Abstract
The genomic lesions that characterize acute lymphoblastic leukemia in childhood include recurrent translocations that result in the expression of fusion proteins that typically involve genes encoding tyrosine kinases, cytokine receptors, and transcription factors. These genetic rearrangements confer phenotypic hallmarks of malignant transformation, including unrestricted proliferation and a relative resistance to apoptosis. In this Minireview, we discuss the molecular mechanisms that link these fusions to the control of cell death. We examine how these fusion genes dysregulate the BCL-2 family of proteins, preventing activation of the apoptotic effectors, BAX and BAK, and promoting cell survival.
Keywords: apoptosis, B-cell lymphoma 2 (Bcl-2) family, fusion protein, leukemia, tyrosine-protein kinase (tyrosine kinase)
Recurrent fusion genes in acute lymphoblastic leukemia
Acute lymphoblastic leukemia (ALL)2 is the most common form of childhood malignancy. Therapy for ALL is one of the great success stories of modern chemotherapy, and overall cure rates are now >90% in developed countries, depending on molecular subtypes and clinical features (1). The extraordinary improvements in outcomes in ALL have unquestionably been driven by the treatment of patients on international collaborative clinical trials (2), which have made it possible to rapidly recruit sufficient numbers of patients to studies of new treatment regimes. The analysis of treatment responses, based on measurement of minimal residual disease, has allowed the early identification of treatment failure or relapse and the consequent adjustment of treatment intensity.
The next revolution in our understanding of ALL biology is being driven by many studies, involving thousands of patient samples, characterizing the genomic landscape of ALL through genome and transcriptome sequencing (3–7). This has led to the recognition of novel molecular subtypes of ALL, defined by the genomic lesions that drive them. Characterization of leukemic genomes provides insight into the key molecular pathways involved in ALL subtypes.
Many recently identified genomic lesions in ALL are fusion genes, arising from chromosomal translocations (8). These include fusions that activate tyrosine kinases, cytokine receptors, and transcription factors. The presence of these fusions has important prognostic and treatment implications. In this Minireview, we consider how these genomic lesions promote resistance to apoptosis in ALL.
Gene fusions in ALL
Chromosomal translocations, resulting in the expression of fusion genes, are a hallmark of B-cell malignancies. This likely arises as fusion partners are mistakenly juxtaposed during periods of genomic editing and recombinase-activating gene (RAG1 and RAG2) activation or somatic hypermutation during B-cell development (9). Recurrent chromosomal translocations have been recognized and detected in ALL, initially by staining of metaphases and microscopy, and more recently by fluorescent in situ hybridization (FISH). The detection of a small number of recurrent translocations is a standard component of ALL diagnosis and risk assessment. For example, t(12;21) ETV6-RUNX1 and t(1;19) TCF3-PBX1 identify ALL associated with excellent outcomes, whereas TCF3-PBX1 is associated with a higher incidence of central nervous system involvement. Other fusions are associated with poor prognosis and are an indication for treatment intensification (8). This includes the mixed lineage leukemia-rearranged (MLL-r) leukemias, which are discussed later.
Fusions activating tyrosine kinases are particularly important in ALL, as they are potentially amenable to treatment with tyrosine kinase inhibitors (TKI's). Imatinib, a small molecule ABL1 inhibitor, has dramatically altered the treatment paradigm for patients with Philadelphia chromosome positive (Ph+) B-ALL and chronic myeloid leukemia (CML) because it encodes the BCR-ABL1 fusion gene (10). The majority of Ph+ patients are now successfully treated with TKI-containing regimens, without the need for hematopoietic stem cell transplant. Philadelphia-like ALL (Ph-like ALL) is characterized by a gene expression profile resembling Ph+ ALL but with the absence of BCR-ABL1. Ph-like ALL includes 10–15% of all pediatric B-ALL cases, and the prevalence in adults peaks in the 16–40-year age group (20%) (4, 11). Ph-like ALL is frequently associated with resistance to frontline chemotherapy and poor clinical outcomes (8, 11, 12). Ph-like ALL is also a fusion-driven disease, and to date, rearrangements involving 13 kinases and cytokine receptors have been identified (4). The kinases activated in Ph-like ALL, which include ABL1, ABL2, and JAK2, can potentially be targeted with specific TKIs (4, 13).
Many ALL fusion genes block apoptosis
A key test that a fusion gene is a true driver of leukemia is its capacity to inhibit apoptosis induced by cytokine deprivation in cytokine-dependent cell lines (14, 15). This test and determining whether expression of the fusion in hematopoietic stem cells recapitulates the primary tumor when cells are transplanted into syngeneic mice are commonly used to establish genes as drivers of leukemia.
Not all leukemia fusions have the same capacity to block cytokine withdrawal-induced cell death. Fusions that activate tyrosine kinases or cytokine receptor signaling often have this function. Others, principally those that involve transcriptional regulators, readily cause leukemia in mice, but leukemic cells expressing such fusion genes remain cytokine-dependent when cultured ex vivo. Understanding the mechanisms by which fusions repress apoptosis has important therapeutic implications, as blocks in apoptosis are a mechanism of chemoresistance, including resistance to therapies targeting fusions (16, 17). The application of drugs that directly target cell death pathways, or inhibit the signaling pathways downstream of activated tyrosine kinases, may significantly enhance the efficacy of standard chemotherapy. The BCL-2 family of apoptosis regulators is key in this process, because they regulate the cell death responses repressed by leukemia fusion oncogenes (18).
BCL-2 family and cancer
Overexpression of members of the BCL-2 protein family that block apoptosis contributes to malignant transformation. This was first recognized as the mechanism of action of the recurrent translocation t(14;18) in follicular lymphoma (19–21). Amplification of pro-survival, BCL-2-like, genes MCL-1 and BCL-XL and deletion of pro-apoptotic genes BOK and PUMA are over-represented in the somatic copy number variations in over 3000 cancer specimens, across 26 human cancers, including ALL (22).
The BCL-2 family has been reviewed in detail elsewhere (15, 23, 24). However, for the purposes of this review, it is worth considering the key molecular events that regulate cellular commitment to apoptosis in cytokine withdrawal models. Both the cytokine signaling pathways on which cells normally depend and the cell survival pathways activated by kinase fusion genes converge on repressing the activation of the intrinsic apoptotic pathway regulated by the BCL-2 family (Fig. 1).
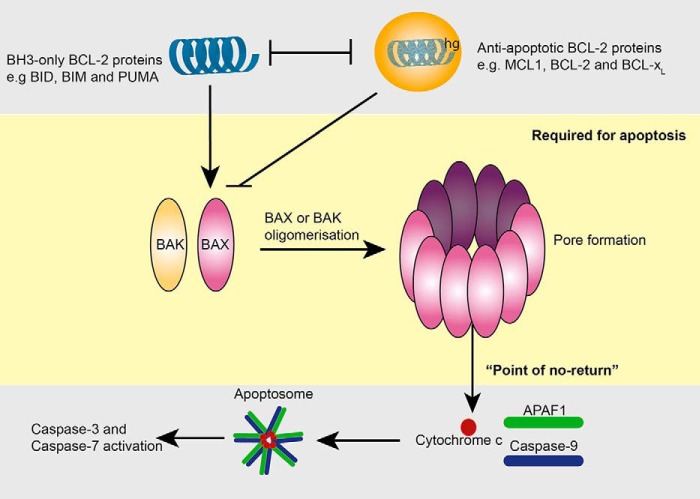
Figure 1.
Regulation of apoptosis by the BCL-2 family of proteins. This schematic shows the interactions between the BCL-2 family members that regulate BAX and BAK activation. The middle (yellow) panel shows that in healthy cells BAX and BAK exist in monomeric form. Following an apoptotic stimulus, they assemble into higher order oligomeric structures, which form a pore in the outer mitochondrial membrane. This results in loss of the mitochondrial outer membrane potential and egress of cytochrome c from mitochondria. This is the “point of no return” or commitment to cell death. Cytochrome c (lower gray panel) together with APAF-1 and caspase-9 form a molecular machine known as the apoptosome. This machine drives activation of caspase-9 and the subsequent activation of other caspases, including the “executioner caspases” caspase-3 and caspase-7. This caspase activation is responsible for the morphological features of apoptosis. The upper gray panel illustrates the key interactions that regulate BAX and BAK activation. The BH3-only proteins, notably BID and BIM, promote activation of BAX and BAK by directly binding to BAX or BAK. Other BH3-only proteins such as PUMA (and also BIM) bind anti-apoptotic BCL-2 proteins at the hydrophobic groove (hg) and inhibit anti-apoptotic function. BCL-2, BCL-xL, and MCL-2 function to directly inhibit BAX and BAK or prevent their activation by binding and blocking the BH3-only proteins.
The BCL-2 family is functionally grouped into proteins that repress apoptosis, BCL-2, BCL-XL, BCL-w, MCL-1, and A1, and the proteins that promote apoptosis (pro-apoptotic) (23). The pro-apoptotic members are further subdivided into two groups, the BAX/BAK subfamily and the BH3-only subfamily, consisting of BIM, BAD, BID, BIK, BMF, PUMA, NOXA, and HRK (25). The pro-survival and BH3-only subgroups regulate the activation of BAX and BAK. Activation of BAX and BAK is the critical step in the commitment to apoptosis, as it is required to trigger mitochondrial outer membrane permeabilization, initiating a cascade of programmed downstream events, including cytochrome c release, formation of the apoptosome by APAF-1, and activation of caspase-9 and the effector caspases, that characterize apoptotic cell death (Fig. 1) (24, 26–28).
The interleukin-3 (IL-3)-dependent cell lines are commonly used to test the capacity of ALL fusions to block apoptosis induced by cytokine deprivation (29). IL-3-dependent cell lines from gene-deleted mice have shown that the presence of at least one of BAX and BAK is absolutely required for cytokine withdrawal-induced apoptosis (30–32). When cytokine is removed from cultures of IL-3-dependent Ba/F3 cells lacking both BAX and BAK, cells remain viable for long periods, although they do not proliferate. When cytokine is restored, cells re-enter the cell cycle and divide again. Demonstrating that some ALL fusions can maintain cell survival in the absence of cytokine provides clear evidence that such leukemia fusion genes directly or indirectly prevent BAX and BAK activation (33, 34).
The molecular mechanisms of BAX and BAK activation are complex and are tightly regulated by the interactions between the BCL-2 protein family members. Mutational and structural analysis shows that BAX and BAK activation is initiated by conformational changes that, for example in the case of BAX, allow translocation and insertion into the outer mitochondrial membrane (35). For both BAX and BAK, the conformational changes expose interaction domains that favor first homodimerization and then the formation of higher order oligomeric structures that ultimately punch a hole in the mitochondrial membrane.
The BH3-only BCL-2 family members initiate BAX and BAK activation in two ways. Some BH3-only proteins directly engage BAX and BAK through an interaction between their BH3 domain and a hydrophobic groove on the surface of BAX and BAK (36, 37). Other BH3-only proteins do not directly bind BAX or BAK, but instead they repress the function of the anti-apoptotic BCL-2 family members through an analogous molecular interaction (37). Anti-apoptotic BCL-2 family members and BAX and BAK share significant structural homology, including the surface of the hydrophobic groove interaction site for the BH3 domain of the BH3-only proteins (25). Anti-apoptotic BCL-2 family proteins can thus “soak up” BH3-only proteins that might otherwise bind BAX or BAK. This is overwhelmed when the abundance of BH3-only proteins increases. The anti-apoptotic BCL-2 family members are also able to directly bind and inhibit activated monomeric BAX and BAK to inhibit apoptosis (24).
Kinase-activating fusions and anti-apoptotic BCL-2 proteins
Fusions involving tyrosine kinases in ALL typically result in the loss of regulatory domains, overexpression of the kinase domain, and the acquisition of coiled-coil domains or helix-loop-helix motifs that facilitate oligomerization and autophosphorylation of the kinase domain (38). This could be through the repression of BH3-only protein expression or increased expression of anti-apoptotic BCL-2 proteins. Both mechanisms may operate simultaneously.
The regulation of MCL-1 protein abundance is important. MCL-1 is absolutely required for normal hematopoietic development (39). The half-life of MCL-1 is short, and it plays a key role in regulating apoptosis in response to cytokine receptor signaling, including interleukin-7 (IL-7)-dependent survival of T- and B-lymphocytes (40). In IL-3 or granulocyte-macrophage colony-stimulating factor (GM-CSF)-dependent hematopoietic cells, MCL-1 undergoes rapid proteasomal degradation soon after cytokine withdrawal (41). When cytokine is restored, MCL-1 expression is rapidly up-regulated. The most compelling evidence that kinase-activating fusions function to maintain MCL-1 expression and repress BH3-only proteins is from studies of the BCR-ABL1 fusion.
BCR-ABL1 shifts the balance of BCL-2 family proteins in favor of cell survival
There are three forms of the BCR-ABL1 fusion gene, with alternate breakpoints in the breakpoint cluster region (BCR) gene. Each is associated with distinct subtypes of leukemia (42). The p210 BCR-ABL1 is the hallmark of CML; p185 is found in adult and pediatric B-ALL; and p230 is associated with neutrophilic CML (38). In Arf−/− B-ALL cells expressing p185 BCR-ABL1, there is selection against silencing of MCL-1 expression, unless apoptosis was blocked by BAX and BAK deletion, or overexpression of BCL-2 or BCL-XL (43, 44). This shows that MCL-1 is required to prevent BAX and BAK activation for the fusion to maintain cell viability. Treatment of these cells with imatinib induced a dose-dependent reduction in MCL-1 protein, but it had no effect on BCL-2 or BCL-XL expression (43). In CML models, BCR-ABL1 appears to maintain MCL-1 expression through the activation of STAT3-dependent transcription. Deletion of STAT3 prevents BCR-ABL1 from initiating CML, and there is a correlation between STAT3 phosphorylation and resistance to imatinib (45, 46). These observations raise the possibility that inhibition of STAT3 phosphorylation, directly or indirectly, could synergize with imatinib treatment.
Other evidence supports repression of pro-apoptotic BH3-only proteins by BCR-ABL1. Imatinib treatment up-regulated expression of BIM and BAD in human CML cell lines (17). There is some indication that the presence of BCR-ABL1 drives ERK-mediated phosphorylation of BIM and subsequent BIM degradation (Fig. 2) (47, 48). However, the functional significance of BIM phosphorylation driven by leukemia fusion genes is not established (49).
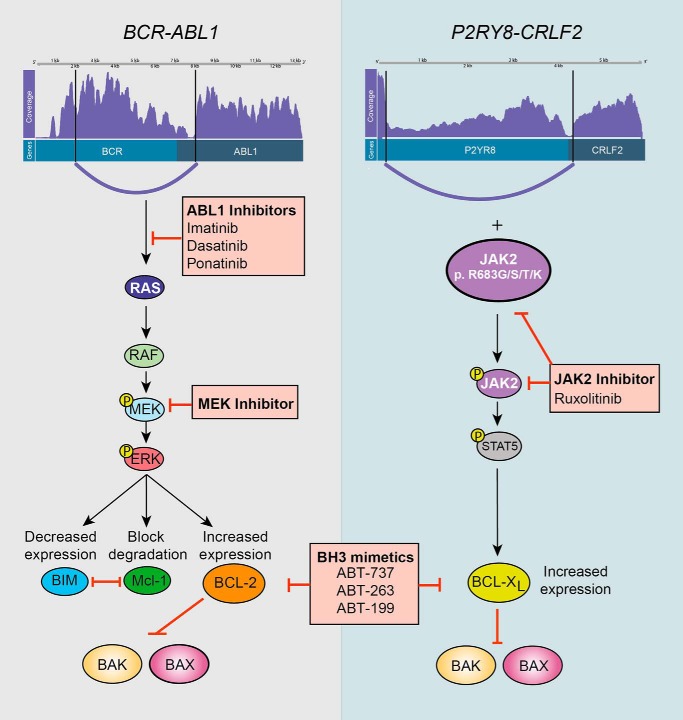
Figure 2.
Mechanisms by which fusion genes promote kinase activation and cell survival and opportunities for therapeutic intervention. This figure shows two common gene rearrangements in ALL, BCR-ABL1 (left panel) and P2RY8-CRLF2 (right panel). The schematics depicting each fusion were produced using a program for visualization of gene rearrangements from RNA sequencing data (Clinker https://github.com/Oshlack/Clinker. Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.). The Clinker images show, from top to bottom, the coverage of reads mapped to each fusion, functional domains, and transcript variants. The breakpoint in each gene and associated fusion is also shown. The pathways show downstream kinases that are activated as a result of each fusion and consequently the impact on expression and/or degradation of mediators of cell survival. Therapeutic interventions at each point are shown in the red-shaded boxes. Constitutive activation of the ABL1 kinase, mediated by BCR-ABL1, promotes activation of the RAS/RAF/MEK/ERK pathway. Tyrosine kinase inhibitors can be used to either target ABL1 (imatinib, dasatinib, or ponatinib) directly or by MEK inhibitors. Phosphorylation of ERK, in turn, decreases BIM expression, blocks MCL-1 degradation, and increases BCL-2 expression. This prevents BAX/BAK activation. Pro-survival proteins BCL-2 and BCL-XL can be therapeutically targeted with BH3-mimetics (ABT-737, ABT-263, or ABT-199). The P2RY8-CRLF2 fusion cooperates with JAK2 mutations, the most common of which, p.R683G/R683S/R683T/R683K, is depicted to promote cell survival. Activated JAK2 can be directly inhibited with ruxolitinib. STAT5 promotes transcription and increased expression of BCL-XL, and perhaps BCL-2 and MCL-1 also. Treatment with BH3-mimetics may increase the capacity of these cells to undergo apoptosis in response to JAK2 inhibition.
The levels of other anti-apoptotic BCL-2 proteins may also be regulated by BCR-ABL1, which may raise the threshold that a death stimulus must reach before apoptosis can proceed. An intriguing line of evidence comes from experiments using BCL-2 inhibitor drugs (or BH3-mimetic drugs). ABT-737 is the prototype inhibitor with broad activity against BCL-2, BCL-XL, and BCL-W, but with no activity against MCL-1 (50). This drug enhanced imatinib cell killing and prolonged the survival of mice transplanted with Ph+ ALL (17, 51). Mechanistically, ABT-737 increases the pool of unbound BH3-only proteins, which in steady state are otherwise bound to and sequestered by BCL-2 and BCL-XL (17). Although ABT-737 treatment increased survival in some imatinib-treated Ph+ ALL xenografts, ABT-263 (navitoclax, identical activity to ABT-737) and ABT-199 (selectively inhibits BCL-2) were ineffective in another Ph+ ALL xenograft model (51, 52). Nevertheless, therapeutic combinations of TKIs and BH3 mimetics are of interest, as anti-apoptotic BCL-2 proteins other than MCL-1 can compensate for the loss of MCL-1 induced by TKIs.
Leonard et al. (53) highlighted the importance of the BCL-2/MCL-1 ratio in sensitivity to venetoclax. The high BCL-2/MCL-1 ratio (in a pediatric Ph+ ALL cell line) mediated sensitivity to venetoclax, whereas low expression of BCL-2 conferred resistance in another CML cell line. Combined treatment with venetoclax and the TKI dasatinib induced the highest levels of cell killing (53). Depletion or inhibition of the total BCL-2 anti-apoptotic protein pool lowers resistance to cell death. Thus, diminished expression of MCL-1, driven by inhibition of BCR-ABL1 kinase, together with direct inhibition of BCL-2 by BH3-mimetic drugs may drive ALL cells to activate their intrinsic apoptosis pathways (48). Similarly, a MEK inhibitor in human Ph+ cell lines was insufficient as a single agent to induce cell killing (48, 54). However, when combined with ABT-263 or ABT-199, cell killing was induced (48). These studies highlight the potential efficacy of combining tyrosine kinase and BCL-2 family inhibition to treat patients with BCR-ABL1-driven leukemia.
JAK2 fusions and the role of BCL-XL
JAK-STAT pathway-activating fusions in ALL are also associated with poor outcomes. Many Janus kinase 2 (JAK2) fusions with different fusion partners have been reported (4). In addition, fusions that increase expression of cytokine receptor-like factor 2 (CRLF2), most commonly with P2RY8 or as an IGH rearrangement, cooperate with activating JAK2 mutations to drive ALL (Fig. 2) (55). How these fusions regulate the apoptotic pathway is less well studied, but activating JAK2 mutations provides some insights. JAK2V617F is the causal mutation of polycythemia vera (PV) and is also found in a range of other myeloproliferative neoplasms. Like JAK2 fusions, JAK2V617F is constitutively activated (56). There is evidence supporting the hypothesis that activated JAK2 signaling leads to elevated expression of BCL-2, BCL-XL, and MCL-1. BCL-2 and BCL-XL expression is increased in PV patients with the JAK2V617F mutation, and higher JAK2V617F expression in erythroid precursor cells from PV patients was associated with increased sensitivity to ABT-737 treatment (57). RNA interference-induced knockdown of BIM decreased sensitivity to JAK2 inhibition, implying that JAK2 signaling also represses BIM, whereas MCL-1 knockdown increased apoptosis in JAK2V617F cells and increased sensitivity to JAK2 inhibition (58). Enforced expression of JAK2V617F increased MCL-1 expression by STAT3-dependent transcription (59). Together, this suggests that JAK2 signaling raises the apoptosis threshold by increasing expression of anti-apoptotic members of the BCL-2 family.
One might then predict synergy between JAK2 inhibitors and BH3-mimetic drugs. The combination of JAK2 inhibitors, such as ruxolitinib or AZD1480, with BH3-mimetics enhanced the limited efficacy of JAK inhibitors as single agents (60, 61). Waibel et al. (62) used an Eμ-TEL-JAK2 (ETV6-JAK2) mouse model of T-ALL to show that up-regulation of BCL-2/BCL-XL and down-regulation of BIM expression promote leukemic cell survival. BCL-2/BCL-XL inhibition (ABT-737), combined with JAK2 inhibition, induced the greatest therapeutic response, compared with either agent alone. Clearly, there are common mechanisms by which BCR-ABL1 fusions and JAK2 fusions maintain cell viability. This may apply to all leukemia fusions activating tyrosine kinase domains. They must all, in some way, regulate the activation of BAX and BAK, although the individual BCL-2 proteins and the pathways that connect the fusions to the apoptosis machinery may vary (63).
ETV6-RUNX1 and cell survival
The ETV6-RUNX1 fusion is present in up to 25% of pediatric B-ALL cases and is associated with favorable outcomes for patients (4, 64). The ETV6-RUNX1 fusion is not necessary to maintain leukemic cell viability, and it is not sufficient alone to cause a leukemia in murine transplant models (65, 66). There is substantial evidence to indicate that the ETV6-RUNX1 fusion can be detected antenatally in pre-leukemic clones. The full manifestation of ETV6-RUNX1-driven ALL therefore requires secondary changes that confer resistance to apoptosis (65).
The erythropoietin receptor (EPOR) is consistently overexpressed in ETV6-RUNX1-positive ALL (67, 68). EPOR is a homodimeric cytokine receptor that may also be expressed as a fusion in Ph-like ALL (4). Normal EPOR signaling requires binding of the EPO ligand to the receptor to induce signal transduction through JAK2 and STAT5 phosphorylation (69). Chromatin immunoprecipitation assays showed the ETV6-RUNX1 fusion bound to the promoter of EPOR, driving EPOR transcription. However, most data suggest that elevated expression of EPOR alone is not sufficient to activate signaling and that EPO ligand is also required. It may be that ETV6-RUNX1 drives unregulated EPOR signaling, STAT5 activation, and elevated BCL-XL expression (70). Another suggested survival mechanism may be a combination of STAT3 activation (which increases BCL-2 and BCL-XL expression) and c-MYC-dependent transcription (71).
Infant MLL-r ALL
Infant MLL-r ALL is often aggressive, and the outcomes remain poor despite intensified chemotherapy or allogeneic hematopoietic stem cell transplantation (74, 75). The MLL (KMT2A) gene is a histone methyltransferase located on chromosome 11q23. MLL-r ALL accounts for 80% of infant ALL, and more than 80 fusion partners have been identified. The most common are AF4, ENL, and AF9 (76, 77). KMT2A functions in a multiprotein transcriptional regulatory complex and is required for normal hematopoiesis (78). Among the key transcription targets regulated by KMT2A are the homeobox (HOX) genes and in particular the HOXA cluster. The transgenic expression of the MLL-AF9 fusion does not cause AML in HoxA9-deficient animals. MLL rearrangements may arise during fetal hematopoiesis and may be detected at birth, prior to the onset of disease (79). Although additional genetic mutations are uncommon in infant MLL-r ALL (3, 80), there are recognized associations with activating PI3K/RAS pathway mutations and FLT3 (79).
The common MLL translocations in infant ALL confer resistance to apoptosis (81). This may result from high BCL-2 expression (82). Although BCL-2 is not required for MLL-AF9 to initiate leukemia in murine models, deletion of BCL-2 delays disease onset and diminishes clonal proliferation (83). MLL-AF4 also specifically up-regulates the BCL-2 expression by DOT1L-mediated H3K79 methylation at the BCL-2 locus (84).
Transcriptional up-regulation of anti-apoptotic BCL-2 proteins by MLL rearrangements may inhibit glucocorticoid-dependent apoptosis. Glucocorticoids form the backbone of ALL treatment protocols. Approximately 30% of infant MLL-r ALL have poor glucocorticoid responses (75, 85). Glucocorticoid-induced apoptosis in lymphocytes proceeds via the BCL-2-regulated pathway and is absolutely dependent on BAX and BAK (86). Stam et al. (88) and others (87) showed increased expression of MCL-1 in prednisolone-resistant pediatric ALL samples, most notably MLL-r ALL. RNAi knockdown of MCL-1 in prednisolone-resistant MLL-r leukemia cells partially restores prednisolone killing (88). Epigenetic silencing of BIM in MLL-r ALL also contributes to glucocorticoid resistance (89, 90).
The role of BCL-2 in the viability of MLL-r leukemia suggests that including BCL-2 inhibition in treatment regimens may be effective. Data from the Pediatric Preclinical Testing Program showed the BH3 mimetic navitoclax has significant anti-tumor activity, particularly in MLL-r ALL (91). Jayanthan et al. (92) also demonstrated that combining ABT-737 with a histone deacetylase inhibitor, proteasome inhibitor, multi-tyrosine kinase inhibitor, and anthracycline had additive effects. Venetoclax was also effective at killing MLL-r ALL leukemic cells. In MLL-r ALL xenografts, venetoclax was associated with a higher response rate of 50%, compared with 26% in non-MLL-ALL xenografts, suggesting that BCL-2 inhibition, in conjunction with chemotherapeutics, is effective in this sub-group (93, 94). These data support the trial of the introduction of BCL-2-inhibitor drugs into therapeutic regimens for MLL-r leukemias (84).
DOT1L is a critical component of the MLL-r transcriptional complex. Phase I trials of small molecule DOT1L inhibitors have shown promising molecular efficacy (95, 96). DOT1L inhibitors also sensitize MLL-r ALL to venetoclax (84), providing additional evidence that BCL-2 proteins play a role in MLL-r leukemia and that BCL-2 inhibition may improve clinical outcomes.
Exploiting other cell death pathways in ALL
Repressing BH3-only protein expression while maintaining expression of anti-apoptotic BCL-2 proteins is a common theme of ALL fusion function. Other genetically programmed cell death pathways, which do not necessarily contribute directly to fusion-driven ALL, may be clinically exploited to bypass blocks in apoptosis. Necroptosis, or programmed necrosis, is activated by diverse extrinsic stimuli but prominently by TNF receptor 1 signaling (97). The key molecules that mediate necroptosis include the RIP kinases (RIPK1 and RIPK3) and the mixed lineage kinase domain-like protein. The inhibitor of apoptosis proteins, cIAP1 and cIAP2, determines whether TNFR1 signaling activates necroptosis. When these are expressed, the necroptosis pathway is blocked (98). IAP inhibitor small molecule drugs bind to the IAPs and cause rapid proteasomal degradation and simultaneous up-regulation of TNF-activated TNFR1 signaling (99). The net effect is autonomous TNFR1-dependent necroptosis. In ALL, the IAP antagonists have shown single agent efficacy in a range of patient-derived xenograft models (100). The mechanism of action of IAP inhibitor drugs exploits pathways not primarily regulated by the leukemia driver fusions but that are intact even in highly resistant tumors.
Conclusion
The mechanisms by which oncogenic fusion genes promote survival converge on the BCL-2 family of proteins and the repression of BAX and BAK activation. These fusions promote cell survival by altering the balance of the BCL-2 family members to favor survival, repressing expression of pro-apoptotic BH3-only proteins, and up-regulating pro-survival BCL-2 family members. For fusions that activate tyrosine kinases, most interest has focused on how the signal transduction pathways initiated by the fusion impact on key anti-apoptotic proteins such as BCL-2 and MCL-1. Transcription factor fusions also regulate the levels of these same BCL-2 family proteins. Fusion-dependent up-regulation of BCL-2 or MCL-1 expression is a mechanism of drug resistance but also a therapeutic opportunity. Although drugs that inhibit BCL-2 (BH3 mimetics) have shown some efficacy in vitro, their use has yet to be implemented into ALL treatment protocols. The promise of this approach will be tested in upcoming clinical trials.
The authors declare that they have no conflicts of interest with the contents of this article.
- ALL
- acute lymphoblastic leukemia
- MLL
- mixed lineage leukemia
- MLL-r
- mixed lineage leukemia-rearranged
- TKI
- tyrosine kinase inhibitor
- CML
- chronic myeloid leukemia
- Ph+
- Philadelphia chromosome positive
- B-ALL
- B-cell acute lymphoblastic leukemia
- EPOR
- erythropoietin receptor
- PV
- polycythemia vera
- BCR
- breakpoint cluster region
- IAP
- inhibitor of apoptosis protein.
References
- 1. Adamson P. C. (2015) Improving the outcome for children with cancer: development of targeted new agents. CA Cancer J. Clin. 65, 212–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pui C. H., Carroll W. L., Meshinchi S., and Arceci R. J. (2011) Biology, risk stratification, and therapy of pediatric acute leukemias: an update. J. Clin. Oncol. 29, 551–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mullighan C. G., Goorha S., Radtke I., Miller C. B., Coustan-Smith E., Dalton J. D., Girtman K., Mathew S., Ma J., Pounds S. B., Su X., Pui C. H., Relling M. V., Evans W. E., Shurtleff S. A., and Downing J. R. (2007) Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446, 758–764 [DOI] [PubMed] [Google Scholar]
- 4. Roberts K. G., Li Y., Payne-Turner D., Harvey R. C., Yang Y. L., Pei D., McCastlain K., Ding L., Lu C., Song G., Ma J., Becksfort J., Rusch M., Chen S. C., Easton J., et al. (2014) Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N. Engl. J. Med. 371, 1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roberts K. G., Morin R. D., Zhang J., Hirst M., Zhao Y., Su X., Chen S. C., Payne-Turner D., Churchman M. L., Harvey R. C., Chen X., Kasap C., Yan C., Becksfort J., Finney R. P., et al. (2012) Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell 22, 153–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mullighan C. G., Zhang J., Harvey R. C., Collins-Underwood J. R., Schulman B. A., Phillips L. A., Tasian S. K., Loh M. L., Su X., Liu W., Devidas M., Atlas S. R., Chen I. M., Clifford R. J., Gerhard D. S., et al. (2009) JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. U.S.A. 106, 9414–9418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Den Boer M. L., van Slegtenhorst M., De Menezes R. X., Cheok M. H., Buijs-Gladdines J. G., Peters S. T., Van Zutven L. J., Beverloo H. B., Van der Spek P. J., Escherich G., Horstmann M. A., Janka-Schaub G. E., Kamps W. A., Evans W. E., and Pieters R. (2009) A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 10, 125–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roberts K. G., and Mullighan C. G. (2015) Genomics in acute lymphoblastic leukaemia: insights and treatment implications. Nat. Rev. Clin. Oncol. 12, 344–357 [DOI] [PubMed] [Google Scholar]
- 9. Papaemmanuil E., Rapado I., Li Y., Potter N. E., Wedge D. C., Tubio J., Alexandrov L. B., Van Loo P., Cooke S. L., Marshall J., Martincorena I., Hinton J., Gundem G., van Delft F. W., Nik-Zainal S., et al. (2014) RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat. Genet. 46, 116–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Druker B. J., Sawyers C. L., Kantarjian H., Resta D. J., Reese S. F., Ford J. M., Capdeville R., and Talpaz M. (2001) Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N. Engl. J. Med. 344, 1038–1042 [DOI] [PubMed] [Google Scholar]
- 11. Roberts K. G., Gu Z., Payne-Turner D., McCastlain K., Harvey R. C., Chen I. M., Pei D., Iacobucci I., Valentine M., Pounds S. B., Shi L., Li Y., Zhang J., Cheng C., Rambaldi A., Tosi M., et al. (2017) High frequency and poor outcome of Philadelphia chromosome-like acute lymphoblastic leukemia in adults. J. Clin. Oncol. 35, 394–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jain N., Roberts K. G., Jabbour E., Patel K., Eterovic A. K., Chen K., Zweidler-McKay P., Lu X., Fawcett G., Wang S. A., Konoplev S., Harvey R. C., Chen I. M., Payne-Turner D., Valentine M., et al. (2017) Ph-like acute lymphoblastic leukemia: a high-risk subtype in adults. Blood 129, 572–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chase A., Bryant C., Score J., Haferlach C., Grossmann V., Schwaab J., Hofmann W. K., Reiter A., and Cross N. C. (2013) Ruxolitinib as potential targeted therapy for patients with JAK2 rearrangements. Haematologica 98, 404–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cambier N., Chopra R., Strasser A., Metcalf D., and Elefanty A. G. (1998) BCR-ABL activates pathways mediating cytokine independence and protection against apoptosis in murine hematopoietic cells in a dose-dependent manner. Oncogene 16, 335–348 [DOI] [PubMed] [Google Scholar]
- 15. Adams J. M., and Cory S. (2007) The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene 26, 1324–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferrao P. T., Frost M. J., Siah S. P., and Ashman L. K. (2003) Overexpression of P-glycoprotein in K562 cells does not confer resistance to the growth inhibitory effects of imatinib (STI571) in vitro. Blood 102, 4499–4503 [DOI] [PubMed] [Google Scholar]
- 17. Kuroda J., Puthalakath H., Cragg M. S., Kelly P. N., Bouillet P., Huang D. C., Kimura S., Ottmann O. G., Druker B. J., Villunger A., Roberts A. W., and Strasser A. (2006) Bim and Bad mediate imatinib-induced killing of Bcr/Abl+ leukemic cells, and resistance due to their loss is overcome by a BH3 mimetic. Proc. Natl. Acad. Sci. U.S.A. 103, 14907–14912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Delbridge A. R., Grabow S., Strasser A., and Vaux D. L. (2016) Thirty years of BCL-2: translating cell death discoveries into novel cancer therapies. Nat. Rev. Cancer 16, 99–109 [DOI] [PubMed] [Google Scholar]
- 19. Fukuhara S., and Rowley J. D. (1978) Chromosome 14 translocations in nonBurkitt lymphomas. Int. J. Cancer 22, 14–21 [DOI] [PubMed] [Google Scholar]
- 20. Tsujimoto Y., Finger L. R., Yunis J., Nowell P. C., and Croce C. M. (1984) Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science 226, 1097–1099 [DOI] [PubMed] [Google Scholar]
- 21. Vaux D. L., Cory S., and Adams J. M. (1988) Bcl-2 gene promotes hematopoietic-cell survival and cooperates with c-Myc to immortalize pre-B-cells. Nature 335, 440–442 [DOI] [PubMed] [Google Scholar]
- 22. Beroukhim R., Mermel C. H., Porter D., Wei G., Raychaudhuri S., Donovan J., Barretina J., Boehm J. S., Dobson J., Urashima M., Mc Henry K. T., Pinchback R. M., Ligon A. H., Cho Y. J., Haery L., et al. (2010) The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cory S., Huang D. C., and Adams J. M. (2003) The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene 22, 8590–8607 [DOI] [PubMed] [Google Scholar]
- 24. Chipuk J. E., Moldoveanu T., Llambi F., Parsons M. J., and Green D. R. (2010) The BCL-2 family reunion. Mol. Cell 37, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Delbridge A. R., and Strasser A. (2015) The BCL-2 protein family, BH3-mimetics and cancer therapy. Cell Death Differ. 22, 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu X., Kim C. N., Yang J., Jemmerson R., and Wang X. (1996) Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 86, 147–157 [DOI] [PubMed] [Google Scholar]
- 27. Ekert P. G., Read S. H., Silke J., Marsden V. S., Kaufmann H., Hawkins C. J., Gerl R., Kumar S., and Vaux D. L. (2004) Apaf-1 and caspase-9 accelerate apoptosis, but do not determine whether factor-deprived or drug-treated cells die. J. Cell Biol. 165, 835–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Henry-Mowatt J., Dive C., Martinou J. C., and James D. (2004) Role of mitochondrial membrane permeabilization in apoptosis and cancer. Oncogene 23, 2850–2860 [DOI] [PubMed] [Google Scholar]
- 29. Warmuth M., Kim S., Gu X. J., Xia G., and Adrián F. (2007) Ba/F3 cells and their use in kinase drug discovery. Curr. Opin. Oncol. 19, 55–60 [DOI] [PubMed] [Google Scholar]
- 30. Wei M. C., Zong W. X., Cheng E. H., Lindsten T., Panoutsakopoulou V., Ross A. J., Roth K. A., MacGregor G. R., Thompson C. B., and Korsmeyer S. J. (2001) Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science 292, 727–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lum J. J., Bauer D. E., Kong M., Harris M. H., Li C., Lindsten T., and Thompson C. B. (2005) Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 120, 237–248 [DOI] [PubMed] [Google Scholar]
- 32. Ekert P. G., Jabbour A. M., Manoharan A., Heraud J. E., Yu J., Pakusch M., Michalak E. M., Kelly P. N., Callus B., Kiefer T., Verhagen A., Silke J., Strasser A., Borner C., and Vaux D. L. (2006) Cell death provoked by loss of interleukin-3 signaling is independent of Bad, Bim, and PI3 kinase, but depends in part on Puma. Blood 108, 1461–1468 [DOI] [PubMed] [Google Scholar]
- 33. Okuda K., Golub T. R., Gilliland D. G., and Griffin J. D. (1996) p210BCR/ABL, p190BCR/ABL, and TEL/ABL activate similar signal transduction pathways in hematopoietic cell lines. Oncogene 13, 1147–1152 [PubMed] [Google Scholar]
- 34. Li S., Ilaria R. L. Jr., Million R. P., Daley G. Q., and Van Etten R. A. (1999) The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J. Exp. Med. 189, 1399–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Czabotar P. E., Westphal D., Dewson G., Ma S., Hockings C., Fairlie W. D., Lee E. F., Yao S., Robin A. Y., Smith B. J., Huang D. C., Kluck R. M., Adams J. M., and Colman P. M. (2013) Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell 152, 519–531 [DOI] [PubMed] [Google Scholar]
- 36. Letai A., Bassik M. C., Walensky L. D., Sorcinelli M. D., Weiler S., and Korsmeyer S. J. (2002) Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell 2, 183–192 [DOI] [PubMed] [Google Scholar]
- 37. Willis S. N., Fletcher J. I., Kaufmann T., van Delft M. F., Chen L., Czabotar P. E., Ierino H., Lee E. F., Fairlie W. D., Bouillet P., Strasser A., Kluck R. M., Adams J. M., and Huang D. C. (2007) Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science 315, 856–859 [DOI] [PubMed] [Google Scholar]
- 38. Greuber E. K., Smith-Pearson P., Wang J., and Pendergast A. M. (2013) Role of ABL family kinases in cancer: from leukaemia to solid tumours. Nat. Rev. Cancer 13, 559–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Opferman J. T., Iwasaki H., Ong C. C., Suh H., Mizuno S., Akashi K., and Korsmeyer S. J. (2005) Obligate role of anti-apoptotic MCL-1 in the survival of hematopoietic stem cells. Science 307, 1101–1104 [DOI] [PubMed] [Google Scholar]
- 40. Opferman J. T., Letai A., Beard C., Sorcinelli M. D., Ong C. C., and Korsmeyer S. J. (2003) Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature 426, 671–676 [DOI] [PubMed] [Google Scholar]
- 41. Chao J. R., Wang J. M., Lee S. F., Peng H. W., Lin Y. H., Chou C. H., Li J. C., Huang H. M., Chou C. K., Kuo M. L., Yen J. J., and Yang-Yen H. F. (1998) mcl-1 is an immediate-early gene activated by the granulocyte-macrophage colony-stimulating factor (GM-CSF) signaling pathway and is one component of the GM-CSF viability response. Mol. Cell. Biol. 18, 4883–4898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Melo J. V. (1996) The diversity of BCR-ABL fusion proteins and their relationship to leukemia phenotype. Blood 88, 2375–2384 [PubMed] [Google Scholar]
- 43. Koss B., Morrison J., Perciavalle R. M., Singh H., Rehg J. E., Williams R. T., and Opferman J. T. (2013) Requirement for antiapoptotic MCL-1 in the survival of BCR-ABL B-lineage acute lymphoblastic leukemia. Blood 122, 1587–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Williams R. T., Roussel M. F., and Sherr C. J. (2006) Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. U.S.A. 103, 6688–6693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bewry N. N., Nair R. R., Emmons M. F., Boulware D., Pinilla-Ibarz J., and Hazlehurst L. A. (2008) Stat3 contributes to resistance toward BCR-ABL inhibitors in a bone marrow microenvironment model of drug resistance. Mol. Cancer Ther. 7, 3169–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Eiring A. M., Page B. D., Kraft I. L., Mason C. C., Vellore N. A., Resetca D., Zabriskie M. S., Zhang T. Y., Khorashad J. S., Engar A. J., Reynolds K. R., Anderson D. J., Senina A., Pomicter A. D., Arpin C. C., et al. (2015) Combined STAT3 and BCR-ABL1 inhibition induces synthetic lethality in therapy-resistant chronic myeloid leukemia. Leukemia 29, 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Steelman L. S., Franklin R. A., Abrams S. L., Chappell W., Kempf C. R., Bäsecke J., Stivala F., Donia M., Fagone P., Nicoletti F., Libra M., Ruvolo P., Ruvolo V., Evangelisti C., Martelli A. M., and McCubrey J. A. (2011) Roles of the Ras/Raf/MEK/ERK pathway in leukemia therapy. Leukemia 25, 1080–1094 [DOI] [PubMed] [Google Scholar]
- 48. Korfi K., Smith M., Swan J., Somervaille T. C., Dhomen N., and Marais R. (2016) BIM mediates synergistic killing of B-cell acute lymphoblastic leukemia cells by BCL-2 and MEK inhibitors. Cell Death Dis. 7, e2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Clybouw C., Merino D., Nebl T., Masson F., Robati M., O'Reilly L., Hübner A., Davis R. J., Strasser A., and Bouillet P. (2012) Alternative splicing of Bim and Erk-mediated Bim(EL) phosphorylation are dispensable for hematopoietic homeostasis in vivo. Cell Death Differ. 19, 1060–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van Delft M. F., Wei A. H., Mason K. D., Vandenberg C. J., Chen L., Czabotar P. E., Willis S. N., Scott C. L., Day C. L., Cory S., Adams J. M., Roberts A. W., and Huang D. C. (2006) The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell 10, 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Scherr M., Elder A., Battmer K., Barzan D., Bomken S., Ricke-Hoch M., Schröder A., Venturini L., Blair H. J., Vormoor J., Ottmann O., Ganser A., Pich A., Hilfiker-Kleiner D., Heidenreich O., and Eder M. (2014) Differential expression of miR-17∼92 identifies BCL2 as a therapeutic target in BCR-ABL-positive B-lineage acute lymphoblastic leukemia. Leukemia 28, 554–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jones L., Carol H., Evans K., Richmond J., Houghton P. J., Smith M. A., and Lock R. B. (2016) A review of new agents evaluated against pediatric acute lymphoblastic leukemia by the Pediatric Preclinical Testing Program. Leukemia 30, 2133–2141 [DOI] [PubMed] [Google Scholar]
- 53. Leonard J. T., Rowley J. S., Eide C. A., Traer E., Hayes-Lattin B., Loriaux M., Spurgeon S. E., Druker B. J., Tyner J. W., and Chang B. H. (2016) Targeting BCL-2 and ABL/LYN in Philadelphia chromosome-positive acute lymphoblastic leukemia. Sci. Transl. Med. 8, 354ra114. [DOI] [PubMed] [Google Scholar]
- 54. Irving J., Matheson E., Minto L., Blair H., Case M., Halsey C., Swidenbank I., Ponthan F., Kirschner-Schwabe R., Groeneveld-Krentz S., Hof J., Allan J., Harrison C., Vormoor J., von Stackelberg A., and Eckert C. (2014) Ras pathway mutations are prevalent in relapsed childhood acute lymphoblastic leukemia and confer sensitivity to MEK inhibition. Blood 124, 3420–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mullighan C. G., Collins-Underwood J. R., Phillips L. A., Loudin M. G., Liu W., Zhang J., Ma J., Coustan-Smith E., Harvey R. C., Willman C. L., Mikhail F. M., Meyer J., Carroll A. J., Williams R. T., Cheng J., et al. (2009) Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat. Genet. 41, 1243–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Quintas-Cardama A., Kantarjian H., Cortes J., and Verstovsek S. (2011) Janus kinase inhibitors for the treatment of myeloproliferative neoplasias and beyond. Nat. Rev. Drug Discov. 10, 318–318 [DOI] [PubMed] [Google Scholar]
- 57. Zeuner A., Pedini F., Francescangeli F., Signore M., Girelli G., Tafuri A., and De Maria R. (2009) Activity of the BH3 mimetic ABT-737 on polycythemia vera erythroid precursor cells. Blood 113, 1522–1525 [DOI] [PubMed] [Google Scholar]
- 58. Rubert J., Qian Z., Andraos R., Guthy D. A., and Radimerski T. (2011) Bim and Mcl-1 exert key roles in regulating JAK2(V617F) cell survival. BMC Cancer 11, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Guo J., Roberts L., Chen Z., Merta P. J., Glaser K. B., and Shah O. J. (2015) JAK2(V617F) Drives Mcl-1 expression and sensitizes Hematologic Cell lines to dual inhibition of JAK2 and Bcl-xL. PLoS One 10, e0114363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schinnerl D., Fortschegger K., Kauer M., Marchante J. R., Kofler R., Den Boer M. L., and Strehl S. (2015) The role of the Janus-faced transcription factor PAX5-JAK2 in acute lymphoblastic leukemia. Blood 125, 1282–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Suryani S., Bracken L. S., Harvey R. C., Sia K. C., Carol H., Chen I. M., Evans K., Dietrich P. A., Roberts K. G., Kurmasheva R. T., Billups C. A., Mullighan C. G., Willman C. L., Loh M. L., Hunger S. P., et al. (2015) Evaluation of the in vitro and in vivo efficacy of the JAK inhibitor AZD1480 against JAK-mutated acute lymphoblastic leukemia. Mol. Cancer Ther. 14, 364–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Waibel M., Solomon V. S., Knight D. A., Ralli R. A., Kim S. K., Banks K. M., Vidacs E., Virely C., Sia K. C., Bracken L. S., Collins-Underwood R., Drenberg C., Ramsey L. B., Meyer S. C., Takiguchi M., et al. (2013) Combined targeting of JAK2 and Bcl-2/Bcl-xL to cure mutant JAK2-driven malignancies and overcome acquired resistance to JAK2 inhibitors. Cell Rep. 5, 1047–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cuesta-Domínguez Á., Ortega M., Ormazábal C., Santos-Roncero M., Galán-Díez M., Steegmann J. L., Figuera Á., Arranz E., Vizmanos J. L., Bueren J. A., Río P., and Fernández-Ruiz E. (2012) Transforming and tumorigenic activity of JAK2 by fusion to BCR: molecular mechanisms of action of a novel BCR-JAK2 tyrosine-kinase. PLoS One 7, e32451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rubnitz J. E., Wichlan D., Devidas M., Shuster J., Linda S. B., Kurtzberg J., Bell B., Hunger S. P., Chauvenet A., Pui C. H., Camitta B., Pullen J., and Children's Oncology Group. (2008) Prospective analysis of TEL gene rearrangements in childhood acute lymphoblastic leukemia: a Children's Oncology Group study. J. Clin. Oncol. 26, 2186–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fischer M., Schwieger M., Horn S., Niebuhr B., Ford A., Roscher S., Bergholz U., Greaves M., Löhler J., and Stocking C. (2005) Defining the oncogenic function of the TEL/AML1 (ETV6/RUNX1) fusion protein in a mouse model. Oncogene 24, 7579–7591 [DOI] [PubMed] [Google Scholar]
- 66. Zaliova M., Madzo J., Cario G., and Trka J. (2011) Revealing the role of TEL/AML1 for leukemic cell survival by RNAi-mediated silencing. Leukemia 25, 313–320 [DOI] [PubMed] [Google Scholar]
- 67. Fine B. M., Stanulla M., Schrappe M., Ho M., Viehmann S., Harbott J., and Boxer L. M. (2004) Gene expression patterns associated with recurrent chromosomal translocations in acute lymphoblastic leukemia. Blood 103, 1043–1049 [DOI] [PubMed] [Google Scholar]
- 68. Inthal A., Krapf G., Beck D., Joas R., Kauer M. O., Orel L., Fuka G., Mann G., and Panzer-Grümayer E. R. (2008) Role of the erythropoietin receptor in ETV6/RUNX1-positive acute lymphoblastic leukemia. Clin. Cancer Res. 14, 7196–7204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Szenajch J., Wcislo G., Jeong J. Y., Szczylik C., and Feldman L. (2010) The role of erythropoietin and its receptor in growth, survival and therapeutic response of human tumor cells from clinic to bench–a critical review. Biochim. Biophys. Acta 1806, 82–95 [DOI] [PubMed] [Google Scholar]
- 70. Torrano V., Procter J., Cardus P., Greaves M., and Ford A. M. (2011) ETV6-RUNX1 promotes survival of early B lineage progenitor cells via a dysregulated erythropoietin receptor. Blood 118, 4910–4918 [DOI] [PubMed] [Google Scholar]
- 71. Mangolini M., de Boer J., Walf-Vorderwülbecke V., Pieters R., den Boer M. L., and Williams O. (2013) STAT3 mediates oncogenic addiction to TEL-AML1 in t(12;21) acute lymphoblastic leukemia. Blood 122, 542–549 [DOI] [PubMed] [Google Scholar]
- 72. Strasser A., Harris A. W., Bath M. L., and Cory S. (1990) Novel primitive lymphoid tumors induced in transgenic mice by cooperation between Myc and Bcl-2. Nature 348, 331–333 [DOI] [PubMed] [Google Scholar]
- 73. Fanidi A., Harrington E. A., and Evan G. I. (1992) Cooperative interaction between C-Myc and Bcl-2 protooncogenes. Nature 359, 554–556 [DOI] [PubMed] [Google Scholar]
- 74. Dreyer Z. E., Hilden J. M., Jones T. L., Devidas M., Winick N. J., Willman C. L., Harvey R. C., Chen I. M., Behm F. G., Pullen J., Wood B. L., Carroll A. J., Heerema N. A., Felix C. A., Robinson B., et al. (2015) Intensified chemotherapy without Sct in infant all: results from COG P9407 (Cohort 3). Pediatr. Blood Cancer 62, 419–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pieters R., Schrappe M., De Lorenzo P., Hann I., De Rossi G., Felice M., Hovi L., LeBlanc T., Szczepanski T., Ferster A., Janka G., Rubnitz J., Silverman L., Stary J., Campbell M., et al. (2007) A treatment protocol for infants younger than 1 year with acute lymphoblastic leukaemia (Interfant-99): an observational study and a multicentre randomised trial. Lancet 370, 240–250 [DOI] [PubMed] [Google Scholar]
- 76. Chessells J. M., Harrison C. J., Kempski H., Webb D. K., Wheatley K., Hann I. M., Stevens R. F., Harrison G., Gibson B. E., and MRC Childhood Leukaemia Working Party. (2002) Clinical features, cytogenetics and outcome in acute lymphoblastic and myeloid leukaemia of infancy: report from the MRC Childhood Leukaemia Working Party. Leukemia 16, 776–784 [DOI] [PubMed] [Google Scholar]
- 77. Yokoyama A., Lin M., Naresh A., Kitabayashi I., and Cleary M. L. (2010) A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell 17, 198–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Muntean A. G., and Hess J. L. (2009) Epigenetic dysregulation in cancer. Am. J. Pathol. 175, 1353–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Andersson A. K., Ma J., Wang J., Chen X., Gedman A. L., Dang J., Nakitandwe J., Holmfeldt L., Parker M., Easton J., Huether R., Kriwacki R., Rusch M., Wu G., Li Y., et al. (2015) The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat. Genet. 47, 330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Andersson A. K., Ma J., Wang J. M., Chen X., Rusch M., Wu G., Easton J., Parker M., Raimondi S. C., Holmfeldt L., Gedman A. L., Song G. C., Becksfort J., Gupta P., Ulyanov A., Payne-Turner D., et al. (2011) Whole genome sequence analysis of 22 MLL rearranged infant acute lymphoblastic leukemias reveals remarkably few somatic mutations: a report from the St Jude Children's Research Hospital–Washington University Pediatric Cancer Genome Project. Blood 118, 33–34 [Google Scholar]
- 81. Kersey J. H., Wang D., and Oberto M. (1998) Resistance of t(4;11) (MLL-AF4 fusion gene) leukemias to stress-induced cell death: possible mechanism for extensive extramedullary accumulation of cells and poor prognosis. Leukemia 12, 1561–1564 [DOI] [PubMed] [Google Scholar]
- 82. Robinson B. W., Behling K. C., Gupta M., Zhang A. Y., Moore J. S., Bantly A. D., Willman C. L., Carroll A. J., Adamson P. C., Barrett J. S., and Felix C. A. (2008) Abundant anti-apoptotic BCL-2 is a molecular target in leukaemias with t(4;11) translocation. Br. J. Haematol. 141, 827–839 [DOI] [PubMed] [Google Scholar]
- 83. Brumatti G., Salmanidis M., Kok C. H., Bilardi R. A., Sandow J. J., Silke N., Mason K., Visser J., Jabbour A. M., Glaser S. P., Okamoto T., Bouillet P., D'Andrea R. J., and Ekert P. G. (2013) HoxA9 regulated Bcl-2 expression mediates survival of myeloid progenitors and the severity of HoxA9-dependent leukemia. Oncotarget 4, 1933–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Benito J. M., Godfrey L., Kojima K., Hogdal L., Wunderlich M., Geng H. M., Marzo I., Harutyunyan K. G., Golfman L., North P., Kerry J., Ballabio E., Chonghaile T. N., Gonzalo O., Qiu Y., et al. (2015) MLL-rearranged acute lymphoblastic leukemias activate BCL-2 through H3K79 methylation And are Sensitive to the BCL-2-specific antagonist ABT-199. Cell Rep. 13, 2715–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Dördelmann M., Reiter A., Borkhardt A., Ludwig W. D., Götz N., Viehmann S., Gadner H., Riehm H., and Schrappe M. (1999) Prednisone response is the strongest predictor of treatment outcome in infant acute lymphoblastic leukemia. Blood 94, 1209–1217 [PubMed] [Google Scholar]
- 86. Bouillet P., Metcalf D., Huang D. C., Tarlinton D. M., Kay T. W., Köntgen F., Adams J. M., and Strasser A. (1999) Proapoptotic Bcl-2 relative bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 286, 1735–1738 [DOI] [PubMed] [Google Scholar]
- 87. Holleman A., Cheok M. H., den Boer M. L., Yang W., Veerman A. J., Kazemier K. M., Pei D., Cheng C., Pui C. H., Relling M. V., Janka-Schaub G. E., Pieters R., and Evans W. E. (2004) Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N. Engl. J. Med. 351, 533–542 [DOI] [PubMed] [Google Scholar]
- 88. Stam R. W., Den Boer M. L., Schneider P., de Boer J., Hagelstein J., Valsecchi M. G., de Lorenzo P., Sallan S. E., Brady H. J., Armstrong S. A., and Pieters R. (2010) Association of high-level MCL-1 expression with in vitro and in vivo prednisone resistance in MLL-rearranged infant acute lymphoblastic leukemia. Blood 115, 1018–1025 [DOI] [PubMed] [Google Scholar]
- 89. Bachmann P. S., Gorman R., Mackenzie K. L., Lutze-Mann L., and Lock R. B. (2005) Dexamethasone resistance in B-cell precursor childhood acute lymphoblastic leukemia occurs downstream of ligand-induced nuclear translocation of the glucocorticoid receptor. Blood 105, 2519–2526 [DOI] [PubMed] [Google Scholar]
- 90. Bachmann P. S., Piazza R. G., Janes M. E., Wong N. C., Davies C., Mogavero A., Bhadri V. A., Szymanska B., Geninson G., Magistroni V., Cazzaniga G., Biondi A., Miranda-Saavedra D., Göttgens B., Saffery R., et al. (2010) Epigenetic silencing of BIM in glucocorticoid poor-responsive pediatric acute lymphoblastic leukemia, and its reversal by histone deacetylase inhibition. Blood 116, 3013–3022 [DOI] [PubMed] [Google Scholar]
- 91. Lock R., Carol H., Houghton P. J., Morton C. L., Kolb E. A., Gorlick R., Reynolds C. P., Maris J. M., Keir S. T., Wu J., and Smith M. A. (2008) Initial testing (stage 1) of the BH3 mimetic ABT-263 by the pediatric preclinical testing program. Pediatr. Blood Cancer 50, 1181–1189 [DOI] [PubMed] [Google Scholar]
- 92. Jayanthan A., Incoronato A., Singh A., Blackmore C., Bernoux D., Lewis V., Stam R., Whitlock J. A., and Narendran A. (2011) Cytotoxicity, drug combinability, and biological correlates of ABT-737 against acute lymphoblastic leukemia cells with MLL rearrangement. Pediatr. Blood Cancer 56, 353–360 [DOI] [PubMed] [Google Scholar]
- 93. Khaw S. L., Suryani S., Evans K., Richmond J., Robbins A., Kurmasheva R. T., Billups C. A., Erickson S. W., Guo Y., Houghton P. J., Smith M. A., Carol H., Roberts A. W., Huang D. C., and Lock R. B. (2016) Venetoclax responses of pediatric ALL xenografts reveal sensitivity of MLL-rearranged leukemia. Blood 128, 1382–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hanna D., and Anderson M. A. (2016) A new approach to high risk pediatric acute lymphoblastic leukemia? Transl. Cancer Res. 1428–1432 [Google Scholar]
- 95. Chen C. W., and Armstrong S. A. (2015) Targeting DOT1L and HOX gene expression in MLL-rearranged leukemia and beyond. Exp. Hematol. 43, 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Daigle S. R., Olhava E. J., Therkelsen C. A., Basavapathruni A., Jin L., Boriack-Sjodin P. A., Allain C. J., Klaus C. R., Raimondi A., Scott M. P., Waters N. J., Chesworth R., Moyer M. P., Copeland R. A., Richon V. M., and Pollock R. M. (2013) Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood 122, 1017–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hitomi J., Christofferson D. E., Ng A., Yao J., Degterev A., Xavier R. J., and Yuan J. (2008) Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell 135, 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Deveraux Q. L., and Reed J. C. (1999) IAP family proteins–suppressors of apoptosis. Genes Dev. 13, 239–252 [DOI] [PubMed] [Google Scholar]
- 99. Vince J. E., Wong W. W., Khan N., Feltham R., Chau D., Ahmed A. U., Benetatos C. A., Chunduru S. K., Condon S. M., McKinlay M., Brink R., Leverkus M., Tergaonkar V., Schneider P., Callus B. A., et al. (2007) IAP antagonists target cIAP1 to induce TNF α-dependent apoptosis. Cell 131, 682–693 [DOI] [PubMed] [Google Scholar]
- 100. Richmond J., Robbins A., Evans K., Beck D., Kurmasheva R. T., Billups C. A., Carol H., Heatley S., Sutton R., Marshall G. M., White D., Pimanda J., Houghton P. J., Smith M. A., and Lock R. B. (2016) Acute sensitivity of Ph-like acute lymphoblastic leukemia to the SMAC-mimetic birinapant. Cancer Res. 76, 4579–4591 [DOI] [PMC free article] [PubMed] [Google Scholar]