Abstract
Cleidocranial dysplasia (CCD) is an autosomal dominant disorder linked to mutations in the Runt-related transcription factor 2, encoded by the RUNX2 gene, which is essential for osteoblast differentiation and skeletal development. Here, we describe a novel nonsense mutation (c.532C>T; p.Q178X) in RUNX2 identified in 3 affected members of a Polish family with CCD. The localization and transcriptional transactivation studies show that the mutated form of the protein has altered the subcellular localization and significantly decreased transactivation properties, respectively. Consequently, our data show that the c.532C>T mutation generates a defective RUNX2 protein and is genetically linked to the CCD phenotype.
Keywords: Cleidocranial dysplasia, Genotype-phenotype correlation, Novel mutation, RUNX2
Cleidocranial dysplasia (CCD) occurs with a prevalence of 1/1,000,000 with no gender or ethnic predilection. It is an autosomal dominant disorder with multiple phenotypes including hypoplastic or aplastic clavicles, failure of cranial suture closure, hypertelorism, dental anomalies, and brachydactyly with hypoplastic distal phalanges (OMIM 119600). Other phenotypes often consist of multiple Wormian bones, hypoplastic pubic bones, scoliosis, and short stature. Additionally, hearing loss and susceptibility to ear infections is implicated in 39% of the cases.
CCD is genetically linked to a mutation in RUNX2 [Mundlos et al., 1997], a transcription factor from the RUNT family critical for the osteochondrogenic differentiation and consequently limb patterning, among others [Komori et al., 1997; Otto et al., 1997].
The human RUNX2 gene maps to chromosome 6p21 [Levanon et al., 1994] and consists of 8 exons encoding a 521-amino acid protein (NCBI: NM_001024630.3; NP_001019801.3). The protein contains 3 activation domains and 1 repression domain. Starting with the N-terminus, the first 19 amino acids and the subsequent QA domain, consisting of glutamine-alanine repeats (23Q and 17A), act as transactivation domains [Thirunavukkarasu et al., 1998]. This is followed by a highly conserved Runt homologous domain (RHD) responsible for DNA binding to a specific DNA motif [Ogawa et al., 1993] and heterodimerization with CBFβ (core-binding factor subunit beta). The latter increases the DNA-binding affinity as well as protects and stabilizes RUNX2 against proteolytic degradation [Kagoshima et al., 1993; Tahirov et al., 2001]. A PRRHRQKLD sequence located adjacently to RHD functions as a nuclear localization signal (NLS) [Thirunavukkarasu et al., 1998]. The PST (proline/serine/threonine)-rich region follows, which contains phosphorylation sites [Pande et al., 2013] and represents the third transactivation domain [Thirunavukkarasu et al., 1998]. Additionally, within PST, the nuclear matrix targeting sequence is present [Kanno et al., 1998; Zaidi et al., 2001]. The last VWRPY pentapeptide sequence functions as a transcriptional repression domain [Levanon et al., 1998; Thirunavukkarasu et al., 1998].
To date, more than 170 RUNX2 mutations have been described, including insertions, deletions, splicing, nonsense, and missense mutations. In this study, we describe a family diagnosed with CCD. The family consists of the mother, her 2 daughters (all affected), and a granddaughter (not affected). Subsequent genetic analysis revealed that all the affected family members carry a novel nonsense mutation c.532C>T (Q178X) in the RUNX2 gene. To investigate the consequences of this mutation on the activity of the protein, transactivation analysis and subcellular localization studies were performed. We demonstrate that the Q178X truncating nonsense mutation generates a 177-amino acid protein lacking part of the RHD motif and devoid of the transcriptional transactivation activity. Furthermore, by carrying out the nuclear localization studies, we showed the impaired subcellular localization of Q178X caused by the absence of the NLS sequence. Overall, we propose that the single base change at position c.532C>T results in a defective RUNX2 protein and is the causative factor of CCD in the investigated family.
Patients and Methods
Patients and DNA Isolation
The proband, her sister, and their mother (all affected) as well as a daughter of the proband (not affected) were recruited by the Children's Orthopedic Clinic and Rehabilitation Department of the Medical University of Lublin, Poland. The 3 affected members of the family show a typical clinical manifestation of CCD (Fig. 1A). Genomic DNA of the family members (affected and unaffected) was isolated from peripheral blood using the MiniBlood Kit (A&A Biotechnology) following the manufacturer's recommendations.
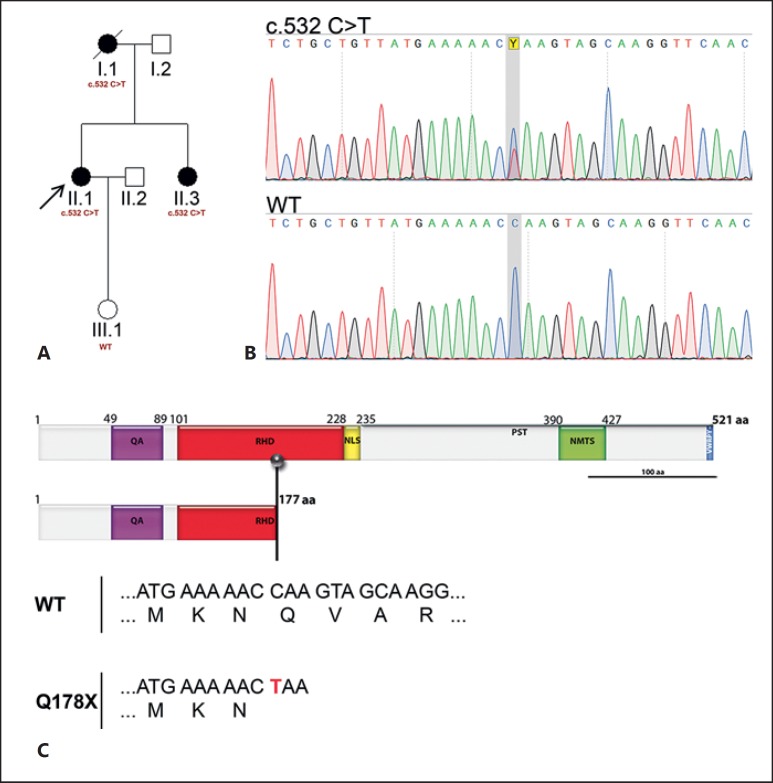
Fig. 1.
A The pedigree of the family. The arrow indicates the proband. B Sangersequence chromatograms of the RUNX2 gene show a single base change at nucleotide position 532 within exon 3 (c.532C>T, p.Q178X) in all affected individuals (I.1, II.1, and II.3). C Schematic representation of RUNX2-WT and the mutant Q178X showing the consequence of the mutation. The mutant allele with a single base change at position 532 (c.532C>T) generates a premature stop codon leading to a shorter protein (p.Q178X). NLS, nuclear localization signal; NMTS, nuclear matrix targeting signal; PST, proline/serine/threonine rich region; QA, polyglutamine and polyalanine domain; RHD, Runt homologous domain; VWRPY, carboxyterminal pentapetide; WT, wild type (III.1).
Sequencing and Mutation Analysis
All 8 exons of the RUNX2 gene from all recruited family members were amplified by PCR reaction with proofreading Vent Polymerase (NEB). Primers used in this study are indicated in Table 1. Next, the sequencing reactions with purification were carried out by an outside contractor (CB DNA, Poznan, Poland). DNA sequences were analyzed using BLAST algorithm (NCBI BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi RRID:SCR_004870) querying NCBI (http://www.ncbi.nih.gov/ RRID:SCR_006472), and Ensembl (http://www.ensembl.org/ RRID:SCR_002344) databases.
Table 1.
Primers used in the study
| Exon | Forward primer (5′→3′) | Reverse primer (5′→3′) | Product size |
|---|---|---|---|
| 1 | TTCTGGATGCCAGGAAAGGC | TTACTGTTTTCATATCCTCACCTCT | 954 bp |
| 2 | GCAGTCGGCCTCATCAAACT | GGGGTTTCTGGGGTTAGAGC | 675 bp |
| 3 | TGCCTGCCTTTGAAATGGGT | CACACCCAGTGAAATTAGGAGC | 616 bp |
| 4 | GAGTCCTGCCTCTTGTCTTTGTTTC | CATTTCCCAAAGCTACGGAGGC | 476 bp |
| 5 | CTGAGAGGGCCAGAGAAGGTGTC | GCATTATTACCGTTCCACCAGG | 734 bp |
| 6 | TGGGTTGCATGTTTCTAAGGCTG | CCAGTTGTCATTCCCTTGCCC | 360 bp |
| 7 | GGCTGTTGCTTCTCCTTCTCTCTTG | CAAGGGTTAAGTGCCATGATGTG | 307 bp |
| 8 | CTGTGGCTTGCTGTTCCTTTATG | CTCTTCTCCCTCCTGAACCTGG | 974 bp |
Site-Directed Mutagenesis
Wild type (WT) human RUNX2 cloned into the eukaryotic expression vector pcDNA3.1(+) was provided by GenScript and used as a template for mutagenesis. Two mutants were generated using the QuikChange Lightning Site-Directed Mutagenesis Kit (STRATAGENE). One of them recreated the mutation found in the proband (c.532C>T resulting in p.Q178X change), and the other one was the previously described mutation (c.391C>G resulting in p.R131G change) that served as a positive control [Kim et al., 2006; Han et al., 2010]. The results of mutagenesis were confirmed by sequencing.
Construction of Hemagglutinin-Tagged Clones
Following mutagenesis, the hemagglutinin (HA)-tagged clones were prepared using the restriction enzyme-based cloning method. Specifically, the WT and mutant clones were cut out using EcoRV and XhoI enzymes (NEB) and ligated into the vector pcDNA3.1-N-3xHA digested with the above-mentioned enzymes. The ligation with T4 Ligase (Thermo Fisher Scientific) was carried out for 2 h at 22°C. This strategy placed the RUNX2 open reading frames in frame with the HA protein tag. Ligation reactions were electroporated into Escherichia coli DH10B cells. The next day, 16 colonies were selected for colony-PCR using TaqNova HS polymerase (Blirt). The clones contacting an insert of desired size were isolated, and their identity was confirmed by Sanger sequencing.
Transfections
HEK293 cells were cotransfected using TurboFect Reagent (Thermo Fisher Scientific) with 1 μg of DNA consisting of: (1) HA-tagged WT, Q178X, or R131G expression plasmid (400 ng); (2) pOSE2-6Luc reporter plasmid (200 ng), containing 6 tandemly linked osteoblast-specific elements (OSE2) in the osteocalcin promoter (mouse OG2 promoter) [Ducy and Karsenty, 1995]; (3) CMV-LacZ (10 ng) for normalization of luciferase assay, and (4) pUC18 plasmid to fill up to 1 μg of total DNA content. Transfections for luciferase assay were performed in triplicate. Following the 24-h incubation, cells were washed once with PBS and used in reporter assays.
Luciferase Assay
Luciferase assays were carried out using the Luciferase Assay System and β-Galactosidase Enzyme Assay System with Reporter Lysis Buffer (Promega) according to the manufacturer's instructions. Transfected HEK cells were lysed, and the luciferase activity was measured according to manufacturer's instructions (Promega). The relative luciferase activity was normalized to the β- galactosidase activity. All measurements were performed using Tecan Infinite M200Pro reader.
Western Blot Analysis
HEK293 cells were transfected with 400 ng of HA-tagged RUNX2-WT, Q178X, or R131G expression vectors using TurboFect Reagent. Next, 24 h after transfection, they were lysed with 40 μl of 2× SDS PAGE sample buffer. The known protein, chick SMOC2, tagged with HA was used as a positive control for tag detection. The negative control consisted of lysate of untransfected HEK cells. The loading control for all the experiments was endogenous β-actin. Lysates were separated with 12% SDS-PAGE gel and then transferred onto a PVDF membrane. Blocking was performed with 5% Blotting-Grade Blocker (Bio-Rad) diluted in a TBST buffer (1× TBS with 0.1% Tween-20), and the immunodetection was performed in the same buffer. Following antibodies were used: Primary Rabbit Anti-HA monoclonal antibody (Cell Signaling Technology Cat No. 3724S, RRID:AB_1549585) 1:1,000, Secondary Bovine Anti-Rabbit IgG HRP-linked antibody (Cell Signaling Technology Cat No. 7074V, RRID:AB_11178390) 1:5,000, Primary β-actin (Santa Cruz Biotechnology Cat No. sc-69879, RRID:AB_1119529) 1:1,000, and Secondary bovine anti-mouse IgG-HRP (Santa Cruz Biotechnology Cat No. sc-2371, RRID:AB_634824) 1:5,000.
Immunoblotting was performed according to Lumi-Light Western Blotting Substrate protocol (Roche) to detect the HRP-conjugated secondary antibodies. Blot was developed in G:Box Chemi XR5 (Syngene).
Immunofluorescence
HEK293 cells were transfected with 500 ng of HA-tagged WT, Q178X or R131G using TurboFect Reagent. Following the 48-h incubation, the cells were methanol fixed (10 min at −20°C) and quenched with 50 mM ammonium chloride in PBS (2 × 5 min). Next, cells were permeabilized with 0.2% Triton X100 (Pierce) and blocked with 0.5% Blocking Reagent Solution (Roche) in PBS for 1 h to prevent the nonspecific protein-protein interactions. Following the 1-h incubation with primary Anti-HA rabbit antibody (Cell Signaling) 1:1,000, and subsequent 1-h incubation with secondary AlexaFluor® 555 Donkey Anti-Rabbit IgG (H+L) (Thermo Fisher Scientific Cat No. A-31572, RRID:AB_2536182) 1:200, the slides were rinsed and mounted using ProLong® Gold Antifade with DAPI (Invitrogen). After the 24-h incubation at room temperature, samples were visualized. The visualization was carried out using a Nikon Ti Eclipse Microscope and the image analysis was done using the software provided by the manufacturer.
Results
Clinical Examination of CCD Patients
All patients diagnosed with CCD show the typical CCD phenotype; however, its manifestation varies among family members (Table 2). The mother of the proband (I.1) (Fig. 1A) has the mildest phenotype including hypoplasia of clavicles, while the proband (II.1) and her sister (II.3) display a more severe CCD, including the lack of clavicles with hypermobility of both shoulders, and enlarged fontanelles. Additionally, II.1 has supernumerary teeth and altered craniofacial features such as a flattened face and a wide nasal bridge. At the age of 6, she was diagnosed with varus deformity of the right hip joint (congenital coxa vara). Her sister (II.3) has a similar phenotype except craniofacial features but with the developmental dislocation of the left hip. The daughter of proband II.1 is healthy without any features associated with CCD. The phenotypes were consistent with phenotypes caused by mutations in the RUNX2 gene.
Table 2.
Phenotypes of family members
| I.1 (mother) | II.3 (sister) | II.1 (proband) | |
|---|---|---|---|
| Facial features | flattened face, wide nasal bridge | ||
| Dental status | ? | ? | multiple supernumerary teeth |
| Enlarged fontanelle | − | + | + |
| Clavicles | hypoplasia | aplasia | aplasia |
| Other features | dislocation of left hip, short stature | varus of right hip joint (coxa vara), short stature | |
Identification of Mutation
We decided to investigate if this family also had defects in the RUNX2 gene. To identify the mutation, we designed primers amplifying all exons of RUNX2 and carried out PCR on the patients' DNA. Sequencing of the PCR products identified a novel mutation in the third exon of all affected patients. The sequencing has unambiguously shown that the patients are heterozygous for the mutation. The newly identified mutation introduced a 1-base codon change at position c.532C>T located within the RHD domain, resulting in a premature stop codon (CAA→TAA) by replacing glutamine with an ochre stop codon [Brenner et al., 1965] (Q178X, Fig. 1B, C, 2B). No mutation was detected in the unaffected family member (III.1), supporting the notion that this novel nonsense mutation segregates with the CCD phenotype in this family.
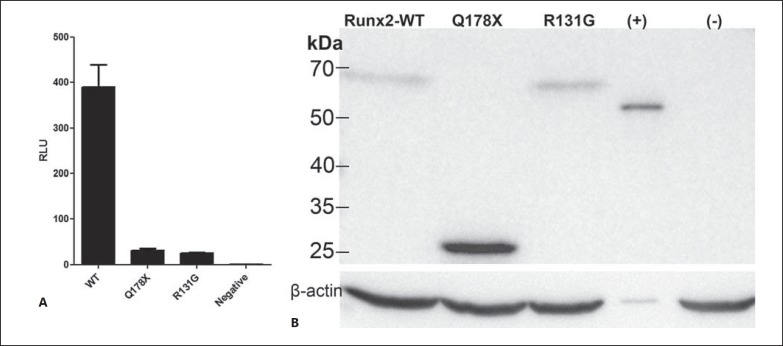
Fig. 2.
A The results of transactivation of the OSE2 reporter plasmid by RUNX2-Q178X WT or control mutation R131G. The results of luciferase assay are presented as relative light units (RLUs) of tested variants normalized to β-galactosidase activity. The graph shows results of a representative experiment. Data represent the average and standard deviation of triplicate samples. B Western blot analysis of the expression levels of RUNX2 mutants used in the reporter assays. The names of the loaded proteins are listed on the top of the gel. (+) denotes Chick SMOC2-HA tag control.
Nonsense Mutation Q178X Results in a Truncated Protein with Defective Activity of the RUNX2 Protein
The newly identified mutation results in production of a truncated RUNX2 protein. Therefore, we tested if this variant was still biologically active by measuring its transactivation activity in a reporter assay. To accomplish that, we used a previously described RUNX2 reporter containing 6 tandemly linked OSE2 from the osteocalcin promoter cloned upstream of a luciferase reporter (pOSE2-6Luc) [Ducy et al., 1997]. The reporter plasmid was cotransfected into HEK293 cells with HA-tagged expression plasmids encoding either the recreated Q178X mutation or the WT RUNX2, or a previously described mutation with a missense change in the RHD domain (R131G) [Han et al., 2010]. To normalize for the transfection efficiency, all samples were cotransfected with CMV-LacZ expression constructs encoding β-galactosidase. Following the cell lysis, we measured the luciferase activity of the OSE2 reporter and normalized it to β-galactosidase activity. We could show that both mutants of RUNX2 displayed a significant reduction of transactivation activity as compared to the WT protein (Fig. 2A). The above-mentioned outcome was confirmed by 4 independent experiments done in triplicate at 1-2 week intervals. The statistical analysis included the non-parametric Kruskal-Wallis (one-way ANNOVA) test. The same results were obtained with plasmids containing untagged WT, Q178X, and R131G (data not shown).
To exclude the possibility that the mutant protein was simply not stable or not translated efficiently and therefore did not elicit any reporter response, we measured the amount of produced protein by Western Blot analysis in lysates of HEK cells transfected with the expression plasmids used in the reporter assays. Indeed, all 3 forms of RUNX2 have been produced at similar levels, with the Q178X form at an even higher efficiency than the WT. This result confirms that the loss of DNA binding resulted from the absence of the C-terminal fragment of the RHD domain and not from the low or nonexpression of the Q178X mutant (Fig. 2B). Thus, we can conclude that the Q178X mutation abolishes the transactivation activity of RUNX2 without affecting the efficiency of translation or protein stability.
Nuclear Localization of RUNX2Q178X
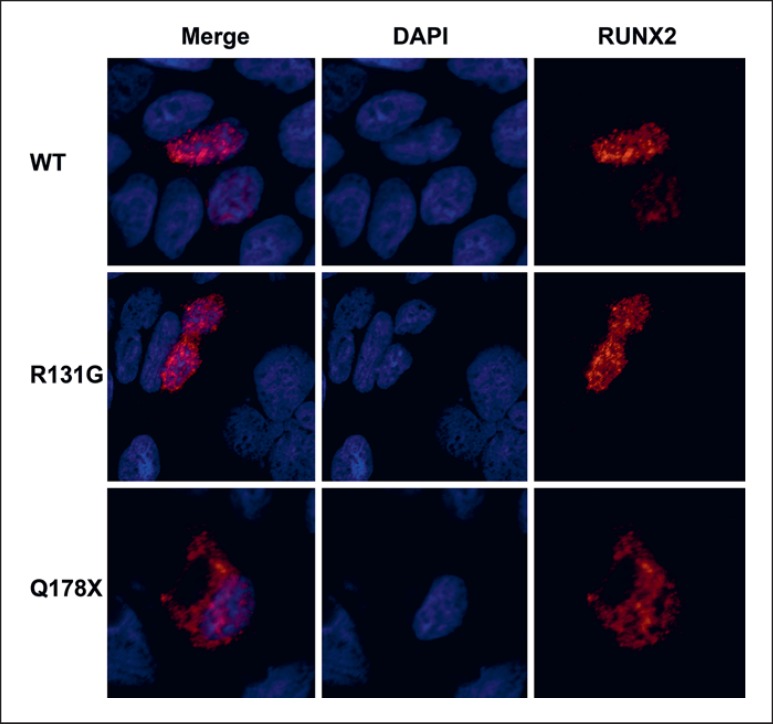
The WT RUNX2 localizes to the nucleus due to the presence of the NLS sequence situated downstream of RHD. The examined c.532C>T mutant (Q178X) lacks the C-terminus of the protein containing the NLS element (Fig. 1C, 2B), suggesting its defect in nuclear transport. To test this, we performed the immunofluorescence for WT, control mutant R131G, and Q178X after transfection to HEK cells. We could confirm that both WT and R131G localized to the nucleus, while the Q178X was present both in the nucleus and cytoplasm (Fig. 3). This result suggests that the truncated RUNX2 protein contains an unidentified nuclear localization motif or that the mechanism of nuclear transport is partially independent on the presence of NLS.
Fig. 3.
Cellular localization of RUNX2-WT, control R131G, and Q178X mutant proteins. The names of the RUNX2 variants are listed on the left. The first column shows the merged images. The second and third columns show the DAPI nuclear staining and AlexaFluor 555 channel for RUNX variants. The WT and control mutant R131G localize in the nucleus; however, the Q178X variant is also present in the cytoplasm.
Discussion
In this study, we performed screening and functional analyses to determine the cause of a familial case of CCD with variable familial clinical presentations. The investigation identified a single base change in the RUNX2 gene at position c.532C>T, causing truncation of the protein in all affected patients. The Q178X mutation has not been described before; although, a previously reported deletion (c.532delC) resulted in a frameshift and premature termination of RUNX2 [Zhang et al., 2000]. However, the authors of this study did not provide experimental evidence of the consequence of that frameshift on the activity of RUNX2.
The discovered Q178X mutation localizes within a highly conserved RHD domain that heterodimerizes with the CBFβ subunit and is responsible for DNA-binding activity [Kagoshima et al., 1993]. Importantly, 3 regions within RHD are involved in DNA-binding properties: the βA'-B loop-spanning region 125V-138, βE'-F loop (R190-S196), and arginine residues at positions R225, R228, and R229 that also are common to the NLS domain [Nagata et al., 1999; Otto et al., 2002]. To determine whether the truncated form of RUNX2-Q178X affects RHD activity, the transactivation studies with luciferase reporter were performed. The detected loss of transactivation properties of Q178X was very likely caused by the reduced DNA-binding activity of RHD and is consistent with other studies demonstrating that changes within RHD result in absent or decreased transactivation of target DNA sequences [Yoshida et al., 2002, 2003; Han et al., 2010].
It has been reported that mutations causing the deletion of NLS, located downstream of RHD, affect the nuclear localization of the RUNX2 protein [Yoshida et al., 2002; Zhang et al., 2010]. Indeed, the Q178X mutant protein was detected in the cytoplasm as well as in the cell nucleus, suggesting that the NLS sequence is required for the efficient nuclear localization of RUNX2.
There is still no clear genotype-phenotype correlation established for cleidocranial dysplasia due to variable penetrance of mutations in RUNX2. Nonsense mutations resulting in a truncated protein are found throughout the gene [Machuca-Tzili et al., 2002; Otto et al., 2002; Tessa et al., 2003; Cunningham et al., 2006; Kim et al., 2006; Xuan et al., 2008a, b; Ott et al., 2010; Hansen et al., 2011; Bufalino et al., 2012; Huang et al., 2013; Anthonappa et al., 2014; Guo et al., 2015] likewise splicing, insertion, or deletion changes. Interestingly, the majority of CCD-causing mutations are present within the Runt domain, and the most frequent are missense changes [Ott et al., 2010] leading to substantial decrease in the transactivation potential of the protein. Moreover, these mutations have been linked to severe dental abnormalities including supernumerary teeth [Yoshida et al., 2002; Bufalino et al., 2012; Lu et al., 2015].
The point mutation analyzed in detail in this study, is located within RHD and, as expected, is correlated with the presence of dental abnormalities in the proband. Interestingly, the proband exhibits the most severe type of CCD in comparison to the other affected members of the family. These familial phenotypic variations have been previously described [Quack et al., 1999; Zhou et al., 1999], and they are considered as a common feature of CCD. We hypothesize that the overall phenotype could be caused by at least 2 mutually nonexclusive events. One of them may be a decrease of the nuclear concentration of WT RUNX2 to 50% (or potentially less). The other event may involve the competition between the truncated Q178X and the WT RUNX2 for protein partners leading to the decrease of the effective concentration of active RUNX2 protein complexes in the cell nucleus. We exclude the possibility that the mutation results in premature RNA degradation due to the nonsense-mediated decay. If that had been the case, we would not have been able to produce the truncated protein without any problems and in large amounts as done.
In summary, we performed screening and functional analyses to determine the cause of the CCD phenotype in patients. In this study, we provide evidence that the newly characterized nonsense mutation Q178X results in defective RUNX2 transcriptional in vitro activity, likely to be responsible for the CCD phenotype.
Statement of Ethics
Informed consent was obtained from the patients. The performed procedures were in accordance with the ethical standards.
Disclosure Statement
The authors declare no conflicts of interest.
Acknowledgments
The authors would like to acknowledge Jakub Czapiński, Michał Kiełbus, and Adrian Odrzywolski (Department of Biochemistry and Molecular Biology, Medical University, Lublin, Poland) for invaluable assistance. This work was supported by a grant from the Polish National Science Centre (2013/11/B/NZ4/03660).
References
- Anthonappa RP, Yan-Hui F, King NM, Rabie AB, You-Qiang S. Novel complex disease allele mutations in cleidocranial dysplasia patients. J Oral Pathol Med. 2014;43:798–800. doi: 10.1111/jop.12198. [DOI] [PubMed] [Google Scholar]
- Brenner S, Stretton AOW, Kaplan S. Genetic code: the “nonsense” triplets for chain termination and their suppression. Nature. 1965;206:994–998. doi: 10.1038/206994a0. [DOI] [PubMed] [Google Scholar]
- Bufalino A, Paranaíba LM, Gouvêa AF, Gueiros LA, Martelli-Júnior H, et al. Cleidocranial dysplasia: oral features and genetic analysis of 11 patients. Oral Dis. 2012;18:184–190. doi: 10.1111/j.1601-0825.2011.01862.x. [DOI] [PubMed] [Google Scholar]
- Cunningham ML, Seto ML, Hing AV, Bull MJ, Hopkin RJ, Leppig KA. Cleidocranial dysplasia with severe parietal bone dysplasia: C-terminal RUNX2 mutations. Birth Defects Res A Clin Mol Teratol. 2006;76:78–85. doi: 10.1002/bdra.20231. [DOI] [PubMed] [Google Scholar]
- Ducy P, Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol Cell Biol. 1995;15:1858–1869. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- Guo YW, Chiu CY, Liu CL, Jap TS, Lin LY. Novel mutation of RUNX2 gene in a patient with cleidocranial dysplasia. Int J Clin Exp Pathol. 2015;8:1057–1062. [PMC free article] [PubMed] [Google Scholar]
- Han MS, Kim HJ, Wee HJ, Lim KE, Park NR, et al. The cleidocranial dysplasia-related R131G mutation in the Runt-related transcription factor RUNX2 disrupts binding to DNA but not CBF-beta. J Cell Biochem. 2010;110:97–103. doi: 10.1002/jcb.22516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen L, Riis AK, Silahtaroglu A, Hove H, Lauridsen E, et al. RUNX2 analysis of Danish cleidocranial dysplasia families. Clin Genet. 2011;79:254–263. doi: 10.1111/j.1399-0004.2010.01458.x. [DOI] [PubMed] [Google Scholar]
- Huang Y, Song Y, Zhang C, Chen G, Wang S, Bian Z. Novel RUNX2 frameshift mutations in Chinese patients with cleidocranial dysplasia. Eur J Oral Sci. 2013;121:142–147. doi: 10.1111/eos.12048. [DOI] [PubMed] [Google Scholar]
- Kagoshima H, Shigesada K, Satake M, Ito Y, Miyoshi H, et al. The Runt domain identifies a new family of heteromeric transcriptional regulators. Trends Genet. 1993;9:338–341. doi: 10.1016/0168-9525(93)90026-e. [DOI] [PubMed] [Google Scholar]
- Kanno T, Kanno Y, Chen LF, Ogawa E, Kim WY, Ito Y. Intrinsic transcriptional activation-inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor alpha subunit revealed in the presence of the beta subunit. Mol Cell Biol. 1998;18:2444–2454. doi: 10.1128/mcb.18.5.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Nam SH, Kim HJ, Park HS, Ryoo HM, et al. Four novel RUNX2 mutations including a splice donor site result in the cleidocranial dysplasia phenotype. J Cell Physiol. 2006;207:114–122. doi: 10.1002/jcp.20552. [DOI] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Levanon D, Negreanu V, Bernstein Y, Bar-Am I, Avivi L, Groner Y. AML1, AML2, and AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics. 1994;23:425–432. doi: 10.1006/geno.1994.1519. [DOI] [PubMed] [Google Scholar]
- Levanon D, Goldstein RE, Bernstein Y, Tang H, Goldenberg D, et al. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc Natl Acad Sci USA. 1998;95:11590–11595. doi: 10.1073/pnas.95.20.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Zeng B, Yu D, Jing X, Hu B, et al. Complex dental anomalies in a belatedly diagnosed cleidocranial dysplasia patient. Imaging Sci Dent. 2015;45:187–192. doi: 10.5624/isd.2015.45.3.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machuca-Tzili L, Monroy-Jaramillo N, González-del Angel A, Kofman-Alfaro S. New mutations in the CBFA1 gene in two Mexican patients with cleidocranial dysplasia. Clin Genet. 2002;61:349–353. doi: 10.1034/j.1399-0004.2002.610505.x. [DOI] [PubMed] [Google Scholar]
- Mundlos S, Otto F, Mundlos C, Mulliken JB, Alysworth AS, et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- Nagata T, Gupta V, Sorce D, Kim WY, Sali A, et al. Immunoglobulin motif DNA recognition and heterodimerization of the PEBP2/CBF Runt domain. Nat Struct Biol. 1999;6:615–619. doi: 10.1038/10658. [DOI] [PubMed] [Google Scholar]
- Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, et al. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AMLI gene. Proc Natl Acad Sci USA. 1993;90:6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott CE, Leschik G, Trotier F, Brueton L, Brunner HG, et al. Deletions of the RUNX2 gene are present in about 10% of individuals with cleidocranial dysplasia. Hum Mutat. 2010;31:E1587–1593. doi: 10.1002/humu.21298. [DOI] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Otto F, Kanegane H, Mundlos S. Mutations in the RUNX2 gene in patients with cleidocranial dysplasia. Hum Mutat. 2002;19:209–216. doi: 10.1002/humu.10043. [DOI] [PubMed] [Google Scholar]
- Pande S, Browne G, Padmanabhan S, Zaidi SK, Lian JB, et al. Oncogenic cooperation between PI3K/Akt signaling and transcription factor Runx2 promotes the invasive properties of metastatic breast cancer cells. J Cell Physiol. 2013;228:1784–1792. doi: 10.1002/jcp.24339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quack I, Vonderstrass B, Stock M, Aylsworth AS, Becker A, et al. Mutation analysis of core binding factor A1 in patients with cleidocranial dysplasia. Am J Hum Genet. 1999;65:1268–1278. doi: 10.1086/302622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirov TH, Inoue-Bungo T, Morii H, Fujikawa A, Sasaki M, et al. Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFbeta. Cell. 2001;104:755–767. doi: 10.1016/s0092-8674(01)00271-9. [DOI] [PubMed] [Google Scholar]
- Tessa A, Salvi S, Casali C, Garavelli L, Digilio MC, et al. Six novel mutations of the RUNX2 gene in Italian patients with cleidocranial dysplasia. Hum Mutat. 2003;22:104. doi: 10.1002/humu.9155. [DOI] [PubMed] [Google Scholar]
- Thirunavukkarasu K, Mahajan M, McLarren KW, Stifani S, Karsenty G. Two domains unique to osteoblast-specific transcription factor Osf2/Cbfa1 contribute to its transactivation function and its inability to heterodimerize with Cbfb. Mol Cell Biol. 1998;18:4197–4208. doi: 10.1128/mcb.18.7.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan D, Li S, Zhang X, Hu F, Lin L, et al. Mutations in the RUNX2 gene in Chinese patients with cleidocranial dysplasia. Ann Clin Lab Sci. 2008a;38:15–24. [PubMed] [Google Scholar]
- Xuan D, Li S, Zhang X, Lin L, Wang C, Zhang J. A novel RUNX2 mutation in cleidocranial dysplasia patients. Biochem Genet. 2008b;46:702–707. doi: 10.1007/s10528-008-9184-4. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Kanegane H, Osato M, Yanagida M, Miyawaki T, et al. Functional analysis of RUNX2 mutations in Japanese patients with cleidocranial dysplasia demonstrates novel genotype-phenotype correlations. Am J Hum Genet. 2002;71:724–738. doi: 10.1086/342717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Kanegane H, Osato M, Yanagida M, Miyawaki T, et al. Functional analysis of RUNX2 mutations in cleidocranial dysplasia: novel insights into genotype-phenotype correlations. Blood Cells Mol Dis. 2003;30:184–193. doi: 10.1016/s1079-9796(03)00020-2. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Javed A, Choi JY, van Wijnen AJ, Stein JL, et al. A specific targeting signal directs Runx2/Cbfa1 to subnuclear domains and contributes to transactivation of the osteocalcin gene. J Cell Sci. 2001;114:3093–3102. doi: 10.1242/jcs.114.17.3093. [DOI] [PubMed] [Google Scholar]
- Zhang C, Zheng S, Wang Y, Zhao Y, Zhu J, Ge L. Mutational analysis of RUNX2 gene in Chinese patients with cleidocranial dysplasia. Mutagenesis. 2010;25:589–594. doi: 10.1093/mutage/geq044. [DOI] [PubMed] [Google Scholar]
- Zhang YW, Yasui N, Kakazu N, Abe T, Takada K, et al. PEBP2αA/CBFA1 mutations in Japanese cleidocranial dysplasia patients. Gene. 2000;244:21–28. doi: 10.1016/s0378-1119(99)00558-2. [DOI] [PubMed] [Google Scholar]
- Zhou G, Chen Y, Zhou L, Thirunavukkarasu K, Hecht J, et al. CBFA1 mutation analysis and functional correlation with phenotypic variability in cleidocranial dysplasia. Hum Mol Genet. 1999;8:2311–2316. doi: 10.1093/hmg/8.12.2311. [DOI] [PubMed] [Google Scholar]