Abstract
Background & Aims
Lamins are nuclear intermediate filament proteins that comprise the major components of the nuclear lamina. Mutations in LMNA, which encodes lamins A/C, cause laminopathies, including lipodystrophy, cardiomyopathy, and premature aging syndromes. However, the role of lamins in the liver is unknown, and it is unclear whether laminopathy-associated liver disease is caused by primary hepatocyte defects or systemic alterations.
Methods
To address these questions, we generated mice carrying a hepatocyte-specific deletion of Lmna (knockout [KO] mice) and characterized the KO liver and primary hepatocyte phenotypes by immunoblotting, immunohistochemistry, microarray analysis, quantitative real-time polymerase chain reaction, and Oil Red O and Picrosirius red staining.
Results
KO hepatocytes manifested abnormal nuclear morphology, and KO mice showed reduced body mass. KO mice developed spontaneous male-selective hepatosteatosis with increased susceptibility to high-fat diet–induced steatohepatitis and fibrosis. The hepatosteatosis was associated with up-regulated transcription of genes encoding lipid transporters, lipid biosynthetic enzymes, lipid droplet-associated proteins, and interferon-regulated genes. Hepatic Lmna deficiency led to enhanced signal transducer and activator of transcription 1 (Stat1) expression and blocked growth hormone–mediated Janus kinase 2 (Jak2), signal transducer and activator of transcription 5 (Stat5), and extracellular signal–regulated kinase (Erk) signaling.
Conclusions
Lamin A/C acts cell-autonomously to maintain hepatocyte homeostasis and nuclear shape and buffers against male-selective steatohepatitis by positively regulating growth hormone signaling and negatively regulating Stat1 expression. Lamins are potential genetic modifiers for predisposition to steatohepatitis and liver fibrosis. The microarray data can be found in the Gene Expression Omnibus repository (accession number: GSE93643).
Keywords: Nonalcoholic Fatty Liver Disease, Laminopathy, Growth Hormone Signaling, Lipodystrophy, Fibrosis
Abbreviations used in this paper: Erk, extracellular signal–regulated kinase; FPLD2, Dunnigan familial partial lipodystrophy; GH, growth hormone; Het, heterozygous; HFD, high-fat diet; Igf1, insulin-like growth factor 1; Jak2, Janus kinase 2; KO, knockout; % liver weight, liver percentage of body mass; NAFLD, nonalcoholic fatty liver disease; ND, normal diet; PBS, phosphate-buffered saline; qPCR, quantitative polymerase chain reaction; Stat, signal transducer and activator of transcription; WT, wild type
Graphical abstract
See editorial on page 441.
Summary.
The nuclear intermediate filament protein lamin A/C acts cell-autonomously in hepatocytes as a positive regulator of growth hormone signaling and maintains hepatocyte homeostasis and nuclear shape. Lamin A/C absence leads to male-selective fatty liver disease and predisposes to nonalcoholic steatohepatitis and fibrosis.
Nuclear lamins are type V intermediate filament proteins that play important roles in maintaining nuclear stability and regulating gene expression, and they also serve as signaling scaffolds at the inner nuclear membrane.1, 2 Lamins are classified based on their isoelectric points and sequence homology as A- or B-type lamins. LMNA (in human beings; Lmna in mice) encodes the alternatively spliced lamins A and C, and lamins B1 and B2 are encoded by LMNB1 and LMNB2, respectively.3, 4, 5
Lamin B1 is expressed ubiquitously, whereas lamin A/C is expressed postnatally in differentiated cells, including cardiac and skeletal muscle, adipocytes, and hepatocytes.6, 7, 8 Lamin A/C binds to chromatin, lamina-associated proteins, and signaling mediators, such as extracellular signal–regulated kinase (Erk), c-Fos, retinoblastoma protein, and sterol regulatory element-binding protein-1c, to regulate gene expression and cell signaling.9, 10, 11 Mutations in lamin A/C cause several human diseases, termed laminopathies, that can affect muscle (eg, Emery–Dreifuss muscular dystrophy), adipose tissue, liver (eg, Dunnigan familial partial lipodystrophy [FPLD2]), bone (eg, mandibuloacral dysplasia), or multiple tissues (eg, Hutchinson–Gilford progeria syndrome), depending on the site of the mutation.12, 13
FPLD2 (OMIM 151660) is characterized by partial lipodystrophy and metabolic syndrome, including insulin resistance, glucose intolerance, and hypertriglyceridemia.14, 15, 16, 17 Hepatosteatosis occurs in many patients with FPLD2 and other lipodystrophies and may progress to steatohepatitis.18, 19
The mechanism by which lipodystrophy-associated lamin mutations promote hepatic steatosis and metabolic syndrome is unclear. One possibility is that lamin A/C is dispensable in hepatocytes and, if so, the liver simply serves as a storage depot for excess fatty acids shunted into the circulation as a result of adipose tissue loss. Alternatively, FPLD2-associated lamin A/C variants in hepatocytes might directly promote fatty acid uptake and/or lipogenesis in the liver.
Previous characterization of laminopathy mouse models focused on adipose and muscle tissues. For example, characterization of mice transgenic for a LMNA mutation that causes FPLD2 focused primarily on its effect on adipose tissue, using the adipocyte protein 2 enhancer, which directs overexpression to adipose tissues, and showed a defect in adipose tissue renewal.20 In addition, assessment of total-body Lmna-deficient mice showed normal fat distribution and metabolism in heterozygous mice. However, complete characterization of a potential FPLD2 phenotype in homozygous mice was prevented by severe muscular dystrophy that became lethal by 8 weeks of age.21, 22
To characterize the role of lamin A/C specifically in the liver, we generated mice carrying hepatocyte-specific deletion of Lmna, using mice carrying a Lmna allele with loxP sites.23 We found that hepatocyte lamin A/C deficiency induced spontaneous liver injury and steatosis and led to markedly increased susceptibility to steatohepatitis upon feeding a high-fat diet (HFD) in a male-specific manner that correlated with the up-regulation of genes encoding fatty acid binding proteins, lipogenic enzymes, and lipid transporters. Notably, lamin A/C deficiency disrupted hepatic growth hormone (GH)-receptor signaling through the Janus kinase 2 (Jak2), Erk, and signal transducer and activator of transcription (Stat)5 axes, leading to dysregulation of downstream Stat5-dependent gene transcription, with up-regulated expression of Stat1 and downstream interferon-regulated genes. Hepatic GH signaling regulates liver metabolism and gender-specific hepatic gene expression in rodents and human beings.24, 25, 26, 27, 28, 29, 30 Our findings show that lamins help maintain hepatocyte homeostasis cell-autonomously and regulate hepatic growth hormone signaling. Lamin A/C genetic variants potentially may contribute to the development and/or progression of nonalcoholic fatty liver disease (NAFLD), which is becoming a major global liver disease.31
Materials and Methods
Antibodies
The antibodies used were as follows: lamin A/C (H-110; Santa Cruz Biotechnology, Santa Cruz, CA); lamin B1 (ab16048; Abcam, Cambridge, UK); phosphorylated Stat5 (9314S), total Stat5 (9363S), phosphorylated Jak2 (3771S), total Erk (clone L34F12), phosphorylated Erk (4370P), total Akt (clone 40D4), total Jak2 (clone D2E12), phosphorylated Akt (4058S), total Stat1 (clone D1K9Y), and phosphorylated Stat1 (clones D4A7 and 58D6) (Cell Signaling Technology, Danvers, MA); pan-actin (Ab-5, 1:2500 dilution; ThermoFisher Scientific, Wayne, MI); and CD45 (clone 30-F11, 1:200 dilution; BD Biosciences, San Jose, CA). All antibodies were used at a 1:1000 dilution unless specified otherwise.
Mouse Experiments
Mouse experiments were performed in accordance with guidelines outlined in the Guide for the Care and Use of Laboratory Animals prepared by the National Academy of Sciences and published by the National Institutes of Health, and with approval from the University of Michigan Institutional Animal Care and Use Committee. C57BL/6 mice with a floxed allele of Lmna23 were crossed to C57BL/6 albumin-Cre mice32 to generate C57BL/6 offspring with hepatocyte-specific deletion of exons 10 and 11 of Lmna and littermate control mice that either lacked the floxed Lmna allele or the albumin-Cre transgene. Both male and female mice were used and ranged in age from 8 to 39 weeks. Mice were fed normal chow or the high-fat Surwit Diet (Research Diets, Inc, New Brunswick, NJ) plus water containing 42 g/L of carbohydrates at 55% fructose and 45% sucrose (Sigma Aldrich, St. Louis, MO) for 14 weeks.33 For GH injection experiments, mice were fasted for 6 hours and then injected intraperitoneally with phosphate-buffered saline (PBS) or GH in PBS (2.5 mg/kg) followed by euthanasia and harvesting of the livers 10 minutes after GH administration. Mice were euthanized by CO2 asphyxiation. Blood was collected by intracardiac puncture, and livers were harvested for paraffin embedding (after fixation in 10% formalin) and immunohistochemistry or were frozen in optimum cutting temperature compound (OCT; ThermoFisher Scientific) for immunofluorescence staining. Mice and whole livers were weighed before liver processing to calculate the liver percentage of body mass (% liver weight). Serum alanine aminotransferase and serum triglyceride values were determined by the Unit for Laboratory Animal Medicine at the University of Michigan.
Microarray Analysis
Total RNA from Lmna wild-type (WT), heterozygous (Het), and knockout (KO) male livers from normal diet (ND)- and HFD-fed mice was isolated using the RNeasy Mini Kit (Qiagen, Hilden, Germany). RNA was hybridized to Mouse Gene ST 2.1 plates (Affymetrix, Santa Clara, CA) using the GeneChip WT PLUS Reagent Kit (Affymetrix).
Immunofluorescence Staining, Immunohistochemistry, and Transmission Electron Microscopy
Six-micron frozen mouse liver sections were fixed in methanol (-20°C, 10 min), washed with PBS, and then permeabilized (22°C, 3 min) with 0.1% Triton X-100 (Sigma Aldrich, St. Louis, MO) in PBS. Sections were blocked with 2% goat serum in 2% bovine serum albumin/PBS for 30 minutes and incubated overnight with a 1:50 dilution of the primary antibody followed by a 1-hour incubation with Alexa Fluor–488 goat anti-rabbit IgG (Life Technologies, Carlsbad, CA) (1:500). Slides were mounted using Prolong Gold Anti-fade Reagent with 4′,6-diamidino-2-phenylindole (ThermoFisher Scientific), and stained sections were visualized with the Axioimager M2 microscope (Zeiss, Pleasanton, CA). For immunohistochemistry, formalin-fixed, paraffin-embedded liver sections were rehydrated via sequential incubations in xylene, ethanol, and water. Sections then were heated in Citra antigen-retrieval solution (Biogenex, Fremont, CA) for 8 minutes, washed, and treated for 15 minutes with 0.6% H2O2 in methanol. Nonspecific antibody binding was blocked using 5% normal goat serum/2.5% bovine serum albumin in PBS (30 minutes). Primary antibody incubation was performed overnight at 4°C at 1:200 dilution in blocking buffer. Detection was performed with the Vectastain ABC rat IgG kit (Vector Laboratories, Burlingame, CA). Slides were developed using 3,3′-diaminobenzidine tetra hydrochloride substrate reagent (Vector Laboratories), counterstained with hematoxylin, dehydrated with ethanol and xylene, and then mounted with coverslips for analysis. Picrosirius red, Oil Red O, and immunohistochemistry staining was visualized using a LMD7000 microscope (Leica, Wetzlar, Germany). Transmission electron microscopy of livers from WT and KO mice was performed as described.34
Sample Fractionation and Immunoblotting
Total tissue homogenates were obtained by dounce homogenization of frozen liver samples in homogenization buffer (0.187 mol/L Tris, 3% sodium dodecyl sulfate, and 5 mmol/L EDTA, pH 6.8). Cytoplasmic and nuclear extracts were obtained from isolated primary hepatocytes and whole livers using NE-PER Nuclear and Cytoplasmic Extraction Reagents (ThermoFisher Scientific). For analysis of GH signaling in liver cytoplasmic extracts, scans of immunoblots were obtained using the ChemiDoc Touch Imaging System (Bio-Rad Laboratories, Hercules, CA), and relative band intensities were estimated using ImageJ version 1.51j8 software (National Institutes of Health, Bethesda, MD). Relative band intensities for phosphorylated Erk, Jak2, Stat5, and Akt were normalized against total Erk, Jak2, Stat5, and Akt, respectively.
Oil Red O Staining and Quantification
Ten-micron frozen liver sections were fixed in ice-cold 10% formalin (10 min), rinsed with water (3×), air dried, and incubated in absolute propylene glycol (Sigma Aldrich) for 5 minutes before staining with 0.5% Oil Red O (Sigma Aldrich) in propylene glycol (60°C, 10 min). Stained slides were washed in 85% propylene glycol and then water before counterstaining with Gill’s 3 Hematoxylin (Sigma Aldrich) for 30–45 seconds. Stained slides then were washed thoroughly with water (3 min) and mounted using glycerol gelatin (Sigma Aldrich). Eight to 10 fields/liver section (10× objective on a Leica LMD7000) were imaged for analysis. The percentage of Oil Red O–stained area was determined using Metamorph Image Analysis Software (Molecular Devices, Sunnyvale, CA).
Hydroxyproline Determination
Hydroxyproline measurement was performed as described previously.35 Absorbance (570 nm) readings were taken using a Synergy 2 Multi-Mode Reader (BioTek, Winooski, VT), and values were normalized to microgram of hydroxyproline per gram of liver weight.
Picrosirius Red Staining
Picrosirius red staining of formalin-fixed paraffin sections of livers from 23-week-old mice was performed by the Unit for Laboratory Animal Medicine at the University of Michigan. Subsequent Picrosirius red staining for 30- to 35-week-old mice was performed as follows: 5-μm paraffin sections were stained in 0.1% Picrosirius red in saturated picric acid for 1 hour, washed twice with 0.5% acetic acid, dehydrated in 3 changes of 100% ethanol, cleared in xylene, and mounted in Permount (ThermoFisher Scientific). Eight to 10 fields/liver section (10× objective using a Leica LMD7000) were imaged for analysis. The percentage of Picrosirius red–stained area was determined using Metamorph Image Analysis Software (Molecular Devices).
Reverse-Transcription Polymerase Chain Reaction
Total RNA was isolated from mouse livers using the RNeasy Mini Kit (Qiagen). Complementary DNA was prepared using the TaqMan Reverse-Transcription Reagents kit (Roche, Branchburg, NJ). Quantitative polymerase chain reaction (qPCR) was performed using Mastercycler realplex (Eppendorf, Hauppauge, NY). The reaction parameters were as follows: 95°C for 3 minutes; 40 cycles of 95°C for 15 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. The relative expression was determined using the 2-ΔΔCt method.36 The list of qPCR primers is included in Table 1.
Table 1.
Oligonucleotides Used in qPCR Analysis
| Primer | Forward | Reverse |
|---|---|---|
| Acta2 | TCAGCGCCTCCAGTTCCT | AAAAAAAACCACGAGTAACAAATCAA |
| Cd36 | GCTGCACCACATATCTACCAAA | AGGATATGGAACCAAACTGAGG |
| Cidea | CAGTCTGCAAGCAACCAAAG | TTGTGCATCGGATGTCGTAG |
| Cidec | AGCTAGCCCTTTCCCAGAAG | CCTTGTAGCAGTGCAGGTCA |
| Col1 | TCTGACTGGAAGAGCGGAGAG | GGCACAGACGGCTGAGTAGG |
| Cyp2a4 | AGCAGGCTACCTTCGACTGG | GCTGCTGAAGGCTATGCCAT |
| Cyp2b13 | GAACTGAGACTACCAGCACCACTCT | TGAGCATGAGCAGGAAACCACT |
| Cyp2b9 | CTGAGACCACAAGCGCCAC | CTTGAGCATGAGCAGGACTCC |
| Cyp39a1 | TTCTGGAACCCTCTTGCAGG | CGTGTTTCCGTCTCCACCAC |
| Cyp7b1 | CAGAAGTTCAGCAGCCGATT | AGTGAGCCACAGAATGCAAA |
| Gstπ | TTGCCGATTACAACTTGCTG | ATGGGACGGTTCACATGTTC |
| Hsd3b5 | CCAGTGTGCCAACATTCATC | AAGTGCCACCATTTTTCAGG |
| Igf1 | AGGCTATGGCTCCAGCATT | GTCTTGGGCATGTCAGTGTG |
| Irf7 | TGATCTTTCCCAGTCCTGCT | TGCCTACCTCCCAGTACACC |
| Mogat1 | TCAAAACGCAGGATTTGGAT | ACAACGGGAAACAGAACCAG |
| Mup3 | TGGGTATTGGTTTTCTATTGCTG | CTCCAAGACAGTGATGTTTTCC |
| Nnmt | GAAGGGACCTGAGAAGGAGGA | AGTACCTGCTTGATTGCACGC |
| Slco1a | CACCTGTTTACTTTGGCGCT | ATGAAGACTGCGGGGAGAAA |
| Stat1 | GTGGAGCCCTACACGAAAAA | ATACTTCCCAAAGGCGTGGT |
| Sult1e1 | TGGACAAACGGTTCACCAAA | GCCTTGCCAAGAACATTTCAA |
| Tgfβ | GGCCAGATCCTGTCCAAACT | TGTTGCGGTCCACCATTAG |
| Themis | TGTGAAGGTGGCTGTGAGAG | CATCTGCAGGCAAACTGCTA |
| Timp1 | TCCCTTGCAAACTGGAGAGT | AGGTGCACAAGCCTGGATTC |
| Tnfa | CGTCAGCCGATTTGCTATCT | CGGACTCCGCAAAGTCTAAG |
| Ubd | CCTTACCCTGAAGGTGGTGA | CTTCCAGCTTCTTTCCGTTG |
| 18S | AAACGGCTACCACATCCAAG | CCTCAAATGGATCCTCGTTA |
Hepatocyte Isolation and GH Treatment
Hepatocytes were isolated from age-matched mice for primary culture as described.37 After 30 minutes (37°C, 5% CO2) to allow for attachment, the cell culture media was replaced (to remove any debris or unattached cells) with fresh media. Primary hepatocytes were serum-starved overnight for 8–12 hours and then stimulated with human growth hormone (Nutropin; Genentech, South San Francisco, CA) in William’s medium E without serum (37°C, 10 min) at the indicated concentrations.
Statistical Analysis
Statistical analysis for microarray data was performed using the Oligo and Limma packages of Bioconductor implemented in the R statistical environment. Expression values for each gene in the microarray was calculated using a robust multi-array average.38 Probe sets with a variance over all samples less than 0.05 were filtered out. P values were adjusted for multiple comparisons using false-discovery rate.39 Statistical analysis of histologic scoring, qPCR, serology, and the percentage of body weight comparisons, body mass comparisons, Oil Red O and Picrosirius red staining, and relative immunoblot band intensities were performed using GraphPad Prism software (GraphPad Software, San Diego, CA). Statistical comparisons were performed using the unpaired t test (2-tailed) or 1-way analysis of variance followed by the Tukey post hoc test.
All authors had access to the study data and have reviewed and approved the final manuscript.
Results
Hepatic Lmna Deficiency Disrupts Nuclear Morphology
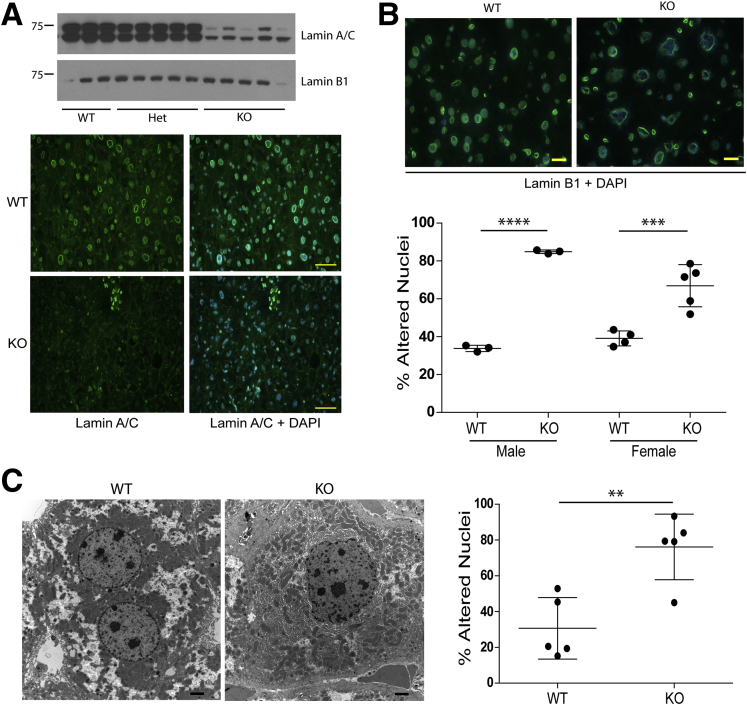
Mice with hepatocyte-specific Lmna deficiency were generated by crossing albumin-Cre transgenic mice with mice carrying a Lmna allele containing loxP sites flanking exons 10 and 11. Livers from mice homozygous for the floxed Lmna allele and positive for the albumin-Cre transgene (designated KO) expressed dramatically lower levels of lamin A/C compared with WT and Het livers (Figure 1A). Staining for lamin B1, which is located at the inner nuclear membrane, showed abnormal nuclear morphology in male and female KO hepatocytes (Figure 1B), which was supported by electron microscopy findings of misshapen nuclei in KO as compared with WT livers (76% vs 31% abnormal nuclei, respectively) (Figure 1C).
Figure 1.
Hepatic Lmna deficiency disrupts nuclear morphology. (A) Whole–cell extracts from Lmna WT, Het, and KO livers were analyzed by immunoblotting using antibodies to lamins A/C and B1. Each lane corresponds to a separate liver. Frozen sections from WT and KO livers were fixed in methanol and stained for lamin A/C followed by 4′,6-diamidino-2-phenylindole (DAPI). Scale bars: 50 μm. (B) Livers of 3 WT and 3 KO males (19–25 weeks of age) and 4 WT and 5 KO females (26–28 weeks of age) were cryosectioned and stained for Lamin B1 and DAPI. Normal-appearing and altered nuclei were counted in a blinded fashion from 5 fields per liver. Error bars represent SD, and statistical significance was determined by 1-way analysis of variance followed by the Tukey post hoc test. ****P < .0001 and ***P = .0005 for WT vs KO males and females, respectively. Scale bars: 20 μm for the representative sections shown. (C) Livers of 5 WT and 5 KO male mice (age, 22–24 wk) were examined by electron microscopy. Normal-appearing and altered nuclei (as defined by the presence of blebs and deformation) were counted in a blinded fashion from an average of 16 fields/liver. Error bars represent SD, and statistical significance (**P < .01) was determined using an unpaired t test (2-tailed). WT nuclei, n = 142; KO nuclei, n = 278. Scale bars: 2 μm for the representative sections shown.
Hepatic Lmna Deficiency Promotes Steatosis and Liver Injury in Male, but Not Female, Mice
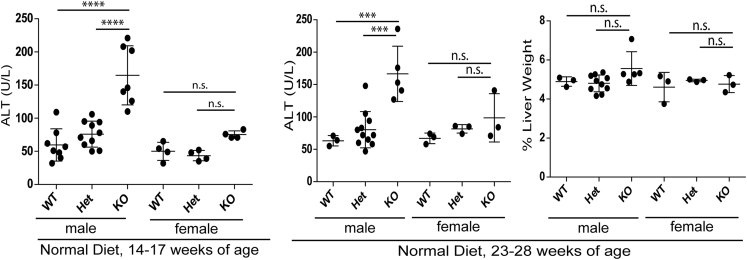
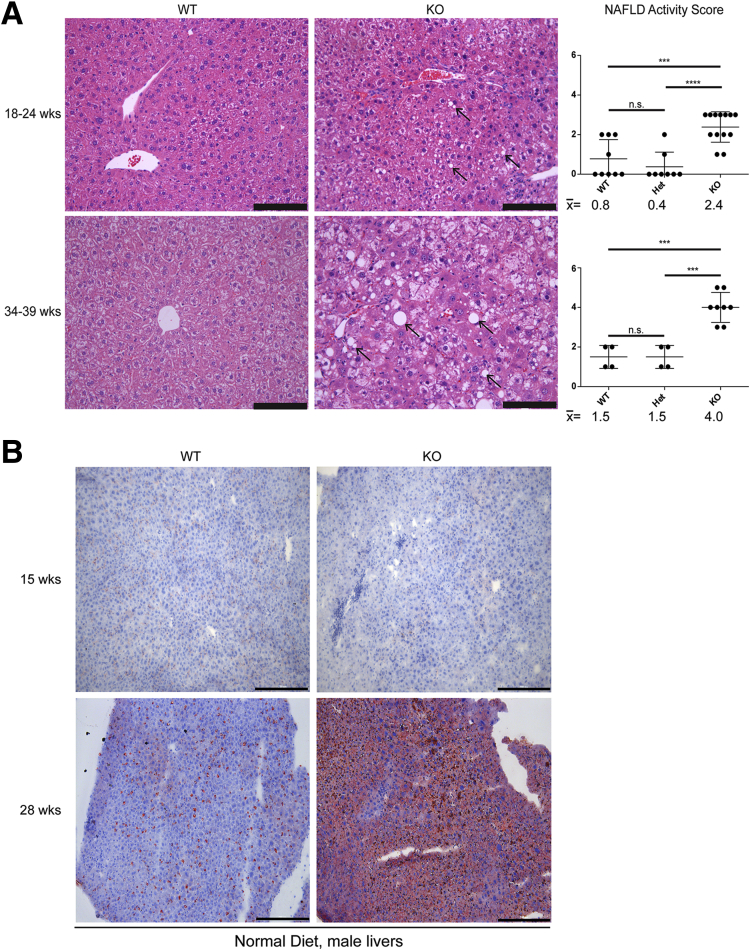
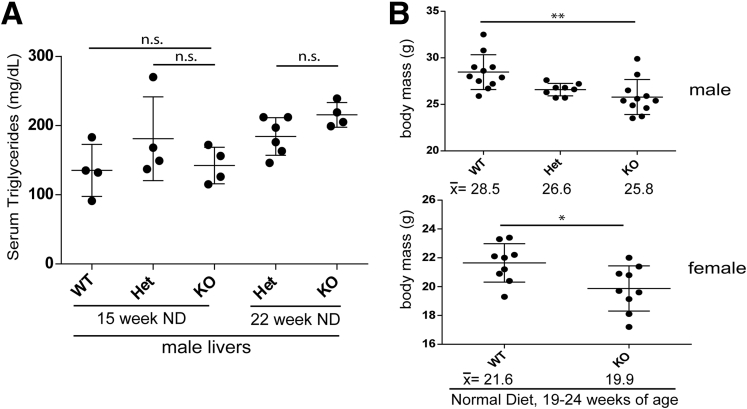
To assess the effect of lamin A/C deficiency on liver injury, mice were fed for 14 weeks with ND, or HFD supplemented with sucrose-fructose in the drinking water. In mice fed ND, lamin A/C absence led to spontaneous, male-selective liver injury by 14–17 weeks of age (Figure 2) and steatosis that progressed with age (Figure 3) with no change in serum triglyceride levels (Figure 4A). The NAFLD activity histologic scoring40 of H&E-stained livers (Figure 3A) showed more prominent liver pathology in male KO livers compared with male WT and Het livers, with significantly higher NAFLD activity scores40 in the KO livers (Figure 3A). No such histologic changes were appreciated in female KO livers under the same conditions (not shown). Both male and female KO animals showed decreased body mass (Figure 4B) without any effect on the % liver weight (Figure 2). The animals showed no gross morphologic changes (not shown).
Figure 2.
Hepatic Lmna deficiency promotes liver injury in male, but not female, mice fed a normal diet. Liver-specific lamin A/C KO leads to increased serum alanine aminotransferase (ALT) levels in male, but not female, mice (ages, 14–17 wk; 23–28 wk) fed normal chow (****P < .0001 for WT vs KO and Het vs KO comparisons at 14–17 weeks, and ***P < .001 for both comparisons at 23–28 weeks). n = 8, 10, and 7 for male WT, Het, and KO mice, respectively, and n = 4 for each of the female WT, Het, and KO mice, respectively, at 14–17 weeks. n = 3, 11, and 5 for male WT, Het, and KO mice, respectively, and n = 3 for each of the female WT, Het, and KO mice, respectively, at 23–28 weeks. For the 23- to 28-week group, the % liver weight is shown. NS, no significant differences in ALT values between genotypes for female livers and no significant difference in the % liver weight between genotypes for both sexes.
Figure 3.
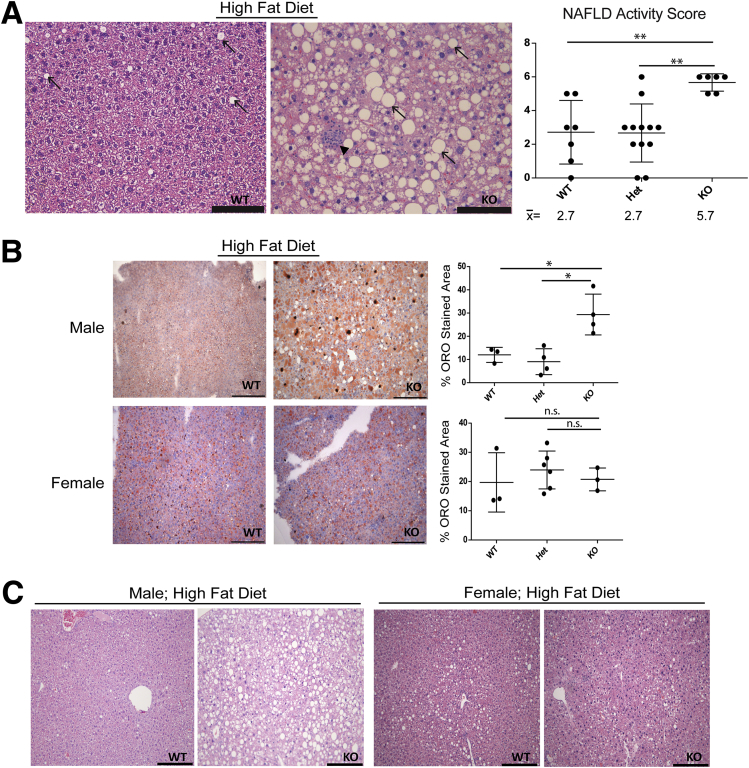
Hepatic Lmna deficiency promotes hepatosteatosis in male mice in an age-dependent manner. (A) Livers from male mice of the indicated genotypes (18–24 and 34–39 weeks of age) were fixed and then stained with H&E. Arrows indicate steatosis. Scale bars: 100 μm. NAFLD activity scores were determined in a blinded fashion for WT (n = 9), Het (n = 8), and KO (n = 13) livers at 18–24 weeks (upper panel) and for WT (n = 4), Het (n = 4), and KO (n = 8) livers at 34–39 weeks (lower panel). Error bars represent the SD. Mean values are shown under each graph. ***P < .001 and ****P < .0001. (B) Frozen liver sections (10 μm) from ND male WT and KO livers at 15 and 28 weeks of age were stained with Oil Red O, showing lipid accumulation in the KO liver. Scale bars: 200 μm.
Figure 4.
Hepatic Lmna deficiency has no effect on serum triglyceride levels but reduces body mass. (A) Hepatocyte-specific Lmna deficiency has no effect on serum triglyceride levels with ND feeding conditions at 15 and 22 weeks of age. (B) Male and female KO mice (19–24 weeks of age) showed reduced body mass compared with their WT counterparts. The mean body mass in grams for each group is shown below each graph. **P < .01 for WT vs KO males, and *P < .05 for WT vs KO females.
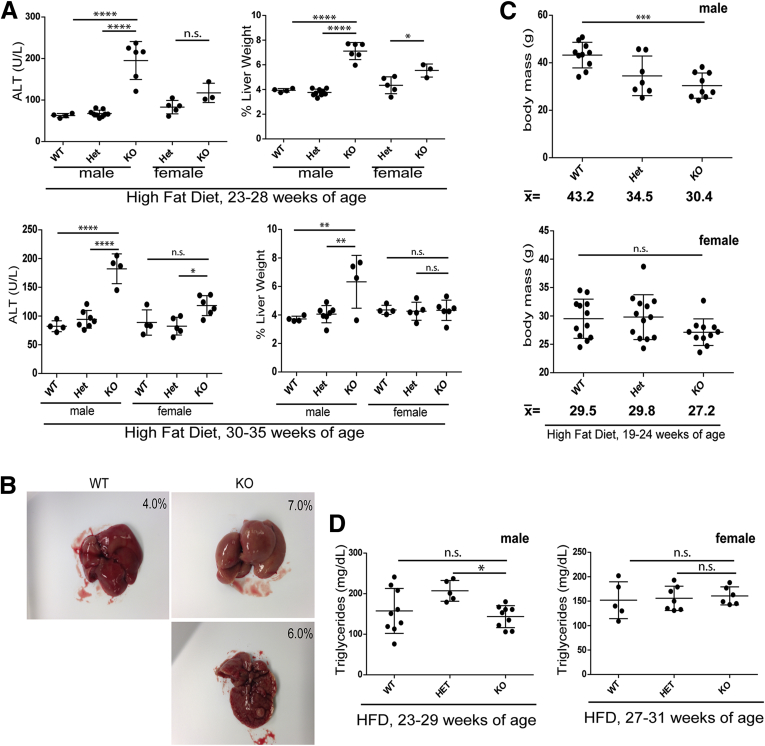
The difference between WT and KO male and female mice became even more apparent after HFD feeding, which led to neutral fat accumulation (based on H&E and Oil Red O staining) (Figure 5), liver injury (Figure 6A), hepatomegaly (Figure 6A and B), nodularity (Figure 6B), and body mass decrease (Figure 6C) in male, but not female, KO mice. H&E staining of male KO livers after HFD feeding (Figure 5A) showed extensive steatotic changes with inflammatory infiltrates and significantly higher NAFLD activity scores compared with WT and Het livers (with all KO livers receiving scores >4, indicating steatohepatitis). Despite the increase in hepatic steatosis in Lmna-deficient male mice, serum triglyceride levels in WT and KO mice were equivalent under both ND and HFD conditions (Figures 4A and 6D).
Figure 5.
Hepatic Lmna deficiency leads to steatohepatitis after HFD feeding. (A) HFD induces steatosis and lobular hepatitis to a much greater degree in KO compared with WT livers based on H&E staining. Scale bars: 100 μm. NAFLD activity scores were determined for WT (n = 7), Het (n = 12), and KO (n = 6) livers from mice ranging in age from 23 to 32 weeks (23–32 weeks for WT, 23–32 weeks for Het, and 23–25 weeks for KO) in a blinded fashion. **P < .01 for WT vs KO and Het vs KO. Arrows indicate steatosis; arrowhead indicates a focus of lobular hepatitis. (B) Frozen liver sections from male and female (WT/Het/KO) livers were stained with Oil Red O (ORO), and the percentage of stained area was calculated as described in the Materials and Methods section (*P < .05). Scale bars: 200 μm. Each point represents data from a separate mouse liver. Male mice were 22–23 weeks of age, and female mice were 27–31 weeks of age. (C) Paraffin sections of livers from HFD-fed male (age, 23 wk) and female (age, 31 wk) WT and KO mice were stained with H&E. Scale bars: 200 μm.
Figure 6.
Hepatic Lmna deficiency promotes liver hypertrophy and nodularity and body mass decrease after HFD feeding in a male-specific manner. (A) HFD led to an increase in alanine aminotransferase (ALT) levels in male, but not female, lamin A/C KO mice for those ages 23–28 and 30–35 weeks. For mice ages 23–28 weeks, n = 4, 9, and 6 for the male WT, Het, and KO mice, respectively; and n = 5 and 3 for female Het and KO, respectively. For mice ages 30–35 weeks, n = 4, 7, and 4 for male WT, Het, and KO, respectively; and n = 4, 5, and 6 for female WT, Het, and KO, respectively. The percentage of liver weights for mice ages 23–28 and 30–35 weeks were more prominently different in KO male livers compared with WT and Het male livers, as compared with WT vs KO female livers. Error bars in all graphs represent SD. Statistical significance was determined using 1-way analysis of variance followed by the Tukey post hoc test (*P < .05, **P < .01, and ****P < .0001). (B) Gross hepatomegaly and nodularity in HFD KO vs WT male livers. Percentages in each panel refer to the percentage of liver weight. (C) Male, but not female, KO mice (19–24 weeks of age) showed reduced body mass compared with their WT counterparts after HFD feeding. The mean body mass for each group is shown below each graph. (***P < .001 for WT vs KO males; NS, no significant difference for WT vs KO females). (D) Serum triglyceride levels for male and female KO mice were equivalent to WT counterparts (*P < .05 for Het vs KO). Males were between 23 and 29 weeks of age, and females were between 27 and 31 weeks of age.
Fatty Acid Metabolism Genes Are Up-Regulated Constitutively in Hepatocytes From Lamin A/C-Deficient Male Livers
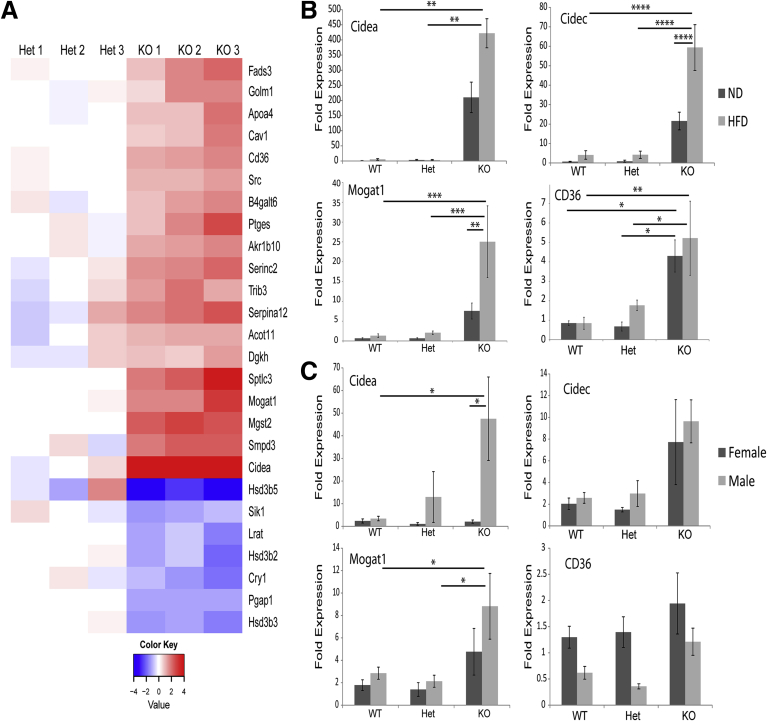
To understand the molecular basis of the male-specific hepatic steatosis phenotype, we performed expression profiling of Lmna WT, Het, and KO male livers from mice fed ND or HFD. Numerous genes encoding proteins involved in lipid storage and metabolism were up-regulated in KO livers (Figure 7A). In fact, the most highly up-regulated gene in male, but not female, livers was Cidea (Figure 7B and C), which encodes a lipid droplet–associated protein that promotes hepatic steatosis.41, 42 The gene encoding cell death-inducing DNA fragmentation factor, a subunit-like effector C, a related lipid droplet–associated protein, also was highly up-regulated in lamin A/C KO male mouse livers after HFD feeding (Figure 7B), although there was no significant difference between male and female KO livers under ND feeding conditions (Figure 7C). We were not able to validate the up-regulation of these genes at the protein level owing to inefficient reactivity or high background of several tested antibodies (not shown). Other notable male KO liver up-regulated genes related to lipid metabolism included Sptlc3, B4galt6, and Serinc2, which encode proteins involved in sphingolipid biosynthesis; and Fads3 and Mogat1, which encode proteins involved in fatty acid and triglyceride biosynthesis (Figure 7A). Mogat1, which encodes an enzyme that converts monoacylglycerol to diacylglycerol and is up-regulated in the livers of both lipodystrophic and ob/ob mice,43, 44 was highly up-regulated in lamin A/C-deficient livers, and the up-regulation was enhanced further after HFD feeding (Figure 7B and C). Similarly, the fatty acid translocase CD36, which is up-regulated in the livers of patients with NAFLD and in mice after HFD feeding,45, 46 was increased in Lmna-deficient male livers under both ND and HFD conditions (Figure 7B).
Figure 7.
Fatty acid metabolism genes are up-regulated in Lmna-deficient livers in a male-specific manner. (A) Lmna Het and KO livers (3 each) from male mice fed ND were compared using Affymetrix microarrays. Het and KO livers were isolated from 26-week-old and 39-week-old mice, respectively. Genes that were significantly different in Het vs KO and carried the gene ontology term fatty acid were grouped together in a heat map (red, high expression; blue, low expression). Cidea, a gene that encodes a lipid binding protein, and Hsd3b5, a male-specific gene involved in steroid metabolism, were the most highly up-regulated and down-regulated genes, respectively, in Lmna KO male livers. (B) qPCR analysis of messenger RNA from WT/Het/KO male livers of mice (20–25 weeks old) fed ND or HFD (*P < .05, **P < .01, ***P < .001, and ****P < .0001); n = 3–4 livers per genotype group for the ND or the HFD feeding. (C) qPCR analysis of messenger RNA from WT/Het/KO livers from male and female mice (13–17 weeks old; n = 3–4 mice/genotype) fed a ND (*P < .05). Error bars in all graphs represent SEM. Statistical significance was determined using 1-way analysis of variance followed by the Tukey post hoc test.
Lmna Deficiency Promotes Steatohepatitis and Fibrosis With Up-Regulation of Proinflammatory Genes
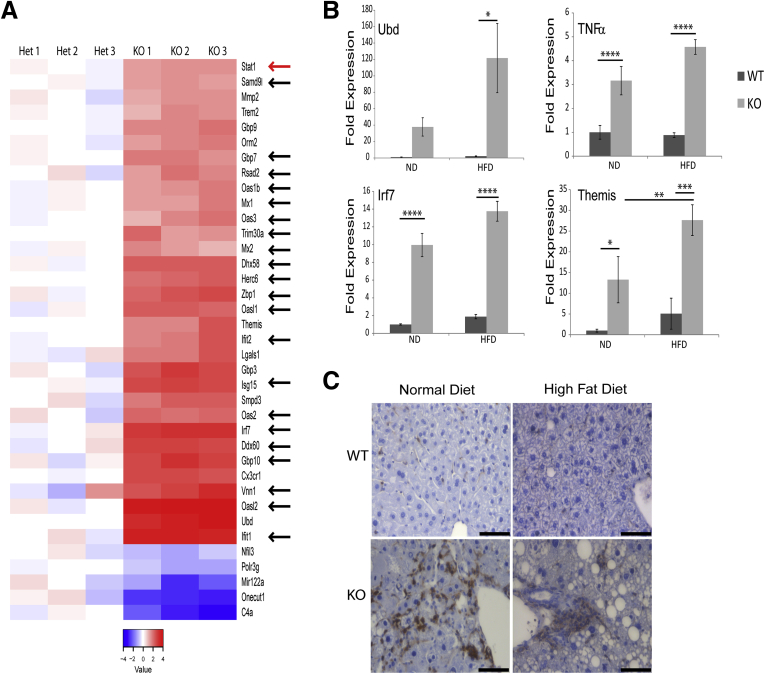
Given that progressive inflammation and fibrosis are thought to be the primary drivers of liver-related morbidity and mortality in human NAFLD, we tested whether lamin A/C KO livers also harbor inflammation and progress from simple steatosis to steatohepatitis and fibrosis. The expression profiling analysis showed that even with ND feeding, the lamin A/C KO livers manifested up-regulation of genes related to immunity and the interferon response (Figure 8A), including Oas1, Oasl2, Oas2, Ifit1, Ifit2, Ddx60, and Dhx58 (which encode interferon-induced antiviral proteins); Irf7, which encodes an interferon-inducible transcription factor that promotes lipid accumulation; the chemokine receptor Cx3cr1; the ubiquitin-like modifier Ubd, which is involved in tumor necrosis factor α–induced nuclear factor-κB activation; and the thymocyte-expressed protein Themis, which is involved in thymocyte selection. A subset of the immune-related genes identified in the microarray, as well as Tnfa, were validated using qPCR of RNA isolated from livers of ND- and HFD-fed male mice (Figure 8B). Notably, all of the analyzed proinflammatory genes were up-regulated in KO livers under both ND and HFD conditions (Figure 8B).
Figure 8.
Hepatocyte-specific Lmna-deficiency promotes liver inflammation. (A) Three Lmna Het and 3 Lmna KO livers from male mice fed a normal diet were compared using Affymetrix microarrays (same as those described in Figure 7). Het and KO livers were isolated from 26-week-old and 39-week-old mice, respectively. Transcripts that were significantly different and carried the gene ontology term immune were grouped together in a heat map (red, high expression; blue, low expression as compared with the mean values for the WT livers). Black arrows denote interferon-regulated genes that are up-regulated in KO livers. The red arrow indicates up-regulated Stat1 expression in KO livers. (B) qPCR analysis of messenger RNA from livers of WT and KO male mice (23–28 weeks old, 5 livers/genotype) fed ND or HFD. Expression of the indicated inflammation-associated genes is shown (*P < .05, **P < .01, ***P < .001, and ****P < .0001). Error bars in all graphs represent SEM. Statistical significance was determined using 1-way analysis of variance followed by the Tukey post hoc test. (C) Immunohistochemical staining of CD45+ cells in WT and KO liver sections from mice fed ND (WT liver: 27 weeks of age; KO liver: 24 weeks of age) or HFD (23 weeks of age for both WT and KO) (representative images are shown; n ≥ 3 mouse livers/genotype/condition). Scale bars: 50 μm.
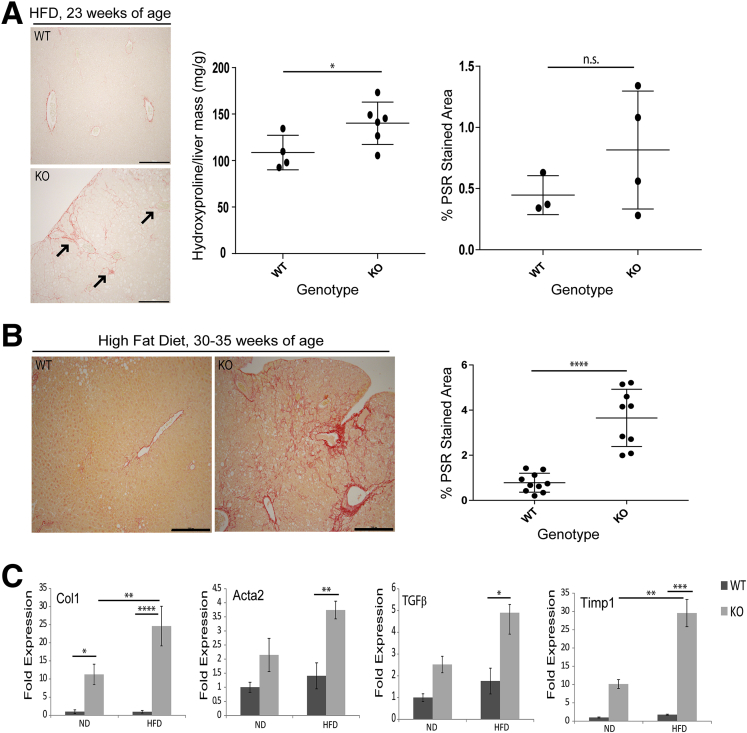
Consistent with the observed increase in transcription of immune-related genes in KO livers, there was increased inflammatory cell infiltration in KO livers under both ND and HFD conditions, as assessed by CD45 staining (Figure 8C), consistent with steatohepatitis. Indeed, we also noted an increase in fibrosis in livers of HFD-fed KO mice, as evidenced by Picrosirius red staining and hydroxyproline measurement (Figure 9A and B), which became more severe with age (Figure 9B) and correlated with up-regulation of fibrosis-related genes in KO livers (Figure 9C). Although up-regulation of inflammation-related genes in KO livers was similar under ND and HFD conditions, up-regulation of fibrosis-related genes in KO livers was much more pronounced after HFD feeding (Figure 9C).
Figure 9.
Hepatic Lmna deficiency promotes liver fibrosis. (A) Picrosirius red staining of paraffin sections of WT and KO livers from HFD-fed mice at 23 weeks of age. Arrows indicate fibrosis. Scale bars: 200 μm. Hydroxyproline measurement of liver extracts is shown (4–6 livers/genotype) (*P = .05). The statistical comparison was performed using the unpaired t test (2-tailed), and error bars represent SD. Quantitation of Picrosirius red–stained area in WT (n = 3) and KO (n = 4) livers from 23-week-old HFD-fed mice was performed as described in the Materials and Methods section. Error bars represent SD. NS indicates no significant difference between WT vs KO. Statistical significance was determined using the unpaired t test (2-tailed). (B) Picrosirius red (PSR) staining of paraffin sections of WT (n = 10) and KO (n = 9) livers from HFD-fed mice between 30 and 35 weeks of age. Scale bars: 200 μm. Quantitation was performed as described in the Materials and Methods section. Statistical significance (****P < .0001) was determined using the unpaired t test (2-tailed), and error bars represent SD. (C) qPCR analysis of messenger RNA comparing WT and KO male livers (23–28 weeks old, 5 livers/genotype) fed ND or HFD for the expression of the indicated fibrosis-associated genes (*P < .05, **P < .01, ***P < .001, and ****P < .0001). Error bars in all qPCR graphs represent SEM. Statistical significance for qPCR analysis was determined using 1-way analysis of variance followed by the Tukey post hoc test.
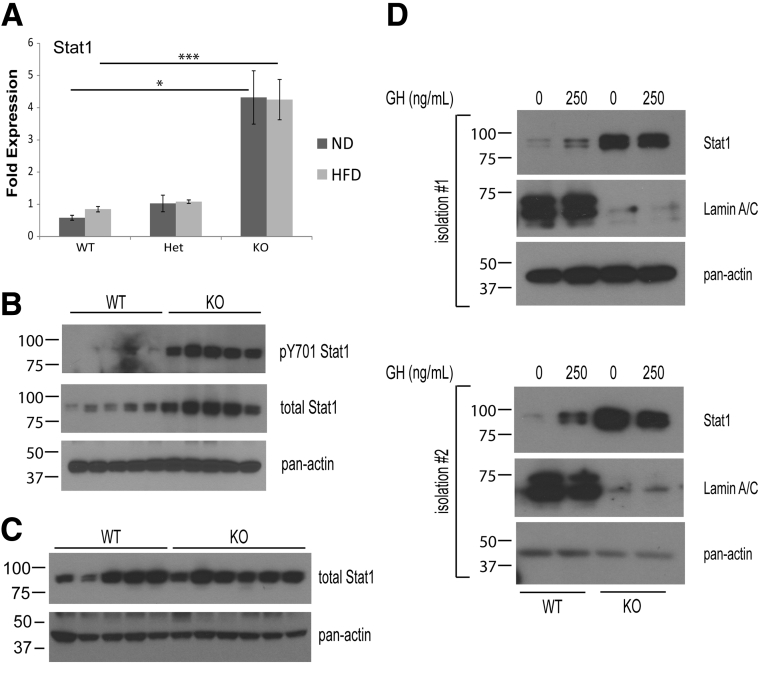
Lmna Deficiency Causes Up-Regulated Stat1 Expression and Phosphorylation in Hepatocytes
Expression profiling analysis indicated that a large proportion of immune genes that are up-regulated in KO livers comprise interferon-regulated genes, suggesting that Lmna deficiency dysregulates interferon α/β receptor signaling (Figure 8A, interferon-regulated genes are denoted with arrows). Stat1, a key transcription factor downstream of the interferon α/β receptor and important in the modulation of interferon-regulated gene expression,47 is up-regulated in KO livers at the messenger RNA and protein levels (Figures 8A and 10A and B) under both ND and HFD feeding. There is also a dramatic increase in Stat1 Y701 phosphorylation in male KO vs WT livers (Figure 10B). In contrast, WT and KO female livers showed equivalent Stat1 protein expression, indicating that the phenotype is gender-specific (Figure 10C). The increase in Stat1 protein levels also was observed in primary hepatocytes from male mice, indicating that Lmna deficiency acts cell-autonomously to increase Stat1 protein in hepatocytes (Figure 10D).
Figure 10.
Hepatic Lmna deficiency causes up-regulation of Stat1 expression and phosphorylation in male liver. (A) Stat1 qPCR analysis of messenger RNA from WT/Het/KO male livers of mice (age, 20–25 wk) fed ND or HFD (*P < .05 and ***P < .001); n = 3–4 livers per genotype group for the ND or the HFD feeding. Error bars represent SEM. Statistical significance was determined using 1-way analysis of variance followed by the Tukey post hoc test. (B) Cytoplasmic liver extracts were prepared from WT and KO male mice (age, 23–28 wk) and then analyzed by phosphorylated Stat1 and Stat1 immunoblotting. (C) Cytoplasmic liver extracts were prepared from WT and KO female mice (age, 23–28 wk) and then analyzed by immunoblotting as indicated. (D) Primary hepatocytes (2 independent isolations are shown) were obtained from WT and KO male mice between 8 and 12 weeks of age, serum-starved for 8–12 hours, treated with 0 or 250 ng/mL of human GH (37°C, 10 min), and then lysed to prepare cytoplasmic extracts for immunoblotting. Note that KO male hepatocytes expressed higher levels of Stat1 than their WT counterparts.
Lmna Deficiency Causes Defects in GH Signaling and Aberrant Regulation of Sexually Dimorphic Genes in Hepatocytes
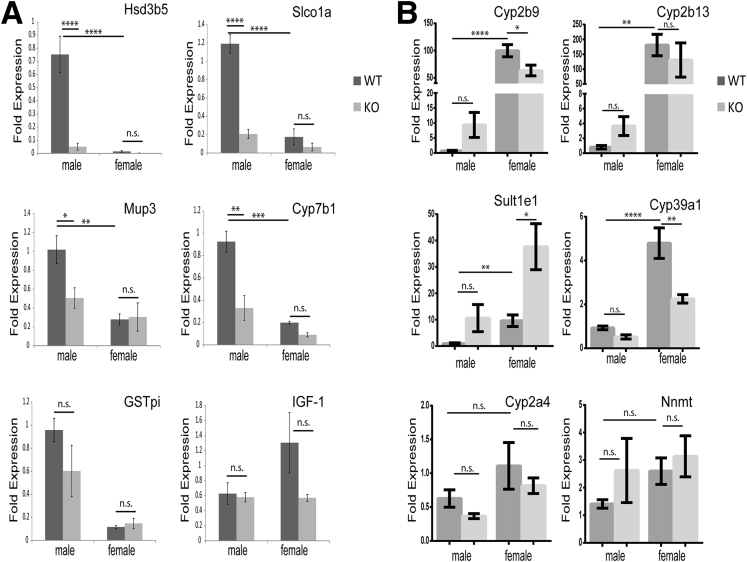
The male specificity of the hepatic Lmna KO phenotype suggested that the male-specific pattern of hepatic gene expression might be disrupted in KO livers. GH signaling is known to regulate sexually dimorphic gene expression in hepatocytes, as evidenced by the induction of male-specific genes by pulsatile GH secretion (in contrast to continuous secretion in females) and resultant hepatocyte activation of Stat5b, and to a lesser extent Stat5a.26, 28, 29, 30 Liver-specific Stat5ab deficiency abolishes growth hormone–mediated induction of male-specific genes, with relative preservation of female-specific gene expression, and de-represses female-specific gene expression in male livers.48, 49, 50 Interestingly, Hsd3b5, a male-specific hepatic gene that is Stat5ab-dependent, was the most highly down-regulated gene in male KO livers (Figure 7A), suggesting that GH/Stat5ab signaling might be altered in Lmna KO livers. We tested this hypothesis by examining the relative expression of Stat5ab-dependent, male-specific genes in WT and KO livers. Similar to Stat5ab-deficient livers, Lmna deficiency greatly reduced male-specific gene expression in male livers, but had no effect on the expression of these genes in females (Figure 11A). Not all Stat5ab-dependent genes were affected to the same degree by Lmna deficiency because liver insulin-like growth factor 1 (Igf-1) transcripts were equivalent in WT and KO livers (Figure 11A), and female-specific genes that are de-repressed by Stat5ab KO in male livers were affected only modestly in male Lmna KO livers (Figure 11B).
Figure 11.
Male-specific gene expression is compromised in Lmna-deficient livers. (A) qPCR analysis of liver messenger RNA from male and female mice (age, 14–17 wk) show that WT male livers express significantly higher levels of male-specific genes than female livers and that Lmna deficiency significantly reduces expression of these genes in male livers. (B) qPCR analysis comparing Lmna WT and KO livers from male and female mice (age, 14–17 wk) for the expression of female-specific hepatic genes (note the marginal effect of Lmna deficiency on the expression of female-specific genes in male KO livers). (A and B) n = 4/genotype for male WT vs KO mice, and n = 3/genotype for female WT vs KO mice. Error bars in all qPCR graphs represent SEM. *P < .05, **P < .01, ***P < .001, and ****P < .0001. Statistical significance for qPCR analysis was determined using 1-way analysis of variance followed by the Tukey post hoc test.
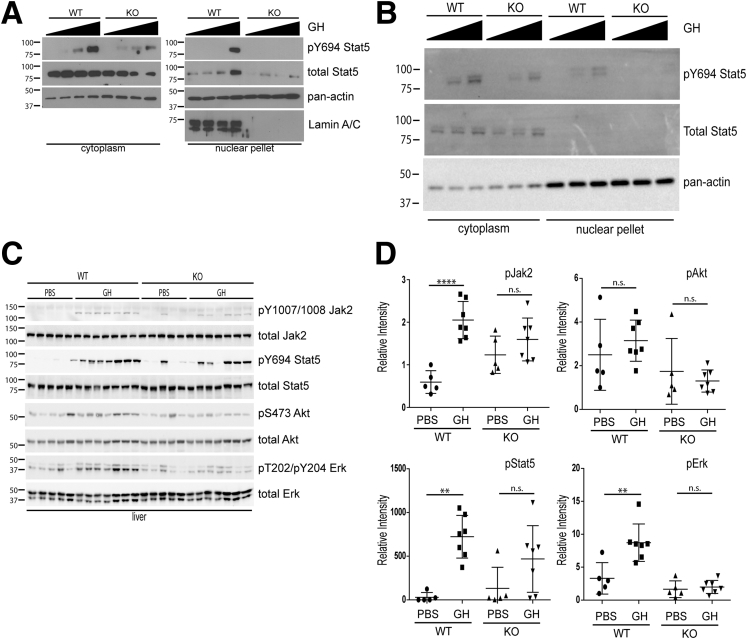
To assess whether Lmna deficiency impacted Stat5 activation via growth hormone signaling, primary hepatocytes from Lmna WT and KO livers were treated with varying concentrations of human GH. KO hepatocytes from male (Figure 12A) and female livers (Figure 12B) showed a decrease in Stat5 Y694 phosphorylation, which corresponded with decreased translocation of Stat5 to the nucleus in KO hepatocytes.
Figure 12.
GH signaling is compromised in Lmna-deficient hepatocytes and livers. (A) Primary hepatocytes were isolated from 8- to 12-week-old WT and KO male mice, serum-starved for 8–12 hours, treated with 0, 25, 50, or 250 ng/mL of human GH (37°C, 10 min), and then lysed to prepare cytoplasmic and nuclear extracts for immunoblotting. Primary hepatocyte isolation experiments were repeated 5 times, and results shown are representative. (B) Primary hepatocytes were isolated from 8- to 12-week-old WT and KO female mice, serum-starved for 8–12 hours, treated with 0, 50, or 250 ng/mL of human GH (37°C, 10 min), and then lysed to prepare cytoplasmic and nuclear extracts for immunoblotting. Primary hepatocyte isolation experiments were repeated 2 times. (C) Cytoplasmic liver extracts were prepared from WT and KO male mice (20–24 weeks old) that were injected intraperitoneally with either PBS or GH (2.5 mg/kg). Livers were harvested 10 minutes after injection. Extracts then were analyzed by immunoblotting as indicated in the Materials and Methods section. (D) Relative band intensities for phosphorylated and total Jak2, Akt, Stat5, and Erk for immunoblots shown in panel C were determined using ImageJ software, and relative band intensities for the phosphorylated proteins were normalized to total Jak2, Akt, Stat5, or Erk. Note that GH induced Jak2, Stat5, and Erk phosphorylation in WT but not KO livers. **P < .01 for WT PBS vs GH for phosphorylated Stat5 and phosphorylated Erk analysis, and ****P < .0001 for WT PBS vs GH for phosphorylated Jak2 analysis. Error bars in all graphs represent SD. Statistical significance was determined using 1-way analysis of variance followed by the Tukey post hoc test.
In addition to primary hepatocytes, we observed decreased induction of Jak2 Y1007/Y1008, Stat5 Y694, and Erk T202/Y204 phosphorylation in male Lmna KO livers after GH administration (Figure 12C and D). In contrast, there was no significant induction of Akt S473 in response to GH in either WT or KO livers, suggesting that Jak2, Stat5, and Erk are the primary pathways downstream of GH that are altered in the hepatocyte-specific lamin A/C KO model (Figure 12C and D).
Discussion
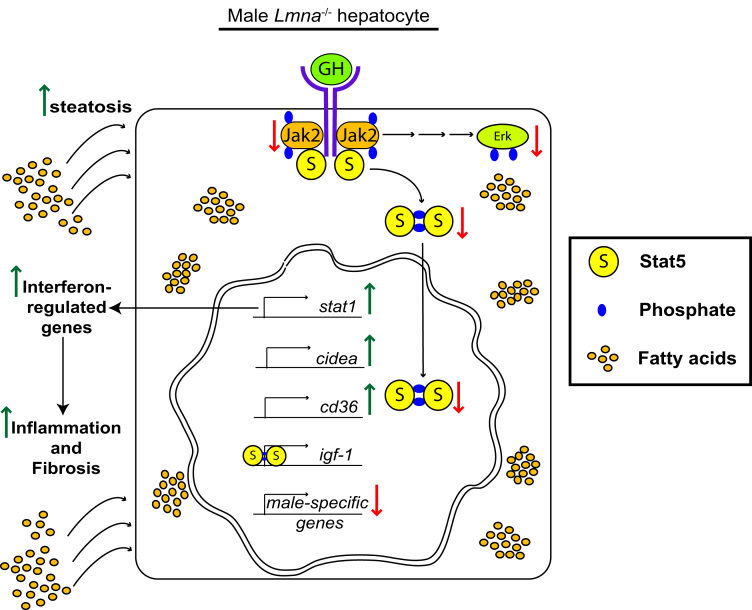
Our data indicate that hepatocyte Lmna deficiency leads to the following: (1) alteration in nuclear shape; (2) male-selective spontaneous steatosis under normal feeding conditions with increased age-associated susceptibility to HFD-induced steatohepatitis and fibrosis; (3) a blunted response to GH in hepatocytes characterized by reduced induction of Jak2, Stat5, and Erk phosphorylation; (4) decreased expression of a subset of Stat5-dependent, GH-induced genes; and (5) increased hepatic Stat1 expression and phosphorylation in a male-specific manner (Figure 13). There are several likely contributors to our observed phenotype, including the marked up-regulation of Cidea and other lipogenic genes in response to lamin A/C absence, given their known function in fatty liver disease development. In addition, the up-regulation of CD36 that we observe meshes well with the known role of CD36 in promoting hepatic steatosis and insulin resistance.45, 51 Notably, CD36 also is up-regulated in hepatocyte-specific GH-receptor–, Jak2-, and Stat5-deficient mice, suggesting that dysregulated hepatic growth hormone signaling might be responsible for CD36 up-regulation in Lmna-deficient hepatocytes because Stat5 previously was shown to repress CD36 transcription directly.52, 53, 54, 55, 56 Similar to hepatocyte-specific Stat5ab-deficient mice, our KO mice showed body mass loss.48, 50 In addition, Lmna-deficient livers showed similar decreases in the expression of male-specific hepatic genes as Stat5ab-deficient livers.48, 50 Both male and female KO hepatocytes showed decreased Stat5 phosphorylation and nuclear translocation in response to GH treatment. The greater severity of the Lmna KO phenotype in male vs female livers may be owing to the greater effect hepatocyte-specific Stat5 deficiency has on male- vs female-specific hepatic gene expression.49, 50
Figure 13.
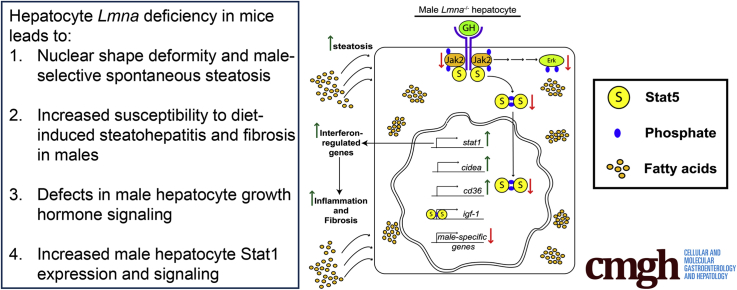
Absence of lamin A/C in hepatocytes interferes with GH signaling in male hepatocytes. The schematic summarizes our findings. Lmna deficiency is responsible for the following: (1) alters nuclear shape; (2) inhibits growth hormone receptor–mediated Jak2, Stat5, and Erk phosphorylation; (3) decreases the expression of Stat5-dependent male-specific genes; (4) increases expression of the Stat5-regulated fatty acid translocator CD36, (5) up-regulates Cidea transcription, and (6) increases Stat1 messenger RNA and protein expression and activation, resulting in the induction of interferon-regulated genes. These alterations lead to hepatocyte storage of excess fatty acids with consequent induction of hepatic inflammation and fibrosis upon feeding with a HFD.
It is important to note that not all aspects hepatic GHR-, Jak2-, and Stat5ab-deficiency are recapitulated in our model. For example, we did not observe a difference in hepatic Igf-1 transcript levels between WT and KO mice, as had been reported in the other models.52, 53, 54, 55, 56 The decreased serum IGF-1 and the resultant increased serum GH levels in hepatic Jak2- and Stat5-deficient mice are thought to increase serum triglyceride levels by GH-induced lipolysis in adipocytes. In these animals, hepatic steatosis was observed by 15 weeks of age. Hepatic Lmna-deficient mice, on the other hand, showed no increase in serum triglyceride levels or hepatic steatosis at 15 weeks in ND-fed animals, and serum triglyceride levels in KO males were equivalent to WT after HFD feeding, despite increased hepatic steatosis. These differences in phenotype likely reflect normal hepatic Igf-1 expression in hepatic Lmna-deficient mice (Figure 11A), despite the dysregulation of Stat5 signaling. Similarly, hepatic Lmna deficiency had a reduced effect on the expression of female-specific genes in male livers compared with hepatic Stat5ab deficiency, in which female-specific genes are de-repressed.49, 50 Taken together, these data indicate that there are qualitative and/or quantitative differences in Stat5 signaling in Lmna-deficient livers that affect male-specific hepatic gene expression, but not all Stat5-dependent genes.
Loss of hepatocyte Stat5 expression results in increased levels of total and Y701 phosphorylated Stat1.53 Increased Stat1 expression and phosphorylation positively regulate interferon α/β receptor signaling.57, 58 These findings support our hypothesis that the increased Stat1 levels, interferon-regulated gene expression, and inflammation in male KO livers likely are the result of dysregulation of Stat5 signaling as a result of Lmna deficiency. Our data are consistent with a model in which lamin A/C represses the activation of steatosis-, inflammation-, and fibrosis-related genes in the liver by positively regulating GH-induced Stat5 signaling (Figure 13).
GH-induced Erk signaling also is compromised in male KO hepatocytes (Figure 12C and D), but the potential role that Erk plays in the Lmna KO liver phenotype is unknown. GH induces Erk phosphorylation through Jak2-mediated phosphorylation of the adaptor protein Shc59 and also through a Jak2-independent/Src-dependent pathway.60, 61 Of note, transgenic mice that express a human LMNA allele that causes dilated cardiomyopathy show increased Erk activation in heart tissue, and pharmacologic inhibition of Erk phosphorylation ameliorates the cardiomyopathy phenotype.62, 63, 64 This is in contrast to our system, in which we see blunted Erk activation in Lmna-deficient livers. It is important to note that expression of a disease-causing lamin A/C variant, with potential gain-of-function effects (with or without loss-of-function effects), could have different cellular consequences compared with absence of lamin A/C.
The male-specific steatohepatitis phenotype in the Lmna KO mice is reminiscent of the gender differences observed in some instances of human NAFLD and in some of the laminopathies. For example, studies from the United States and China have indicated a higher prevalence of NAFLD in men than in premenopausal women, and there also have been variations among different ethnicities.65, 66 However, in the case of FPLD2, female patients are affected more prominently than males in terms of the diabetes phenotype,16 but the liver phenotype was not assessed in the studied cohort. Our findings are highly relevant to human fatty liver disease, in which relatively few genetic variants that contribute to disease development and progression have been defined.67 In this context, we posit that lamin A/C variants may contribute to fatty liver disease progression. There is already clear evidence to support this hypothesis because patients with FPLD2 caused by lamin A/C mutation also develop hepatosteatosis.68 However, given the multi-organ involvement of this familial lipodystrophy, it has not been clear whether the liver disease is related to direct lamin-related effects on the hepatocyte or a secondary effect of the metabolic syndrome. The findings described herein help answer this question by showing that hepatocyte-specific ablation of lamin A/C in mice leads to fatty liver disease and also provide support for lamins playing a role in normal hepatocyte homeostasis.
Acknowledgments
The authors thank Bradley Nelson for assistance with electron microscopy, Craig Johnson for assistance with microarray analysis, and Stephen Lentz for assistance with Metamorph analysis.
Footnotes
Author contributions Raymond Kwan, Graham Brady, Lei Yin, Xin Tong, Ram Menon, Colin Stewart, and Bishr Omary were responsible for the study concepts and design; Raymond Kwan, Graham Brady, Maria Brzozowski, Sujith V. Weerasinghe, Hope Martin, Min-Jung Park, and Makayla Brunt were responsible for the experiments and procedures; and Raymond Kwan, Graham Brady, and Bishr Omary were responsible for writing and revision of the article.
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by National Institutes of Health grants R01 DK47918 (M.B.O.); R01 DK099593 (L.Y.); National Institutes of Health training grant T32 DK094775 (G.F.B.); and an institutional National Institutes of Health grant (P30 DK34933) to the University of Michigan.
References
- 1.Butin-Israeli V., Adam S.A., Goldman A.E., Goldman R.D. Nuclear lamin functions and disease. Trends Genet. 2012;28:464–471. doi: 10.1016/j.tig.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gruenbaum Y., Foisner R. Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu Rev Biochem. 2015;84:131–164. doi: 10.1146/annurev-biochem-060614-034115. [DOI] [PubMed] [Google Scholar]
- 3.Biamonti G., Giacca M., Perini G., Contreas G., Zentilin L., Weighardt F., Guerra M., Della Valle G., Saccone S., Riva S., Falaschi A. The gene for a novel human lamin maps at a highly transcribed locus of chromosome 19 which replicates at the onset of S-phase. Mol Cell Biol. 1992;12:3499–3506. doi: 10.1128/mcb.12.8.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin F., Worman H.J. Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J Biol Chem. 1993;268:16321–16326. [PubMed] [Google Scholar]
- 5.Lin F., Worman H.J. Structural organization of the human gene (LMNB1) encoding nuclear lamin B1. Genomics. 1995;27:230–236. doi: 10.1006/geno.1995.1036. [DOI] [PubMed] [Google Scholar]
- 6.Rober R.A., Sauter H., Weber K., Osborn M. Cells of the cellular immune and hemopoietic system of the mouse lack lamins A/C: distinction versus other somatic cells. J Cell Sci. 1990;95:587–598. doi: 10.1242/jcs.95.4.587. [DOI] [PubMed] [Google Scholar]
- 7.Rober R.A., Weber K., Osborn M. Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animal: a developmental study. Development. 1989;105:365–378. doi: 10.1242/dev.105.2.365. [DOI] [PubMed] [Google Scholar]
- 8.Stewart C., Burke B. Teratocarcinoma stem cells and early mouse embryos contain only a single major lamin polypeptide closely resembling lamin B. Cell. 1987;51:383–392. doi: 10.1016/0092-8674(87)90634-9. [DOI] [PubMed] [Google Scholar]
- 9.Gruenbaum Y., Medalia O. Lamins: the structure and protein complexes. Curr Opin Cell Biol. 2015;32:7–12. doi: 10.1016/j.ceb.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Harr J.C., Luperchio T.R., Wong X., Cohen E., Wheelan S.J., Reddy K.L. Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J Cell Biol. 2015;208:33–52. doi: 10.1083/jcb.201405110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solovei I., Wang A.S., Thanisch K., Schmidt C.S., Krebs S., Zwerger M., Cohen T.V., Devys D., Foisner R., Peichl L., Herrmann H., Blum H., Engelkamp D., Stewart C.L., Leonhardt H., Joffe B. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013;152:584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 12.Burke B., Stewart C.L. Functional architecture of the cell's nucleus in development, aging, and disease. Curr Top Dev Biol. 2014;109:1–52. doi: 10.1016/B978-0-12-397920-9.00006-8. [DOI] [PubMed] [Google Scholar]
- 13.Worman H.J., Bonne G. “Laminopathies”: a wide spectrum of human diseases. Exp Cell Res. 2007;313:2121–2133. doi: 10.1016/j.yexcr.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garg A. Acquired and inherited lipodystrophies. N Engl J Med. 2004;350:1220–1234. doi: 10.1056/NEJMra025261. [DOI] [PubMed] [Google Scholar]
- 15.Shackleton S., Lloyd D.J., Jackson S.N., Evans R., Niermeijer M.F., Singh B.M., Schmidt H., Brabant G., Kumar S., Durrington P.N., Gregory S., O'Rahilly S., Trembath R.C. LMNA, encoding lamin A/C, is mutated in partial lipodystrophy. Nat Genet. 2000;24:153–156. doi: 10.1038/72807. [DOI] [PubMed] [Google Scholar]
- 16.Vigouroux C., Magre J., Vantyghem M.C., Bourut C., Lascols O., Shackleton S., Lloyd D.J., Guerci B., Padova G., Valensi P., Grimaldi A., Piquemal R., Touraine P., Trembath R.C., Capeau J. Lamin A/C gene: sex-determined expression of mutations in Dunnigan-type familial partial lipodystrophy and absence of coding mutations in congenital and acquired generalized lipoatrophy. Diabetes. 2000;49:1958–1962. doi: 10.2337/diabetes.49.11.1958. [DOI] [PubMed] [Google Scholar]
- 17.Speckman R.A., Garg A., Du F., Bennett L., Veile R., Arioglu E., Taylor S.I., Lovett M., Bowcock A.M. Mutational and haplotype analyses of families with familial partial lipodystrophy (Dunnigan variety) reveal recurrent missense mutations in the globular C-terminal domain of lamin A/C. Am J Hum Genet. 2000;66:1192–1198. doi: 10.1086/302836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ajluni N., Meral R., Neidert A.H., Brady G.F., Buras E., McKenna B., DiPaola F., Chenevert T.L., Horowitz J.F., Buggs-Saxton C., Rupani A.R., Thomas P.E., Tayeh M.K., Innis J.W., Omary M.B., Conjeevaram H., Oral E.A. Spectrum of disease associated with partial lipodystrophy (PL): lessons from a trial cohort. Clin Endocrinol (Oxf) 2017;86:698–707. doi: 10.1111/cen.13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ludtke A., Genschel J., Brabant G., Bauditz J., Taupitz M., Koch M., Wermke W., Worman H.J., Schmidt H.H. Hepatic steatosis in Dunnigan-type familial partial lipodystrophy. Am J Gastroenterol. 2005;100:2218–2224. doi: 10.1111/j.1572-0241.2005.00234.x. [DOI] [PubMed] [Google Scholar]
- 20.Wojtanik K.M., Edgemon K., Viswanadha S., Lindsey B., Haluzik M., Chen W., Poy G., Reitman M., Londos C. The role of LMNA in adipose: a novel mouse model of lipodystrophy based on the Dunnigan-type familial partial lipodystrophy mutation. J Lipid Res. 2009;50:1068–1079. doi: 10.1194/jlr.M800491-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cutler D.A., Sullivan T., Marcus-Samuels B., Stewart C.L., Reitman M.L. Characterization of adiposity and metabolism in Lmna-deficient mice. Biochem Biophys Res Commun. 2002;291:522–527. doi: 10.1006/bbrc.2002.6466. [DOI] [PubMed] [Google Scholar]
- 22.Sullivan T., Escalante-Alcalde D., Bhatt H., Anver M., Bhat N., Nagashima K., Stewart C.L., Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang A.S., Kozlov S.V., Stewart C.L., Horn H.F. Tissue specific loss of A-type lamins in the gastrointestinal epithelium can enhance polyp size. Differentiation. 2015;89:11–21. doi: 10.1016/j.diff.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Jaffe C.A., Ocampo-Lim B., Guo W., Krueger K., Sugahara I., DeMott-Friberg R., Bermann M., Barkan A.L. Regulatory mechanisms of growth hormone secretion are sexually dimorphic. J Clin Invest. 1998;102:153–164. doi: 10.1172/JCI2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaffe C.A., Turgeon D.K., Lown K., Demott-Friberg R., Watkins P.B. Growth hormone secretion pattern is an independent regulator of growth hormone actions in humans. Am J Physiol Endocrinol Metab. 2002;283:E1008–E1015. doi: 10.1152/ajpendo.00513.2001. [DOI] [PubMed] [Google Scholar]
- 26.MacLeod J.N., Pampori N.A., Shapiro B.H. Sex differences in the ultradian pattern of plasma growth hormone concentrations in mice. J Endocrinol. 1991;131:395–399. doi: 10.1677/joe.0.1310395. [DOI] [PubMed] [Google Scholar]
- 27.Moller N., Jorgensen J.O. Effects of growth hormone on glucose, lipid, and protein metabolism in human subjects. Endocr Rev. 2009;30:152–177. doi: 10.1210/er.2008-0027. [DOI] [PubMed] [Google Scholar]
- 28.Udy G.B., Towers R.P., Snell R.G., Wilkins R.J., Park S.H., Ram P.A., Waxman D.J., Davey H.W. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci U S A. 1997;94:7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waxman D.J., Pampori N.A., Ram P.A., Agrawal A.K., Shapiro B.H. Interpulse interval in circulating growth hormone patterns regulates sexually dimorphic expression of hepatic cytochrome P450. Proc Natl Acad Sci U S A. 1991;88:6868–6872. doi: 10.1073/pnas.88.15.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waxman D.J., Ram P.A., Pampori N.A., Shapiro B.H. Growth hormone regulation of male-specific rat liver P450s 2A2 and 3A2: induction by intermittent growth hormone pulses in male but not female rats rendered growth hormone deficient by neonatal monosodium glutamate. Mol Pharmacol. 1995;48:790–797. [PubMed] [Google Scholar]
- 31.Younossi Z., Henry L. Contribution of alcoholic and nonalcoholic fatty liver disease to the burden of liver-related morbidity and mortality. Gastroenterology. 2016;150:1778–1785. doi: 10.1053/j.gastro.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 32.Postic C., Shiota M., Niswender K.D., Jetton T.L., Chen Y., Moates J.M., Shelton K.D., Lindner J., Cherrington A.D., Magnuson M.A. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 33.Kohli R., Kirby M., Xanthakos S.A., Softic S., Feldstein A.E., Saxena V., Tang P.H., Miles L., Miles M.V., Balistreri W.F., Woods S.C., Seeley R.J. High-fructose, medium chain trans fat diet induces liver fibrosis and elevates plasma coenzyme Q9 in a novel murine model of obesity and nonalcoholic steatohepatitis. Hepatology. 2010;52:934–944. doi: 10.1002/hep.23797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singla A., Griggs N.W., Kwan R., Snider N.T., Maitra D., Ernst S.A., Herrmann H., Omary M.B. Lamin aggregation is an early sensor of porphyria-induced liver injury. J Cell Sci. 2013;126:3105–3112. doi: 10.1242/jcs.123026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strnad P., Tao G.Z., Zhou Q., Harada M., Toivola D.M., Brunt E.M., Omary M.B. Keratin mutation predisposes to mouse liver fibrosis and unmasks differential effects of the carbon tetrachloride and thioacetamide models. Gastroenterology. 2008;134:1169–1179. doi: 10.1053/j.gastro.2008.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Weerasinghe S.V., Ku N.O., Altshuler P.J., Kwan R., Omary M.B. Mutation of caspase-digestion sites in keratin 18 interferes with filament reorganization, and predisposes to hepatocyte necrosis and loss of membrane integrity. J Cell Sci. 2014;127:1464–1475. doi: 10.1242/jcs.138479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 39.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 40.Kleiner D.E., Brunt E.M., Van Natta M., Behling C., Contos M.J., Cummings O.W., Ferrell L.D., Liu Y.C., Torbenson M.S., Unalp-Arida A., Yeh M., McCullough A.J., Sanyal A.J. Nonalcoholic Steatohepatitis Clinical Research Network. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 41.Puri V., Ranjit S., Konda S., Nicoloro S.M., Straubhaar J., Chawla A., Chouinard M., Lin C., Burkart A., Corvera S., Perugini R.A., Czech M.P. Cidea is associated with lipid droplets and insulin sensitivity in humans. Proc Natl Acad Sci U S A. 2008;105:7833–7838. doi: 10.1073/pnas.0802063105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou L., Xu L., Ye J., Li D., Wang W., Li X., Wu L., Wang H., Guan F., Li P. Cidea promotes hepatic steatosis by sensing dietary fatty acids. Hepatology. 2012;56:95–107. doi: 10.1002/hep.25611. [DOI] [PubMed] [Google Scholar]
- 43.Agarwal A.K., Tunison K., Dalal J.S., Yen C.E., Farese R.V., Jr., Horton J.D., Garg A. Mogat1 deletion does not ameliorate hepatic steatosis in lipodystrophic (Agpat2-/-) or obese (ob/ob) mice. J Lipid Res. 2016;57:616–630. doi: 10.1194/jlr.M065896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hall A.M., Soufi N., Chambers K.T., Chen Z., Schweitzer G.G., McCommis K.S., Erion D.M., Graham M.J., Su X., Finck B.N. Abrogating monoacylglycerol acyltransferase activity in liver improves glucose tolerance and hepatic insulin signaling in obese mice. Diabetes. 2014;63:2284–2296. doi: 10.2337/db13-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koonen D.P., Jacobs R.L., Febbraio M., Young M.E., Soltys C.L., Ong H., Vance D.E., Dyck J.R. Increased hepatic CD36 expression contributes to dyslipidemia associated with diet-induced obesity. Diabetes. 2007;56:2863–2871. doi: 10.2337/db07-0907. [DOI] [PubMed] [Google Scholar]
- 46.Miquilena-Colina M.E., Lima-Cabello E., Sanchez-Campos S., Garcia-Mediavilla M.V., Fernandez-Bermejo M., Lozano-Rodriguez T., Vargas-Castrillon J., Buque X., Ochoa B., Aspichueta P., Gonzalez-Gallego J., Garcia-Monzon C. Hepatic fatty acid translocase CD36 upregulation is associated with insulin resistance, hyperinsulinaemia and increased steatosis in non-alcoholic steatohepatitis and chronic hepatitis C. Gut. 2011;60:1394–1402. doi: 10.1136/gut.2010.222844. [DOI] [PubMed] [Google Scholar]
- 47.McNab F., Mayer-Barber K., Sher A., Wack A., O'Garra A. Type I interferons in infectious disease. Nat Rev Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baik M., Yu J.H., Hennighausen L. Growth hormone-STAT5 regulation of growth, hepatocellular carcinoma, and liver metabolism. Ann N Y Acad Sci. 2011;1229:29–37. doi: 10.1111/j.1749-6632.2011.06100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clodfelter K.H., Holloway M.G., Hodor P., Park S.H., Ray W.J., Waxman D.J. Sex-dependent liver gene expression is extensive and largely dependent upon signal transducer and activator of transcription 5b (STAT5b): STAT5b-dependent activation of male genes and repression of female genes revealed by microarray analysis. Mol Endocrinol. 2006;20:1333–1351. doi: 10.1210/me.2005-0489. [DOI] [PubMed] [Google Scholar]
- 50.Holloway M.G., Cui Y., Laz E.V., Hosui A., Hennighausen L., Waxman D.J. Loss of sexually dimorphic liver gene expression upon hepatocyte-specific deletion of Stat5a-Stat5b locus. Endocrinology. 2007;148:1977–1986. doi: 10.1210/en.2006-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson C.G., Tran J.L., Erion D.M., Vera N.B., Febbraio M., Weiss E.J. Hepatocyte-specific disruption of CD36 attenuates fatty liver and improves insulin sensitivity in HFD-fed mice. Endocrinology. 2016;157:570–585. doi: 10.1210/en.2015-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barclay J.L., Nelson C.N., Ishikawa M., Murray L.A., Kerr L.M., McPhee T.R., Powell E.E., Waters M.J. GH-dependent STAT5 signaling plays an important role in hepatic lipid metabolism. Endocrinology. 2011;152:181–192. doi: 10.1210/en.2010-0537. [DOI] [PubMed] [Google Scholar]
- 53.Cui Y., Hosui A., Sun R., Shen K., Gavrilova O., Chen W., Cam M.C., Gao B., Robinson G.W., Hennighausen L. Loss of signal transducer and activator of transcription 5 leads to hepatosteatosis and impaired liver regeneration. Hepatology. 2007;46:504–513. doi: 10.1002/hep.21713. [DOI] [PubMed] [Google Scholar]
- 54.Fan Y., Menon R.K., Cohen P., Hwang D., Clemens T., DiGirolamo D.J., Kopchick J.J., Le Roith D., Trucco M., Sperling M.A. Liver-specific deletion of the growth hormone receptor reveals essential role of growth hormone signaling in hepatic lipid metabolism. J Biol Chem. 2009;284:19937–19944. doi: 10.1074/jbc.M109.014308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi S.Y., Martin R.G., Duncan R.E., Choi D., Lu S.Y., Schroer S.A., Cai E.P., Luk C.T., Hopperton K.E., Domenichiello A.F., Tang C., Naples M., Dekker M.J., Giacca A., Adeli K., Wagner K.U., Bazinet R.P., Woo M. Hepatocyte-specific deletion of Janus kinase 2 (JAK2) protects against diet-induced steatohepatitis and glucose intolerance. J Biol Chem. 2012;287:10277–10288. doi: 10.1074/jbc.M111.317453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sos B.C., Harris C., Nordstrom S.M., Tran J.L., Balazs M., Caplazi P., Febbraio M., Applegate M.A., Wagner K.U., Weiss E.J. Abrogation of growth hormone secretion rescues fatty liver in mice with hepatocyte-specific deletion of JAK2. J Clin Invest. 2011;121:1412–1423. doi: 10.1172/JCI42894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ivashkiv L.B., Donlin L.T. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tassiulas I., Hu X., Ho H., Kashyap Y., Paik P., Hu Y., Lowell C.A., Ivashkiv L.B. Amplification of IFN-alpha-induced STAT1 activation and inflammatory function by Syk and ITAM-containing adaptors. Nat Immunol. 2004;5:1181–1189. doi: 10.1038/ni1126. [DOI] [PubMed] [Google Scholar]
- 59.Xu J., Keeton A.B., Franklin J.L., Li X., Venable D.Y., Frank S.J., Messina J.L. Insulin enhances growth hormone induction of the MEK/ERK signaling pathway. J Biol Chem. 2006;281:982–992. doi: 10.1074/jbc.M505484200. [DOI] [PubMed] [Google Scholar]
- 60.Barclay J.L., Kerr L.M., Arthur L., Rowland J.E., Nelson C.N., Ishikawa M., d'Aniello E.M., White M., Noakes P.G., Waters M.J. In vivo targeting of the growth hormone receptor (GHR) box1 sequence demonstrates that the GHR does not signal exclusively through JAK2. Mol Endocrinol. 2010;24:204–217. doi: 10.1210/me.2009-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu T., Ling L., Lobie P.E. Identification of a JAK2-independent pathway regulating growth hormone (GH)-stimulated p44/42 mitogen-activated protein kinase activity. GH activation of Ral and phospholipase D is Src-dependent. J Biol Chem. 2002;277:45592–45603. doi: 10.1074/jbc.M201385200. [DOI] [PubMed] [Google Scholar]
- 62.Muchir A., Pavlidis P., Decostre V., Herron A.J., Arimura T., Bonne G., Worman H.J. Activation of MAPK pathways links LMNA mutations to cardiomyopathy in Emery-Dreifuss muscular dystrophy. J Clin Invest. 2007;117:1282–1293. doi: 10.1172/JCI29042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muchir A., Wu W., Worman H.J. Mitogen-activated protein kinase inhibitor regulation of heart function and fibrosis in cardiomyopathy caused by lamin A/C gene mutation. Trends Cardiovasc Med. 2010;20:217–221. doi: 10.1016/j.tcm.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu W., Muchir A., Shan J., Bonne G., Worman H.J. Mitogen-activated protein kinase inhibitors improve heart function and prevent fibrosis in cardiomyopathy caused by mutation in lamin A/C gene. Circulation. 2011;123:53–61. doi: 10.1161/CIRCULATIONAHA.110.970673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Browning J.D., Szczepaniak L.S., Dobbins R., Nuremberg P., Horton J.D., Cohen J.C., Grundy S.M., Hobbs H.H. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40:1387–1395. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- 66.Xu C., Yu C., Ma H., Xu L., Miao M., Li Y. Prevalence and risk factors for the development of nonalcoholic fatty liver disease in a nonobese Chinese population: the Zhejiang Zhenhai Study. Am J Gastroenterol. 2013;108:1299–1304. doi: 10.1038/ajg.2013.104. [DOI] [PubMed] [Google Scholar]
- 67.Anstee Q.M., Seth D., Day C.P. Genetic factors that affect risk of alcoholic and nonalcoholic fatty liver disease. Gastroenterology. 2016;150:1728–1744 e7. doi: 10.1053/j.gastro.2016.01.037. [DOI] [PubMed] [Google Scholar]
- 68.Guenantin A.C., Briand N., Bidault G., Afonso P., Bereziat V., Vatier C., Lascols O., Caron-Debarle M., Capeau J., Vigouroux C. Nuclear envelope-related lipodystrophies. Semin Cell Dev Biol. 2014;29:148–157. doi: 10.1016/j.semcdb.2013.12.015. [DOI] [PubMed] [Google Scholar]