Abstract
Background
Curcumin (diferuloylmethane) has chemopreventive and therapeutic properties against many types of tumors, both in vitro and in vivo. Previous reports have shown that curcumin exhibits anti‐invasive activities, but the mechanisms remain largely unclear.
Methods
In this study, both microRNA (miRNA) and messenger RNA (mRNA) expression profiles were used to characterize the anti‐metastasis mechanisms of curcumin in human non‐small cell lung cancer A549 cell line.
Results
Microarray analysis revealed that 36 miRNAs were differentially expressed between the curcumin‐treated and control groups. miR‐330‐5p exhibited maximum upregulation, while miR‐25‐5p exhibited maximum downregulation in the curcumin treatment group. mRNA expression profiles and functional analysis indicated that 226 differentially expressed mRNAs belonged to different functional categories. Significant pathway analysis showed that mitogen‐activated protein kinase, transforming growth factor‐β, and Wnt signaling pathways were significantly downregulated. At the same time, axon guidance, glioma, and ErbB tyrosine kinase receptor signaling pathways were significantly upregulated. We constructed a miRNA gene network that contributed to the curcumin inhibition of metastasis in lung cancer cells. let‐7a‐3p, miR‐1262, miR‐499a‐5p, miR‐1276, miR‐331‐5p, and miR‐330‐5p were identified as key microRNA regulators in the network. Finally, using miR‐330‐5p as an example, we confirmed the role of miR‐330‐5p in mediating the anti‐migration effect of curcumin, suggesting the importance of miRNAs in the regulation of curcumin biological activity.
Conclusion
Our findings provide new insights into the anti‐metastasis mechanism of curcumin in lung cancer.
Keywords: Curcumin, gene expression profile, lung cancer, metastasis, miRNA expression profile
Introduction
Lung cancer is the major cause of cancer‐related mortality worldwide. The outcome of chemotherapeutic agents long used in clinical treatment remains disappointing. Phytochemicals, such as curcumin (diferuloylmethane), an active component of the spice turmeric (Curcuma longa), have encouraging anti‐cancer effects. Curcumin shows chemopreventive and therapeutic properties against many tumors, both in vitro and in vivo.1, 2, 3, 4 A plethora of molecular targets such as Jun N‐terminal kinase (JNK), c‐Jun/AP‐1, P53, and nuclear factor κB, and signaling pathways such as phosphoinositide 3‐kinase (PI3K)/protein kinase B (Akt)/mammalian target of rapamycin (mTOR) are modulated by curcumin, resulting in the inhibition of cell proliferation, invasion, metastasis, and angiogenesis, and the induction of apoptosis.5, 6, 7 Previous studies from our laboratory have shown that curcumin inhibits lung cancer cell proliferation and induces cell apoptosis through the lysosomal and reactive oxygen species‐dependent mitochondrial signaling pathways.8 Recently, studies have further focused on the anti‐invasion and anti‐metastasis effect of curcumin in many types of cancers. Zhu et al. reported that curcumin could induce apoptosis and inhibit invasion through mitogen‐activated protein kinase (MAPK) and matrix metalloproteinase (MMP) signaling in human monocytic leukemia SHI‐1 cells.9 Wang et al. demonstrated that curcumin suppresses growth and invasion and induces apoptosis in glioma cells by inhibiting the Skp2 pathway.10 In lung cancer, the protein kinase C alpha (PKCα)/NADPH oxidase 2 (Nox‐2)/reactive oxygen species (ROS)/activating transcription factor‐2 (ATF‐2) pathway was involved in the anti‐invasion effect of curcumin.11 Although more and more molecular targets and pathways are being discovered, the mechanisms of curcumin on lung cancer metastasis remain largely unknown.
MicroRNAs (miRNAs) are a class of ~22 nt endogenous, non‐coding RNAs generated from a larger stem loop structure, which can be expressed in a cell‐specific or tissue‐specific manner, influencing messenger RNA (mRNA) stability and translation. miRNAs are reported to be involved in many biological processes, including migration, invasion, epithelial to mesenchymal transition (EMT), and metastasis. For example, miR‐21 is reported to be involved in growth, invasion, and chemoresistant or radioresistant non‐small‐cell lung cancer. miR‐21 overexpression downregulates phosphatase and tensin homolog (PTEN) expression levels in A549 cells.12 miR‐483‐5p is upregulated in human lung adenocarcinoma, which is correlated with the progression of lung cancer.13 Recently, several miRNAs, such as miR‐135a, miR‐146a, and miR‐200, have been shown to be involved in the EMT of lung cancer.14, 15, 16 Furthermore, studies have shown that miR‐29a and miR‐200 inhibit proliferation and invasion in lung cancer cells in vitro, and this function might be mediated through the post‐transcriptional regulation of several proteins or abrogate the capacity of cells to undergo EMT.17, 18 These results show the important roles of miRNAs in anti‐lung cancer metastasis. Although a large number of miRNAs have been described in lung cancer, few global miRNA analyses have been performed to examine the inhibitory effects of curcumin on lung cancer metastasis.
In this study, by integrating miRNA and gene expression data from A549 cells and their curcumin‐treated counterparts, we revealed the miRNA‐mRNA interaction networks potentially involved in curcumin‐inhibited lung cancer metastasis. Moreover, we identified five key miRNAs involved in the inhibition of A549 cell migration and invasion by curcumin. These findings provide new insights into the mechanism of curcumin regulated miRNAs and mRNA expression profiles for anti‐metastasis.
Methods
Reagents and cell culture
Curcumin was purchased from Sigma‐Aldrich (St. Louis, MO, USA). The A549 human non‐small cell lung cancer cell line was obtained from the cell bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in RPMI‐1640 medium containing 10% fetal bovine serum at 37°C with 5% v/v CO2.
Methyl thiazolyl tetrazolium (MTT) assay
The anti‐proliferation effect of curcumin on A549 cells was determined by methyl thiazolyl tetrazolium (MTT) assay. Briefly, the cells were seeded in quintuplicate at a density of 5 × 103 cells per well in a 96‐well plate. After 24 hours of culture, curcumin was added to each well and incubated for 12, 24, and 48 hours. The cells were then washed with phosphate buffered saline (PBS) and 20 μL MTT solution (5 mg/mL) was added. After four hours of incubation at 37°C, 150 μL isopropanol was added. The absorbance of MTT‐formazan was measured at 570 nm.
microRNA (miRNA) transfections
GenePharma Biotechnology (Shanghai, China) chemically synthesized the 2′‐O‐methyl (2′‐O‐Me) oligonucleotides. The sequences were as follows: miR‐330‐5p mimics: (forward) 5′‐UCUCUGGGCCUGUGUCUUAGGC‐3′, (reverse) 5′‐CUAAGACACAGGCCCAGAGAUU‐3′; miR‐330‐5p inhibitor small interfering RNA (siRNA): 5′‐GCCUAAGACACAGGCCCAGAGA‐3′, negative control (NC): (forward) 5′‐UUCUCCGAACGUGUCACGUTT‐3′, (reverse) 5′‐ACGUGACACGUUCGGAGAATT‐3′. A549 cells at 60–70% confluence were transfected with miR‐330‐5p mimics, miR‐330‐5p inhibitor siRNA, or NC using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol.
Migration and invasion assay
For the wound‐healing assays, transfected and non‐transfected A549 cells were allowed to grow to 80–90% confluence in 24‐well plates. A pipette tip was used to scrape the cell monolayer to induce the wound. After washing with PBS, the cells were exposed to indicated concentrations of curcumin. At the next two time points (0 and 24 hours), the wound closure was photographed under a Leica inverted microscope (Leica Microsystems, Wetzlar, Germany) and transwell assay was carried out. In brief, 5 × 104 cells were plated on the transwell upper chamber (in a 24‐well plate) and coated with 10 μg Matrigel/well in serum‐free medium containing 0.1% bovine serum albumin and 10 μM curcumin; 10% fetal bovine serum medium in the lower chamber served as the chemoattractant. After 48 hours, non‐invading cells were removed with cotton swabs. The invading cells were observed and photographs were taken under optical microscope. All experiments were carried out in triplicate.
Real‐time PCR array analysis
Total RNA was isolated from cultured A549 and A549 treated with 10 μM curcumin. Reverse transcription was performed to generate cDNAs. Samples were loaded onto 384‐well microfluidic cards (TaqMan Array Human MicroRNA A+B Card Set v3) using a 7900HT RT‐PCR System (Life Technologies, Gaithersburg, MD, USA). Experimental data were then analyzed using SDS 2.2.2 software (Applied Biosystems, Foster City, CA, USA) and the relative miRNA expression values were calculated using U6 small nuclear RNA as an endogenous control according to the ΔΔCt method. A Ct value < 40 was defined as the limit of detection of the individual assays.19
Gene microarray analysis
Total RNA was isolated from A549 and curcumin (10 μM, 24 hours) treated A549 cells. A total of 100 ng of total RNA was reverse transcribed to cDNA, which was used as template in the following reaction. The biotinylated cDNA (5 μg) was then fragmented and hybridized to a GeneChip Human Gene 1.0 ST Array (Affymetrix Inc., Santa Clara, CA, USA). All microarray experiments were performed in triplicate. GeneChips were then scanned using GeneChip Scanner 3000 7G Plus 2 and Command Console Software (AGCC) version 1.0 (Affymetrix Inc.). Raw gene expression data in the generated CEL files were then normalized using the Robust MultiChip Averaging (RMA) algorithm.20
Integrated analysis of significant trends of miRNA and messenger RNA (mRNA)
The relationships between differentially expressed miRNAs and mRNAs were further analyzed. The predicted mRNA targets of each of the miRNAs that were differentially regulated were obtained using TargetScan version 5.1 (Whitehead Institute for Biological Research, Cambridge, UK). The intersection genes between the miRNA target genes and the different expressing genes were analyzed by spike‐triggered covariance analysis. Corresponding miRNAs were analyzed in the next step.
Gene ontology (GO) and pathway analysis
Gene ontology (GO) analysis was applied to analyze the main function of the differential expression genes according to the Gene Ontology database, the key functional classification of the National Center for Biotechnology Information. Specifically, two‐side Fisher's exact and χ2 tests were used to classify the GO category, and the false discovery rate (FDR) was calculated to correct the P value. Pathway analysis was used to determine the significant pathway of the differential genes according to KEGG, Biocarta, and Reatome databases. Fisher's exact and χ2 tests were then used to select the significant pathway, and the threshold of significance was defined by P value and FDR.
miRNA gene network
To build a miRNA gene network, we counted the differential expression values between miRNAs and genes, according to their interactions in the Sanger miRNA database. The adjacency matrix of microRNA and genes A = [ai,j] was made by attributing relationships between genes and microRNA, where ai,j represents the weight of the relationship between gene i and microRNA j. In the miRNA gene network, the circles represent genes and their interactions are represented by the edges. The center of the network was represented by degree, which is the contribution of one miRNA to the genes around it or the contribution of one gene to the miRNAs around it. The key miRNAs and genes in the network displayed the greatest degrees.
Statistical analysis
All statistical analyses were performed using SPSS version 13.0 (SPSS Inc., Chicago, IL, USA). Data were expressed as mean ± standard deviation. The statistical difference of data between groups was analyzed by Student's t test. Differences were considered significant when P < 0.05.
Results
Low curcumin concentrations inhibit migration and invasion but not proliferation in lung cancer A549 cells
In a previous study, we found that curcumin inhibited lung cancer 801D cell proliferation in concentration‐dependent manner. Low concentrations of curcumin did not affect 801D cell proliferation, but significantly suppressed cell migration and invasion.21 To further confirm the effects of curcumin in lung cancer cells, A549 cells were treated with various concentrations of curcumin (10, 20, 40 μM) and the cell viabilities of each group were tested by MTT assay. As shown in Figure 1a, curcumin treatment resulted in time and dose dependent loss of cell viability. However, lower concentrations of curcumin (10 μM) were shown to have a weak effect on lung cancer cell growth within 24 hours of treatment. We then measured the effect of curcumin on the invasive ability of A549 cells. A Boyden chamber coated with Matrigel was used in a dosage experiment. The results showed that the number of invaded cells was significantly reduced in 10 μM in the curcumin treated group (Fig 1b). The inhibitory effect of curcumin (10 μM) on A549 cell migration was also confirmed by wound‐healing assay (Fig 1c). These data demonstrated that lower concentrations of curcumin significantly inhibited the invasion and migration of A549 cells. As a result, 10 μM curcumin was used for migration and invasion experiments and microarray analysis.
Figure 1.
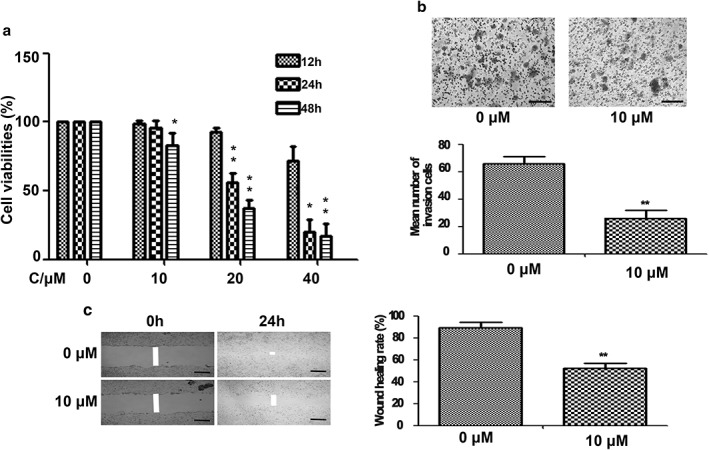
The effect of curcumin on A549 lung cancer cells. (a) Curcumin treatment resulted in a time and dose dependent loss of cell viabilities. Columns, mean from three different experiments with three duplicates; bars, standard error (*P < 0.05 and **P < 0.01 vs. respective 0 μM curcumin group). Curcumin significantly inhibited (b) A549 cell invasion and (c) migration at 10 μM curcumin after 24 hours of treatment. Scale bar = 200 μm.
Curcumin treatment alters miRNA expression profiles
To understand the mechanisms of how curcumin inhibited migration and invasion in A549 cells, we treated the A549 cells with 10 μM of curcumin for 24 hours, extracted the total RNA, and profiled miRNA levels on microarrays (TaqMan Array Human MicroRNA A+B Card Set v3, Life Technologies). When we set the cut‐off as a fold change > 2.0 and P < 0.05 between the curcumin‐treated and control groups, 36 differentially expressed miRNAs were identified (Table S1). The miR‐330‐5p exhibited maximum upregulation (fold change 142.34) and the miR‐25‐5p exhibited maximum downregulation (fold change 0.005 [200 times downregulated]) in the curcumin‐treated compared to the control group. These data suggest that in A549 cell migration and invasion, curcumin may modulate the expression of some key miRNAs.
Curcumin treatment alters mRNA expression profile
The total RNA samples were also used for mRNA profiling. We analyzed the global gene transcription profiles of the lung cancer cells treated with 10 μM of curcumin. We found that 86 genes were upregulated and 140 genes were downregulated by curcumin when using fold change > 2 and P < 0.01 as a cut‐off (data not shown). Functional annotation of the differentially expression genes was conducted using the National Institutes of Health DAVID database. The results showed that specific enriched functional categories of GO included cell adhesion (such as sialic acid binding Ig‐like lectin 16), cell fate determination (such as GATA binding protein 3), and positive regulation of gene expression (such as leucine rich repeat containing 32).
Functional (GO) analysis of the differential miRNA regulated target genes
TargetScan 5.1 was used to predict potential mRNAs with targets of 36 differentially expressed miRNAs. GO analysis was performed on the list of predicted target mRNAs that corresponded with altered mRNAs after curcumin treatment. The results showed that the top GO category of downregulated mRNAs was multicellular organismal development, while the top GO category of upregulated mRNAs was genes regulating sodium ion transport (Fig 2).
Figure 2.
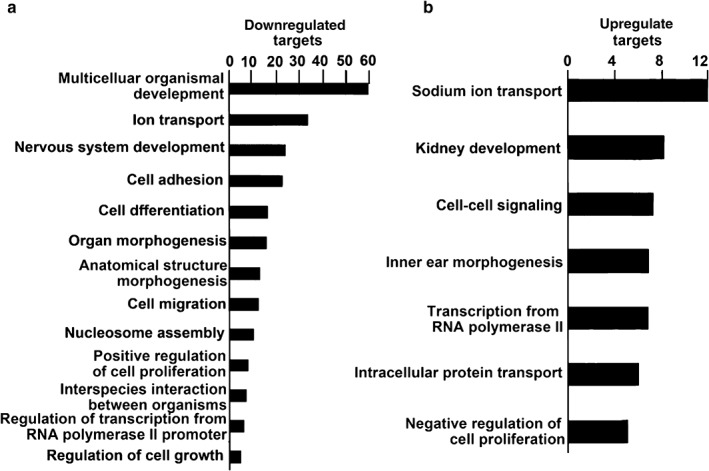
Gene ontology analysis identified potential messenger RNA functional categories regulated by microRNAs between the curcumin‐treated and control groups. Gene ontology analysis was performed on messenger RNAs predicted to be targets of micro RNAs. Three technical and one biological replicate was used for each experiment, except methyl thiazolyl tetrazolium assay, which had five biological replicates included. Only categories with a P value < 0.05 are included. The negative log of the P value is plotted on the x‐axis.
Signal transduction pathway analysis of differential miRNA target genes
Significant pathway analysis of the negatively related intersections of the target genes was performed using methods similar to the abovementioned gene function analysis. We found that the upregulated miRNA target genes were involved in 27 signal pathways, while the downregulated miRNA target genes were involved in 16 (P < 0.05, FDR < 0.05). Significantly downregulated pathways included MAPK, transforming growth factor‐β (TGF‐β), and Wnt signaling pathways (Fig S1). Significantly upregulated pathways included Axon guidance, glioma, and ErbB signaling pathways. These results were similar to those found using GO analysis, indicating that differential miRNA target genes are involved in many cellular pathways, promoting lung cancer occurrence and development.
Integrated analysis of the miRNA and mRNA expression profiles in the curcumin‐treated A549 cells
It is well known that miRNAs induce mRNA degradation and regulate protein expression by post‐transcriptional silencing. Therefore, miRNA levels should be inversely correlated to the levels of target gene transcripts. To understand the mechanisms of curcumin in suppressing the migration and invasion of lung cancer cells, we analyzed the potential association of differentially expressed miRNAs with mRNAs. Our results revealed that 14 out of 75 upregulated miRNAs (20%) had 100 downregulated mRNA targets, while 20 out of 74 downregulated miRNAs (27%) had 104 upregulated target mRNAs in the curcumin‐treated A549 cells. Furthermore, curcumin inhibited the migration and invasion of A549 cells via a network formed by these miRNAs and mRNAs. Thus, miRNAs might play key roles in this process. Further, we focused on seven miRNA regulators that have higher degrees in the miRNA‐mRNA network and might have key regulation roles in curcumin‐inhibited lung cancer metastasis (Fig 3).
Figure 3.
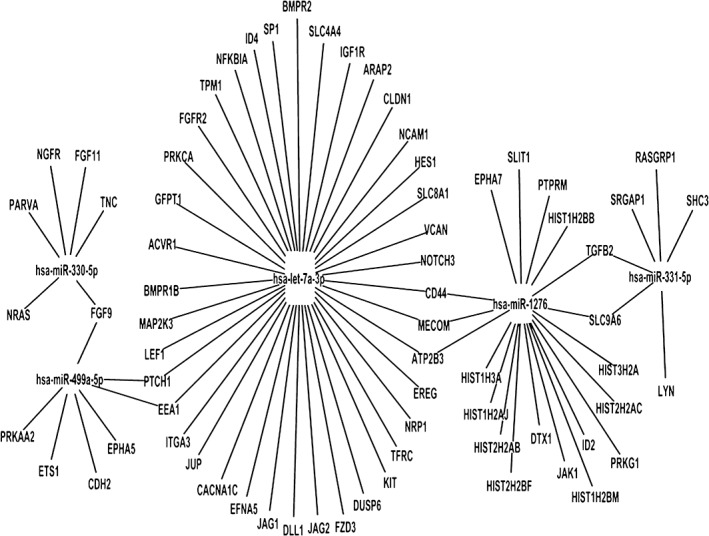
Differential regulatory network and key microRNA (miRNA) regulators of lung cancer metastasis. Hsa‐miR‐330‐5p, hsa‐miR‐1276, and hsa‐miR‐331‐5p are upregulated miRNAs, while hsa‐miR‐499a‐5p and hsa‐let‐7a‐3p are downregulated miRNAs in curcumin‐treated A549 cells.
miR‐330‐5p mimics significantly inhibited A549 cell migration
Because miR‐330‐5p exhibited maximum upregulation in curcumin treated A549 cells, we performed miRNA mimic transfection and wound‐healing assays to evaluate the inhibitory effect of miR‐330‐5p on A549 cell migration. Our results showed that miR‐330‐5p mimic transfection significantly inhibited A549 cell migration, and the effect was similar to that exhibited by curcumin. When the miR‐330‐5p inhibitor was used, it partly counteracted the inhibitory effect of curcumin on A549 cell migration (Fig 4). These results suggest that key miRNAs exist and are necessary for curcumin to have an effect on anti‐lung cancer migration.
Figure 4.
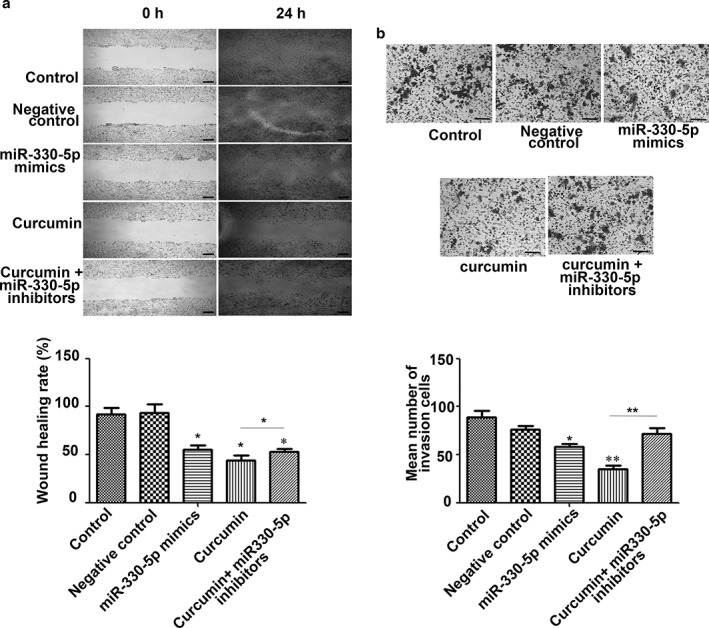
microRNA (miR)‐330‐5p is involved in the (a) anti‐migration and (b) invasion effects of curcumin. Compared to the control and negative control, the miR‐330‐5p mimic significantly inhibited the migration and invasion of lung cancer A549 cells, which was similar to the effect of curcumin. Furthermore, miR‐330‐5p inhibitors can partly counteract the inhibitory effect of curcumin on A549 cell migration and invasion. Scale bar = 200 μm. Columns, mean from three different experiments with three duplicates; bars, standard error (*P < 0.05 and **P < 0.01 vs. control or indicated control).
Discussion
Curcumin is a natural polyphenol compound that has potent anti‐proliferative and anti‐metastasis effects on various cancers.22, 23 In this study, we found that relatively low concentrations of curcumin significantly inhibited the migration of lung cancer A549 cells. Twenty‐one miRNAs were upregulated, whereas 15 miRNAs were downregulated with curcumin treatment. Integrated microRNA and gene expression profiling reveals that curcumin increased the expression of 86 mRNAs and decreased the expression of 140 mRNAs. Using miR‐330‐5p as an example, we showed that screened miRNAs and their targeted genes were involved in curcumin‐mediated lung cancer cell migration. Previous studies have shown that miR‐330‐5p is highly expressed and acts as tumor suppressor in a variety of tumors, including pancreatic cancer, glioblastoma, and cutaneous malignant melanoma.24, 25, 26
In our study, the KEGG pathway and GO enrichment analysis showed that many signaling pathways participated in lung cancer metastasis. To narrow the scope and evaluate the most significant candidates, future studies could focus on miRNAs and their target genes in the intersection of pathways in cancer, such as the MAPK, TGF‐β, and Wnt signaling pathways. miR‐302b‐3p and let‐7f‐2‐3p might be key regulators of the MAPK and Wnt signaling pathways, respectively. The classic MAPK pathway consists of RAS, RAF, MEK1/2, and ERK1/2, which sequentially relay proliferative signals generated at cell surface receptors through cytoplasmic signaling into the nucleus.27 Numerous reports indicate that the MAPK pathway plays a major role in tumor progression and invasion, while inhibition of MAPK signaling reduces invasion.28 Notably, our data revealed that the MAPK signaling events were downregulated by curcumin and their expressions were likely regulated by miR‐190a, miR‐34a‐5p, miR‐330‐5p, miR‐338‐3p, miR‐1276, miR‐544a, miR‐331‐5p, and miR‐302b‐3p. Likewise, the Wnt signaling pathway includes the canonical or Wnt/beta‐catenin pathway in which Wnt proteins bind to “frizzled” receptors, which leads to downstream activation of gene transcription by beta‐catenin. In our study, Wnt signaling events were downregulated by curcumin and were likely targeted by miR‐544a, miR‐302b‐3p, miR‐335‐5p, and miR‐338‐3p. These data suggest a positive regulatory role of miRNAs for their target genes by some alternative indirect mechanisms, particularly for those genes involved in cell migration. We are interested in further investigating how curcumin modulates the transcription of these miRNAs and their predicted target genes in lung cancer cells.
More importantly, five key classifying modules were identified, whose target gene members were enriched in KEGG pathways: miR‐330‐5p, miR‐331‐5p, miR‐499a‐5p, miR‐1276, and let‐7a‐3p modules. Their predicted functions have been reported to be associated with tumor metastasis activity. We predicted that miR‐330‐5p targeted PARVA, miR‐331‐5p targeted TGFB2, miR‐499a‐5p targeted CDH2, miR‐1276 targeted CD44, and let‐7a‐3p targeted BMPR2. In previous studies, downregulation of the miR‐330 family miRNA has been reported in various human malignant diseases, and the role of miR‐330 family miRNA in tumor suppression has also been examined.29, 30 miR‐1276 has been reported to be a NF‐κB target miRNA in tumor necrosis factor α‐stimulated HepG2 cells. Bioinformatic analysis revealed that two potential target genes of miR‐1276, BMP2 and CASP9, were also enriched in lung cancer.31 miR‐331‐5p was found to be involved in doxorubicin resistance in K562 leukemia cells.32 In addition, miR499a was increased in hepatitis B virus‐mediated hepatocellular carcinoma cell proliferation and migration by targeting MAPK6.33 Very few studies have reported the role of let‐7a‐3p in carcinogenesis and none have examined miRNAs in curcumin‐anti‐metastasis.34, 35 Our results may provide some clues enabling further functional characterization of the role of these miRNAs and their target genes in this process.
miR‐330‐5p is a member of the miR‐330 family; however, the expression level and downstream target genes of miR‐330‐5p, as well as its biological roles in lung cancer, are still unknown. In our study, miR‐330‐5p exhibited maximum upregulation in the curcumin treatment group and had higher degrees in our miRNA‐mRNA network. It is exciting to speculate that miR‐330‐5p may represent the first non‐coding miRNA to be related to curcumin treatment in lung cancer. miR‐330‐5p mimic transfection significantly inhibited A549 cell migration, while miR‐330‐5p inhibitor transfection attenuated curcumin anti‐migration on lung cancer cells. These results suggest that curcumin manipulates the levels of key microRNAs, such as miR‐330‐5p, and may help to potentiate the anti‐cancer and anti‐metastatic effects of curcumin.
In conclusion, using miRNA and mRNA expression profiling, we demonstrated that the expression of a number of miRNAs and their predicted target genes was altered in A549 cells in response to curcumin. Analysis of the differentially expressed mRNAs and miRNAs revealed a signaling network that could mediate curcumin‐anti‐metastasis activities in A549 cells. These findings provide new insights into the mechanism of curcumin‐anti‐metastasis.
Disclosure
No authors report any conflict of interest.
Supporting information
Figure S1. microRNA (miRNA) target genes were involved in many cellular pathways. Significant pathways targeted by (a) upregulated miRNAs and (b) downregulated miRNAs. The vertical axis is the pathway category, and the horizontal axis is the negative log of the P value.
Table S1. Thirty‐six differentially expressed microRNA (miRNAs) between the curcumin‐treated and control groups.
Acknowledgments
This work was supported by the Medical Scientific Research Foundation of Zhejiang Province (201485114), the Public Welfare Project of the Science and Technology Department of Zhejiang Province (2013C33209, 2014C33277) and the Science and Technology Plan Project of Hangzhou City (20130633B29, 20140633B40).
Contributor Information
Qing‐yong Chen, Email: cqyong117@163.com.
Sheng‐lin Ma, Email: mashenglin@medmail.com.cn.
References
- 1. Chen Q, Wang Y, Xu K et al Curcumin induces apoptosis in human lung adenocarcinoma A549 cells through a reactive oxygen species‐dependent mitochondrial signaling pathway. Oncol Rep 2010; 23: 397–403. [PubMed] [Google Scholar]
- 2. Li L, Braiteh FS, Kurzrock R. Liposome‐encapsulated curcumin: in vitro and in vivo effects on proliferation, apoptosis, signaling, and angiogenesis. Cancer 2005;104:1322–31. [DOI] [PubMed] [Google Scholar]
- 3. Kim SY, Jung SH, Kim HS. Curcumin is a potent broad spectrum inhibitor of matrix metalloproteinase gene expression in human astroglioma cells. Biochem Biophys Res Commun 2005;337:510–6. [DOI] [PubMed] [Google Scholar]
- 4. Singh M, Singh N. Curcumin counteracts the proliferative effect of estradiol and induces apoptosis in cervical cancer cells. Mol Cell Biochem 2011;347: 1–11. [DOI] [PubMed] [Google Scholar]
- 5. Yu T, Li J, Qiu Y, Sun H. 1‐phenyl‐2‐decanoylamino‐3‐morpholino‐1‐propanol (PDMP) facilitates curcumin‐induced melanoma cell apoptosis by enhancing ceramide accumulation, JNK activation, and inhibiting PI3K/AKT activation. Mol Cell Biochem 2012;361: 47–54. [DOI] [PubMed] [Google Scholar]
- 6. Ye M, Zhang J, Zhang J, Miao Q, Yao L, Zhang J. Curcumin promotes apoptosis by activating the p53‐miR‐192‐5p/215‐XIAP pathway in non‐small cell lung cancer. Cancer Lett 2015;357:196–205. [DOI] [PubMed] [Google Scholar]
- 7. Jin H, Qiao F, Wang Y, Xu Y, Shang Y. Curcumin inhibits cell proliferation and induces apoptosis of human non‐small cell lung cancer cells through the upregulation of miR‐192‐5p and suppression of PI3K/Akt signaling pathway. Oncol Rep 2015;34:2782–9. [DOI] [PubMed] [Google Scholar]
- 8. Chen QY, Shi JG, Yao QH et al Lysosomal membrane permeabilization is involved in curcumin‐induced apoptosis of A549 lung carcinoma cells. Mol Cell Biochem 2012;359:389–98. [DOI] [PubMed] [Google Scholar]
- 9. Zhu GH, Dai HP, Shen Q, Ji O, Zhang Q, Zhai YL. Curcumin induces apoptosis and suppresses invasion through MAPK and MMP signaling in human monocytic leukemia SHI‐1 cells. Pharm Biol 2016; 54:1303–11. [DOI] [PubMed] [Google Scholar]
- 10. Wang L, Ye X, Cai X et al Curcumin suppresses cell growth and invasion and induces apoptosis by down‐regulation of Skp2 pathway in glioma cells. Oncotarget 2015;6:18027–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fan Z, Duan X, Cai H et al Curcumin inhibits the invasion of lung cancer cells by modulating the PKCα /Nox‐2/ROS/ATF‐2/MMP‐9 signaling pathway. Oncol Rep 2015; 34:691–8. [DOI] [PubMed] [Google Scholar]
- 12. Liu ZL, Wang H, Liu J, Wang ZX. MicroRNA‐21 (miR‐21) expression promotes growth, metastasis, and chemo‐ or radioresistance in non‐small cell lung cancer cells by targeting PTEN. Mol Cell Biochem 2013;372:35–45. [DOI] [PubMed] [Google Scholar]
- 13. Song Q, Xu Y, Yang C et al miR‐483‐5p promotes invasion and metastasis of lung adenocarcinoma by targeting RhoGDI1 and ALCAM. Cancer Res 2014;74:3031–42. [DOI] [PubMed] [Google Scholar]
- 14. Shi H, Ji Y, Zhang D, Liu Y, Fang P. MiR‐135a inhibits migration and invasion and regulates EMT‐related marker genes by targeting KLF8 in lung cancer cells. Biochem Biophys Res Commun 2015;465:125–30. [DOI] [PubMed] [Google Scholar]
- 15. Park DH, Jeon HS, Lee SY et al MicroRNA‐146a inhibits epithelial mesenchymal transition in non‐small cell lung cancer by targeting insulin receptor substrate 2. Int J Oncol 2015;47:1545–53. [DOI] [PubMed] [Google Scholar]
- 16. Kim JS, Kurie JM, Ahn YH. BMP4 depletion by miR‐200 inhibits tumorigenesis and metastasis of lung adenocarcinoma cells. Mol Cancer 2015;14: 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Muniyappa MK, Dowling P, Henry M et al MiRNA‐29a regulates the expression of numerous proteins and reduces the invasiveness and proliferation of human carcinoma cell lines. Eur J Cancer 2009;45:3104–18. [DOI] [PubMed] [Google Scholar]
- 18. Gibbons DL, Lin W, Creighton CJ et al Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR‐200 family expression. Genes Dev 2009;23:2140–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) method. Methods 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- 20. Irizarry RA, Hobbs B, Collin F et al Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003;4:249–64. [DOI] [PubMed] [Google Scholar]
- 21. Chen QY, Zheng Y, Jiao DM et al Curcumin inhibits lung cancer cell migration and invasion through Rac1‐dependent signaling pathway. J Nutr Biochem 2014;25:177–85. [DOI] [PubMed] [Google Scholar]
- 22. Ji C, Cao C, Lu S et al. Curcumin attenuates EGF‐induced AQP3 up‐regulation and cell migration in human ovarian cancer cells. Cancer Chemother Pharmacol 2008;62:857–65. [DOI] [PubMed] [Google Scholar]
- 23. Lin SS, Lai KC, Hsu SC et al Curcumin inhibits the migration and invasion of human A549 lung cancer cells through the inhibition of matrix metalloproteinase‐2 and ‐9 and vascular endothelial growth factor (VEGF). Cancer Lett 2009;285:127–33. [DOI] [PubMed] [Google Scholar]
- 24. Tréhoux S, Lahdaoui F, Delpu Y et al Micro‐RNAs miR‐29a and miR‐330‐5p function as tumor suppressors by targeting the MUC1 mucin in pancreatic cancer cells Biochim Biophys Acta 2015;1853:2392–403. [DOI] [PubMed] [Google Scholar]
- 25. Feng L, Ma J, Ji J, Ji H, Liu Y, Hu W. MiR‐330‐5p suppresses glioblastoma cell proliferation and invasiveness through targeting ITGA5. Biosci Rep 2017; pii: BSR20170019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26. Su BB, Zhou SW, Gan CB, Zhang XN. MiR‐330‐5p regulates tyrosinase and PDIA3 expression and suppresses cell proliferation and invasion in cutaneous malignant melanoma. J Surg Res 2016;203:434–40. [DOI] [PubMed] [Google Scholar]
- 27. Krueger JS, Keshamouni VG, Atanaskova N, Reddy KB. Temporal and quantitative regulation of mitogen‐activated protein kinase (MAPK) modulates cell motility and invasion, Oncogene 2001;20:4209–18. [DOI] [PubMed] [Google Scholar]
- 28. Choudhury GG, Karamitsos C, Hernandez J, Gentilini A, Bardgette J, Abboud HE. PI‐3‐kinase and MAPK regulate mesangial cell proliferation and migration in response to PDGF. Am J Physiol 1997;273:F931–38. [DOI] [PubMed] [Google Scholar]
- 29. Lee KH, Chen YL, Yeh SD et al MicroRNA‐330 acts as tumor suppressor and induces apoptosis of prostate cancer cells through E2F1‐mediated suppression of Akt phosphorylation. Oncogene 2009;28:3360–70. [DOI] [PubMed] [Google Scholar]
- 30. Hodzic J, Giovannetti E, Diosdado B, Adema AD, Peters GJ. Regulation of deoxycytidine kinase expression and sensitivity to gemcitabine by micro‐RNA 330 and promoter methylation in cancer cells. Nucleosides Nucleotides Nucleic Acids 2011;30:1214–22. [DOI] [PubMed] [Google Scholar]
- 31. Zhou F, Wang W, Xing Y, Wang T, Xu X, Wang J. NF‐κB target microRNAs and their target genes in TNFα‐stimulated HeLa cells. Biochim Biophys Acta 2014;1839:344–54. [DOI] [PubMed] [Google Scholar]
- 32. Feng DD, Zhang H, Zhang P et al Down‐regulated miR‐331‐5p and miR‐27a are associated with chemotherapy resistance and relapse in leukaemia. J Cell Mol Med 2011;15:2164–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xiang Z, Wang S, Xiang Y. Up‐regulated microRNA499a by hepatitis B virus induced hepatocellular carcinogenesis via targeting MAPK6. PLoS ONE 2014;9:e111410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ji H, Chen M, Greening DW et al Deep sequencing of RNA from three different extracellular vesicle (EV) subtypes released from the human LIM1863 colon cancer cell line uncovers distinct miRNA‐enrichment signatures. PLoS ONE 2014;9:e110314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Murray MJ, Bailey S, Raby KL et al Serum levels of mature microRNAs in DICER1‐mutated pleuropulmonary blastoma. Oncogenesis 2014;3:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. microRNA (miRNA) target genes were involved in many cellular pathways. Significant pathways targeted by (a) upregulated miRNAs and (b) downregulated miRNAs. The vertical axis is the pathway category, and the horizontal axis is the negative log of the P value.
Table S1. Thirty‐six differentially expressed microRNA (miRNAs) between the curcumin‐treated and control groups.


