Abstract
In the last few decades, different methods for the detection of genomic imbalances, such as the microdeletion syndromes, were developed. The 22q11.2 deletion syndrome (22q11.2DS) is the most common microdeletion syndrome and presents wide clinical heterogeneity. The aim of this study was to describe 4 unusual cases of genomic imbalances found in individuals with suspected microdeletion syndromes. Different methods were necessary to complete the diagnosis and to obtain information for genetic counseling. The study was retrospective and descriptive. From August 2014 to December 2015, 39 individuals were assessed using FISH and/or MLPA; in 15 cases, chromosomal microarray (CMA) analysis was carried out. Of 39 registered individuals, we found deletions in the 22q11.2 region in 10 individuals (8 individuals with 22q11.2DS and 2 individuals presenting with atypical deletions in the 22q11.2 region: 1 distal deletion and 1 central deletion). In one case with a typical 22q11.2 deletion, a familial balanced translocation was detected. In another case without a 22q11.2 deletion, a 6p duplication concomitant with a 9p deletion was detected by CMA. Clinical data are reported and diagnostic investigations are discussed. Essential aspects for the understanding of different diagnostic techniques of genomic imbalances are considered, and the 4 cases described underline the complexity and the difficulties involved in the diagnostic process. The approach is informative for clinical practice and may be applied in other contexts of genomic imbalance investigation in microdeletion syndromes.
Keywords: Balanced translocation, Deletion 9p, 22q11.2 deletion syndrome, Duplication 6p, Genomic imbalance, Microdeletion syndromes
The great advance of knowledge and technologies in the area of genetics and molecular biology has allowed the development of different methods for the detection of genomic imbalances (GI). These are defined as losses or gains of DNA segments [Stofanko et al., 2013; Vieira et al., 2013; McDonald-McGinn et al., 2015]. Some GIs are recurrent and known as microdeletion syndromes, some of them already well established and clinically recognizable, e.g., the 22q11.2 deletion syndrome (22q11.2DS; OMIM 188400]. This stands out as the most common microdeletion syndrome, with an incidence of 1/4,000–1/5,000 births [Swillen et al., 2000]. The main features of this syndrome include congenital cardiac malformations, palatal abnormalities, common facial dysmorphisms, immunodeficiency, neonatal hypocalcemia, learning disability, and development delay [Fernández et al., 2009; Monteiro et al., 2013; Vieira et al., 2015], indicating the need of healthcare specialists.
Because of clinical heterogeneity, 22q11.2DS is not always simple to detect [Monteiro et al., 2013]. Moreover, depending on the genetic test used, a negative result may not completely disregard the diagnosis, requiring further investigation in cases of persistent clinical suspicion. For many years, FISH was considered as a “gold standard” for detecting this microdeletion [Fernández et al., 2005; Oh et al., 2007; Sgardioli et al., 2015]. Currently, new targeted techniques such as MLPA, analysis of polymorphic DNA markers, real-time PCR, and quantitative fluorescence PCR have been used in screening 22q11.2DS [Swillen et al., 2000; Jalali et al., 2008; Vieira et al., 2013]. Each one has advantages and limitations (GeneTests; https://www.genetests.org/).
In addition to these, chromosomal microarray analysis (CMA) technique, aCGH, or SNP array detects GIs throughout the whole genome using a single test. Currently, this is indicated as a first-tier test for diagnostic screening of patients with multiple congenital anomalies and/or intellectual disabilities [Miller et al., 2010]. CMA is widely available in genetic laboratories and has been the preferred choice for the diagnosis of a 22q11.2 deletion in some countries [McDonald-McGinn and Sullivan, 2011]. This technique, however, does not allow the identification of balanced chromosomal aberrations or low-percentage mosaicism, both being the main complicating factors for genetic counseling [Bi et al., 2013].
Considering different options for 22q11.1DS diagnosis, this study presents some aspects in understanding the different techniques for the investigation of GIs and reports 4 cases that showed the complexity and difficulties involved in the process of diagnosis and, therefore, requiring different techniques.
Patients and Methods
From August 2014 to December 2015, 39 individuals were registered in the Brazilian Database of Craniofacial Anomalies/22q11.2 Deletion Syndrome (http://www.fcm.unicamp.br/fcm/en/cranio-face-brasil/projeto-cranio-face-brasil). For clinical evaluation, a standard protocol based on clinical criteria for suspicion of 22q11.1DS was used proposed by Monteiro et al. [2013]. The biological samples were tested by FISH and/or MLPA techniques for 22q11.2 deletion screening, and 15 cases were also tested by CMA.
Cultured peripheral blood lymphocytes were used for FISH technique as well as for G-banding. Genomic DNA extracted from peripheral blood with the NucleoSpin® Blood XL kit (Macherey-Nagel GmbH & Co. KG) was used for MLPA and CMA, according to the manufacturer's instructions.
Fluorescence in situ Hybridization
Probes used for the 22q11.2 region were DiGeorge/VCFS TUPLE1 + 22q13.3 Deletion Probe Combination (Cytocell Aquarius®). Hybridization and washing procedures were carried out according to the manufacturer's instructions. One hundred interphase nuclei per sample were analyzed, using a BX51-BF-II/BX2 fluorescence microscope (Olympus®) with appropriate filters, and the images were recorded with the FISHView software (Applied Spectral Imaging®).
Multiplex Ligation-Dependent Probe Amplification
The P250-B2 DiGeorge (MRC-Holland MLPA®) kit was used according to the manufacturer's protocol. Results were analyzed with the GeneMapper® software (Applied Biosystems™) and data were processed in a Microsoft® Excel spreadsheet, elaborated specifically for this kit by the National Genetics Reference Laboratory-Manchester (http://www.ngrl.org.uk/Manchester/projects/informatics/mlpa).
Chromosomal Microarray Analysis
This technique was carried out with the Cytoscan 750K or Cytoscan HD (Affymetrix®) according to the manufacturer's instructions. Analysis of the results was performed using the Chromosome Analysis Suite software, version 3.1.0.15 (r9069) (Affymetrix).
Results
Among the 39 individuals registered in the period of this study (14 males and 25 females), ages ranged from 4 months to 23 years. The main clinical signs which led to suspicion are described in Table 1.
Table 1.
Main clinical features found in 39 cases with 22q11DS suspicion
| Clinical features | Frequency, % |
|---|---|
| Congenital heart disease | 69.2 |
| Palatal abnormalities | 64.1 |
| Facial dysmorphisms | 74.4 |
| Immunological changes | 35.9 |
| Development changes | 71.8 |
| Hypoacusis | 23.1 |
| Ophthalmologic changes | 30.8 |
| Neurological changes | 12.8 |
| Abnormalities in the urinary tract | 20.5 |
| Abnormalities in the gastrointestinal tract | 30.8 |
| Skeletal abnormalities | 56.4 |
Using MLPA and/or FISH techniques, we found deletions in the 22q11.2 region in 10 individuals (25.6%), 8/10 (20.5%) with the common 3-Mb deletion and 2/10 (5.1%) with atypical 22q11.2 deletions. In 4 cases, we found unusual results. Among these, in 2 cases, karyotyping and CMA revealed abnormalities in other genomic regions; in one case, MLPA revealed an atypical distal 22q11.2 deletion, and in another case, CMA revealed an atypical central 22q11.2 deletion in a patient without clinical suspicion. These 4 cases are described as follows.
Case 1
The proband was referred for genetic testing at 2 years and 9 months, presenting with congenital heart disease and dysmorphisms. At birth, the girl's weight was 2,210 g (<3rd percentile [<P3]), height 46 cm (P3), and her head circumference was 30 cm (<P3). She had recurrent pneumonia and gastroesophageal reflux as well as motor development and speech delay (she walked with 23 months and uttered first words at 27 months).
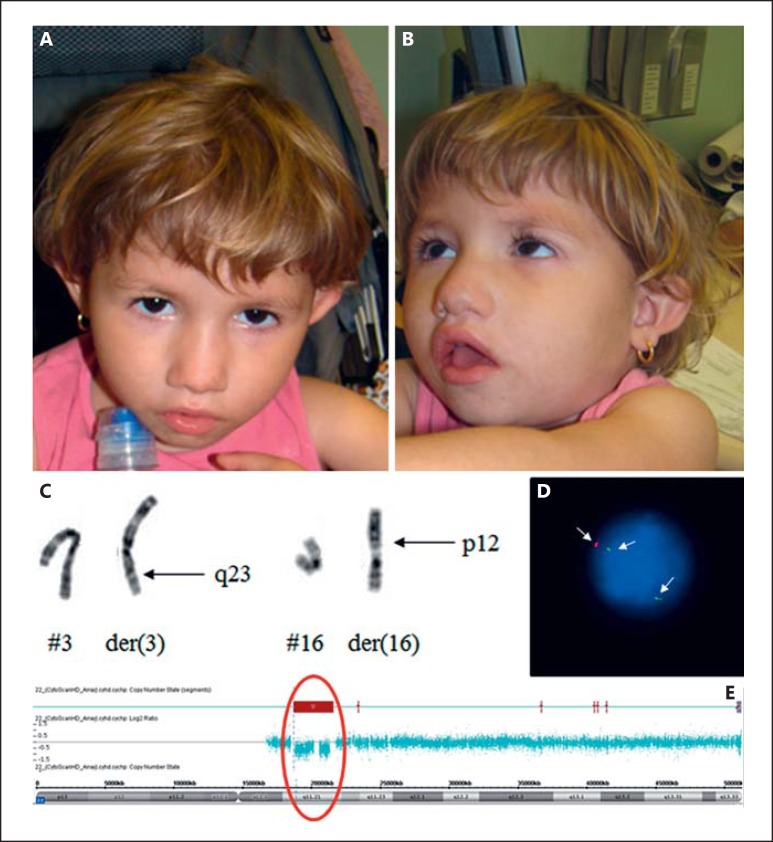
At the age of 2 years and 9 months, her weight was 9,530 g (<P3), height 81.5cm (<P3), and head circumference 45 cm (<P3). Clinical evaluation revealed a tubular nose, retrognathia, question mark left ear, retracted columella, short nasal filter, thin lips, bifid uvula, widely spaced nipples, long and proximally placed thumbs, and an accessory ligament on the fifth finger (Fig. 1A, B). The patient also had a nasal voice, speech and motor delay, behavior disorder, agitation, and sleep difficulties. An echocardiogram revealed small muscular interventricular communication and atrial septal defect. Nasofibroscopy revealed velopharyngeal insufficiency. The G-banded karyotype showed an apparently balanced translocation between chromosomes 3 and 16: 46,XX,t(3;16)(q23;p12)[20] (Fig. 1C). CMA revealed a 22q11.2 deletion of 3 Mb: arr[GRCh37] 22q11.21(18916842_21798907)×1 (Fig. 1E), confirmed by FISH (Fig. 1D). Other GIs were not found by CMA, suggesting that the translocation is balanced. The same translocation was found in the father's karyotype.
Fig. 1.
Patient 1 at the age of 2 years and 9 months. A, B Clinical pictures of the proband showing hooded eyelids, tubular nose, downturned oral commissures, question mark ear, retracted columella, and retrognathia. C Chromosomes 3 and 16 showing an apparently balanced translocation: 46,XX,t(3;16)(q23;p12)[20]. D Interphase FISH showing the 22q11.2 deletion. E CMA hybridization profile of chromosome 22 showing a common deletion of 3 Mb in region q11.2: arr[GRCh37] 22q11.21 (18916842_21798907)×1.
Case 2
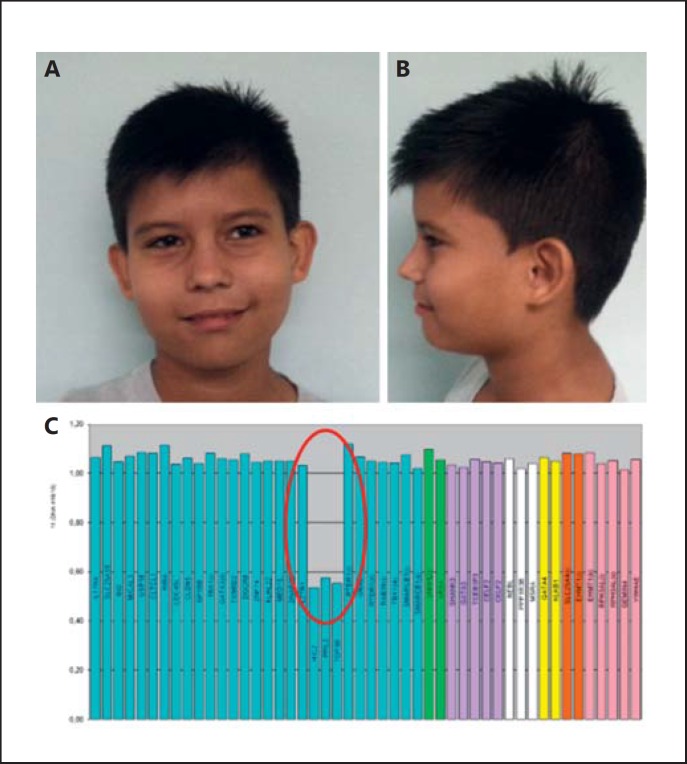
This patient was referred for genetic testing at 9 years and 11 months with subtle facial dysmorphisms (Fig. 2A, B), congenital heart disease, and a nasal voice. Medical records revealed surgically corrected hypospadia. At birth, the boy's weight was 2,500 g (P3), height 47 cm (P3), and his head circumference was 34 cm (P3). Motor development was adequate; however, he evolved with attention deficit hyperactivity disorder and dyslexia. At 11 years, his weight was 48.8 kg (P97), height 147 cm (P75), and head circumference 56 cm (P75–97). Dysmorphology evaluation revealed fifth finger clinodactyly and flat feet. An echocardiogram revealed a ventricular septal defect, and abdominal ultrasound showed a bilateral inguinal hernia. X-ray of the spine showed an S dorsolumbar curve, scoliosis, and lordosis. The G-banded karyotype was normal. MLPA technique identified an atypical distal 22q11.2 deletion, including HIC2, PPIL2, and TOP3B probes (Fig. 2C).
Fig. 2.
Patient 2 at 9 years and 11 months. A, B Facial features of the proband showing subtle dysmorphisms such as epiblepharon and a thin upper lip. C MLPA results showing an atypical 22q11.2 deletion, including HIC2, PPIL2, and TOP3B probes.
Case 3
This patient was referred for genetic testing at 2 years and 1 month. At birth, the boy's weight was 2,890 g (P10–25), height 46 cm (<P3), and head circumference 37 cm (P75–97). Motor development was delayed; he sat up at 9 months, walked and spoke monosyllables at 2 years of age.
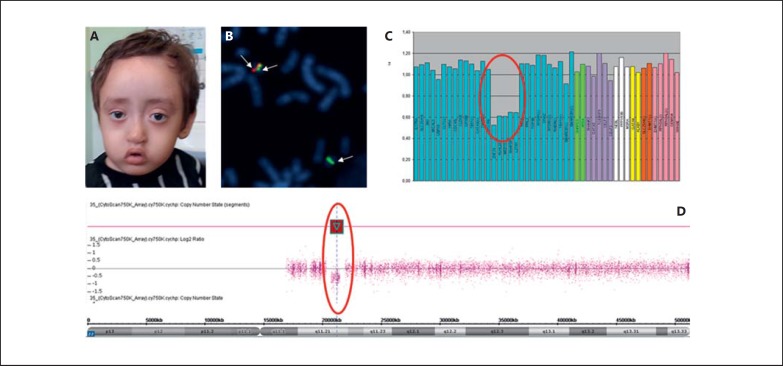
At 2 years and 1 month, his weight was 7,600 g (<P3), height 68 cm (<P3), and his head circumference was 45 cm (P50). Dysmorphology evaluation revealed a high forehead and frontal hair whorl, thickened metopic region, lateral third rarefaction of eyebrows, shallow orbits and supraorbital ridge, short and slightly oblique palpebral fissure, short columella, short nasal filter, hypoplasia of the nasal wing and anteverted nostrils, everted upper lip, and downturned oral commissures (Fig. 3A) as well as a high palate, long fingers, bilateral single transverse palmar crease, absence of distal transverse flexion creases, overlapping and long toes, limited extension of the elbows, dimpled chin, knees, and elbows. His voice was considered dysphonic. Nasofibroscopy revealed an adhesion in the middle and early third of the vocal folds. An echocardiogram detected a patent foramen ovale and physiological pulmonary branch stenosis. Abdominal ultrasound showed left-sided pyelocalyceal dilation. Bone inventory evidenced a left radioulnar synostosis and an abnormal position of the left fibula. Due to multiple congenital anomalies, without a specific etiologic suspicion, investigation initiated by karyotyping, which had normal results, and CMA identified an atypical central deletion of approximately 600 kb in the 22q11.2 region: arr[GRCh37] 22q11.21(20716876_21800471)×1 (Fig. 3D), confirmed by FISH with BAC probes (Fig. 3B) and MLPA (Fig. 3C).
Fig. 3.
Patient 3 at the age of 2 years and 1 month. A Clinical features of the proband showing a high forehead, eyebrows with lateral enlargement, shallow orbits and supraorbital ridges, slightly downslanting palpebral fissures, malar hypoplasia, short nasal filter, and an everted upper lip with downturned oral commissures. B FISH analysis of BAC clones showing 2 signals in the control region 22q22.3 (RP11-876A20) marked in green and a red signal in the region 22q11.2 (RP11-1058B20), indicating a partial deletion of the long arm of chromosome 22 in region 11.21. C MLPA results showing an atypical 22q11.2 deletion. D CMA hybridization profile of chromosome 22 showing an atypical 22q11.2 deletion of approximately 600 kb: arr[GRCh37] 22q11.21(20716876_21800471)×1.
Case 4
This 3-year-old patient was referred for genetic testing because of multiple congenital anomalies. At birth, the girl's weight was 2,430 g (P3), height 46 cm (P3), and her head circumference was 32 cm (<P3). Neuropsychomotor development was delayed (she walked at 24 months, spoke words and phrases at 26 and 36 months, respectively, and acquired complete control of the bladder and anal sphincters at 6 years).
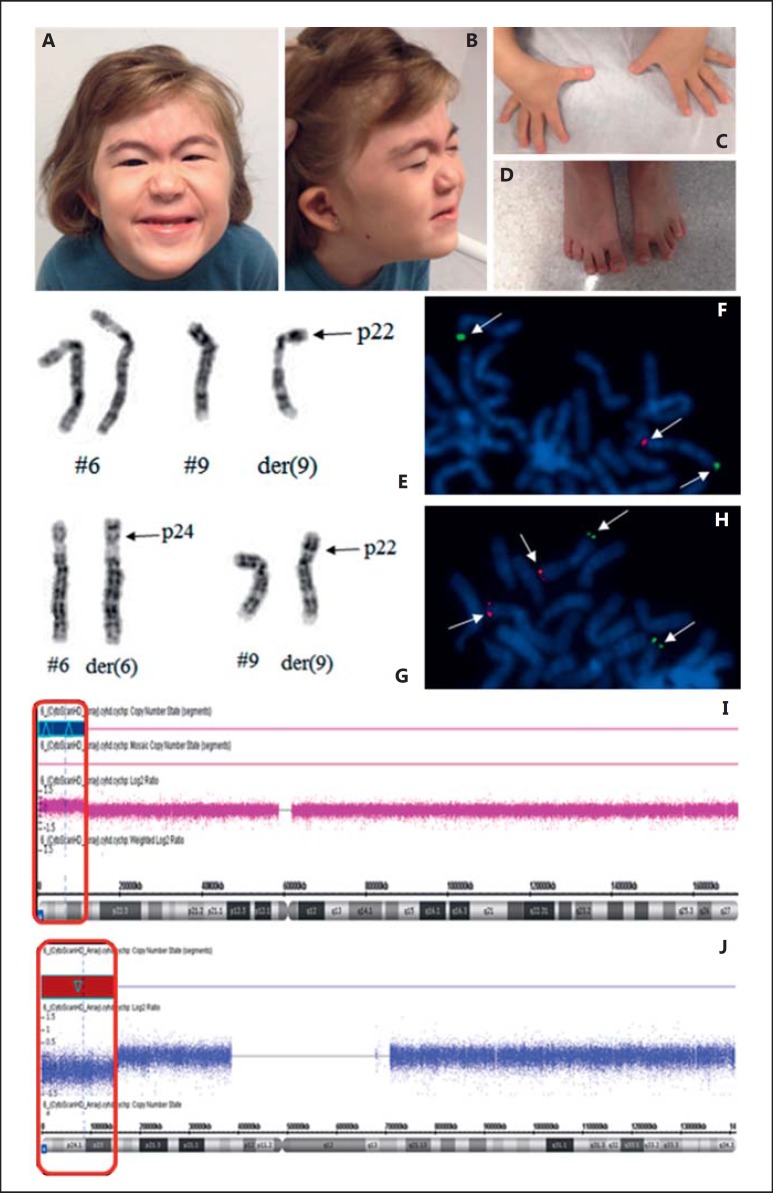
Dysmorphology evaluation at 8 years of age showed slight thickening of metopic suture; facial asymmetry without craniosynostosis; asymmetric, low-set, and dysmorphic ears; port-wine stain on the glabellar area; arched, thin, and laterally enlarged eyebrows; mild synophrys; ocular hypertelorism; short palpebral fissures; convergent strabismus following abducens nerve palsy (cranial nerve VI palsy); a depressed nasal bridge and root as well as a nasal tip with a square shape; small mouth; long and faint nasal filter; everted lower lip; malocclusion; slightly wide central upper incisor; high and narrow anterior palate; posterior palatal fissure; short webbed neck; ligament laxity in hands; slightly clubbed thumbs; shortening of the right fourth and fifth metatarsals and the left third to fifth metatarsals as well as overlapping of the left fourth and third toes (Fig. 4C). She also presented with motor and language delay, nasal voice, intellectual disability, attention deficit hyperactivity disorder, and somatic growth deficit. At 9 years old, her weight was 21 kg (P3), height 108.5 cm (<P3), head circumference 51.8 cm (P50); dysmorphisms were still present. MRI of the skull identified a mild reduction in the thickness of the corpus callosum between body and splenium. Abdominal and pelvic ultrasounds revealed left renal agenesis, confirmed by renal scintigraphy, excretion urography, and abdominal CT. Echocardiogram and skull X-rays showed normal results. Following complete evaluation and registration in the Brazilian Database on Craniofacial Anomalies/22q11.2 Deletion Syndrome, the patient fulfilled the clinical criteria proposed by Monteiro et al. [2013] for investigation of 22q11.2DS. The G-banded karyotype was normal, and FISH was negative for 22q11.2 deletion. CMA identified a duplication of approximately 11.5 Mb in the short arm of chromosome 6: arr[GRCh37] 6p25.3p24.1(156974_11629164)×3 (Fig. 4I) and a deletion of approximately 14.9 Mb in the short arm of chromosome 9: arr[GRCh37] 9p24.3p22.3(203861_15125789)×1 (Fig. 4J). Karyotyping was repeated and revealed a derivative chromosome 9: 46,XX,der(9)t(6;9)(p25.3;p24.3)[20], confirming CMA results. The maternal karyotype showed a balanced translocation: 46,XX,t(6;9)(p25.3;p24.3)[20] (Fig. 4E). Karyotype results were confirmed by FISH (Fig. 4G).
Fig. 4.
Patient 4 at 8 years of age. A-D Clinical pictures of the proband. The facial appearance shows mild asymmetry, arched eyebrows with mild lateral enlargement, mild synophrys, epiblepharon, a port wine stain on the glabellar area, short palpebral fissures, convergent strabismus to the left, low nasal bridge, square nasal tip, faint nasal filter (A), as well as low-set and dysmorphic ears, everted lower lip, and a short neck (B). The patient's hands show slightly clubbed thumbs (C), and her feet show shortening of the right 4th and 5th metatarsals and the left 3rd-5th metatarsals as well as an overlapping of the left 3rd and 4th toes (D). E Chromosomes 6 and 9 showing the derivative chromosome 9: 46,XX,der(9) t(6;9)(p25.3;p24.3)[20]. F FISH analysis of BAC clones of a partial metaphase with a signal for the probe of the 9p24 region (RP11-48M17) marked in red and 2 signals for the probe of the 9q34 region (RP11-644H13) in green (indicated by arrows), revealing a partial deletion of the short arm of chromosome 9 in region 24. G Chromosomes 6 and 9 of the mother showing a balanced translocation: 46,XX,t(6;9) (p25.3;p24.3)[20]. H FISH analysis of BAC clones of a partial metaphase with 2 signals for the probe of the 9p24 region (RP11-48M17) marked in red and 2 signals for the probe in the region 9q34 (RP11-644H13) in green (indicated by arrows); however, this result shows that the region 9p24 is located in another chromosome and confirms the translocation revealed by karyotyping. I CMA hybridization profile of chromosome 6 of the patient showing a duplication of approximately 11.5 Mb in the short arm of chromosome 6: arr[GRCh37] 6p25.3p24.1(156974_11629164)×3. J CMA hybridization profile of chromosome 9 of the patient showing a deletion of approximately 14.9 Mb in the short arm of chromosome 9: arr[GRCh37] 9p24.3p22.3(203861_15125789)×1.
Discussion
With technological advancement and the current availability of different genetic testing methods to investigate GIs, it is necessary to be informed about their applications, advantages, and limitations, allowing a fast and accurate diagnosis of patients with clinical phenotypes possibly caused by a GI.
22q11.2DS is suitable to illustrate this approach because of its high prevalence and clinical heterogeneity [Vieira et al., 2015]. Its clinical suspicion may be difficult; therefore, accurate and precise clinical methods are fundamental in achieving diagnosis, treatment, and management. Follow-up care with a multidisciplinary team, according to each patient's needs, as well as adequate genetic counseling for the families are necessary [Oskarsdóttir et al., 2004; Oh et al., 2007].
Patients with clinical suspicion of 22q11.2DS are often seen by several medical specialties and health professionals. Lack of understanding about limitations of each diagnostic technique by these professionals may lead to difficulties in the interpretation of the clinical results. In this study, 4 cases exemplify the necessity of a combined diagnostic approach, using appropriate and complementary techniques.
In case 1, karyotyping was the first test performed and revealed a reciprocal translocation apparently balanced and inherited. Although this chromosomal abnormality probably does not have impact on the phenotype of the patient, it could be interpreted as causative by nonspecialists, delaying the diagnosis. On the other hand, this translocation revealed the possibility of new unbalanced rearrangements in the couple's offspring recommending genetic counseling. CMA revealed a 3-Mb microdeletion in the 22q11.2 region, which was confirmed by FISH. This technique was used for diagnosis and also to detect other GIs possibly associated with the chromosomal translocation. It was performed since approximately 30% of the cases harboring reciprocal translocations present GIs associated to the breakpoints [Baptista et al., 2008]. Thus, different approaches were necessary for a complete diagnosis and also for providing clarification for the family. The identification of a chromosomal translocation would not have been possible using only CMA or MLPA, which would only have detected the 22q11.2 deletion. One of the limitations of these techniques is the inability to identify balanced rearrangements [Bi et al., 2013].
In case 2, with a normal karyotype, investigation for 22q11.2DS was performed by MLPA, revealing an atypical 22q11.2 distal deletion. This microdeletion could not be identified by FISH with the probe commonly used for the proximal 22q11.2 region, confirming that MLPA is quite effective in detecting atypical deletions involving smaller and variable regions within the 22q11.2 region [Fernández et al., 2005; Jalali et al., 2008; Molck et al., 2013].
In case 3, considering the presence of multiple congenital anomalies and a normal karyotype, CMA was the approach initially chosen, which revealed an atypical central deletion of approximately 600 kb, confirmed by MLPA. This case demonstrates the wide clinical heterogeneity of deletions in 22q11.2 region.
In case 4, with a previously confirmed normal karyotype and no detected deletion in the above-mentioned region tested by FISH, CMA identified a duplication of approximately 11 Mb in the short arm of chromosome 6 and a deletion of approximately 14,9 Mb in the short arm of chromosome 9. Because of the large chromosome segments involved, usually visible by karyotyping, G-banding was repeated, which confirmed CMA results, and revealed that the mother had an apparently balanced translocation. In this case, the first karyotype was normal (false negative), probably because the regions involved have similar lengths and the banding patterns make diagnosis more difficult. Furthermore, the cytogenetic resolution of the previous test may not have been adequate, which is a technical limitation [Bi et al., 2013].
Case 4 demonstrates the spectrum of clinical findings in different genetically determined conditions. Some studies report patients with a 9p deletion presenting with facial asymmetry, hypertelorism, short palpebral fissure, strabismus, asymmetric and dysmorphic ears, wide nasal bridge, long nasal filter, high palate, malocclusion, and psychomotor retardation [Freitas et al., 2011; Recalcati et al., 2012; Spazzapan et al., 2016]. Whereas patients with a 6p duplication are reported having language disorders, motor delay, and intellectual deficit [Vermeesch et al., 2004]. There are only 2 cases reported in the literature with 6p duplication concomitant with 9p deletion. In both cases, the patients present alterations as described above, including a short neck as well as hand and foot abnormalities [Eden et al., 1985; Lytle et al., 1989]. All these clinical features are present in patient 4, and many of these signs have already been described in 22q11.2DS patients, which restate the clinical variability of this condition.
The present study reinforces the difficulties to request investigation and to diagnose the cases with 22q11.2DS suspicion, and describes the application of different techniques that may be used for investigating microdeletion syndromes. Targeted approaches, such as FISH, real-time PCR, analysis of polymorphic DNA markers, and quantitative fluorescent PCR, though well established and fast, are only effective in detecting common deletions [Fernández et al., 2005; Oh et al., 2007; Molck et al., 2013; Vieira et al., 2013; Poirsier et al., 2016]. Thus, negative results may lead to false negative diagnosis, since atypical 22q11.2 deletions may occur [Beaujard et al., 2009; Molck et al., 2013].
In this context, MLPA is a more effective technique for the diagnosis of 22q11.2DS, and identifying typical/atypical deletions, these in smaller size and/or variable regions within the 22q11.2 region [Vorstman et al., 2006; Molck et al., 2013, 2015]. MLPA detected 4 deletions of 3 Mb and 1 atypical distal deletion that would not have been detected by FISH. In addition, it confirmed 1 atypical central 22q11.2 deletion, initially detected by CMA. However, MLPA is also a targeted approach and limited to the detection of approximately 48 genomic regions. In this way, GIs outside the 22q11 region or at loci not represented in the MLPA kit, as well as balanced chromosomal aberrations, cannot be detected.
In the US and in European countries, CMA is an effective widely used method detecting 22q11.2 and other GIs [McDonald-McGinn and Sullivan, 2011]. However, an overall agreement has not yet been found for its use in detecting microdeletion syndromes. In general, a targeted approach, such as FISH or MLPA, is only used when there is a very strong suspicion of 22q11.2DS. In Brazil, FISH and MLPA have been used for specific screening of 22q11.2 deletion, using CMA just for cases with negative FISH and MLPA results, mainly because CMA is still too expensive [Jehee et al., 2011; Vieira et al., 2013]. This approach has been adopted as a low-cost strategy by our research group.
The geneticist may carry out the research and individualized genetic counseling of all cases, which frequently requires the evaluation of the parents. In cases of unconfirmed and maintained suspicion, other strategies should be considered by the genetics expert.
It is worth mentioning that in many cases in spite of technological advancement, karyotyping should not be discarded, especially for the parents. It can be the key tool for genetic counseling as seen in 2 of the 4 cases presented in this study [Sgardioli et al., 2015].
Understanding the complexity of the 22q11.2DS diagnostic approach, from clinical suspicion to diagnostic confirmation - and its specific challenges - make the present article relevant for various health professionals. They will be in the front line, indicating and interpreting findings, always aware of limitations and clinical consequences of each result.
Statement of Ethics
This study was approved by the Ethics Committee Board of the University of Campinas, as stated in report CAAE16525913. 1.0000.5404. All participants or their legal guardians signed the informed consent form.
Disclosure Statement
The authors declare no conflicts of interest.
Acknowledgments
The authors thank the patients and their families for their cooperation. We thank the biologist Nilma Lúcia Viguetti Campos. This study was supported by the State of São Paulo Research Foundation (FAPESP), grant 2012/51799-6 and the National Council for Scientific and Technological Development (CNPq), grant 460422/2014-6). V.L.G.-d.-S.-L. is supported by CNPq, grant 304455/2012-1.
References
- Baptista J, Mercer C, Prigmore E, Gribble SM, Carter NP, et al. Breakpoint mapping and array CGH in translocations: comparison of a phenotypically normal and an abnormal cohort. Am J Hum Genet. 2008;82:927–936. doi: 10.1016/j.ajhg.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaujard MP, Chantot S, Dubois M, Keren B, Carpentier W, et al. Atypical deletion of 22q11.2: detection using the FISH TBX1 probe and molecular characterization with high-density SNP arrays. Eur J Med Genet. 2009;52:321–327. doi: 10.1016/j.ejmg.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Bi W, Borgan C, Pursley AN, Hixson P, Shaw CA, et al. Comparison of chromosome analysis and chromosomal microarray analysis: what is the value of chromosome analysis in today's genomic array era? Genet Med. 2013;15:450–457. doi: 10.1038/gim.2012.152. [DOI] [PubMed] [Google Scholar]
- Eden MS, Thelin JW, Michalski K, Mitchell JA. Partial trisomy 6p and partial monosomy 9p from a de novo translocation 46,XY, −9, +DER(9)T(6:9)(p211:p24) Clin Genet. 1985;28:375–384. doi: 10.1111/j.1399-0004.1985.tb02210.x. [DOI] [PubMed] [Google Scholar]
- Fernández L, Lapunzina P, Arjona D, López Pajares I, García-Guereta L, et al. Comparative study of three diagnostic approaches (FISH, STRs and MLPA) in 30 patients with 22q11.2 deletion syndrome. Clin Genet. 2005;68:373–378. doi: 10.1111/j.1399-0004.2005.00493.x. [DOI] [PubMed] [Google Scholar]
- Fernández L, Nevado J, Santos F, Heine-Suñer D, Martinez-Glez V, et al. A deletion and a duplication in distal 22q11.2 deletion syndrome region. Clinical implications and review. BMC Med Genet. 2009;10:48. doi: 10.1186/1471-2350-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas ÉL, Gribble SM, Simioni M, Vieira TP, Silva-Grecco RL, et al. Maternally inherited partial monosomy 9p (pter → p24.1) and partial trisomy 20p (pter → p12.1) characterized by microarray comparative genomic hybridization. Am J Med Genet A. 2011;155A:2754–2761. doi: 10.1002/ajmg.a.34168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalali GR, Vorstman JA, Errami A, Vijzelaar R, Biegel J, et al. Detailed analysis of 22q11.2 with a high density MLPA probe set. Hum Mutat. 2008;29:433–440. doi: 10.1002/humu.20640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehee FS, Takamori JT, Medeiros PF, Pordeus AC, Latini FR, et al. Using a combination of MLPA kits to detect chromosomal imbalances in patients with multiple congenital anomalies and mental retardation is a valuable choice for developing countries. Eur J Med Genet. 2011;54:e425–432. doi: 10.1016/j.ejmg.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Lytle C, Wade J, Farrier A, Flohrschutz F, 3rd, Hecht B, Allanson J. Duplication 6p and deletion 9p. J Med Genet. 1989;26:64–66. doi: 10.1136/jmg.26.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald-McGinn DM, Sullivan KE. Chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome) Medicine (Baltimore) 2011;90:1–18. doi: 10.1097/MD.0b013e3182060469. [DOI] [PubMed] [Google Scholar]
- McDonald-McGinn DM, Sullivan KE, Marino B, Philip N, Swillen A, et al. 22q11.2 deletion syndrome. Nat Rev Dis Primers. 2015;1:15071. doi: 10.1038/nrdp.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molck MC, Vieira TP, Sgardioli IC, Simioni M, Dos Santos AP, et al. Atypical copy number abnormalities in 22q11.2 region: report of three cases. Eur J Med Genet. 2013;56:515–520. doi: 10.1016/j.ejmg.2013.07.002. [DOI] [PubMed] [Google Scholar]
- Molck MC, Vieira TP, Simioni M, Sgardioli IC, dos Santos AP, et al. Distal 22q11.2 microduplication combined with typical 22q11.2 proximal deletion: a case report. Am J Med Genet A. 2015;167A:215–220. doi: 10.1002/ajmg.a.36809. [DOI] [PubMed] [Google Scholar]
- Monteiro FP, Vieira TP, Sgardioli IC, Molck MC, Damiano AP, et al. Defining new guidelines for screening the 22q11.2 deletion based on a clinical and dysmorphologic evaluation of 194 individuals and review of the literature. Eur J Pediatr. 2013;172:927–945. doi: 10.1007/s00431-013-1964-0. [DOI] [PubMed] [Google Scholar]
- Oh AK, Workman LA, Wong GB. Clinical correlation of chromosome 22q11.2 fluorescent in situ hybridization analysis and velocardiofacial syndrome. Cleft Palate Craniofac J. 2007;44:62–66. doi: 10.1597/05-192. [DOI] [PubMed] [Google Scholar]
- Oskarsdóttir S, Vujic M, Fasth A. Incidence and prevalence of the 22q11 deletion syndrome: a population-based study in Western Sweden. Arch Dis Child. 2004;89:148–151. doi: 10.1136/adc.2003.026880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirsier C, Besseau-Ayasse J, Schluth-Bolard C, Toutain J, Missirian C, et al. A French multicenter study of over 700 patients with 22q11 deletions diagnosed using FISH or aCGH. Eur J Hum Genet. 2016;24:844–851. doi: 10.1038/ejhg.2015.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recalcati MP, Bellini M, Norsa L, Ballarati L, Caselli R, et al. Complex rearrangement involving 9p deletion and duplication in a syndromic patient: genotype/phenotype correlation and review of the literature. Gene. 2012;502:40–45. doi: 10.1016/j.gene.2012.04.030. [DOI] [PubMed] [Google Scholar]
- Sgardioli IC, Vieira TP, Simioni M, Monteiro FP, Gil-da-Silva-Lopes VL. 22q11. 2 Deletion syndrome: laboratory diagnosis and TBX1 and FGF8 mutation screening. J Pediatr Genet. 2015;4:017–022. doi: 10.1055/s-0035-1554976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spazzapan P, Arnaud E, Baujat G, Nizon M, Malan V, et al. Clinical and neuroradiological features of the 9p deletion syndrome. Childs Nerv Syst. 2016;32:327–335. doi: 10.1007/s00381-015-2957-2. [DOI] [PubMed] [Google Scholar]
- Stofanko M, Gonçalves-Dornelas H, Cunha PS, Pena HB, Vianna-Morgante AM, Pena SD. Simple, rapid and inexpensive quantitative fluorescent PCR method for detection of microdeletion and microduplication syndromes. PLoS One. 2013;8:e61328. doi: 10.1371/journal.pone.0061328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swillen A, Vogels A, Devriendt K, Fryns JP. Chromosome 22q11 deletion syndrome: update and review of the clinical features, cognitive-behavioral spectrum, and psychiatric complications. Am J Med Genet. 2000;97:128–135. doi: 10.1002/1096-8628(200022)97:2<128::aid-ajmg4>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Vermeesch JR, Thoelen R, Fryns JP. A familial complex chromosome translocation resulting in duplication of 6p25. Ann Genet. 2004;47:275–280. doi: 10.1016/j.anngen.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Vieira TP, Sgardioli IC, Gil-da-Silva-Lopes VL. Genetics and public health: the experience of a reference center for diagnosis of 22q11.2 deletion in Brazil and suggestions for implementing genetic testing. J Community Genet. 2013;4:99–106. doi: 10.1007/s12687-012-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira TP, Monteiro FP, Sgardioli IC, Souza J, Fett-Conte AC, et al. Clinical features in patients with 22q11.2 deletion syndrome ascertained by palatal abnormalities. Cleft Palate Craniofac J. 2015;52:411–416. doi: 10.1597/13-233. [DOI] [PubMed] [Google Scholar]
- Vorstman JA, Jalali GR, Rappaport EF, Hacker AM, Scott C, Emanuel BS. MLPA: a rapid, reliable, and sensitive method for detection and analysis of abnormalities of 22q. Hum Mutat. 2006;27:814–821. doi: 10.1002/humu.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]