Abstract
Chromosomal mosaicism, which represents a diagnostic challenge for detection and interpretation, has been described in several genetic conditions. It can contribute to a large phenotypic variation in diseases. At analysis of a well-characterized cohort of 714 patients with neurodevelopmental disorders (NDDs) of unknown etiology using a high-resolution chromosomal microarray platform, we found 2 cases (0.28%) of low-level mosaicism and defined a previously detected extra chromosome in a third patient. Two of the cases were mosaics for segmental imbalances (a partial trisomy 3q26.1q27.3 and a partial monosomy 18q21.2qter with 14.6 and 20% mosaic ratios in lymphocytes, respectively), and 1 was a mosaic for an entire chromosome (trisomy 14, mosaic ratio 20%). Our diagnostic yield is in line with the ratios previously published in patients with intellectual disability. Notably, the partial trisomy 3q26.1q27.3 case is an example of a rare and unusual class of a rearranged neocentric ring chromosome, which can neither be categorized in class I, nor in class II of such rearrangements. Our cases further elucidate the phenotypes related to the aberrations of the specific chromosome segments observed and underline the important role of low-level mosaics in the pathogenesis of NDDs of unknown etiology even in the absence of clinical signs of mosaicism.
Keywords: Deletion 18q, Microarray, Mosaicism, Neocentromere, Neurodevelopmental disorders
Chromosomal mosaicism is defined as the presence of 2 or more chromosomally distinct cell lines in one individual. Mosaics are often difficult to detect due to reasons such as subtle phenotypic abnormalities, technical limitations, and tissue specificity. Advances in chromosomal microarray analysis (CMA) have made this method a powerful genome-wide analysis tool able to detect a wide range of aberrations, including mosaics of low level [Ballif et al., 2006; Cheung et al., 2007; Pham et al., 2014]. It is well-known that the standard analysis of 15 cells obtained by traditional cytogenetics only provides a diagnostic accuracy of ≥20% for mosaicism of an entire chromosome [Hook, 1977]. Detection limits of CMA have been determined at 5% for whole chromosomes and 20% for segmental mosaics [Ballif et al., 2006; Conlin et al., 2010; Scott et al., 2010; Hoang et al., 2011]. It has been suggested that for the detection of segmental low-level mosaics (<10%), CMA coupled with conventional chromosome studies on more than 15 cells should be performed [Bi et al., 2013].
In this work, we report on our results concerning chromosomal mosaicism obtained in a cohort of 714 patients with a wide range of isolated or syndromic neurodevelopmental disorders (NDDs) and their phenotypical description.
Patients and Methods
Patient Cohort
The cohort was previously reported [Asadollahi et al., 2014]. Briefly, excluding patients with large-scale chromosomal aberrations (copy number variants >10 Mb) or clinically recognized recurrent microaberration syndromes, we investigated 714 patients with NDDs with or without further congenital anomalies by CMA analysis during a period of 3 years.
Extraction of DNA
DNA extraction was performed from peripheral blood using a chemagen-automated device according to the manufacturer's instruction (Perkin Elmer, Baesweiler, Germany).
Microarray Analysis
DNA was analyzed with Affymetrix Genome-Wide Human SNP Array 6.0 (1.8 million markers; 79 patients), Affymetrix Cytogenetics 2.7 (2.7 million markers; 423 patients), or CytoScan HD (2.6 million markers; 212 patients; Affymetrix Inc., Santa Clara, CA, USA). Array hybridization was performed according to the manufacturer's protocols. Data analysis was performed with the Chromosome Analysis Suite (ChAS) software (Affymetrix). For the mosaic analysis, no filter was applied.
Fluorescence in situ Hybridization
Fluorescence in situ hybridization (FISH) analyses were performed on metaphase preparations or on interphase nuclei using commercial BAC probes (Illumina, San Diego, CA, USA) according to standard protocols. An α-satellite DNA probe specific for all human centromeres (Kreatech, Amsterdam, Netherlands) and an α-satellite chromosome 3-specific probe (Cytocell, Cambridge, UK) were used to characterize the neocentromere ring chromosome (NRC) 3.
Results
Low-Level Mosaicism
In 3/714 individuals (0.4%), we detected a low-level mosaicism (CMA copy number state deviation <0.25). Two cases showed segmental mosaicism: 1 mosaic duplication of 19 Mb in 3q26.1q27.3 and 1 mosaic deletion of 20.7 Mb in 18q21.2qter. The third case was a mosaic trisomy of the entire chromosome 14. All of these findings explained the respective patient's phenotype. Informed consent for detailed publication of the phenotypes was obtained for the 2 cases with unusual segmental aberrations, but not for the third case with a chromosome 14 mosaic. For this reason, no clinical information and no description are given for the latter case.
Case 1 (ID1392): Mosaic Duplication of 3q26.1q27.3 in a Patient with Mild Intellectual Disability and Postaxial Polydactyly of the Hands
This patient was a 14-year-old boy with mild intellectual disability (ID), bilateral postaxial polydactyly of the hands, and mild dysmorphic facial features. He was born after an uneventful pregnancy with normal growth measurements (weight: 3,445 g, length: 48 cm). After birth, bilateral postaxial polydactyly of the hands, mild upslanting palpebral fissures, and folding of the upper ear helices were noted. At the age of 6 months, he was able to sit. At the age of 18 months, he could walk with support, and a developmental assessment showed a mild global developmental delay (DD) with a developmental quotient of 60 in all domains (cognition, language, motor, and social). Fine motor problems and frequent toe walking occurred (improved by physiotherapy). He had also undergone surgery for inguinal hernia and cryptorchidism.
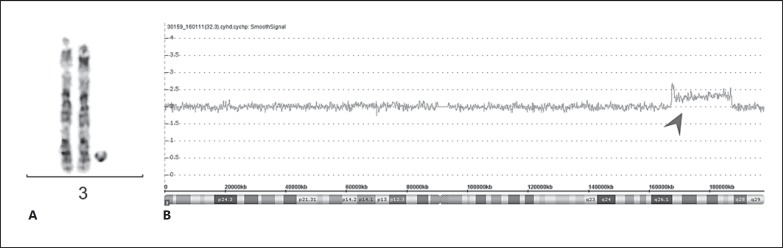
At referral to our center at the age of 14 years for genetic assessment, his weight, height, and head circumference were 50.2 kg (57th centile), 154.9 cm (18th centile), and 56 cm (81st centile), respectively. Physical examination revealed a coarse face with full lips and a bulbous nasal tip, prominent incisors, a high palate, additional interphalangeal flexion creases on both small fingers, which also showed mild clinodactyly, scars from surgery of postaxial polydactyly in both hands, and relatively broad feet with big toes as well as bilateral mild 2–3 syndactyly. He was at the onset of puberty with some axillary and genitalia hair growth. He attended a special needs school but was able to read and write. He showed no signs of mosaicism such as body asymmetry or pigmentary changes. Standard karyotyping performed at the age of 1 year had revealed a small supernumerary marker chromosome (SMC) in 30 and 45% of the analyzed metaphases from cultivated lymphocytes and skin fibroblasts, respectively. The origin of the SMC at that time could not be identified, but was assumed to be extra material from chromosome 16 after microdissection and FISH mapping. CMA performed 13 years later using the Affymetrix 2.7 array on a new blood sample allowed the characterization of the SMC and revealed a 19-Mb duplication of chromosome 3q26.1q27.3 (hg19, chr3:167,268,921–186,249,954) in mosaic form (CMA copy number state: 2.3), encompassing 151 genes in total (Fig. 1). FISH analysis using the BAC probe RP11-436A20 specific for the region 3q26.33 and a whole chromosome 3 painting confirmed the finding of a SMC in 14.6% and in 50% of the analyzed metaphases from newly sampled cultivated lymphocytes and skin fibroblasts, respectively. Of note, FISH analysis using an alphoid probe specific for chromosome 3 and one for all human centromeres resulted in no detectable signals on the marker chromosome, indicating an analphoid SMC. On the other hand, since the mosaic ratio remained unchanged in fibroblasts and comparable in lymphocytes through the years as well as the fact that the subtelomeric sequences are missing, it is likely that this SMC represents an interesting example of NRC. The location of kinetochore formation, however, was not studied with immunofluorescence and antibodies to centromere protein CENP. The NRC was absent in parental blood, and we therefore assume that it occurred de novo, although germinal mosaics cannot be excluded.
Fig. 1.
Mosaic duplication of 3q26.1q27.3 in a patient with mild intellectual disability and postaxial polydactyly of the hands. A G-banding of the 2 normal chromosomes 3 and the neocentric ring chromosome. B Screenshot of microarray raw data. The arrowhead indicates the smooth signal of markers, indicative of the interstitial mosaic duplication in 3q26.1q27.3, slightly above 2. Chromosomal position and cytobands are given at the bottom. CMA copy number state: 2.3.
Case 2 (ID64988): Mosaic Deletion of 18q21.2qter in a Patient with DD and Atresia of the Auditory Canal
This patient was born after an uneventful pregnancy by emergency cesarean section because of breech presentation. Birth growth measurements were normal, but he was hypotonic and exhibited feeding problems. After birth, atresia of the right external auditory canal with a preauricular tag and, subsequently, at the age of 6 months, a bilateral hearing impairment was diagnosed. He was able to sit at age 8.5 months and walk independently at 21 months. He was referred for genetic assessment at the age of 2.5 years because of global DD, muscular hypotonia, mild facial dysmorphic features, and a bilateral hearing impairment. Weight, height, and head circumference were 13 kg (50th centile), 98 cm (97th centile), and 50 cm (25–50th centile), respectively. Physical examination revealed a preauricular tag on the right side, long eye lashes, a broad mouth with everted lips, small fingers with broad short distal phalanges, bilateral clinodactyly of the fifth toes, tender translucent skin, fine hair, and cryptorchidism on the right side. He showed no signs of mosaicism such as body asymmetry or pigmentary changes. He could climb stairs, but had dystonic movements. He had very limited speech with a vocabulary of about 5 words. Brain MRI at the age of 2 years showed delayed myelination. A broad developmental assessment at the age of 3 years showed a severe global DD (developmental quotient of 25) in all domains (cognition, language, and motor) and some autistic behaviors.
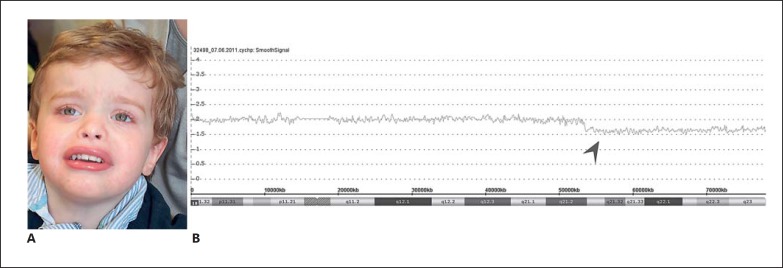
CMA performed with the Affymetrix 2.7 array on blood revealed a 25.2-Mb deletion of chromosome 18q21.2qter (hg19, chr18:53,277,087–78,077,248) in mosaic form, encompassing 97 genes in total (CMA copy number state: 1.7; Fig. 2). FISH analysis confirmed the mosaic deletion in 20% of the cultivated lymphocytes (normal signals in 43 metaphases and 5 interphases, abnormal signals in 11 metaphases and 1 interphase) and in 81% of the cells from a buccal swab.
Fig. 2.
Mosaic deletion of 18q21.2qter in a patient with developmental delay and atresia of auditory canal. A Photograph of the patient at the age of 2½ years. Note the broad forehead, hypertelorism, flat nasal root, epicanthal folds, upturned nares, and everted lips. B Screenshot of microarray raw data. The arrowhead indicates the smooth signal of markers, indicative of the mosaic terminal on chromosome 18, slightly below 2. Chromosomal position and cytobands are given at the bottom. CMA copy number state: 1.7.
Discussion
In a cohort of 714 cases with NDDs with or without further congenital anomalies, we detected chromosomal aberrations in 3 cases (0.4%). All of these mosaics were low level in lymphocytes, and 2 of them remained undetected using classical cytogenetic techniques (mosaic trisomy 14 [not described in detail as mentioned above] and mosaic partial deletion on chromosome 18, i.e., case 2). The third mosaic case, a small SMC, later identified as small NRC, was detected by conventional cytogenetics, but the precise identification of the material had been elusive until CMA analysis was performed. The incidence we observed is in line with what has been previously reported (0.2–1%) in patients with ID [Ballif et al., 2006; Cheung et al., 2007; Conlin et al., 2010; Bruno et al., 2011; Hoang et al., 2011]. Our findings underline the important role of low-level mosaics in the pathogenesis of NDDs of unknown etiology. In particular, a mosaic deletion of 18q in lymphocytes is a rare genetic abnormality with a highly variable phenotype, which represents a diagnostic challenge for clinicians and underlines the importance of CMA testing in children with atresia of the auditory canal. This feature is indeed recurrent in the 18q22.3q23 deletion [Veltman et al., 2003], a syndrome that has been documented in more than 100 cases and occurs in approximately 1/40,000 live births [Cody et al., 1999, 2009]. Other common features include DD/ID, short stature, hypotonia, craniofacial abnormalities, especially midface hypoplasia, hearing impairment, growth hormone deficiencies, abnormalities of the feet, and genital hypoplasia [Kline et al., 1993]. In 2007, a phenotypic map of chromosome 18 was created based on information from 29 patients defining critical regions for microcephaly (region 18q21.33), short stature (18q12.1q12.3, 18q21.1q21.33, and 18q22.3q23), white matter disorders (18q22.3q23), growth hormone insufficiency (18q22.3q23), congenital aural atresia (18q22.3), and cognitive delay [Feenstra et al., 2007]. While mild ID was usually identified with deletions distal to 18q21.33, severe ID was usually observed with deletions proximal to 18q21.31 [Feenstra et al., 2007]. Similar to other reported patients with larger terminal 18q deletions, our patient suffers from severe DD, hypotonia and cryptorchidism, but has neither short stature, nor microcephaly. He shows a very distinct wide hypotonic mouth. Recently, a case of a mosaic deletion resembling the one in our patient has been reported in the literature. The patient exhibited polydactyly of the feet, hypospadia, cardiovascular abnormalities, and eye anomalies, such as bilateral microcornea with dense opacification and unilateral iris as well as chorioretinal coloboma [Galvin et al., 2015]. None of these features were present in our case. Of note, a broad phenotypic ocular spectrum was reported previously in 7 patients with deletion of the regions 18q21.3qter or 18q22.2qter [Kline et al., 1993]. In general, it is assumed that the size of the deletion correlates with the phenotype severity [Cody et al., 2009], but in this case, the mosaic ratio of aberrant cells and their localization may as well play a role.
SMCs without detectable alphoid DNA represent a rare class of rearranged marker chromosomes. They occur with an incidence of 0.14–0.72/1,000 newborns and are not easily identifiable by G-banding or by FISH [Nielsen and Wohlert, 1991]. SMCs are referred to as neocentric marker chromosomes or NRCs, when an ectopic centromere is able to assemble a functional kinetochore that originates occasionally from non-centromeric regions of the chromosome, which rescues the chromosome fragments and restores their ability to segregate efficiently [Marshall et al., 2008].
To date, more than 100 neocentromeres have been characterized in humans [Marshall et al., 2008; Alonso et al., 2010; Klein et al., 2012], and they show remarkable diversity in chromosome position and DNA sequences [Burrack and Berman, 2012].
The formation of a neocentromere is generally associated with a chromosomal rearrangement, such as an inverted duplication of a distal chromosome segment leading to partial tetrasomy (class I) or a balanced chromosomal rearrangement into linear and circular marker chromosomes after an interstitial deletion (class II/McClintock mechanism). Class I rearrangements represent the most common mechanism for the formation of SMCs, and the majority of SMCs reported in the literature affects the distal part of chromosome 3q, which appears to have a high propensity for neocentromere formation [Marshall et al., 2008]. The long arm of chromosome 3, such as 3q26.2 and 3q27.2, is indeed a hotspot, for which other neocentromeres have been reported. Interestingly, the evolution of chromosome 3 in primates has been studied, and it has been shown that the region 3q26 was a centromeric region in a common ancestor of the Old World monkeys (Cercopithecidae) approximately 25–40 MYA. This suggests the possibility of reactivation of longstanding latent centromeres and that there is an inherent potential of these regions to form centromeres [Ventura et al., 2004].
The marker in our case of a chromosome 3-derived neocentromeric chromosome can neither be classified in class I, nor in class II rearrangements, since our patient has 2 normal copies of chromosome 3 and a small additional mitotically stable segment, leading to a partial trisomy 3q in mosaic. An unusual and similar type of neocentromeric formation has already been described for chromosome 1 in one case [Spiegel et al., 2003], but, to the best of our knowledge, it has never been reported for chromosome 3.
Neocentromeric formation of the distal region 3q has been associated with hyper- or hypopigmentation, hypotonia, preauricular pits, accessory nipples, DD, and seizures. In one male with a marker chromosome 3q26.2qter, the postaxial polydactyly has been reported together with asymmetric cerebral ventricles, a duplicated right kidney, and right pulmonary artery stenosis [Teshima et al., 2000]. An acentric marker chromosome involving the region 3q27 has also been reported in a young man with normal intelligence and hyperpigmentation, but no dysmorphism [Portnoi et al., 1999]. Similar to the phenotypic features already described in the literature, our patient shows mild ID and postaxial polydactyly, but no abnormal skin pigmentation. The large variability of the clinical findings in patients with SMCs occurring in the distal region 3q is due to the different genetic content, type of rearrangement, tissue distribution, and the mosaic ratio.
Of note, neither case 1 nor case 2 showed typical signs of chromosomal mosaicism such as body asymmetry or segmental pigmentary anomalies.
In summary, identifying low-level mosaics remains a diagnostic challenge for cytogenetic and molecular testing, and their detection is of utmost importance for an accurate etiological diagnosis and genetic counseling.
Statement of Ethics
Informed consent was obtained for the 2 described cases. All procedures performed were in accordance with the ethical standards.
Disclosure Statement
The authors declare no conflicts of interest.
References
- Alonso A, Hasson D, Cheung F, Warburton PE. A paucity of heterochromatin at functional human neocentromeres. Epigenetics Chromatin. 2010;3:6. doi: 10.1186/1756-8935-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadollahi R, Oneda B, Joset P, Azzarello-Burri S, Bartholdi D, et al. The clinical significance of small copy number variants in neurodevelopmental disorders. J Med Genet. 2014;51:677–688. doi: 10.1136/jmedgenet-2014-102588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballif BC, Rorem EA, Sundin K, Lincicum M, Gaskin S, et al. Detection of low-level mosaicism by array CGH in routine diagnostic specimens. Am J Med Genet A. 2006;140:2757–2767. doi: 10.1002/ajmg.a.31539. [DOI] [PubMed] [Google Scholar]
- Bi W, Borgan C, Pursley AN, Hixson P, Shaw CA, et al. Comparison of chromosome analysis and chromosomal microarray analysis: what is the value of chromosome analysis in today's genomic array era? Genet Med. 2013;15:450–457. doi: 10.1038/gim.2012.152. [DOI] [PubMed] [Google Scholar]
- Bruno DL, White SM, Ganesamoorthy D, Burgess T, Butler K, et al. Pathogenic aberrations revealed exclusively by single nucleotide polymorphism (SNP) genotyping data in 5000 samples tested by molecular karyotyping. J Med Genet. 2011;48:831–839. doi: 10.1136/jmedgenet-2011-100372. [DOI] [PubMed] [Google Scholar]
- Burrack LS, Berman J. Neocentromeres and epigenetically inherited features of centromeres. Chromosome Res. 2012;20:607–619. doi: 10.1007/s10577-012-9296-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung SW, Shaw CA, Scott DA, Patel A, Sahoo T, et al. Microarray-based CGH detects chromosomal mosaicism not revealed by conventional cytogenetics. Am J Med Genet A. 2007;143A:1679–1686. doi: 10.1002/ajmg.a.31740. [DOI] [PubMed] [Google Scholar]
- Cody JD, Ghidoni PD, DuPont BR, Hale DE, Hilsenbeck SG, et al. Congenital anomalies and anthropometry of 42 individuals with deletions of chromosome 18q. Am J Med Genet. 1999;85:455–462. doi: 10.1002/(sici)1096-8628(19990827)85:5<455::aid-ajmg5>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Cody JD, Heard PL, Crandall AC, Carter EM, Li J, et al. Narrowing critical regions and determining penetrance for selected 18q-phenotypes. Am J Med Genet A. 2009;149A:1421–1430. doi: 10.1002/ajmg.a.32899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlin LK, Thiel BD, Bonnemann CG, Medne L, Ernst LM, et al. Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Hum Mol Genet. 2010;19:1263–1275. doi: 10.1093/hmg/ddq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feenstra I, Vissers LE, Orsel M, van Kessel AG, Brunner HG, et al. Genotype-phenotype mapping of chromosome 18q deletions by high-resolution array CGH: an update of the phenotypic map. Am J Med Genet A. 2007;143A:1858–1867. doi: 10.1002/ajmg.a.31850. [DOI] [PubMed] [Google Scholar]
- Galvin JA, LeBoyer RM, Michelotti M, Monte MA, Elner VM, Mian SI. Mosaic chromosome 18q partial deletion syndrome with bilateral full-thickness corneal disease: surgical intervention and histopathology. Ophthalmic Genet. 2015;36:75–78. doi: 10.3109/13816810.2013.833633. [DOI] [PubMed] [Google Scholar]
- Hoang S, Ahn J, Mann K, Bint S, Mansour S, et al. Detection of mosaicism for genome imbalance in a cohort of 3,042 clinical cases using an oligonucleotide array CGH platform. Eur J Med Genet. 2011;54:121–129. doi: 10.1016/j.ejmg.2010.10.010. [DOI] [PubMed] [Google Scholar]
- Hook EB. Exclusion of chromosomal mosaicism: tables of 90%, 95% and 99% confidence limits and comments on use. Am J Hum Genet. 1977;29:94–97. [PMC free article] [PubMed] [Google Scholar]
- Klein E, Rocchi M, Ovens-Raeder A, Kosyakova N, Weise A, et al. Five novel locations of neocentromeres in human: 18q22.1, Xq27.1 approximately 27.2, Acro p13, Acro p12, and heterochromatin of unknown origin. Cytogenet Genome Res. 2012;136:163–166. doi: 10.1159/000336648. [DOI] [PubMed] [Google Scholar]
- Kline AD, White ME, Wapner R, Rojas K, Biesecker LG, et al. Molecular analysis of the 18q- syndrome - and correlation with phenotype. Am J Hum Genet. 1993;52:895–906. [PMC free article] [PubMed] [Google Scholar]
- Marshall OJ, Chueh AC, Wong LH, Choo KH. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet. 2008;82:261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J, Wohlert M. Chromosome abnormalities found among 34,910 newborn children: results from a 13-year incidence study in Arhus, Denmark. Hum Genet. 1991;87:81–83. doi: 10.1007/BF01213097. [DOI] [PubMed] [Google Scholar]
- Pham J, Shaw C, Pursley A, Hixson P, Sampath S, et al. Somatic mosaicism detected by exon-targeted, high-resolution aCGH in 10,362 consecutive cases. Eur J Hum Genet. 2014;22:969–978. doi: 10.1038/ejhg.2013.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoi MF, Boutchnei S, Bouscarat F, Morlier G, Nizard S, et al. Skin pigmentary anomalies and mosaicism for an acentric marker chromosome originating from 3q. J Med Genet. 1999;36:246–250. [PMC free article] [PubMed] [Google Scholar]
- Scott SA, Cohen N, Brandt T, Toruner G, Desnick RJ, Edelmann L. Detection of low-level mosaicism and placental mosaicism by oligonucleotide array comparative genomic hybridization. Genet Med. 2010;12:85–92. doi: 10.1097/GIM.0b013e3181cc75d0. [DOI] [PubMed] [Google Scholar]
- Spiegel M, Hickmann G, Senger G, Kozlowski P, Bartsch O. Two new cases of analphoid marker chromosomes. Am J Med Genet A. 2003;116A:284–289. doi: 10.1002/ajmg.a.10916. [DOI] [PubMed] [Google Scholar]
- Teshima I, Bawle EV, Weksberg R, Shuman C, Van Dyke DL, Schwartz S. Analphoid 3qter markers. Am J Med Genet. 2000;94:113–119. doi: 10.1002/1096-8628(20000911)94:2<113::aid-ajmg3>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Veltman JA, Jonkers Y, Nuijten I, Janssen I, van der Vliet W, et al. Definition of a critical region on chromosome 18 for congenital aural atresia by arrayCGH. Am J Hum Genet. 2003;72:1578–1584. doi: 10.1086/375695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Weigl S, Carbone L, Cardone MF, Misceo D, et al. Recurrent sites for new centromere seeding. Genome Res. 2004;14:1696–1703. doi: 10.1101/gr.2608804. [DOI] [PMC free article] [PubMed] [Google Scholar]