH transcription factor ATOH1 [27], following sonication, SDS (Sigma #L6026) was removed from lysates with the SDS-Out SDS Precipitation kit (Thermo-Fisher Scientific #20308) following the manufacturer's instructions. A total of 5 ng of ChIP DNA or input DNA were used to generate libraries for next generation sequencing using the NEBNext Ultra DNA Library Prep Kit (NEB catalog #E7370). Libraries were sequenced with a paired-end 75-bp read protocol on an Illumina HiSeq 2,500 machine at the Broad Institute Genomics Platform (https://www.broadinstitute.org/scientific-community/science/platforms/genomics/genomics-platform), generating an average of 31.2 million paired-end reads (62.4 million total reads). Quality checking of raw sequence reads was performed by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). All reads passed our criteria, as FastQC revealed that the lower quartile of per-base quality across the reads in all the fastq files were above 20 on the Phred scale. Sequence reads were next aligned to the Ensembl GRCh37 (version 71) human reference genome using the BWA-MEM alignment algorithm (version 0.7.5a-r418) at its default settings [28]. Alignments were then coordinate sorted, flagged for PCR duplicates, and indexed using PicardTools (version 1.95) (https://github.com/broadinstitute/picard). Uniquely mapped reads were extracted using SAMtools suite (version 0.1.18) [29]. These uniquely mapped reads (with duplicates removed) were used to detect TCF4 isoform 10 ChIP peaks with the “callpeak” function of MACS v2.1.0 (https://github.com/taoliu/MACS/), using input DNA reads as the control with a false discovery rate q-value threshold of q <0.05 (read file format = bam); 70 peaks in total were called. Peaks arising in regions of high background (∼50- to 100-fold above baseline signal) apparent in Input DNA samples were removed, leaving a total of 37 “high confidence” peaks. Genes containing, or near to, the peaks were identified by visual inspection of peak locations relative to GENCODE comprehensive genome annotation of human genome build hg19 using the UCSC genome browser (https://genome.ucsc.edu/index.html). Some peaks were intergenic (i.e., >50 kb from any transcript); in these cases, we identified the closest gene. Finally, we determined that none of the resulting “high confidence” list of peaks fell in common ChIP-seq contaminant nonspecific background regions defined as the ENCODE “blacklist” (https://sites.google.com/site/anshulkundaje/projects/blacklists). Finally, TCF4 ChIP-seq peak sequences were analyzed by MEME-ChIP (http://meme-suite.org/tools/meme-chip), using default settings, with the exception of allowing detection of an unlimited number of motifs, to identify ab initio novel DNA sequence motifs enriched in these peaks. The list of genes containing or nearest to TCF4 isoform 10 ChIP peaks was compared with the list of genes regulated by TCF4 RNAi in 8330-8 human NPC-derived neurons to identify genes that were hits in both assays.
Results
Multiple protein products are encoded by the TCF4 gene, arising from transcription initiation at multiple promoters, as well as alternative splicing [30]. Currently, the NCBI lists 12 RefSeq TCF4 transcripts (http://www.ncbi.nlm.nih.gov/gene/6925); UCSC Genes (http://genome.ucsc.edu/) and Ensembl (http://www.ensembl.org) list 20 and 48 transcripts, respectively. An alignment of the RefSeq transcripts with functional domain annotation is shown in Figure 1a and online supplementary Figure 1. All RefSeq transcripts contain the C-terminal exons that encode the bHLH DNA binding and dimerization domains. The transcripts differ in whether they contain N-terminal exons encoding transcriptional regulatory domains that have been defined as activating or repressing in reporter gene studies [30, 31]. While most of these transcripts have been detected in human brain [30], a detailed quantitation of their expression levels in neuronal precursor cells and neurons has not yet been reported.
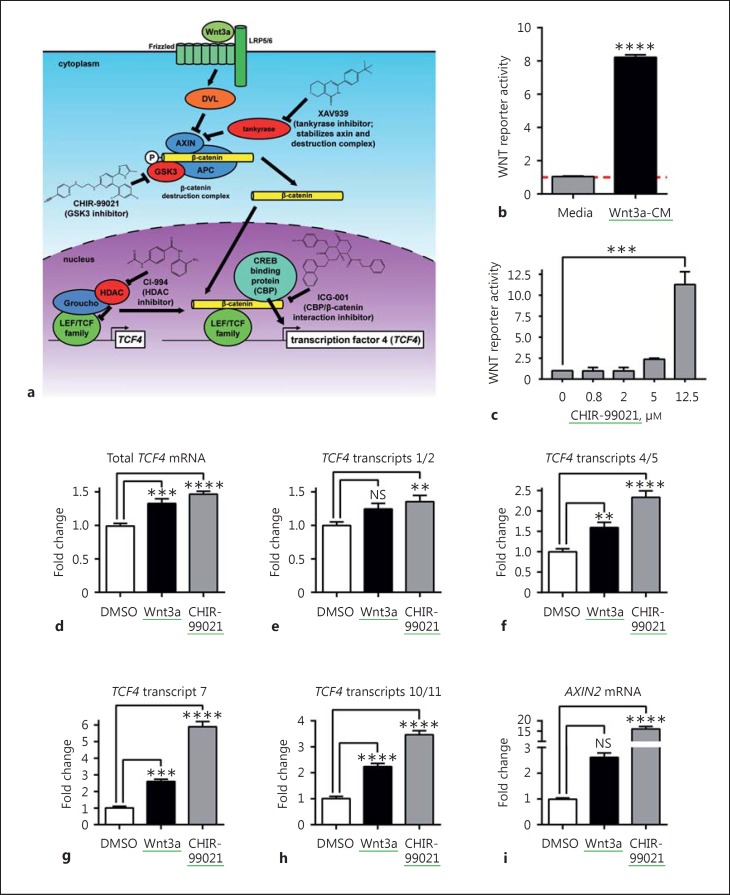
Fig. 1.
Specific TCF4 transcript isoforms are upregulated upon differentiation of human NPCs into neurons. a Exon structure of the human TCF4 gene and NCBI RefSeq transcripts. Gene structure is annotated according to human genome build hg19. Exons are shown as uniquely colored horizontal bars. Wider portions of bars indicate coding regions; narrower regions of bars indicate 5′ and 3′ UTRs. Locations of primer binding sites for TaqMan assays for individual transcripts or sets of transcripts are indicated as vertical black bars connected by a horizontal line. b Immunofluorescent staining of dendrites and axons in neurons differentiated for 3 weeks. Scale bar indicates 100 µm. c Western blot showing emergence of synaptic protein expression following 3–6 weeks of neuronal differentiation. d-i Regulation of the indicated TCF4 transcript levels during differentiation of human NPCs into neurons with n = 3 replicates per time point. One-way ANOVA followed by Dunnett's multiple comparisons test was performed. *, **, ***, and **** indicate p < 0.05, 0.01, 0.001, and 0.0001, respectively, for comparisons of transcript levels in neurons vs. NPCs. NS indicates not statistically significant. Error bars display standard error of the mean.
To enable detection of individual TCF4 transcripts, or subsets of transcripts, wherever possible given the limited amount of individual transcript-specific stretches of sequence, we designed quantitative real time-PCR (qPCR) TaqMan assays for the 12 NCBI RefSeq TCF4 transcripts (based on human genome build GRCh37/hg19; we were unable to identify sufficient transcript-specific sequence to develop real-time PCR assays for transcripts 6 and 9). In human NPC and 3- and 6-week differentiated neuron cDNA libraries, all of these qPCR primer/probe sets generated PCR products at the expected sizes based on the reference genome build (online suppl. Fig. 2), except for transcript 3, which was not detectable by our assay. In addition, the qPCR assay for TCF4 transcripts 1/2 also generated an additional band that corresponds to a size expected for a novel TCF4 transcript annotated in Gencode (Comprehensive Gene Annotation Set version 19) as ENST00000565908.2. Besides these TCF4 transcript-specific assays, we also utilized a TaqMan qPCR assay that detects mRNA sequence common to all 12 TCF4 RefSeq transcripts (total TCF4 mRNA levels).
After validating the panel of mRNA expression assays in human NPC and neurons, we next determined expression levels of total and the specific TCF4 transcripts in human iPSC-derived NPCs while in a proliferative, self-renewing state, as well as over a 6-week neurodevelopmental time course initiated by the removal of EGF/FGF growth factors that activates an intrinsic program of neuronal differentiation into postmitotic neurons [24]. After an initial 3 weeks of differentiation, the human NPCs had developed a highly branched, neuronal morphology and expressed dendritic and axonal markers (MAP2 and SMI312, respectively; Fig. 1b). Over the course of 6 weeks of differentiation, the neurons also initiated expression of synaptic proteins (Fig. 1c). We initially used our panel of TCF4 transcript-specific TaqMan assays to detect the effect of neuronal differentiation on TCF4 transcript expression in human NPCs. As summarized in Figure 1d, e, f, g, h, i, while neuronal differentiation did not produce a change in the total levels of TCF4 mRNA, use of the TCF4 transcript-specific mRNA expression assays revealed that specific TCF4 transcripts were in fact significantly changed. Most notably, TCF4 transcripts 1/2 were slightly downregulated, while TCF4 transcripts 4/5, 7, 8, and 10/11 were significantly upregulated. TCF4 transcripts 3 and 12 were undetectable in both cellular states in our assays (not shown). Of all of these effects, upregulation of transcripts 10/11 by neuronal differentiation was by far the most pronounced.
Having characterized TCF4 expression in the form of various transcripts in human iPSC-derived NPCs and neurons, we next sought to determine the signaling pathways controlling TCF4 expression in human NPCs. In particular, given previous reports in nonneuronal cells of TCF4 being under control of canonical WNT/β-catenin signaling [21, 22], we first tested whether different TCF4 transcripts responded to modulation of the canonical WNT signaling pathway as illustrated in Figure 2a. To first stimulate WNT signaling, we tested activation of the pathway at the level of stimulation of the Frizzled family of G-protein-coupled receptors through the secreted glycoprotein Wnt3a, and also by inhibition of GSK3 (glycogen synthase kinase 3), a negative regulator of canonical WNT signaling that phosphorylates β-catenin to promote its ubiquitin-dependent degradation via the destruction complex composed of AXIN/APC (adenomatosis polyposis coli) and PP2A (protein phosphatase 2A)/CK1 (casein kinase 1) [32, 33, 34]. As a source of Wnt3a that we had already validated as a regulator of canonical WNT signaling in human NPCs, we used Wnt3a conditioned media (Wnt3a-CM [25]). For GSK3 inhibition, we used CHIR-99021, a potent, and highly selective, ATP-competitive inhibitor. Using our established WNT pathway signaling reporter gene system [25], we were able to first confirm that both of these stimulators were active in our current batch of human NPCs: Wnt3a-CM (5%) produced an 8-fold increase (Fig. 2b), while CHIR-99021 (12.5 μM) produced more than a 10-fold increase in WNT reporter gene activity (Fig. 2c). Using similar treatment conditions, we next tested the effect of both Wnt3a-CM and CHIR-99021 on TCF4 transcript levels in human NPCs. As shown in Figure 2d, e, f, g, h, 24-h treatment with Wnt3a or CHIR-99021 upregulated total TCF4 mRNA in human NPCs ∼1.4-fold, whereas transcripts 1/2, 4/5, 7, and 10/11 were upregulated ∼1.4- to 6-fold. Notably, 3 of the TCF4 transcripts (4/5, 7, and 10/11) upregulated by WNT signaling were also upregulated by neuronal differentiation, including most significantly TCF4 transcript 10/11 (also known as ITF2A; SEF2-1A; TCF4-A) encoding a ∼55-kDa short form of TCF4. Although TCF4 transcript 8 expression increased upon neuronal differentiation, it was unresponsive to either Wnt3a-CM or CHIR-99021 treatment (data not shown). Taken together, these data indicate that WNT signaling regulates the expression of a subset of TCF4 transcripts in human NPCs.
Fig. 2.
TCF4 transcript levels are regulated by the WNT signaling pathway. a Model showing the canonical WNT signaling pathway and modes of action of chemical modulators of this pathway. b, c 5% Wnt3a-conditioned media (Wnt3a-CM), or the WNT pathway activator (GSK3 inhibitor) CHIR-99021, were applied to human NPCs (8330-8) carrying a luciferase reporter under the control of multiple TCF/LEF binding sites. Wnt3a-CM results in ∼8-fold induction of luciferase expression (b), and CHIR-99021 causes ∼10-fold induction of luciferase expression (c), compared to luciferase expression in cells treated with media alone. Unpaired, two-tailed t test was performed. *** and **** indicate p < 0.001 and 0.0001, respectively. d-i Regulation of TCF4 and AXIN2 transcript levels in human NPCs by 24-h treatments with Wnt3a (10% Wnt3a-CM) or CHIR-99021 (10 µM) with n = 3 replicates per treatment. One-way ANOVA followed by Sidak's multiple comparisons test was performed. **, ***, and **** indicate p < 0.01, 0.001, and 0.0001, respectively, for comparisons of transcript levels in treated human NPCs vs. control (DMSO). NS indicates not statistically significant. Error bars display standard error of the mean. Green underline = activators.
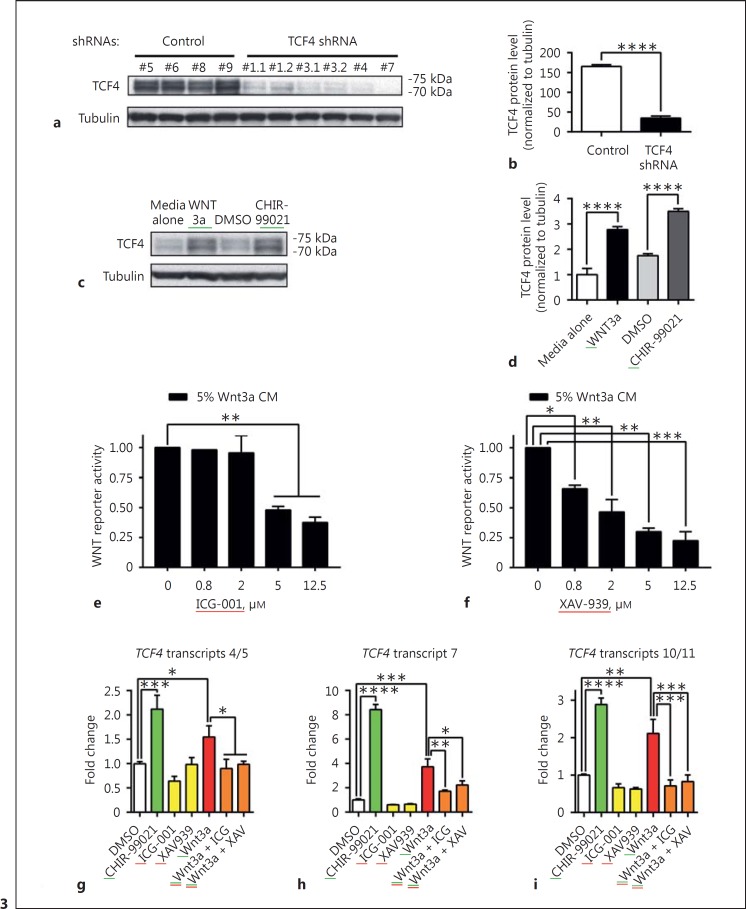
We next sought to test whether WNT signaling regulates TCF4 expression at the protein level. Most commercially available TCF4 antibodies recognize multiple Western blot bands, which may represent different TCF4 gene products or posttranslational modification states. However, the existence of two related genes (TCF3 and TCF12) with high sequence homology to TCF4, each giving rise to similar numbers of distinct transcripts, creates uncertainty about the identity of each band being detected. Thus, to enable specific identification of TCF4 bands detected by Western blotting, we screened for and identified a set of lentiviral vectors that each express a short hairpin RNA (shRNA) designed to induce stable and long-term TCF4 gene silencing through RNA interference (RNAi) in both dividing human NPCs and nondividing human neurons. As shown in Figure 3a, Western blot analysis of NPC samples expressing four distinct control or four distinct TCF4 shRNAs revealed two bands running between the 70- to 75-kDa markers corresponding roughly to the expected molecular weight of full-length TCF4 isoforms. The multiple TCF4 shRNAs produced on average a ∼79% decrease in these bands compared to control shRNAs (Figure 3b). The downregulation of these bands by shRNAs specific for TCF4 indicates that these bands reflect detection of TCF4 protein products by the Western blot antibody. Further supporting this conclusion, we have found that these two Western blot bands are completely absent in lysates from human iPSCs in which TCF4 expression has been eliminated by CRISPR-mediated knockout [Haggarty, in preparation]. Having confirmed the identity of the main TCF4 bands by Western blotting, human NPCs were again treated with Wnt3a-CM (10%) or CHIR-99021(10 μM) for 24 h, and samples were analyzed by Western blots. Both Wnt3a and CHIR-99021 upregulated TCF4 protein levels (Fig. 3c); quantification of TCF4 bands indicated a ∼3- to 3.5-fold upregulation was achieved by Wnt3a and CHIR-99021 treatments (Fig. 3d; p < 0.0001 in both cases). Thus, WNT signaling through GSK3 inhibition upregulates TCF4 mRNA and protein expression in human NPCs.
Fig. 3.
WNT pathway stimulation upregulates TCF4 protein level in human NPCs; WNT pathway antagonists suppress the upregulation of specific TCF4 transcripts induced by WNT pathway stimulation. a Western blot analysis of TCF4 protein levels in human NPC lysates from stable lines expressing 4 different control shRNAs (labeled 5, 6, 8, or 9) or 4 different TCF4 shRNAs (labeled 1.1/1.2 [replicates of one shRNA], 3.1/3.2 [replicates of one shRNA], 4, or 7). See Materials and Methods for sources and sequences of shRNAs. Two bands running between the 70- and 75-kDa markers are shown. b Graph illustrating that these bands are decreased in cells expressing TCF4 shRNAs by ∼79%, when signals from cell lines expressing all TCF4 shRNAs were normalized to tubulin levels and averaged and compared to the average control line signal, confirming the identity of these bands as TCF4. Unpaired, two-tailed t test performed. **** indicates p < 0.0001. c, d 24-h treatments with Wnt3a (10% Wnt3a-CM) or CHIR-99021 (10 μM) upregulate TCF4 protein levels ∼3- to 3.5-fold; TCF4 band signals were normalized to tubulin levels and then normalized to the media alone mean value with n = 3 replicates per treatment (representative blot shown in c; quantitation of all data in d). One-way ANOVA followed by Sidak's multiple comparisons test was performed. **** indicates p < 0.0001. Western blot images in panels a and c were created with different exposure times - longer for a and shorter for c - in order to avoid artifacts due to under- or overexposure of the films, which might have obscured signals for the treatment lanes (TCF4 shRNA in a and WNT3a/CHIR-99021 for c). This has caused the levels of TCF4 protein in control conditions to appear to differ between these two panels, but we do not believe this apparent difference in control levels is real. e, f WNT pathway antagonists ICG-001 and XAV-939 produce dose-dependent inhibition of reporter gene activation induced by 5% Wnt3a-CM. WNT reporter activity was calculated as the fold change in luciferase expression of compound-treated cells over DMSO-treated cells. One-way ANOVA followed by Sidak's multiple comparisons test performed. * and ** indicate p < 0.1 and 0.01, respectively, for the indicated comparisons. g-i The WNT pathway antagonists ICG-001 (25 μM) and XAV939 (10 μM) block Wnt3a-mediated (10% Wnt3a-CM) upregulation of TCF4 transcripts 4/5, 7, and 10/11 (24-h treatments) with n = 3 replicates per treatment. g One-way ANOVA followed by uncorrected Fisher's LSD test was performed. * indicates p < 0.1 for the indicated comparisons. h, i One-way ANOVA followed by Sidak's multiple comparisons test was performed. *, **, and *** indicate p < 0.1, 0.01, and 0.001, respectively, for the indicated comparisons of TCF4 transcript levels in treated NPCs. Error bars display standard error of the mean. Green underline = activators; red underline = inhibitors.
Having shown that WNT pathway activation leads to an increase in TCF4 mRNA and protein expression, we sought to better understand the underlying molecular mechanisms. Activation of canonical WNT signaling is known to regulate the expression of target genes through β-catenin accumulation, nuclear translocation, and binding of β-catenin to DNA-bound members of the TCF/LEF family of High-mobility group (HMG) transcription factors, for example TCF7L2 (RefSeq gene ID 6934; not to be confused with TCF4 itself [32, 35, 36]). This binding of β-catenin displaces co-repressor complexes that normally prevent gene expression in the absence of WNT pathway activation through epigenetic silencing [37, 38]. Therefore, we first looked at the evidence for the presence of TCF/LEF binding sites within the transcripts' start sites that were regulated by Wnt3a-CM and GSK3 inhibitor treatment. Since global ChIP-seq studies have revealed that β-catenin binding can occur at canonical TCF/LEF binding sites (T/AT/ACAAAG) in gene promoters within 2.5 kb of transcript start sites [39, 40], we examined 2.5 kb of genomic sequence upstream of the identified WNT-responsive TCF4 transcripts as well as publicly available TCF7L2 ChIP data from HEK293 cells collected for the ENCODE project (https://genome.ucsc.edu/ENCODE/) by the Farnham laboratory. Indeed, as summarized in online suppl. Table 2, several potential TCF/LEF family binding sites were identified in each of the WNT-responsive transcripts; most notably, the highly WNT responsive TCF4 transcripts 10/11 encoding the ∼55 kDa short form of TCF4 (ITF2A; SEF2-1A; TCF4-A) contained 4 sites and had ChIP-seq evidence for TCF7L2 binding. These data are consistent with the hypothesis that upregulation of TCF4 transcripts upon WNT pathway activation operates through a β-catenin-dependent transcriptional mechanism in human NPCs.
To experimentally test the hypothesis that WNT signaling through β-catenin is essential for activation of TCF4 mRNA expression, we sought next to determine whether inhibition of WNT signaling by interfering with β-catenin accumulation or activity would lead to a repression of basal or WNT-induced increases of TCF4 mRNA levels. For this, we chose two distinct pharmacological mechanisms known to block canonical WNT signaling. The first involved treatment with XAV-939, which reduces β-catenin stability by inhibiting tankyrase 1/2. Tankyrase is a poly-ADP ribosylase that destabilizes AXIN and prevents its interaction with β-catenin within the destruction complex; thus, inhibiting tankyrase has the effect of stabilizing AXIN and causing enhanced degradation of β-catenin [41]. The second was ICG-001, which inhibits the interaction of β-catenin with the histone acetyltransferase CBP (CREB-binding protein [42]), blocking β-catenin's transcriptional activation activity (Fig. 2a).
Using our established WNT signaling reporter gene system [25], in which stimulators are active (Fig. 2b, c), we found that the inhibitors of WNT signaling were also active in our human NPCs (Fig. 3e, f). For ICG-001 and XAV-939, dose-dependent inhibitory activities were detected only when the pathway was stimulated, i.e. in the presence of Wnt3a-CM (5%). To complement our demonstration of Wnt3a and CHIR-99021 upregulation of TCF4 expression (Fig. 2d, e, f, g, h), we tested whether the WNT pathway inhibitors ICG-001 and XAV-939 downregulate TCF4 expression. NPCs were treated with ICG-001 (25 μM) or XAV-939 (10 μM) in the absence or presence of 10% Wnt3a-CM for 24 h, and samples were collected and analyzed by qPCR for TCF4 transcripts 4/5, 7, and 10/11, the transcripts responsive to Wnt3a regulation as well as neuronal differentiation (Fig. 1, 2). As observed in the WNT reporter gene assay, downregulation of TCF4 transcript expression by ICG-001 and XAV-939 was observed in the presence of Wnt3a-CM (10%), with these compounds producing an approximately 50% reduction of transcripts 4/5 and 7, and a 75% reduction of transcripts 10/11 from the levels observed in the presence of Wnt3a (Fig. 3g, h, i). Thus, antagonizing the WNT pathway at the level of disrupting β-catenin/CBP interaction with ICG-001 or through tankyrase inhibition with XAV-939 downregulates TCF4 mRNA expression.
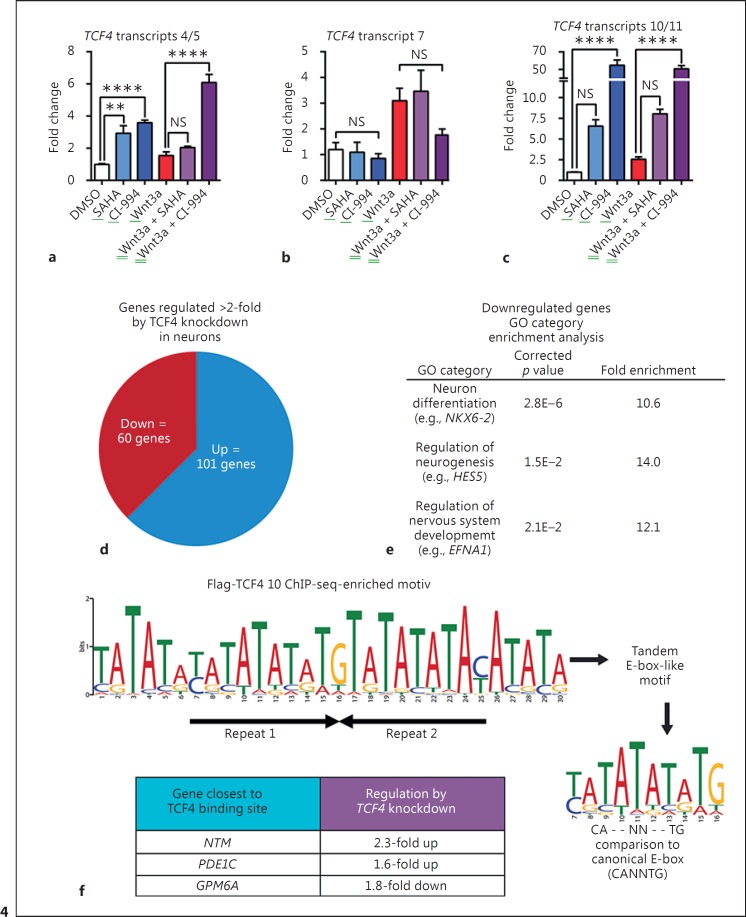
Given the ability of ICG-001 to disrupt the co-activator activity of the β-catenin/CBP complex that has histone acetyltransferase (HAT) activity and repress TCF4 mRNA expression, along with evidence for histone modifying enzymes as playing a key role in canonical WNT signaling [38, 43], we sought to test whether selective inhibition of HDACs that generally oppose co-activator complexes containing HATs would have the opposite effect as ICG-001 and potentiate TCF4 mRNA expression. In the same reporter assays in which both ICG-001 and XAV-939 dose-dependently antagonized the effect of WNT pathway stimulation, the broadly acting HDAC class I, IIb, and IV inhibitor SAHA (suberoyl hydroxamic acid; vorinostat) and the more selective HDAC class I inhibitor CI-994, were found to strongly synergize with Wnt3a-CM (5%) to activate the WNT signaling reporter system in human NPCs (online suppl. Fig. 3). In contrast, there was essentially no transcriptional induction with these agents in the absence of Wnt3a. To determine whether these synergistic effects could be translated into regulation of TCF4 mRNA expression, we next treated human NPCs with CI-994 (10 μM) or SAHA (1 μM) in the absence or presence of Wnt3a-CM (10%). As shown in Figure 4a, b, c, even in the absence of Wnt3a, TCF4 transcripts 4/5 and 10/11, but not transcript 7, were found to be upregulated by CI-994 and SAHA treatment. When effective, we observed stronger induction by the class I HDAC inhibitor CI-994 than the more broadly acting HDAC inhibitor SAHA. These data provide further evidence indicating that TCF4 transcripts 4/5 and 10/11 are regulated by the WNT pathway and implicate a mechanism that may involve the loss of repressive epigenetic regulation by class I HDAC co-repressor complexes antagonizing the effects of β-catenin/CBP co-activator complexes.
Fig. 4.
HDAC inhibitors upregulate specific TCF4 transcript in human NPCs. ChIP-seq identification of TCF4 isoform 10 target genes. a-c Upregulation of TCF4 transcripts by 24-h treatments with CI994 (10 μM) or SAHA (1 μM) alone or in the presence of Wnt3a (10% Wnt3a-CM) with n = 3 replicates per treatment. One-way ANOVA followed by Sidak's multiple comparisons test was performed. **, ***, and **** indicate p < 0.01, 0.001, and 0.0001, respectively, for comparisons of transcript levels in treated NPCs vs. control (DMSO). NS indicates not statistically significant. Error bars in this figure display standard error of the mean. d Pie chart indicating the total number of genes up- or downregulated at least 2-fold by shRNA knockdown of TCF4 in 8330 neurons. e The top three most statistically significantly enriched GO categories for genes downregulated at least 2-fold by shRNA-mediated TCF4 knockdown. f TCF4 binding motif enriched in ChIP DNA fragments pulled down by anti-Flag antibody in FLAG-TCF4 isoform 10 expressing differentiated 8,330 neurons. Two tandem inverted repeats are indicated by arrows. The table below the motif lists three examples of candidate TCF4 target genes that scored as hits both in our TCF4 isoform 10 ChIP-seq assay and in our test of genes regulated by TCF4 knockdown. Note that corrected p values for regulation of PDE1C and GPM6A by TCF4 knockdown were only nominally significant (p = 0.057 and 0.08, respectively). Green underline = activators.
In order to fully understand the consequences of TCF4 haploinsufficiency in PTHS, and to characterize the effectiveness of pharmacological upregulation of TCF4 expression, it is necessary to identify TCF4 target genes. These genes may have aberrant expression levels in PTHS patients, and then a goal of pharmacological upregulation of TCF4 expression could be to normalize the expression of these genes. As a first step in identifying TCF4 target genes, we detected gene expression changes caused by shRNA-mediated knockdown of TCF4 in human neurons differentiated by growth factor withdrawal for 6 weeks using genome-wide microarray analysis. The full set of transcriptome changes meeting our statistical significance criteria (fold change ≥ ±1.3, p < 0.05) are listed in online supplementary Table 4. Among genes whose expression was changed more than 2-fold (online suppl. Table 5), 60 were downregulated, and 101 were upregulated (Fig. 4d). These data suggest that TCF4 can act as both an activator and a repressor of gene transcription in human neurons; this is consistent with the existence of both activation and repression domains within the N-terminal region of TCF4 (online suppl. Fig. 1 [30, 31]). Interestingly, GO category enrichment analysis of the 60 genes downregulated by at least 2-fold found the greatest degree of statistically significant enrichment for sets of genes involved in neuronal development and differentiation (Fig. 4e). Examples of some of these genes are the homeobox transcription factor NKX6-2 (downregulated 2.1-fold; role in neurodevelopment, e.g. [44]); the bHLH transcription factor BHLHE22 (downregulated 4.6-fold; role in neurodevelopment, e.g. [45]); the bHLH transcription factor HES5 (downregulated 3.2-fold; role in neurodevelopment, e.g. [46]); and the ephrin EFNA1 (downregulated 2.6-fold; role in neurodevelopment, e.g. [47]). These data provide further support for the notion that TCF4 regulates the expression of genes that direct neuronal differentiation and development [48].
Genes that have altered expression levels induced by TCF4 knockdown could be direct targets of TCF4, or alternatively could be regulated as downstream secondary effects mediated by other primary TCF4 target genes. To begin to identify direct target genes of TCF4, we utilized ChIP-seq analysis, specifically to detect genomic binding sites for TCF4 isoform 10, one of the short isoforms robustly upregulated by differentiation as well as WNT pathway activators and CI-994 (Fig. 1, 2, 3, 4). We generated a stable NPC line overexpressing Flag-tagged TCF4 isoform 10, differentiated these NPC into neurons, and used an anti-Flag antibody in a ChIP assay to generate a library of genomic TCF4-binding DNA fragments. Sequencing of this library and employment of a stringent enriched DNA fragment peak detection procedure (with input DNA reads as the control and the false discovery rate q-value threshold of q <0.05; see Materials and Methods for details) identified a set of “high confidence” genomic TCF4 binding sites across the human genome (online suppl. Table 6); genes nearest to these binding peaks were determined by visual inspection of GENCODE comprehensive genome annotation of genome build hg19 using the UCSC genome browser (https://genome.ucsc.edu/index.html). To first gain confidence in the specificity of our peak calling procedure, we first analyzed the sequences of these TCF4 binding sites using MEME-ChIP (http://meme-suite.org/tools/meme-chip), a DNA motif discovery and enrichment analysis algorithm [49]. This algorithm detected an extremely highly enriched motif (p = 1.3E-198) that resembles two tandem repeats of the canonical TCF4 binding E-box (CANNTG), with the difference being a 3× “TA” motif replacing the “NN” in the middle of the canonical E-box, and with the second repeat inverted with respect to the first repeat (Fig. 4f). This unbiased computational analysis (as compared to analyses that have started with predefined E-box consensus sequences) provides strong evidence that the genomic DNA sequences we identified are true TCF4 isoform 10 binding sites. Nonetheless, further validation using ChIP assays that measure binding of endogenous TCF4 isoform 10 should be performed once antibodies specific for this isoform become available in the future.
Amongst this set of high-confidence TCF4 isoform 10 genomic binding sites, we noted that the nearest genes (i.e., candidate TCF4 isoform 10 target genes) included a number of gene encoding synaptic protein that have previously been reported to be regulated by E-box binding transcription factors. These include GRIA1 (glutamate receptor 1), which is a member of the alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate receptor family of ionotropic glutamate receptors that are critical to synaptic plasticity [50, 51]; and DLG2 (disks large homolog 2), which is a critical postsynaptic protein scaffold of excitatory synapses that interacts with NMDA receptor subunits and potassium channels [52, 53]. Of further note, GRIA1 is amongst the genes within the 108 loci associated with schizophrenia risk by GWAS studies with a p value of 1.06 × 10−10[54]. We also note that the dosage-sensitive, transcription factor SOX5 (SRY-related HMG-box 5) is amongst the candidate TCF4 isoform 10 target genes, as mutations in this gene that cause haploinsufficiency are known to lead to intellectual disability and developmental delay [55, 56]. Besides coding genes, we note that TCF4 isoform 10 peaks were identified in genes encoding multiple noncoding RNA transcripts, including AC108142.1, RP11-138I17.1, and two microRNAs, MIR4300 and MIR548O2, suggesting a role for TCF4 in regulation of multiple types of genomic elements.
Cross-referencing the list of genes nearest to high confidence TCF4 isoform 10 binding sites (online suppl. Table 6) to the set of genes whose expression levels were altered upon the loss-of-function of TCF4 in differentiated human neurons (online suppl. Table 4) revealed a short list of genes with nearby genomic TCF4 binding sites that were regulated by TCF4 knockdown. Many of these genes we note play critical roles in the nervous system, such as synaptogenesis (NTM[57]), postsynaptic density scaffolding and signaling (DLG2; e.g., [58]), and axon growth and guidance (GPM6A [59]). Additionally, many of these TCF4 target genes have been implicated in genetic studies of autism, intellectual disability, or psychiatric disease (NTM [60]; PDE1C[61]; DLG2[62]; GPM6A [54, 63], suggesting that their aberrant regulation caused by haploinsufficiency of TCF4 may at least in part be involved in the pathophysiology of PTHS. Importantly, each of these genes now provides a functional assay to compare signatures of PTHS patient-derived neurons and the effects of experimental therapeutics as discovered here. Moreover, these genes may represent a link between PTHS and other neuropsychiatric disorders that share an overlapping molecular and cellular pathophysiology.
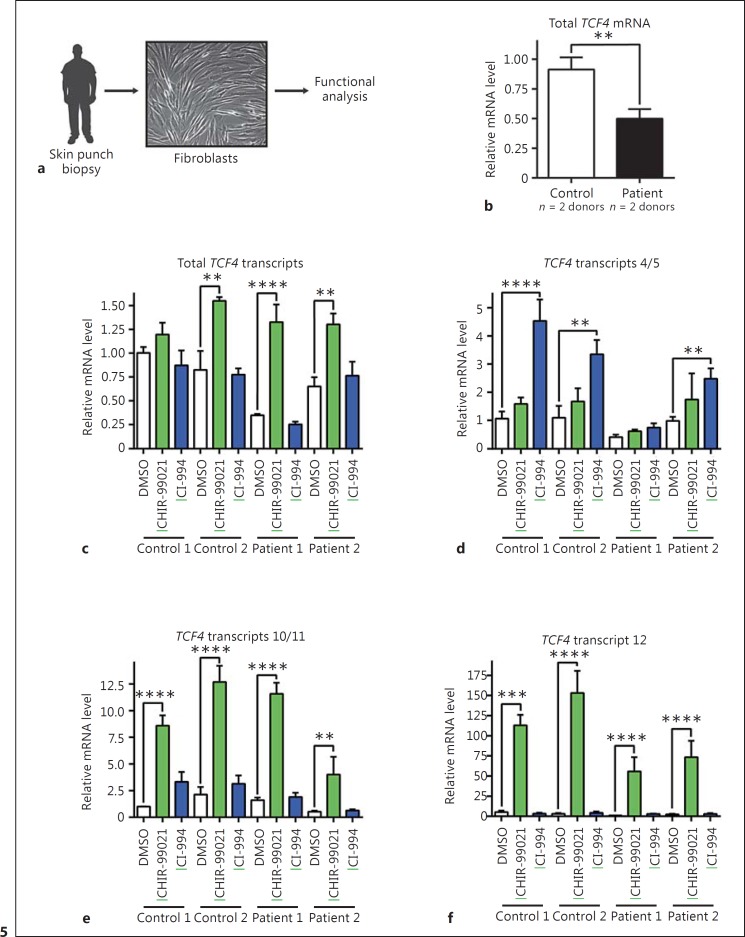
One potential route for the development of therapies for PTHS patients is to identify small-molecule drugs that can upregulate the expression of TCF4 in patient cells to mitigate genetic haploinsufficiency. As a first test of this idea, we obtained dermal fibroblasts from 2 PTHS patients that are heterozygous for TCF4 locus splice site variants (these variants occur in splice sites of two exons that are common to all TCF4 transcripts; unpubl. data), as well as 2 healthy donors (Fig. 5a). Given that splice site variants can produce premature stop codons and loss of mRNA due to non-sense-mediated decay, we first determined if TCF4 mRNA was expressed at a reduced level in the PTHS patient fibroblasts using the same qPCR assays used to measure total TCF4 mRNA levels in our human NPCs. As shown in Figure 5b, indeed, total TCF4 mRNA levels were 46% lower in the patient fibroblasts versus control fibroblasts from healthy donors. We then evaluated the effectiveness of WNT signaling modulators and HDAC inhibitors on induction of TCF4 mRNA expression by treating human primary fibroblasts with CHIR-99021 and CI-994. Stimulation of these control and patients' fibroblasts in primary culture with CHIR-99021 significantly induced increased expression of total TCF4 mRNA and also the WNT-responsive transcripts 10/11 but not TCF4 transcripts 4/5 (Fig. 5c, d, e). In addition, CHIR-99021 upregulated TCF4 transcript 12 that was not previously detected in human NPCs (Fig. 5f). Interestingly, CI-994 was more effective than CHIR-99021 at induction of TCF4 transcripts 4/5 (Fig. 5d), though less effective for other TCF4 transcripts. These data indicate that various TCF4 transcripts respond differently to WNT signaling potentiators and HDAC inhibitors, presumably reflecting different regulatory mechanisms controlling these transcripts.
Fig. 5.
The WNT pathway stimulator CHIR-99021 and HDAC inhibitor CI-994 reverse deficiencies in expression of specific TCF4 transcript in human Pitt-Hopkins patient-derived fibroblasts. a Human fibroblast explant cultures were generated from skin punch biopsies collected from PTHS patients or healthy control donors. b Decreased expression of total TCF4 mRNA in fibroblasts derived and cultured from PTHS patient skin biopsies vs. healthy control donors. Total TCF4 mRNA levels were averaged across 2 control or 2 PTHS patient fibroblast lines (n = 3 replicates per line), and normalized to GAPDH mRNA levels. Unpaired, two-tailed t test was performed. ** indicates p < 0.01. c-f Upregulation of TCF4 transcripts in control and Pitt-Hopkins patient fibroblasts by 24-h treatments with CHIR-99021 (10 μM) or CI-994 (10 μM) with n = 3 replicates per treatment. One-way ANOVA followed by Sidak's multiple comparisons test was performed. **, ***, and **** indicate p < 0.01, 0.001, and 0.0001, respectively, for comparisons of transcript levels in treated fibroblasts vs. control (DMSO). Error bars in this figure display standard error of the mean. Green underline = activators.
Discussion
As a transcription factor in which dominant mutations can cause cognitive deficits in humans, the bHLH family member TCF4 provides a critical link between the molecular mechanisms of transcriptional regulation, neuroplasticity, and cognition. Here, we have performed the first detailed quantitative analysis of expression of individual TCF4 transcripts in human iPSC-derived NPCs and differentiated neurons leading to the demonstration of neurodevelopmental regulation of the TCF4 locus and pointing to the dynamic changes in specific TCF4 isoforms that track with neuronal differentiation. In addition, we have identified the WNT/β-catenin signaling pathway as a robust stimulator of the expression of specific TCF4 transcripts. Excitingly, we were able to induce upregulation of TCF4 mRNA levels in PTHS patients' cells by stimulation of the WNT pathway and class I HDAC inhibitor treatment. Thus, our data suggest the future possibility of drug treatments for TCF4 haploinsufficiency in Pitt-Hopkins via WNT pathway activators and selective modulation of chromatin regulatory complexes.
Future work will be required to elucidate the mechanism of Wnt3a-mediated upregulation of TCF4 transcripts. Activation of WNT signaling can upregulate expression of target genes through the canonical β-catenin-TCF/LEF transcriptional pathway (e.g., [32]). ChIP-seq studies have shown that β-catenin binding can occur at canonical TCF/LEF binding sites (T/AT/ACAAAG) in gene promoters within 2.5 kb of transcript start sites [39, 40]. Indeed, examination of 2.5 kb of genomic sequence upstream of TCF4 transcripts' start sites reveals several potential TCF/LEF binding sites that might regulate each of the WNT-responsive transcripts that we identified (online suppl. Table 2).
Recent reports of mutations in the CTNNB1 gene that encodes β-catenin provide further strong support of the overall notion that the WNT/β-catenin/TCF4 pathway is critical for human cognition. Indeed, recurrent disruptive mutations in CTNNB1 were found in targeted large-scale resequencing studies in the study of autism spectrum disorders [64]. Moreover, a new intellectual disability and neurodevelopment syndrome was recently reported with the discovery of a total of 20 individuals (from 19 different families) with de novo, heterozygous, loss-of-function mutations in CTNNB1[65, 66, 67]. In addition to intellectual disability, affected individuals with CTNNB1 haploinsufficiency were described to commonly show motor delay, severe speech impairment, microcephaly, and abnormal muscle tone (truncal hypotonia and distal hypertonia/spasticity) with characteristic facial features consisting of abnormal nasal and lip structures [65, 66, 67]. Since a number of these features are reminiscent of the clinical diagnosis of PTHS, given our findings suggesting the direct regulation of TCF4 in a WNT/β-catenin-dependent manner, this suggests that both haploinsufficiency disorders may share a common molecular pathogenesis, with the prediction from our work that individuals with reduced β-catenin function will have decreased TCF4 expression.
A critical unanswered question for PTHS is whether the pathophysiology of patients is caused by TCF4 haploinsufficiency during development, or also in the fully developed adult [2]. Likewise, at the cellular level, it is unclear whether TCF4 haploinsufficiency causes dysfunction in NPCs, neurons, both, or other cell types. Indeed, TCF4 has been shown to play roles in all of these developmental stages and brain cell types: e.g., in neuronal migration in the developing cortex [68]; in synaptic plasticity during memory formation in the mature brain [69]; in neurogenesis [70]; and in excitability of mature cortical neurons [15]. Our data suggest that it may be possible to reverse TCF4 haploinsufficiency by drug therapy during development or in brain regions with continual production of NPCs (i.e., in the hippocampus). We also measured TCF4 transcript expression in differentiated human neurons generated by one specific method (described in Sheridan et al. [24]) from iPSC-derived NPCs, and we found that responsiveness to WNT pathway stimulators was greatly reduced or absent (not shown). Further work will be required to determine whether TCF4 expression levels can be upregulated by WNT pathway stimulators using different treatment paradigms (e.g., longer than 24 h), in differentiated human neuron types not present in our cultures as new methods of producing these neurons become available (e.g., [71, 72]), and in differentiated neurons in rodent in vivo models.
With the generation of PTHS patient-specific iPSCs from the fibroblast lines generated and analyzed here [Haggarty, in preparation], it will be possible in the near future to test whether these same pharmacological mechanisms can also rescue reduced TCF4 expression in the context of human NPCs and differentiated neurons. Based upon our observation of cell type-specific and developmental stage-specific regulation of specific TCF4 transcripts, it will be important to not only assess total TCF4 levels in such models, but also specific transcripts, their change over neurodevelopment, and their response to pharmacological treatments in patient-derived cells. Overall, in terms of understanding the function of TCF4 in the human CNS, our findings point to the importance of considering TCF4 function and its target genes, not just in tumor and peripheral cells, but in physiologically relevant cell types that are likely to express relevant bHLH-binding partners and components of the chromatin and transcriptional regulatory machinery interacting with TCF4 to govern the expression of its target genes. Finally, besides cellular assays, a further key next step will also be to test whether WNT pathway stimulators can reverse defects in rodent models of PTHS during development and in adulthood. Encouragingly, pharmacological WNT pathway activators can reduce cognitive deficits in mouse models of other neurodevelopmental disorders [73, 74]. Furthermore, lithium salts used to treat bipolar disorder, as well as valproate used to also treat bipolar disorder and epilepsy, are examples of FDA-approved, brain-penetrant medications with known ability to stimulate the WNT pathway, suggesting the feasibility of identifying therapeutic agents targeting WNT signaling that have safety profiles commensurate with chronic administration.
To fully understand whether pharmacological upregulation of TCF4 expression could work as a PTHS therapeutic approach, it will be necessary to determine whether upregulation of all, or only some, TCF4 transcripts could produce beneficial effects. A recent genetic study of a PTHS patient [19] potentially sheds some light on this question. Maduro et al. [19] reports a complex genomic translocation within the TCF4 locus that, despite causing the loss of certain TCF4 transcripts, results in an overall elevation of expression of total TCF4 mRNA levels. Intriguingly, assessment of individual TCF4 transcripts revealed that the short TCF4 transcripts corresponding to TCF4 transcripts 7–12 in our studies were unregulated in peripheral blood samples, whereas typical Pitt-Hopkins cases examined in this study with damaging mutations in one TCF4 allele showed the expected loss of TCF4 mRNA expression. Since the individuals studied by [19] exhibited mild autosomal dominant intellectual disability, rather than the full constellation of symptoms of PTHS, these findings suggest that the upregulation of the short informs of TCF4 may be at least partially protective of full PTHS. Further studies of additional cases of milder forms of PTHS and cases of intellectual disability with TCF4 mutations at the level of individual TCF4 transcripts, as exemplified by our study, is critical; indeed, our study may yield a human genetic proof-of-concept for therapeutic strategies aiming to enhance TCF4 transcription in an allele-specific manner.
In schizophrenia patients with disease risk-associated genetic variation at the TCF4 locus, it is entirely unknown whether this variation causes under- or overexpression of TCF4, or perhaps even a complex, cell-type dependent pattern of aberrant expression. Similarly, it is unclear whether WNT pathway overactivation, or underactivation, or both, occur in autism spectrum disorders [23]. However, overexpression of TCF4 in mice causes cognitive and sensorimotor gating impairments [16]. These data have led to the hypothesis that TCF4 may be overexpressed in schizophrenia patients' brains [2]. If this hypothesis is correct, our data suggest that it may be possible to attempt to dial down TCF4 overexpression in patients via pharmacological agents that inhibit the WNT pathway, or stimulate HDACs/inhibit HATs. Interestingly, data from animal models of neurodegenerative dementias have also suggested a potential therapeutic role for HDAC activators in those disorders [75]. As future investigations reveal the nature of the pathological consequences of disease risk-associated genetic variation at the TCF4 locus, it will be exciting to work towards development of pharmacological agents to reverse the pathology.
As for the case of the known TCF4 target genes CNTNAP2 and NRXN1[76], which are mutated in an autosomal-recessive manner to cause Pitt-Hopkins-like syndrome 1 and 2, respectively [77], identification of the other CNS targets of TCF4 may also help elucidate a better understanding of convergent pathophysiology and related clinical syndromes. For this, it will be necessary to comprehensively identify the full set of TCF4 target genes, taking into account the many isoforms of TCF4 (different isoforms may regulate different target genes), as well as brain cell type-specific factors (TCF4 may target different genes in different brain cell types). To begin this process, we used a tagged version of the TCF4 isoform 10 to selectively map the targets of this one short TCF4 isoform, which our studies revealed was the WNT pathway and neurodevelopmentally regulated with higher expression in postmitotic neurons than proliferative NPCs. While this strategy has the advantage of allowing a single isoform of TCF4 to be investigated, the overexpression of this one isoform may cause artifactual detection of binding events due to nonphysiological levels of the Flag-TCF4 isoform 10 being expressed. However, there are currently no alternatives as antibodies specific for TCF4 isoform 10 do not exist. Importantly, despite these caveats of our strategy, unbiased, computational analysis of the entire set of DNA motifs found within the genomic regions bound by TCF4 isoform 10 revealed a highly significant (p = 1.3E-198) enrichment of a motif resembling two tandem repeats of the canonical TCF4 binding E-box (CANNTG), which is consistent with current models of the mechanisms through which members of the bHLH family of transcription factors regulate gene expression. Moreover, the small set of genes that we identified by our stringent analysis of the peak enrichment include genes that play critical roles in the development and functioning of the CNS, and many have been implicated in neuropsychiatric disorders by various genetic studies. It is important, however, to keep in mind that our computational analysis intersecting TCF4 target genes identified by ChIP-seq with microarray analysis of genes dysregulated by TCF4 knockdown by RNAi likely underestimates the true number of genes regulated by TCF4 isoform 10 due to false negatives from noise limitations inherent to microarray technology. As such, future studies that intersect TCF4 ChIP-seq datasets with global transcriptome studies from RNA sequencing will help to overcome these technical limitations of our current study. Moreover, while focusing on TCF4 isoform 10 with the use of an overexpression of a tagged transgene had the advantage of allowing us to isolate potential genomic binding sites of a specific TCF4 isoform, we likely missed critical TCF4 target genes that this form of TCF4 does not bind. In addition, overexpressing TCF4 might have caused changes in cell state leading to nonphysiological binding of TCF4 to sites in the genome. Thus, further understanding of the cis-acting regulatory elements that recruit TCF4 and the chromatin modifying and remodeling complexes that control downstream target gene expression will benefit in the future from the use of well-validated TCF4 antibodies that detect specific endogenous TCF4 proteins.
During preparation of this paper, the Sweatt laboratory [69] reported that chronic (11-day) systemic administration of SAHA, the HDAC inhibitor we show here enhances expression of multiple TCF4 transcripts, was able to normalize deficits in hippocampal long-term potentiation and memory in a mouse model of TCF4 haploinsufficiency. Correlated with these functional effects of HDAC inhibition in the TCF4 haploinsufficient mouse, SAHA administration significantly increased the expression of multiple TCF4 transcripts (Tcf4-004, Tcf4-007, Tcf4-011, and Tcf4-014) in the hippocampal CA1 region. Notably, Tcf4-007 is the mouse ortholog of human TCF4 transcripts 10/11 that we also found to be upregulated by SAHA treatment in our human neuronal cells (Fig. 4c). Moreover, consistent with our identification that class I HDAC inhibition through CI-994 is sufficient to enhance TCF4 expression, this study also demonstrated that selective inhibition of the class I HDAC2 using antisense oligonucleotides infused into the brain is sufficient to rescue memory deficits in the TCF4 haploinsufficient mice [69]. While HDAC2 inhibition is likely to affect multiple genes and proteins critical to neuroplasticity based upon our and others' published findings [78, 79, 80, 81], given its critical role in and cognition, these results provide critical in vivo evidence of the potential for targeting the epigenetic regulation of TCF4 as novel therapeutics for PTHS and other cognitive disorders involving dysfunction of TCF4. In addition, in future rodent model experiments, it would be exciting to test whether WNT pathway agonists can upregulate TCF4 in vivo. A related interesting question that could also be addressed is whether endogenously released WNT proteins upregulate TCF4 expression under physiological states in vivo.
In summary, having validated here a new set of pharmacological tools and molecular mechanisms for regulating TCF4 gene expression through the WNT/β-catenin pathway and epigenetic mechanisms involving opposing regulation by class I HDAC complexes that are capable of normalizing reduced TCF4 expression in PTHS patient-derived cells, our study provides critically needed insight into potential therapeutic targets for this rare neurodevelopmental disorder. Furthermore, through our identification of novel TCF4 target genes using ChIP-seq and a global transcriptome study, our study further highlights the critical function that TCF4 plays in the CNS as an integrator of signaling pathways that are fundamental to neurodevelopment and neuroplasticity. Future work will be required to determine whether expression of any of these novel TCF4 neuronal target genes are dysregulated in the context of PTHS and schizophrenia patients, and much additional work will be required to comprehensively catalog the full set of TCF4 target genes in a brain cell type-specific manner. Ultimately, these efforts when integrated with findings emerging from other model systems [15, 69] will lead to the elucidation of pathological mechanisms due to altered TCF4 function and help identify pharmacological therapies for treatment and ideally prevention of neuropsychiatric disorders.
Disclosure Statement
The authors declare no conflict of interest.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgements
We thank members of the Haggarty, Gusella, and Talkowski laboratories for technical assistance and helpful discussions along with families within the Pitt-Hopkins community who generously donated tissue used in this study. Funding was provided by the Pitt-Hopkins Research Foundation, Brace Cove Health Fund, Harvard Stem Cell Institute, Stanley Center for Psychiatric Research, National Institute of Mental Health (5R01MH095088), and the National Institute of General Medical Sciences (Developmental Genome Anatomy Project, 5P01GM061354).
References
- 1.Navarrete K, Pedroso I, De Jong S, Stefansson H, Steinberg S, Stefansson K, et al. TCF4 (e2-2; ITF2): a schizophrenia-associated gene with pleiotropic effects on human disease. Am J Med Genet B Neuropsychiatr Genet. 2013;162b:1–16. doi: 10.1002/ajmg.b.32109. [DOI] [PubMed] [Google Scholar]
- 2.Sweatt JD. Pitt-Hopkins syndrome: intellectual disability due to loss of TCF4-regulated gene transcription. Exp Mol Med. 2013;45:e21. doi: 10.1038/emm.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flora A, Garcia JJ, Thaller C, Zoghbi HY. The E-protein Tcf4 interacts with Math1 to regulate differentiation of a specific subset of neuronal progenitors. Proc Natl Acad Sci USA. 2007;104:15382–15387. doi: 10.1073/pnas.0707456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Persson P, Jogi A, Grynfeld A, Pahlman S, Axelson H. HASH-1 and E2-2 are expressed in human neuroblastoma cells and form a functional complex. Biochem Biophys Res Commun. 2000;274:22–31. doi: 10.1006/bbrc.2000.3090. [DOI] [PubMed] [Google Scholar]
- 5.Ravanpay AC, Olson JM. E protein dosage influences brain development more than family member identity. J Neurosci Res. 2008;86:1472–1481. doi: 10.1002/jnr.21615. [DOI] [PubMed] [Google Scholar]
- 6.Pitt D, Hopkins I. A syndrome of mental retardation, wide mouth and intermittent overbreathing. Aust Paediatr J. 1978;14:182–184. doi: 10.1111/jpc.1978.14.3.182. [DOI] [PubMed] [Google Scholar]
- 7.Whalen S, Heron D, Gaillon T, Moldovan O, Rossi M, Devillard F, et al. Novel comprehensive diagnostic strategy in Pitt-Hopkins syndrome: clinical score and further delineation of the TCF4 mutational spectrum. Hum Mutat. 2012;33:64–72. doi: 10.1002/humu.21639. [DOI] [PubMed] [Google Scholar]
- 8.Zweier C, Peippo MM, Hoyer J, Sousa S, Bottani A, Clayton-Smith J, et al. Haploinsufficiency of TCF4 causes syndromal mental retardation with intermittent hyperventilation (Pitt-Hopkins syndrome) Am J Hum Genet. 2007;80:994–1001. doi: 10.1086/515583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ripke S, Neale BM, Corvin A, Walters JT, Farh KH, Holmans PA, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Donnell L, Soileau B, Heard P, Carter E, Sebold C, Gelfond J, et al. Genetic determinants of autism in individuals with deletions of 18q. Hum Genet. 2010;128:155–164. doi: 10.1007/s00439-010-0839-y. [DOI] [PubMed] [Google Scholar]
- 11.Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brockschmidt A, Filippi A, Charbel Issa P, Nelles M, Urbach H, Eter N, et al. Neurologic and ocular phenotype in Pitt-Hopkins syndrome and a zebrafish model. Hum Genet. 2011;130:645–655. doi: 10.1007/s00439-011-0999-4. [DOI] [PubMed] [Google Scholar]
- 13.D'Rozario M, Zhang T, Waddell EA, Zhang Y, Sahin C, Sharoni M, et al. Type I bHLH proteins Daughterless and Tcf4 restrict neurite branching and synapse formation by repressing neurexin in postmitotic neurons. Cell Rep. 2016;15:386–397. doi: 10.1016/j.celrep.2016.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grubisic V, Kennedy AJ, Sweatt JD, Parpura V. Pitt-Hopkins mouse model has altered particular gastrointestinal transits in vivo. Autism Res. 2015;8:629–633. doi: 10.1002/aur.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rannals MD, Hamersky GR, Page SC, Campbell MN, Briley A, Gallo RA, et al. Psychiatric risk gene transcription factor 4 regulates intrinsic excitability of prefrontal neurons via repression of SCN10a and KCNQ1. Neuron. 2016;90:43–55. doi: 10.1016/j.neuron.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brzozka MM, Radyushkin K, Wichert SP, Ehrenreich H, Rossner MJ. Cognitive and sensorimotor gating impairments in transgenic mice overexpressing the schizophrenia susceptibility gene Tcf4 in the brain. Biol Psychiatry. 2010;68:33–40. doi: 10.1016/j.biopsych.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 17.Sepp M, Pruunsild P, Timmusk T. Pitt-Hopkins syndrome-associated mutations in TCF4 lead to variable impairment of the transcription factor function ranging from hypomorphic to dominant-negative effects. Hum Mol Genet. 2012;21:2873–2888. doi: 10.1093/hmg/dds112. [DOI] [PubMed] [Google Scholar]
- 18.Kharbanda M, Kannike K, Lampe A, Berg J, Timmusk T, Sepp M. Partial deletion of TCF4 in three generation family with non-syndromic intellectual disability, without features of Pitt-Hopkins syndrome. Eur J Med Genet. 2016;59:310–314. doi: 10.1016/j.ejmg.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Maduro V, Pusey BN, Cherukuri PF, Atkins P, du Souich C, Rupps R, et al. Complex translocation disrupting TCF4 and altering TCF4 isoform expression segregates as mild autosomal dominant intellectual disability. Orphanet J Rare Dis. 2016;11:62. doi: 10.1186/s13023-016-0439-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mulligan KA, Cheyette BN. Wnt signaling in vertebrate neural development and function. J Neuroimmun Pharmacol. 2012;7:774–787. doi: 10.1007/s11481-012-9404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolligs FT, Nieman MT, Winer I, Hu G, Van Mater D, Feng Y, et al. ITF-2, a downstream target of the Wnt/TCF pathway, is activated in human cancers with beta-catenin defects and promotes neoplastic transformation. Cancer Cell. 2002;1:145–155. doi: 10.1016/s1535-6108(02)00035-1. [DOI] [PubMed] [Google Scholar]
- 22.Feng Y, Lee N, Fearon ER. TIP49 regulates beta-catenin-mediated neoplastic transformation and T-cell factor target gene induction via effects on chromatin remodeling. Cancer Res. 2003;63:8726–8734. [PubMed] [Google Scholar]
- 23.Kalkman HO. A review of the evidence for the canonical Wnt pathway in autism spectrum disorders. Mol Autism. 2012;3:10. doi: 10.1186/2040-2392-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheridan SD, Theriault KM, Reis SA, Zhou F, Madison JM, Daheron L, et al. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS One. 2011;6:e26203. doi: 10.1371/journal.pone.0026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao WN, Cheng C, Theriault KM, Sheridan SD, Tsai LH, Haggarty SJ. A high-throughput screen for Wnt/beta-catenin signaling pathway modulators in human iPSC-derived neural progenitors. J Biomol Screen. 2012;17:1252–1263. doi: 10.1177/1087057112456876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang JL, Shamah SM, Sun AX, Waldman ID, Haggarty SJ, Perlis RH. Label-free, live optical imaging of reprogrammed bipolar disorder patient-derived cells reveals a functional correlate of lithium responsiveness. Transl Psychiatry. 2014;4:e428. doi: 10.1038/tp.2014.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flora A, Klisch TJ, Schuster G, Zoghbi HY. Deletion of Atoh1 disrupts Sonic Hedgehog signaling in the developing cerebellum and prevents medulloblastoma. Science. 2009;326:1424–1427. doi: 10.1126/science.1181453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sepp M, Kannike K, Eesmaa A, Urb M, Timmusk T. Functional diversity of human basic helix-loop-helix transcription factor TCF4 isoforms generated by alternative 5′ exon usage and splicing. PLoS One. 2011;6:e22138. doi: 10.1371/journal.pone.0022138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herbst A, Kolligs FT. A conserved domain in the transcription factor ITF-2B attenuates its activity. Biochem Biophys Res Commun. 2008;370:327–331. doi: 10.1016/j.bbrc.2008.03.081. [DOI] [PubMed] [Google Scholar]
- 32.Oliva CA, Vargas JY, Inestrosa NC. Wnts in adult brain: from synaptic plasticity to cognitive deficiencies. Front Cell Neurosci. 2013;7:224. doi: 10.3389/fncel.2013.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stamos JL, Weis WI. The beta-catenin destruction complex. Cold Spring Harbor Perspect Biol. 2013;5:a007898. doi: 10.1101/cshperspect.a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yost C, Torres M, Miller JR, Huang E, Kimelman D, Moon RT. The axis-inducing activity, stability, and subcellular distribution of beta-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- 35.Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, et al. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 36.van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, et al. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 37.Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, et al. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- 38.Willert K, Jones KA. Wnt signaling: is the party in the nucleus? Genes Dev. 2006;20:1394–1404. doi: 10.1101/gad.1424006. [DOI] [PubMed] [Google Scholar]
- 39.Bottomly D, Kyler SL, McWeeney SK, Yochum GS. Identification of β-catenin binding regions in colon cancer cells using ChIP-Seq. Nucleic Acids Res. 2010;38:5735–5745. doi: 10.1093/nar/gkq363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe K, Biesinger J, Salmans ML, Roberts BS, Arthur WT, Cleary M, et al. Integrative ChIP-seq/microarray analysis identifies a CTNNB1 target signature enriched in intestinal stem cells and colon cancer. PLoS One. 2014;9:e92317. doi: 10.1371/journal.pone.0092317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 42.Emami KH, Nguyen C, Ma H, Kim DH, Jeong KW, Eguchi M, et al. A small molecule inhibitor of beta-catenin/CREB-binding protein transcription (corrected) Proc Natl Acad Sci USA. 2004;101:12682–12687. doi: 10.1073/pnas.0404875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arce L, Pate KT, Waterman ML. Groucho binds two conserved regions of LEF-1 for HDAC-dependent repression. BMC Cancer. 2009;9:159. doi: 10.1186/1471-2407-9-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sousa VH, Miyoshi G, Hjerling-Leffler J, Karayannis T, Fishell G. Characterization of Nkx6-2-derived neocortical interneuron lineages. Cereb Cortex. 2009;19((suppl 1)):i1–i10. doi: 10.1093/cercor/bhp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joshi PS, Molyneaux BJ, Feng L, Xie X, Macklis JD, Gan L. Bhlhb5 regulates the postmitotic acquisition of area identities in layers II-V of the developing neocortex. Neuron. 2008;60:258–272. doi: 10.1016/j.neuron.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hatakeyama J, Bessho Y, Katoh K, Ookawara S, Fujioka M, Guillemot F, et al. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004;131:5539–5550. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- 47.Martinez A, Soriano E. Functions of ephrin/Eph interactions in the development of the nervous system: emphasis on the hippocampal system. Brain Res Brain Res Rev. 2005;49:211–226. doi: 10.1016/j.brainresrev.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 48.Forrest MP, Hill MJ, Quantock AJ, Martin-Rendon E, Blake DJ. The emerging roles of TCF4 in disease and development. Trends Mol Med. 2014;20:322–331. doi: 10.1016/j.molmed.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 49.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chater TE, Goda Y. The role of AMPA receptors in postsynaptic mechanisms of synaptic plasticity. Front Cell Neurosci. 2014;8:401. doi: 10.3389/fncel.2014.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin CH, Lee EH. JNK1 inhibits GluR1 expression and GluR1-mediated calcium influx through phosphorylation and stabilization of Hes-1. J Neurosci. 2012;32:1826–1846. doi: 10.1523/JNEUROSCI.3380-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fujita A, Kurachi Y. SAP family proteins. Biochem Biophys Res Commun. 2000;269:1–6. doi: 10.1006/bbrc.1999.1893. [DOI] [PubMed] [Google Scholar]
- 53.Wilke SA, Hall BJ, Antonios JK, Denardo LA, Otto S, Yuan B, et al. NeuroD2 regulates the development of hippocampal mossy fiber synapses. Neural Dev. 2012;7:9. doi: 10.1186/1749-8104-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ripke S. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lamb AN, Rosenfeld JA, Neill NJ, Talkowski ME, Blumenthal I, Girirajan S, et al. Haploinsufficiency of SOX5 at 12p12.1 is associated with developmental delays with prominent language delay, behavior problems, and mild dysmorphic features. Hum Mutat. 2012;33:728–740. doi: 10.1002/humu.22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schanze I, Schanze D, Bacino CA, Douzgou S, Kerr B, Zenker M. Haploinsufficiency of SOX5, a member of the SOX (SRY-related HMG-box) family of transcription factors is a cause of intellectual disability. Eur J Med Genet. 2013;56:108–113. doi: 10.1016/j.ejmg.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 57.Hashimoto T, Maekawa S, Miyata S. IgLON cell adhesion molecules regulate synaptogenesis in hippocampal neurons. Cell Biochem Funct. 2009;27:496–498. doi: 10.1002/cbf.1600. [DOI] [PubMed] [Google Scholar]
- 58.Sato Y, Tao YX, Su Q, Johns RA. Post-synaptic density-93 mediates tyrosine-phosphorylation of the N-methyl-D-aspartate receptors. Neuroscience. 2008;153:700–708. doi: 10.1016/j.neuroscience.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mita S, de Monasterio-Schrader P, Funfschilling U, Kawasaki T, Mizuno H, Iwasato T, et al. Transcallosal projections require glycoprotein M6-dependent neurite growth and guidance. Cereb Cortex. 2015;25:4111–4125. doi: 10.1093/cercor/bhu129. [DOI] [PubMed] [Google Scholar]
- 60.Minhas HM, Pescosolido MF, Schwede M, Piasecka J, Gaitanis J, Tantravahi U, et al. An unbalanced translocation involving loss of 10q26.2 and gain of 11q25 in a pedigree with autism spectrum disorder and cerebellar juvenile pilocytic astrocytoma. Am J Med Genet A. 2013;161a:787–791. doi: 10.1002/ajmg.a.35841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gamage TH, Misceo D, Fannemel M, Frengen E. A balanced de novo inv(7)(p14.3q22.3) disrupting PDE1C and ATXN7L1 in a 14-year old developmentally delayed boy. Eur J Med Genet. 2013;56:361–364. doi: 10.1016/j.ejmg.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 62.Egger G, Roetzer KM, Noor A, Lionel AC, Mahmood H, Schwarzbraun T, et al. Identification of risk genes for autism spectrum disorder through copy number variation analysis in Austrian families. Neurogenetics. 2014;15:117–127. doi: 10.1007/s10048-014-0394-0. [DOI] [PubMed] [Google Scholar]
- 63.El-Kordi A, Kastner A, Grube S, Klugmann M, Begemann M, Sperling S, et al. A single gene defect causing claustrophobia. Transl Psychiatry. 2013;3:e254. doi: 10.1038/tp.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O'Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dubruc E, Putoux A, Labalme A, Rougeot C, Sanlaville D, Edery P. A new intellectual disability syndrome caused by CTNNB1 haploinsufficiency. Am J Med Genet A. 2014;164a:1571–1575. doi: 10.1002/ajmg.a.36484. [DOI] [PubMed] [Google Scholar]
- 66.Kuechler A, Willemsen MH, Albrecht B, Bacino CA, Bartholomew DW, van Bokhoven H, et al. De novo mutations in beta-catenin (CTNNB1) appear to be a frequent cause of intellectual disability: expanding the mutational and clinical spectrum. Hum Genet. 2015;134:97–109. doi: 10.1007/s00439-014-1498-1. [DOI] [PubMed] [Google Scholar]
- 67.Tucci V, Kleefstra T, Hardy A, Heise I, Maggi S, Willemsen MH, et al. Dominant beta-catenin mutations cause intellectual disability with recognizable syndromic features. J Clin Invest. 2014;124:1468–1482. doi: 10.1172/JCI70372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen T, Wu Q, Zhang Y, Lu T, Yue W, Zhang D. Tcf4 controls neuronal migration of the cerebral cortex through regulation of Bmp7. Front Mol Neurosci. 2016;9:94. doi: 10.3389/fnmol.2016.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kennedy AJ, Rahn EJ, Paulukaitis BS, Savell KE, Kordasiewicz HB, Wang J, et al. Tcf4 regulates synaptic plasticity, DNA methylation, and memory function. Cell Rep. 2016;16:2666–2685. doi: 10.1016/j.celrep.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmidt-Edelkraut U, Daniel G, Hoffmann A, Spengler D. Zac1 regulates cell cycle arrest in neuronal progenitors via Tcf4. Mol Cell Biol. 2014;34:1020–1030. doi: 10.1128/MCB.01195-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheng C, Fass DM, Folz-Donahue K, MacDonald ME, Haggarty SJ. Highly expandable human iPS cell-derived neural progenitor cells (NPC) and neurons for central nervous system disease modeling and high-throughput screening. Curr Protoc Hum Genet. 2017;92:21.8.1–21.8.21. doi: 10.1002/cphg.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mines MA, Yuskaitis CJ, King MK, Beurel E, Jope RS. GSK3 influences social preference and anxiety-related behaviors during social interaction in a mouse model of fragile X syndrome and autism. PLoS One. 2010;5:e9706. doi: 10.1371/journal.pone.0009706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo W, Murthy AC, Zhang L, Johnson EB, Schaller EG, Allan AM, et al. Inhibition of GSK3beta improves hippocampus-dependent learning and rescues neurogenesis in a mouse model of fragile X syndrome. Hum Mol Genet. 2012;21:681–691. doi: 10.1093/hmg/ddr501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim D, Frank CL, Dobbin MM, Tsunemoto RK, Tu W, Peng PL, et al. Deregulation of HDAC1 by p25/Cdk5 in neurotoxicity. Neuron. 2008;60:803–817. doi: 10.1016/j.neuron.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Forrest M, Chapman RM, Doyle AM, Tinsley CL, Waite A, Blake DJ. Functional analysis of TCF4 missense mutations that cause Pitt-Hopkins syndrome. Hum Mutat. 2012;33:1676–1686. doi: 10.1002/humu.22160. [DOI] [PubMed] [Google Scholar]
- 77.Zweier C, de Jong EK, Zweier M, Orrico A, Ousager LB, Collins AL, et al. CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in Drosophila. Am J Hum Genet. 2009;85:655–666. doi: 10.1016/j.ajhg.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kilgore M, Miller CA, Fass DM, Hennig KM, Haggarty SJ, Sweatt JD, et al. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer's disease. Neuropsychopharmacology. 2010;35:870–880. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fass DM, Reis SA, Ghosh B, Hennig KM, Joseph NF, Zhao WN, et al. Crebinostat: a novel cognitive enhancer that inhibits histone deacetylase activity and modulates chromatin-mediated neuroplasticity. Neuropharmacology. 2013;64:81–96. doi: 10.1016/j.neuropharm.2012.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Graff J, Joseph NF, Horn ME, Samiei A, Meng J, Seo J, et al. Epigenetic priming of memory updating during reconsolidation to attenuate remote fear memories. Cell. 2014;156:261–276. doi: 10.1016/j.cell.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data