Abstract
Synaptic vesicles (SVs) form distinct pools at synaptic terminals, and this well-regulated separation is necessary for normal neurotransmission. However, how the SV cluster, in particular synaptic compartments, maintains normal neurotransmitter release remains a mystery. The presynaptic protein Neurexin (NRX) plays a significant role in synaptic architecture and function, and some evidence suggests that NRX is associated with neurological disorders, including autism spectrum disorders. However, the role of NRX in SV clustering is unclear. Here, using the neuromuscular junction at the 2–3 instar stages of Drosophila larvae as a model and biochemical imaging and electrophysiology techniques, we demonstrate that Drosophila NRX (DNRX) plays critical roles in regulating synaptic terminal clustering and release of SVs. We found that DNRX controls the terminal clustering and release of SVs by stimulating presynaptic F-actin. Furthermore, our results indicate that DNRX functions through the scaffold protein Scribble and the GEF protein DPix to activate the small GTPase Ras-related C3 Botulinum toxin substrate 1 (Rac1). We observed a direct interaction between the C-terminal PDZ-binding motif of DNRX and the PDZ domains of Scribble and that Scribble bridges DNRX to DPix, forming a DNRX–Scribble–DPix complex that activates Rac1 and subsequently stimulates presynaptic F-actin assembly and SV clustering. Taken together, our work provides important insights into the function of DNRX in regulating SV clustering, which could help inform further research into pathological neurexin-mediated mechanisms in neurological disorders such as autism.
Keywords: actin, cell adhesion, Drosophila, synapse, vesicles
Introduction
Neurons are the basic unit of the nervous system, and they communicate with each other through synapses. After being captured by our sensory organs, neural signals pass between synapses in the form of neurotransmitters. As the vehicles of neurotransmitters, synaptic vesicles (SV)2 are essential for neurotransmission. SVs can be divided into three distinct pools according to their localization and function. The SVs adjacent to the active zone and ready to be released are referred to as the ready release pool. The second pool of vesicles is the exo/endo-cycling pool, which is also found close to release sites and supplies the ready release pool. Finally, the pool located away from the active zone, and that contains the majority of SVs, is referred to as the reserve pool and is considered to be a storage pool (1, 2). Actin is a major part of the cytoskeleton that is required for maintaining the architecture of synapses as well as contributing to their function (3, 4), and a significant amount of evidence has shown that the localization, translocation, and release of SVs can be altered by disturbing the polymerization of pre-synaptic actin (5, 6).
The synapse is a highly specialized structure, and synaptogenesis is a highly complex process. Previous studies have shown that synaptic adhesive molecules play important roles in synaptogenesis and neurotransmission (7), and a number of synaptic cell adhesion molecules, including Neuroligins and Neurexins have been identified over the past few decades. Neurexin was first recognized as a receptor for α-latrotoxin, a black widow spider venom component that triggers massive neurotransmitter release (8). There are three neurexin genes in mammals, each of which has two promoters generating α-Neurexin and β-Neurexins, whereas there is only one neurexin-1 gene in Drosophila (dnrx) (9). Recent studies both in Drosophila and mammals showed that Neurexin plays a significant role in synaptic architecture and function, and there is evidence suggesting that neurexin is associated with autism spectrum disorders (ASDs) (10, 11). The complexity and redundancy of the neurexin genes in mammals motivated us to focus on simpler model systems, such as Drosophila, to investigate the in vivo function of DNRX.
Neurexin has been shown to bind to several molecules, including the presynaptic scaffolding proteins Mint (12), CASK (13, 14), and LRRTM2 (15, 16). Recently, DNRX also has been demonstrated to interact with the N-ethylmaleimide sensitive factor to regulate short-term synaptic depression and to interact with Spinophilin to maintain active zone architecture (17, 18). A typical trans-synaptic complex is formed by the heterophilic interaction of presynaptic Neurexins and postsynaptic Neuroligins (19, 20), and these complexes have attracted much attention as scaffolding complexes that not only maintain the normal structure of the synapse but also function in passing signals across the synapses. It is thus clear that Neurexin is a multifunctional molecule. Despite the identification and characterization of these proteins that functionally associated with DNRX, our understanding of the pathways including DNRX that control synaptic function are still incomplete, with other partners and mechanisms yet to be uncovered and analyzed.
In this study, we have investigated the role of DNRX in the cluster and release of SVs at synaptic terminals. We demonstrate that the effect of DNRX is mediated by presynaptic F-actin and there is a direct interaction between the C-terminal PDZ-binding motif of DNRX and the PDZ domains of the tumor suppressor protein Scribble. Furthermore, we find that Scribble bridges DNRX to DPix, forming a DNRX–Scribble–DPix complex to activate Rac1 and affect presynaptic F-actin assembly and SV clustering. Taken together, these studies provide novel insight into the mechanisms underlying the regulation of neurotransmitter release by DNRX.
Results
The clustering and release of SVs at the synaptic terminal is altered in dnrx mutants
To better understand the role of Neurexin at the synapse, we investigated whether there are some additional defects after ablating dnrx. Previous studies both in mammals and Drosophila have shown that Neurexin affects neurotransmitter release, and that the excitatory junction potential (EJP) amplitude and miniature EJP (mEJP) frequency are altered in the neurexin mutant (21, 22). Therefore, we examined whether the localization and membrane fusion of SVs would be affected in a dnrx mutant. As a marker of synaptic activity, we used Synaptotagmin (SYT), which is a synaptic vesicle membrane protein and calcium sensor that is efficiently transported to presynaptic terminals and is expressed in specific regions of the synapse (23), to indicate SV.
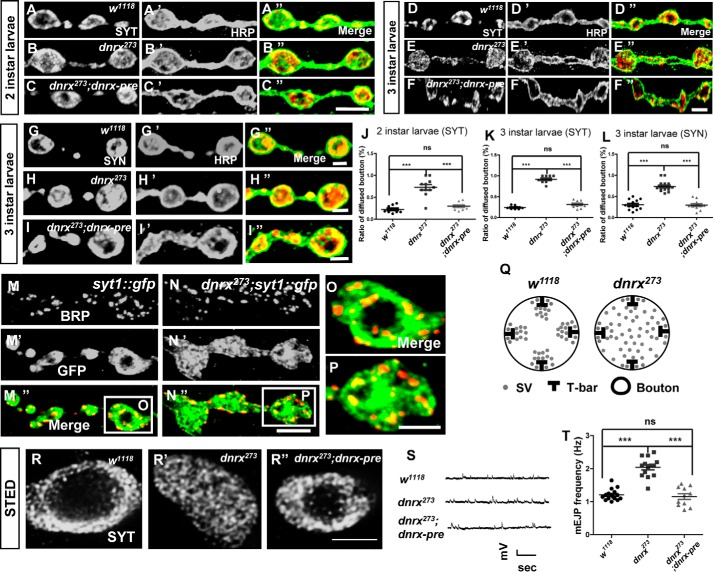
To assess the defects of SVs in dnrx mutants, we observed the synaptic terminal SV distribution of Drosophila melanogaster neuromuscular junction (NMJ) at different development stages of larvae from 2 to 3 instar. We found that loss of DNRX resulted in an ∼200% increase in the ratio of SYT-diffused boutons when compared with controls. Just as we expected, this defect could be fully rescued by overexpressing full-length dnrx cDNA within the pre-synapse in the dnrx mutant at 2 instar larvae (Fig. 1, A–C″ and J). When it turned to 3 instar larvae we got the same result for losing of DNRX resulted in an ∼300% increase in the ratio of SYT-diffused boutons when compared with controls (Fig. 1, D–F″ and K). Synapsin (SYN) is also a SV protein, which is localized at the SV membrane and can be used as another SV marker. We observed that the distribution of synaptic terminal SVs is diffused in dnrx mutant (Fig. 1, G–I″ and L). To be able to show the relative position of active zone (AZ) and SVs at the same time, we employed the syt::gfp knock-in fly with an integrated dnrx mutant background. Confocal images of third instar larvae NMJ boutons double labeled with active zone protein Bruchpilot (BRP) and GFP showed that loss of DNRX disrupted the distribution of terminal SVs (Fig. 1, M–P). The representative diagrammatic sketch illustrates the distribution of SVs in the dnrx mutant (Fig. 1Q). To make our measurements more precise, we turned to stimulated emission depletion (STED) microscopy to visualize the terminal distribution of SVs, and we observed the same phenotype (Fig. 1, R–R″). To determine whether the number of SVs and synapses were changed in dnrx mutants, we detected the number and density of AZ in wild-type and dnrx mutant and the immunostaining results showed that the number of AZ of single bouton is increased, whereas the AZ density of single bouton is decreased in dnrx mutant (supplemental Fig. S1, A–D). Moreover, we analyzed the total protein levels of SYN and BRP in adult heads with antibodies, and there were no differences between dnrx mutants and wild-type flies (supplemental Fig. S2, A–C).
Figure 1.
DNRX is necessary for synaptic terminal aggregation and release of SVs. A–C″, synaptic boutons of wild-type (A–A″), dnrx mutant (B–B″), and pre-synaptic rescue (C–C″) at two instar larvae stage double stained for SYT (red) and HRP (green), which labels the pre-synaptic SVs and neuronal membrane, respectively. D–F″, synaptic boutons of wild-type (D–D″), dnrx mutant (E–E″), and pre-synaptic rescue (F–F″) at third instar larvae stage double stained for SYT (red) and HRP (green), which labels the pre-synaptic SVs and neuronal membrane, respectively. G–I″, synaptic boutons of wild-type (G–G″), dnrx mutant (H–H″), and pre-synaptic rescue (I–I″) at third instar larvae stage double stained for SYN (red) and HRP (green), which labels the pre-synaptic SVs and neuronal membrane, respectively. J, quantification of the SYT-diffused boutons ratio at two instar larvae stage in muscle 4 shows that dnrx mutants have disturbed the distribution of SVs and this phenotype can be rescued by pre-synaptic DNRX. K, quantification of the SYT-diffused boutons ratio at the third instar larvae stage in muscle 4 shows that dnrx mutants have disturbed the distribution of SVs and this phenotype can be rescued by pre-synaptic DNRX. L, quantification of the SYN-diffused boutons ratio at third instar larvae stage in muscle 4 shows that dnrx mutants have disturbed the distribution of SVs and this phenotype can be rescued by pre-synaptic DNRX. M–N″, confocal images of third instar larvae NMJ boutons double labeled with anti-BRP (red) and anti-GFP (green) in syt::gfp (M–M″), and dnrx273,syt::gfp mutants (N–N″) showing that loss of DNRX disrupts the distribution of terminal SVs. O–P, amplified confocal images of single third instar larvae NMJ bouton double labeled with anti-Brp (red) and anti-GFP (green) in syt::gfp (O), and dnrx273,syt::gfp (P) showing the dispersed distribution of SVs in dnrx mutants. Q, representative diagrammatic sketch of SVs in single bouton of wild-type and dnrx mutant. R–R″, STED images of third instar larvae single bouton of wild-type (R), dnrx mutant (R′), and pre-synaptic rescue (R″) labeled with SYT, showing the SVs were diffused in dnrx mutant and can be rescued by pre-synaptic DNRX. S, representative traces of spontaneous responses of indicated genotypes. T, quantification of mEJP frequency of the indicated genotypes, showing that the mEJP frequency was increased in dnrx mutant and the DNRX could fully rescue the defects at pre-synapse. Data are mean ± S.E. ***, p < 0.001; **, p < 0.01; and *, p < 0.05. ns, not significant. Two-tailed Student's t tests were used to compare genotypes. Scale bar, 5 (A–C″), 5 (D–F″), 2 (G–G″), 2 (H–H″), 2 (I–I″), 5 (M–N″), 2.5 (O–P), and 2.5 μm (R–R″).
The defect of SV localization was reminiscent of SV release. Thus, we also explored whether the release of SVs was altered in dnrx mutants. We observed profound abnormalities in the spontaneous release of SVs in the dnrx mutant and found that it can be restored to a normal level after inducing full-length DNRX within the pre-synapse in the dnrx mutant (Fig. 1, S–T). Collectively, these results suggest that DNRX is essential for the proper aggregation and release of SVs at synaptic terminals.
The effects of DNRX on SV aggregation and release depend on presynaptic F-actin
Studies in mammals have suggested that cell adhesion complexes can localize presynaptic SVs by regulating local actin polymerization (24). Actin is one of the most prominent cytoskeletal molecules at synapses, and it is especially abundant at presynaptic terminals and postsynaptic dendritic spines (25). Moreover, pharmacological studies revealed a role for subsynaptic distribution of F-actin in SV mobilization and exocytosis (26). Studies in mammals have reported that treatment with latrunculin-A (an actin monomer sequestering agent) completely disrupts the actin arrangement and interferes with SV dynamics (4, 26). Conspicuously, actin participates in a regulatory mechanism at the synaptic terminals that inhibits the fusion of SVs at the active zone (26, 27). These observations indicate that DNRX might affect the distribution and release of SVs by regulating the presynaptic actin arrangement.
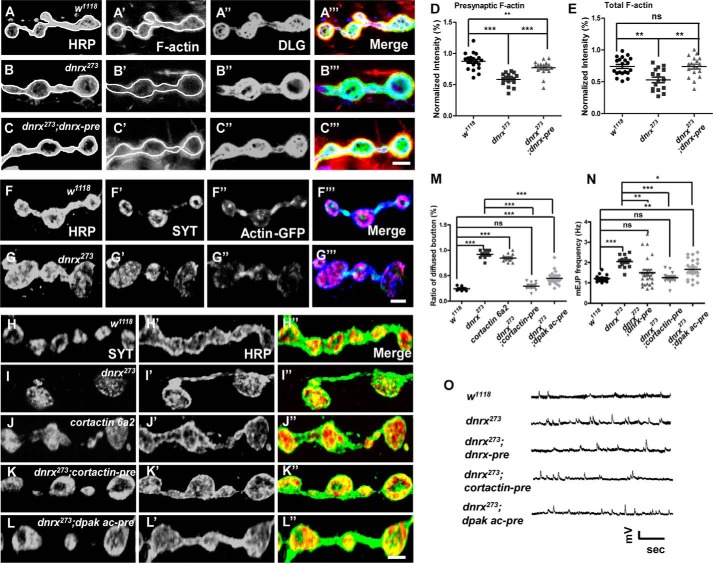
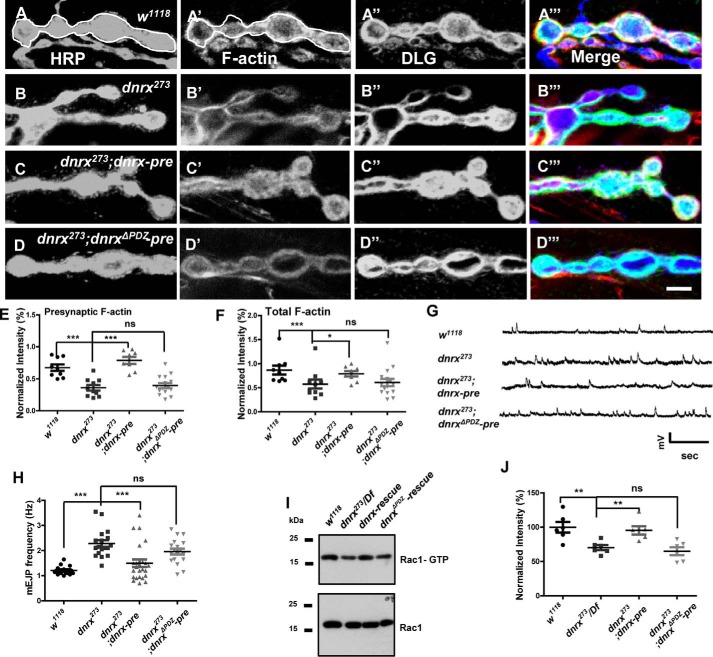
To determine whether the impact of DNRX on SV localization depends on presynaptic F-actin, we used Texas Red-conjugated phalloidin and fluorescently tagged actin (GFP-actin) to determine the distribution and amount of F-actin. We found that the fluorescence intensity of F-actin was significantly reduced in dnrx mutants and that this defect could be restored to normal levels after introducing full-length DNRX at the pre-synapse (Fig. 2, A–C‴). Furthermore, we measured the fluorescence intensity of F-actin in relationship to neuronal plasma membrane (HRP) and Discs large (DLG), which can be used to reflect the pre-synaptic and total F-actin, respectively. We found that both pre-synaptic and total F-actin were decreased in the dnrx mutant and could be rescued by introducing full-length DNRX (Fig. 2, D and E). GFP-tagged actin has previously been used to observe the localization of actin at synaptic terminals, and previous research has shown that these bright spots most likely correspond to F-actin (28). When we focus on the distribution of presynaptic F-actin, GFP-tagged actin has been confirmed to be particularly useful. We found that the presynaptic bright spots decreased in the dnrx mutant compared with wild-type (Fig. 2, F and G‴), suggesting that DNRX positively regulates F-actin recruitment to the synapse.
Figure 2.
DNRX regulates SV clustering and release dependent on presynaptic F-actin. A–C‴, confocal images of third instar larvae NMJ type Ib boutons at muscles 12/13 triple labeled with Texas Red phalloidin (red), anti-DLG (green), and anti-HRP (blue) in wild-type (A), dnrx mutants (B), ok6>dnrx rescue (C). The phalloidin signal contained in white circles correspond to HRP, largely reflecting the presynaptic F-actin. D and E, summary graph of relative fluorescence intensity of F-actin correspond to HRP (D) and DLG (E) showing that the F-actin fluorescence intensities were significantly reduced in dnrx mutant and can be restored to normal levels. F–G‴, representative confocal images of third instar larvae NMJ labeled with anti-HRP (blue), anti-SYT (red), and anti-GFP (green) in wild-type and dnrx mutant, respectively. The light green spot represents the pre-synaptic F-actin decreased in the dnrx mutant. H–L″, synaptic boutons of wild-type (H–H″), dnrx mutant (I–I″), cortactin mutant (J–J″), and pre-synaptic overexpress Cortactin (K–K″), and the active form of DPak (L–L″) in the dnrx mutant double stained for SYT (red) and HRP (green), which labels pre-synaptic SVs and neuronal membrane, respectively. M, quantification of SYT-diffused bouton ratio shows presynaptic branched F-actin can fully rescue the diffused distribution of SVs in dnrx mutant. N, quantification of mEJP frequency of the indicated genotypes, showing that mEJP frequency was increased in dnrx mutant and the Cortactin, the active form of DPak could rescue the defects at pre-synapse. O, representative traces of spontaneous responses of the indicated genotypes. Data are mean ± S.E. ***, p < 0.001; **, p < 0.01; and *, p < 0.05. ns, not significant. Two-tailed Student's t tests were used to compare genotypes. Scale bar, 5 (A–C‴), 5 (F–G‴), and 5 μm (H–L″).
To investigate the role of pre-synaptic F-actin in DNRX-regulated clustering of SVs at synaptic terminals, we adjusted the polymerization of actin at the pre-synapse by controlling the expression of Cortactin and the active form of DPak (Drosophila Pak). Cortactin is an actin-associated protein that promotes the polymerization of actin by stabilizing Arp2/3 complexes (29), whereas Pak regulates G-actin nucleation through Limk/Coffilin signaling (30). Our results suggested that the defects in SV clustering at presynaptic terminals were rescued by expressing Cortactin and the activated form of DPak in neurons using motor neuron-specific Gal4 (OK6-Gal4) (Fig. 2, H–L″ and M), although the rescue extent was different. It may involve distinct F-actin-regulating forms by these two proteins.
To further test the mechanism through which DNRX modulates the spontaneous release of SVs, we attempted to rescue the abnormality in the spontaneous release of SVs in the dnrx mutant. The result showed that Cortactin and activated DPak could rescue the abnormality (Fig. 2, N–O). In addition, we also measured the protein level of Cortactin and Arp2 in the dnrx mutant, and we saw no effect of DNRX on the amount of Arp2 or Cortactin (supplemental Fig. S2, A, D, and E). Taken together, these results suggest that the effect of DNRX on SV aggregation and release depends on presynaptic F-actin.
DNRX is co-localized with Scribble in the nervous system
Scribble is a cytoplasmic multimodular protein, and a previous study showed that mammalian Scribble regulates epithelial cell adhesion and migration (31). The Drosophila homologue of Scribble is located at the basolateral membrane and the septate junctions of epithelia (32). Recently, Scribble has been found to play essential roles in the nervous system (33, 34) and in active forgetting (35), a process in which dnrx was found to be involved (36). Loss of function of Scribble causes numerous defects in the nervous system, including alterations in synaptic architecture and function. DNRX is abundantly expressed in neurons in the central nervous system and plays important roles in associative learning in Drosophila (9, 20), and DNRX can also be detected, and it functions in the Drosophila NMJ (21, 37, 38). Thus we hypothesized that DNRX functions together with Scribble.
We used a GFP knock-in fly to mark endogenous Scribble, and to determine whether DNRX and Scribble are co-localized in the nervous system. At the embryonic stage, the immunostaining showed that DNRX was highly co-localized with Scribble in the developing nervous system (Fig. 3, A–A″). We also found that DNRX and Scribble were co-localized in the central nervous system at the third instar larval stage, and they were abundantly expressed and highly co-localized in the mushroom body. Moreover, they were co-localized in the ventral nerve cord with different expression levels (Fig. 3, B–B″). Previously, we found that DNRX could also be detected in a small subset of peripheral nervous system neurons (9, 38). Because of the low sensitivity of the DNRX antibody, we used motor neuron-specific Gal4 (OK6-Gal4) to drive DNRX expression and found that DNRX and Scribble were co-localized at NMJs of Drosophila (Fig. 3, C–C‴). Finally, we also observed a similar distribution of DNRX and Scribble in the heads of adult Drosophila, such as antennal lobe and mushroom body with different expression levels (Fig. 3, D–D″). However, in epithelial cells, we could only detect the signal from Scribble (Fig. 3, E–E‴). Collectively, these results show that DNRX and Scribble are specifically co-localized in the nervous system at all developmental stages of Drosophila.
Figure 3.
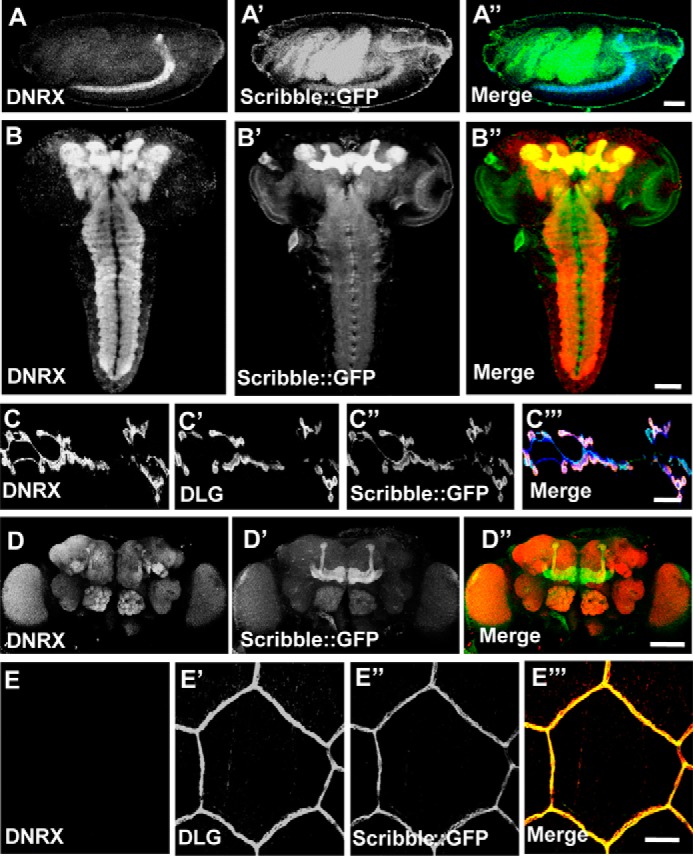
DNRX is co-localized with Scribble in the nervous system. A–A″, staining for scribble::gfp knock-in fly embryo with anti-DNRX (blue) and GFP is the spontaneous green fluorescence, showing that DNRX and Scribble are co-localized in the central nervous system. B–B″, third instar larvae brains of scribble::gfp staining with anti-DNRX (red) and GFP is the spontaneous green fluorescence, showing that DNRX and Scribble are co-localized in the central nervous system especially concentrated at mushroom body. C–C‴, confocal images of the third instar larvae neuromuscular junction at muscles 6/7 staining with anti-DNRX (blue), anti-DLG (red), and GFP is the spontaneous green fluorescence in scribble::gfp integrated with the DNRX overexpression fly, showing that DNRX and Scribble were co-localized in neuromuscular junction. D–D″, scribble::gfp knock-in fly adult brain staining with anti-DNRX (red) and GFP are the spontaneous green fluorescence, showing that DNRX and Scribble are co-localized in the central nervous system. E–E‴, epithelial cell staining with anti-DNRX, anti-DLG (red), and GFP is the spontaneous green fluorescence, showing that DNRX and Scribble are not co-localized at epithelial cells. Scale bar, 50 (A–A″), 50 (B–B″), 20 (C–C‴), 100 (D–D″), and 20 μm (E–E‴).
DNRX physically interacts with Scribble PDZ domains through C-terminal PDZ-binding motif
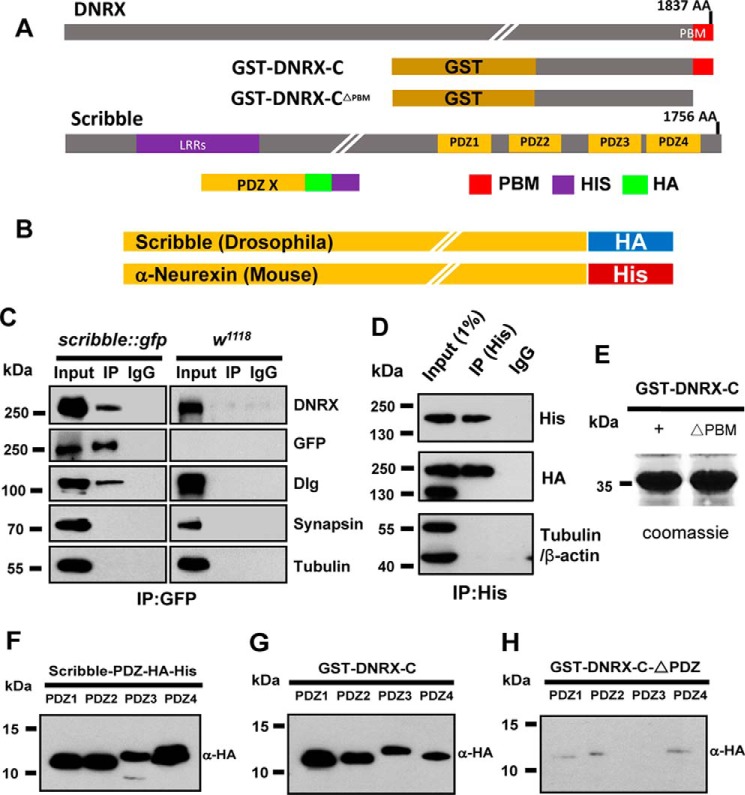
Previous studies have shown that Scribble belongs to the LAP protein family and consists of 16 leucine-rich repeats and 4 PDZ domains (Fig. 4A). The 4 PDZ domains have been shown to interact with several molecules that function both in neurons and epithelial cells. In mammals, it has been shown that Scribble can interact with the C-terminal PDZ-binding site of β-catenin to localize SVs to synapses through the PDZ domain (39). In addition, it has been shown that Scribble can form a complex with βPix to regulate actin arrangements and SV assembly (40). Recently, it has been found that NOS1AP associates with Scribble and regulates dendritic spine development (41).
Figure 4.
DNRX binds with the PDZ domains of Scribble via the C-terminal PDZ-binding motif. A, schematic representation of the protein structure of DNRX and Scribble. The polypeptide structure of the GST-fused DNRX C-terminal section with and without the C-terminal PDZ-binding motif and the PDZ domains of Scribble with HA and HIS tags. B, schematic graph of full-length HA-tagged Scribble of Drosophila and HIS-tagged α-Neurexin of mouse constructs. C, immunoprecipitated (IP) from whole Drosophila adult brain extracts with GFP nano-antibody fused beads in scribble::gfp and wild-type strains, respectively. Anti-GFP antibody precipitated the DNRX from the lysates, whereas the control IgG and wild-type sample did not. Suggesting DNRX physically interacts with Scribble in vivo. Input is 1%. D, Western blots of anti-His immunoprecipitates using protein lysate of HEK293 cells that can express the proteins of HA-Scribble and HIS-α-Neurexin, showing that HA-Scribble was coimmunoprecipitated with HIS-α-Neurexin in mouse. E, Coomassie Brilliant Blue for the purified DNRX C-terminal protein and deleted final 7-amino acid protein. F, Western blot validation for the purified 4 PDZ domain polypeptides of Scribble. G and H, GST pulldown with the DNRX C-terminal protein and deleted a very C-terminal 7-amino acid protein, respectively. Incubating with 4 PDZ domains of Scribble. Western blot results showed DNRX directly binds with all the PDZ domains of Scribble.
To test whether there is a physical interaction between DNRX and Scribble, we used lysates from adult heads for co-immunoprecipitation assay, and found that anti-GFP antibody was able to immunoprecipitate DNRX (Fig. 4C). This provided evidence that DNRX can form a complex with Scribble in vivo. To validate a conserved interaction between Scribble and Neurexin, we cotransfected murine α-Neurexin with a HIS tag and Drosophila Scribble with an HA tag (Fig. 4B) into HEK293 cells. After inducing and extracting these two proteins we incubated them together. Co-immunoprecipitation experiments showed that Drosophila Scribble physically interacts with murine α-Neurexin (Fig. 4D). Therefore, the physical interaction between Neurexin and Scribble in the central nervous system is conserved at least between Drosophila and mouse.
To further confirm the interaction between DNRX and Scribble, we performed a GST pulldown assay. We constructed the C-terminal portion of DNRX with or without the PDZ-binding motif (Fig. 4E), and we generated four polypeptides of each Scribble PDZ domain with HA and HIS tags (Fig. 4F). Our GST pulldown results showed that the full C-terminal portion of DNRX was able to efficiently pulldown each of the four Scribble PDZ domains (Fig. 4G). However, deletion of the very C-terminal seven amino acids containing the PDZ-binding motif of DNRX significantly reduced the affinity between DNRX and Scribble (Fig. 4H). These results strongly support the idea that the very C-terminal PDZ-binding motif of DNRX directly binds to the Scribble PDZ domains in vitro.
The interaction between DNRX and Scribble is required for the aggregation of SVs
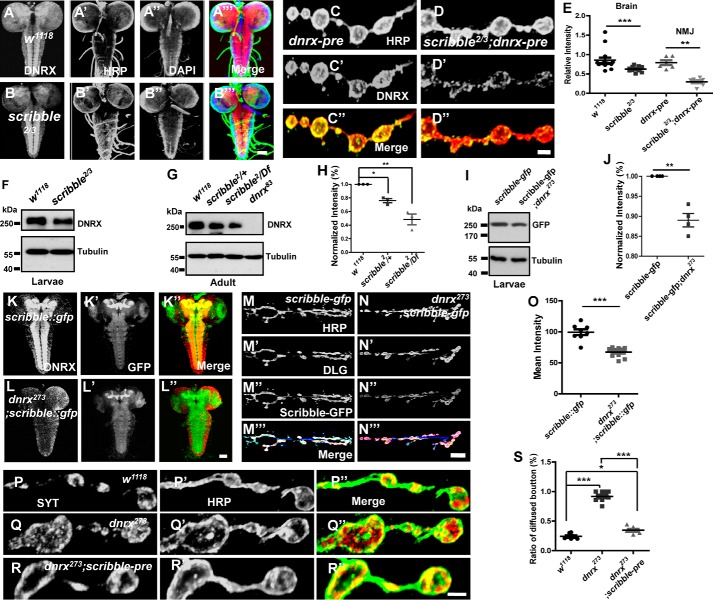
Because the data above showed that DNRX could directly bind to Scribble through the PDZ-binding motif, we hypothesized that a functional relationship exists between these two proteins. To test this, we measured the expression pattern of DNRX in scribble-null mutants. Surprisingly, the immunostaining results both in the brain and NMJ showed that the level of DNRX was significantly reduced compared with controls (Fig. 5, A–D″ and E), and Western blots confirmed that the protein level of DNRX was clearly decreased in scribble mutants compared with wild-type (Fig. 5, F–H) in a dose-dependent manner.
Figure 5.
DNRX interacts with Scribble and sustains the terminal SV aggregation. A–B‴, representative images of larvae brains of the indicated genotypes were stained with DNRX (red), HRP (green), and DAPI (blue), showing the level of DNRX was decreased in scribble mutant. C–D″, representative images of larvae NMJ of the indicated genotypes were stained with DNRX (green) and HRP (red), showing the level of DNRX was decreased in the scribble mutant. E, quantification of the fluorescence intensity for DNRX both in brain and NMJ of the indicated genotypes. F and G, Western blot analysis of protein lysates prepared from heads using anti-DNRX antibody showing that the level of DNRX was reduced in scribble mutant both in larvae (F) and adult (G) stages. H, quantitative analysis of data for Western blot analysis of the indicated genotypes for adult flies. I, Western blot analysis of protein lysates prepared from heads using anti-GFP antibody showing that the level of Scribble was reduced in the dnrx mutant in larvae stage. J, quantitative analysis for Western blots of the indicated genotypes. K–L″, representative images of larvae brain of the indicated genotypes stained with DNRX (red) and GFP (green), showing the level of Scribble was decreased in dnrx mutant. M–N‴, representative images of larvae NMJ of the indicated genotypes were stained with HRP (blue), DLG (red), and GFP (green), showing the level of Scribble was decreased in dnrx mutant. O, quantification of the fluorescence intensity for Scribble::GFP of the indicated genotypes. P–R″, synaptic boutons of wild-type (P–P″), dnrx mutant (Q–Q″), and pre-synaptic overexpress Scribble in dnrx mutant (R–R″) double stained for SYT (red) and HRP (green), which labels the pre-synaptic SVs and neuronal membrane, respectively. S, quantification of the ratio of SYT-diffused boutons in muscle 4 shows that dnrx mutants have disturbed the terminal cluster of SVs and this phenotype can be rescued by pre-synaptic Scribble. Data are mean ± S.E. ***, p < 0.001; **, p < 0.01; and *, p < 0.05. Two-tailed Student's t tests were used to compare genotypes. Scale bar, 50 (A–B‴), 5 (C–D″), 50 (K–L″), 20 (M–N‴), and 5 μm (P–R″).
We also sought to determine whether DNRX has an impact on the level and localization of Scribble at the larval NMJ. Because there are no antibodies available against Scribble, and Scribble is expressed at both the pre- and post-synapse (33, 41), we used the GFP knock-in fly to reflect the endogenous Scribble. We observed that the level of GFP fused Scribble in the dnrx mutant was decreased both in the central nervous system and NMJ when compared with controls (Fig. 5, I–O). We performed qRT-PCR to determine whether DNRX affects the transcription of Scribble, but we found no statistical difference in the mRNA level of Scribble between wild-type and dnrx mutant (supplemental Fig. S3A). Thus DNRX regulates Scribble mostly at the protein level and it is possible that DNRX and Scribble are stabilized by each other. Together, these results suggest that DNRX regulates the recruitment of Scribble to the synapse and that the levels of DNRX and Scribble in the synapse are stabilized by each other.
To better understand the effect of Scribble on SV distribution we assessed the terminal SV in scribble mutant and we found that SVs are diffused in bouton after ablating Scribble (supplemental Fig. S3, B–C″). To further determine the relationship between DNRX and Scribble, we next investigated the functional significance of Scribble in DNRX-dependent aggregation of SVs. We independently overexpressed Scribble in the dnrx mutant background and the immunostaining results showed that the ratio of SYT-diffused boutons was decreased when compared with the dnrx mutant and was nearly restored to wild-type level (Fig. 5, P–R″ and S). In summary, these results suggest that DNRX interacts with Scribble and that this complex is required for sustaining normal SV clustering.
DNRX interacts with DPix via Scribble and modulates the activity of Rac1 to sustain the aggregation and release of SVs
Mammalian Scribble forms a tight complex with βPix (40), which is a GEF that is involved in the pathways that regulate actin polymerization (42). Based on this, we hypothesized that DNRX regulates presynaptic F-actin through DPix. There is only one DPix in Drosophila, whereas two isoforms, αPix and βPix, are found in mammals (43). To reveal the biological role of DPix in DNRX-dependent SV distribution and release, we prepared GFP-fused DPix transgenic flies and overexpressed GFP-DPix in pan-neurons driven by Elav-Gal4. DNRX co-immunoprecipitated with DPix in the Drosophila central nervous system (Fig. 6A), and this suggested that Scribble might act as a bridge between DNRX and DPix. This hypothesis was supported by another co-immunoprecipitation experiment in which DNRX was not observed in the scribble mutants using a GFP antibody (Fig. 6B). Therefore, DNRX interacts with DPix through Scribble in vivo (Fig. 6C), and this indicates that DNRX might regulate presynaptic F-actin via the GEF (DPix).
Figure 6.
DNRX interacts with DPix through Scribble and regulates the activity of Rac1, and maintains the terminal SV aggregation and release. A, Western blots of anti-GFP immunoprecipitates using protein lysates of adult Drosophila heads showing immunoprecipitated proteins for DNRX and DPix in vivo. Input is 1%. B, Western blots of anti-GFP immunoprecipitates using protein lysate of adult Drosophila heads showing loss of Scribble, DNRX could not immunoprecipitate with DPix in vivo. Input is 1%. C, schematic representation of DNRX interacts with DPix through Scribble and activates Rac1. D, Western blots showing the level of Rac-GTP and total Rac1 in adult heads of wild-type, dnrx mutant. E, summary graph showing that the activated Rac1 was decreased after knocking out DNRX. F, Western blots showing the level of Rac-GTP and total Rac1 in adult heads of elav dcr/+, dnrx, and scribble knocking down flies. G, summary graph showing that the activated Rac1 was decreased after knocking down the expression of DNRX and Scribble using the RNAi method. H, summary graph showing that the mean fluorescence intensity of DPix-GFP was decreased both in NMJ and VNC in dnrx mutant when compared with wild-type controls. I and J″, representative images of larvae VNC of the indicated genotypes were stained with DNRX (red) and GFP (green), showing DNRX regulates the amount of DPix. K and L‴, representative images of larvae NMJ type Ib boutons of the indicated genotypes were stained with HRP (blue), DLG (red), and GFP (green), showing DNRX regulates the amount of DPix. M–O″, confocal images of third instar larvae NMJ type Ib boutons of the indicated genotypes double labeled with anti-SYT (red) and anti-HRP (green), which labels the pre-synaptic SVs and neuronal membrane, respectively. P, quantification of the ratio of the SYT-diffused boutons in muscle 4 shows that in dnrx mutants the distribution of SVs has been disturbed and this phenotype can be rescued by pre-synaptic DPix. Q, representative traces of spontaneous responses of the indicated genotypes. R, quantification of mEJP frequency of indicated genotypes, showing that mEJP frequency was increased in the dnrx mutant and the Scribble and DPix could rescue the defects at pre-synapse. Data are mean ± S.E., ***, p < 0.001; **, p < 0.01; and *, p < 0.05. ns, not significant. Two-tailed Student's t tests were used to compare genotypes. Scale bars, 50 (I–J″), 5 (K-L‴), and 5 μm (M–O″).
To further confirm the role of DNRX in presynaptic actin arrangement, we measured the level of activated Rac1, which has been shown to positively promote the polymerization of actin (44, 45). A previous study has shown that Rac1 is locally activated in dendritic spines and that this local activation of Rac1 is regulated by βPix, a Rac GEF (46). Our results showed that DPix associates with DNRX and Scribble. Therefore, we hypothesized that DNRX regulates the activity of Rac1 through its interactions with DPix and thus that the DNRX–Scribble–DPix complex promotes presynaptic actin polymerization. To directly test this, we used GST-fused Pak-PBD, a domain of Pak that can specifically bind to Rac1 and efficiently pulldown activated Rac1 (Rac1-GTP) after treating with GTPrs. Compared with wild-type, less Rac1-GTP was pulled down in the dnrx mutant (Fig. 6, D and E). In addition, reducing the expression of DNRX and Scribble in the central nervous system using a RNAi method inhibited the activity of Rac1 (Fig. 6, F and G). To further confirm whether DNRX regulates actin polymerization and SV release via activating Rac1, we got the dominant-negative form of Rac1 (Rac1DN) and integrated it into the dnrx mutant background. The immunostaining and electrophysiology results showed that Rac1DN could not rescue the defects of F-actin assembly and SV release in dnrx mutant (supplemental Fig. S5). Similarly, we measured another GTPase signaling protein, Cdc42, which can also regulate actin cytoskeleton like Rac1 (47, 48), however, the results showed that the level of activated Cdc42 has no significant difference between wild-type and dnrx mutant although it has a decreased tendency in dnrx mutant (supplemental Fig. S6).
To further determine whether DNRX can affect the distribution and level of pre-synaptic DPix, as mentioned above, the lack of DPix antibody and the fact that DPix is localized at both the pre- and postsynapse (49) forced us to use motor neuron-specific Gal4 (OK6-Gal4) to drive DPix in the dnrx mutant and wild-type background separately. Surprisingly, we found that the level of DPix was significantly decreased in the dnrx mutant compared with wild-type background both in the NMJ and VNC (Fig. 6, I–L‴), and the fluorescence intensity measurements validated this phenotype (Fig. 6H). To determine whether the level of DPix mRNA was also changed in the dnrx mutant, we performed qRT-PCR and found that the mRNA level of DPix had a downward tendency, but the differences compared with controls were not statistically significant (supplemental Fig. S3A). Collectively, these results highlight the essential role of DNRX in affecting GEF through Scribble and support the hypothesis that DNRX regulates presynaptic F-actin by activating Rac1.
To determine whether SV distribution is altered in dpix knockdown flies, we examined the terminal SVs in dpix RNAi driven by elav-gal4 and we found that SVs are diffused in bouton after knocking down the expression of DPix (supplemental Fig. S3, B–D″). To further confirm the functional significance of DPix in DNRX-dependent aggregation of SVs, we next independently overexpressed DPix in the dnrx mutant background and the immunostaining result showed that the ratio of SYT-diffused boutons could be restored to the wild-type level (Fig. 6, M–O″ and P). However, the defect in which SVs are retained in the axon in the dnrx mutant could not be rescued by Scribble–DPix (supplemental Fig. S4, A–D″). To better understand the effect of Scribble–DPix on DNRX, we assessed the spontaneous release of SVs and found that Scribble–DPix can rescue the abnormality in the dnrx mutant (Fig. 6, Q–R).
The PDZ-binding motif is essential for the effect of DNRX on F-actin assembly and SV release
To determine the specific motif of DNRX that might be involved in the regulation of F-actin and SV, we subsequently constructed the dnrxΔPDZ transgenic fly, which lacks the C-terminal 7 amino acid residues according to the above result. To determine whether the PDZ-binding motif of DNRX is required for F-actin assembly, we first examined the F-actin intensity in wild-type, dnrx mutant, dnrx rescue, and dnrxΔPDZ rescue lines. The immunostaining results showed that the fluorescence intensity of F-actin was significantly reduced in dnrx mutants and this defect could not be restored to normal levels after introducing DNRXΔPDZ at presynapse (Fig. 7, A–D‴). Furthermore, we measured the fluorescence intensity of F-actin in relationship to neuronal plasma membrane (HRP) and Discs large (DLG). The data showed that DNRXΔPDZ had no effect on the regulation of F-actin (Fig. 7, E and F). Next, to examine the association of the DNRX PDZ-binding motif with SV release, we performed electrophysiology experiments and the result also showed that DNRXΔPDZ could not rescue the increased frequency of SV release observed in the dnrx mutant (Fig. 7, G and H). Finally, to better understand the effect of DNRX on the activity of Rac1, we assessed the activated Rac1 level after inducing DNRXΔPDZ in the dnrx mutant background and we found that DNRXΔPDZ still could not rescue the defect of Rac1 activity (Fig. 7, I and J). Altogether, our data consistently indicate that the PDZ-binding motif of DNRX is essential for regulating F-actin assembly and SV release. Therefore, our results support a model that DNRX binds to DPix via Scribble and regulates F-actin polymerization by activating Rac1, and subsequently control SV clustering and release at synaptic terminals (Fig. 8).
Figure 7.
The PDZ-binding motif is essential for the effect of DNRX on F-actin assembly and SV release. A–D‴, representative images of third instar larvae NMJ type Ib boutons at muscles 12/13 triple labeled with Texas Red phalloidin (red), anti-DLG (green), and anti-HRP (blue) in wild-type (A), dnrx mutants (B), dnrx rescue (C), and dnrxΔPDZ rescue (D). The phalloidin signal contained in white circles corresponds to HRP, and largely reflects the presynaptic F-actin. E and F, summary graph of the relative fluorescence intensity of F-actin corresponding to HRP (E) and DLG (F) showing that F-actin fluorescence intensities were significantly reduced in dnrx mutant and could not be rescued by inducing DNRXΔPDZ at presynapse. G, representative traces of spontaneous responses of the indicated genotypes. H, quantification of mEJP frequency of the indicated genotypes, showing that mEJP frequency was increased in the dnrx mutant and that DNRXΔPDZ could not rescue the defects. I, Western blots showing the level of Rac-GTP and total Rac1 in adult heads of wild-type, dnrx mutant, dnrx rescue, and dnrxΔPDZ rescue. J, summary graph showing that the activated Rac1 was decreased after knocking out DNRX and this defect can be rescued by inducing the full-length of DNRX at presynapse, however, driving DNRXΔPDZ at presynapse in the dnrx mutant had no rescue effect. Data are mean ± S.E. ***, p < 0.001; **, p < 0.01; and *, p < 0.05. ns, not significant. Two-tailed Student's t tests were used to compare genotypes. Scale bar, 5 μm (A–D‴).
Figure 8.
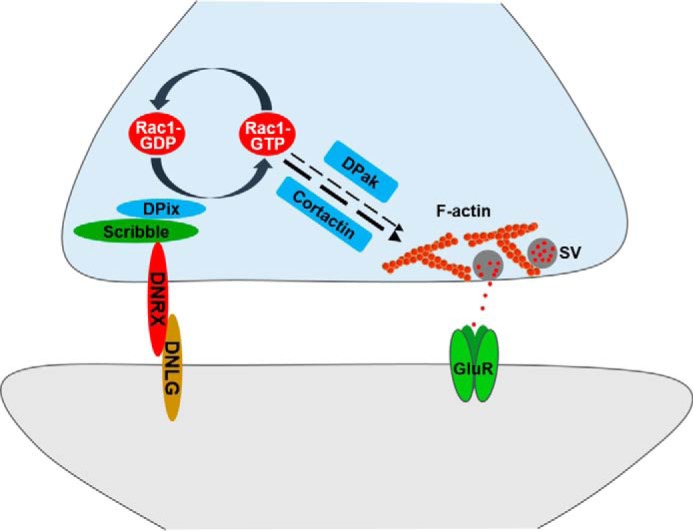
The model shows that DNRX regulates SV aggregation and release via presynaptic F-actin by modulating Scribble–DPix. Presynaptic DNRX interacts with Scribble–DPix to regulate the activity of Rac1, the activated Rac1 (Rac1-GTP) then affects actin polymerization to finally modulate the SVs localization and release.
Discussion
The brain is one of the most complicated organs and synapse is the functional unit of the brain. A large number of genes and proteins are involved in regulating the function of the brain. Mutations in synapse-associated genes cause neurologic disease. It would thus be desirable to explore the fundamental function of these genes in vivo and it may help attenuate the suffering of the patient. Considering the genetic techniques are available to manipulate, rapid life cycle, low cost, and a large number of genes are conserved between flies and vertebrates, it makes Drosophila emerging as an outstanding model to identify the biological basis for disease. Here, we use drosophila NMJ as a model to study and reveal the vital function of the cell adhesion molecule DNRX at synapse. Our study provides new mechanistic insights into the SV cluster and release regulation, which conceptually advances current knowledge of DNRX function at synapse.
DNRX and Scribble are essential for nervous system
Neurexin is a highly conserved cell adhesion molecule that is predominantly localized at the presynaptic terminal. Previous studies have shown that Neurexin plays a significant role in synaptic architecture and function. In addition, accumulating evidence has implicated Neurexin in ASDs. ASDs are neurodevelopmental disorders characterized by deficits in communication and social interaction as well as restricted interests and repetitive and stereotypic patterns of behavior (10, 20, 50, 51). However, the precise function and underlying molecular mechanisms of Neurexin in both normal physiology and ASDs remain unclear. Drosophila Scribble is a cytoplasmic scaffolding protein that was first recognized as a tumor suppressor that regulates epithelial cell adhesion and migration in mammals (31, 32). Recently, it has been shown that Scribble is also localized in the nervous system both in invertebrate and vertebrate animals (33, 41), and plays a role in synaptic plasticity and animal behavior, including learning, memory, social behavior, and olfactory behavior (52). However, how Scribble functions at the synapse remains unknown. In this study, we have revealed that DNRX interacts directly with the Scribble PDZ domains through the very C-terminal PDZ-binding motif to regulate presynaptic F-actin and SVs.
F-actin is highly enriched at synaptic terminals and is vital for SV traffic, localization, and release
For decades, F-actin emerged as the major cytoskeleton identified in presynaptic nerve terminals. Considering the especial location of F-actin at the internal space of presynaptic nerve terminals, it raised a hypothesis that F-actin regulates synaptic vesicle localization and release. Previous studies from several groups provided consistent evidence that after disrupting the polymerization of actin, pre-synapse affected SV traffic, localization, and neurotransmitter release (4, 5, 28). Moreover, cortical actin has been identified as a barrier for vesicles during the process of moving to the active zone in the presynaptic terminal (27). However, whether and how F-actin participates in the regulation of SV at synapse is still controversial. Therefore, further investigation will be necessary to determine the function of F-actin at synapse and especially for advances in our understanding of the relationship with SV. By now the function of F-actin at synapse is still poorly understood. To better understand the effect of F-actin on SV we assessed the cluster and release of SV after ablating the actin-associated genes and found the obvious defects of SV cluster and release. To further test this possibility, we need to perform additional experiments to elaborate the vital role of F-actin in SV regulation in future.
The function of DNRX in regulating SVs via presynaptic F-actin
SV distribution and dynamics are essential for normal neural signal transmission and for synaptic plasticity both at peripheral and central synapses (53). Disruptions in SV function will lead to various forms of neurological disorders. The process of synaptic vesicle priming, docking, and fusion with the presynaptic membrane has been investigated extensively, and numerous molecules involved in this process have been identified. These include synaptotagmins, synapsins, synaptobrevins, and Munc18. However, how SV cluster in particular compartments and their role in regulating neurotransmitter release at the presynaptic terminal are unclear. The results from this study provide compelling evidences that DNRX plays an essential role in the distribution and release of SVs.
In this study, our data suggest the amount of F-actin is significantly reduced. Moreover, presynaptic Cortactin or the active form of DPak, key regulators of the actin cytoskeleton, are able to rescue the defects in SV distribution and spontaneous release frequency in the dnrx mutant. These results illustrate the essential role of presynaptic actin in regulating SVs localization and release. Our results are consistent with the recent findings showing that Arp2/3 complex-mediated actin regulation is important for presynaptic neurotransmitter release (54).
Scribble is required for DNRX associates with DPix
Previous work has shown that Scribble is a scaffolding, tumor suppressor protein that through its PDZ domains interacts with a number of proteins, including β-catenin, βPix, and NOS1AP. Although NOS1AP can bind directly to the fourth Scribble PDZ domain (41), βPix has binding affinity to all four PDZ domains of Scribble (40). In the present study, we use co-immunoprecipitation to show that DNRX can form a complex with Scribble in the Drosophila nervous system in vivo. We also show that DNRX and Scribble are co-localized in both central and peripheral nervous system during embryonic, larval, and adult stages. Importantly, these two proteins are highly expressed in the mushroom body of Drosophila, suggesting a key role in learning and memory. We also show that DNRX can directly bind to all four Scribble PDZ domains through its C-terminal PDZ-binding motif, similar to what is observed for βPix. These interaction results suggest that there may be competitions and cross-effect between DNRX and βPix. These interactions may also relate to the mutual effect on the protein level of Scribble and DNRX. Because the mRNA level of Scribble is not altered in the dnrx mutant, it is possible that when the DNRX or Scribble was absent, the complex becomes destabilized and subsequently degraded. Further experiments are needed to address this possibility.
DNRX–Scribble–DPix regulates presynaptic F-actin by activating Rac1
What is the functional consequence of the Scribble and DNRX interaction? The present study provides evidence that Scribble may act as a bridge between DNRX and DPix to regulate the actin cytoskeleton. βPix is a GEF specific to Rac1/Cdc2, a key mediator of actin reorganization in response to various stimuli. In the mammalian system, Rac1 is locally activated in dendritic spines, and this spatial restricted activation is regulated by Pix (46). In the present study, we show that the active form of Rac1 (Rac1-GTP) is reduced in the dnrx mutant, dnrx, and scribble knockdown flies compared with wild-type for the protein level of DPix in these lines were decreased, supporting the idea that the DNRX and Scribble interaction activates Rac1. Recent studies show that Rac1 plays a critical roles in animal behavior, particularly in the process of forgetting (36). In addition, Scribble has recently been reported to activate forgetting through Rac1 in Drosophila (35). Taken together, we suggest that DNRX may also be involved in forgetting by regulating the Rac1 signaling pathway. Indeed, it has been demonstrated that Rac1 activation is defective in multiple autism-related gene mutations, including the dnrx gene, and that Rac1 has been proposed to be a converging node linked to ASD (36). Thus, the present study demonstrating that DNRX interacts with Scribble and DPix to regulate Rac1 provides direct mechanistic insight into not only the fundamental mechanisms underlying the roles of the neuroligin–neurexin complex in actin-mediated presynaptic regulation, but also the pathological mechanism of ASD.
Experimental procedures
cDNA clones
The Scribble, DPix, Cortactin, and DNRX full-length cDNA were obtained from DGRC.
Fly stocks
The following fly strains were used in this study. 1) The dnrx mutant allele dnrx273 was generated as a gift from Dr. Manzoor A. Bhat, and Df(3R)5C1 (a deficiency that removes dnrx) were introduced into the TM6B fly line that was obtained from the Bloomington Stock Center. The dnrx mutant allele dnrx83 was generated by our lab (9). UAS-dnrx and dnrxΔPDZ transgenic flies were generated by cloning the entire dnrx full-length cDNA and dnrx full-length cDNA without the terminal region of the PDZ-binding motif into the pUAST-attB vector for germ line transformation. 2) Mutations in scribble, scribble2, and scribble3 were generated as a gift from Norbert Perrimon, and Df(3R)Tl-x flies, which are missing the scribble region, were obtained from the Bloomington Stock Center. UAS-scribble-gfp and UAS-dpix-gfp transgenic flies were generated by cloning the entire scribble, dpix full-length cDNA with GFP cDNA C-terminal into the pUAST-attB vector for germ line transformation. 3) The Gal4 lines, Elav-Gal4, Elav dcr-Gal4, and OK6-Gal4 were obtained from the Bloomington Stock Center. 4) The UAS-dnrx RNAi, UAS-scribble RNAi, UAS-dpix RNAi, UAS-actin-GFP transgenic fly, and scribble::gfp (07683) were purchased from the model animal center of D. melanogaster Tsinghua University. The uas-pak, usa-racL89 (dominant-nagtive rac), and tsr mutant flies were obtained from the Bloomington Stock Center. 5) The wild-type (WT) D. melanogaster strain used in this study was w1118. 6) All transgenic flies and gene Integrated flies were identified (supplemental Fig. S7). All the flies were raised at 25 °C on standard medium.
Western blotting analysis
Western blotting analysis was performed as described previously (9). In brief, adult heads and larvae muscles were homogenized with RAPI buffer and centrifuged at 12,000 rpm at 4 °C for 10 min, and the supernatant was collected. The protein lysates were electrophoresed on an SDS-PAGE gel and electrotransferred onto polyvinylidene difluoride membranes. Immobilized proteins on the membrane were probed with the primary antibody and then incubated with HRP-conjugated secondary antibodies. The primary antibodies used in this study were rabbit polyclonal anti-DNRX (55); mouse monoclonal anti-GFP, mouse anti-Arp2, and mouse anti-Cortactin from Santa Cruz Biotechnology; mouse monoclonal anti-α-tubulin and anti-β-actin were from Sigma; mouse anti-DLG, mouse anti-Brp, and mouse anti-Synapsin antibodies from the Developmental Studies Hybridoma Bank; rabbit polyclonal anti-Cdc42 from Thermo Scientific.
Immunostaining
Preparation and antibody staining for whole mount embryos, wandering third-instar larvae, and adult. Briefly, embryonic samples were washed three times in 0.3% PBST (PBS + 0.3% Triton X-100) rapidly, adding 50% commercial bleach diluted in water for decorticating, blocked for 1 h in the blocking solution, and incubated with primary antibodies at 4 °C overnight. For larvae and adult NMJ and head staining, larvae and adult were dissected in Ca2+-free HL3 saline (70 mm NaCl, 5 mm KCl, 20 mm MgCl2, 10 mm NaHCO3, 5 mm trehalose, 115 mm sucrose, and 5 mm Hepes, pH 7.2) and fixed in 4% paraformaldehyde/PBS for 30 min. After dissection and fixation, samples were stained with antibodies. The primary antibodies used in this study were as follows: purified mouse antiserum against DNRX (A4, 1:100) (14), mouse anti-SYT (DSHB, 1:50), mouse anti-SYN (DSHB, 1:50), mouse anti-DLG (DSHB, 1:50), mouse anti-GFP (DSHB, 1:50), and rabbit anti-HRP (DSHB, 1:500), Texas Red®-X Phalloidin (Molecular Probes, 1:10) and Alexa Fluor 488-, Alexa Fluor 555-, or Alexa Fluor 647-conjugated secondary antibodies (Invitrogen, 1:500). Samples were imaged at room temperature using a LSM 700 (Carl Zeiss) confocal microscope at 1,024 × 1,024 pixels using a ×63 (NA 1.4, oil immersion; Carl Zeiss) objective for boutons staining, ×40 (NA 0.95, dry; Carl Zeiss) objective for whole NMJ staining, ×20 (NA 0.8, dry; Carl Zeiss) for epithelial cells staining and ×10 (NA 0.45, dry; Carl Zeiss) for brain, VNC, and embryo staining. All images were initially acquired through Zen 2010 software (Carl Zeiss). Fluorescent intensities for F-actin, DNRX, Scribble::GFP, and DPix-GFP were measured using Image J software (National Institutes of Health). Brightness and contrast were adjusted using AimImageBrowser software (Carl Zeiss).
STED imaging
For STED imaging the Alexa Fluor 555-conjugated secondary antibodies were used (Invitrogen, 1:500). Samples were imaged at room temperature using a STED microscope of TCS SP8 (Leica, Germany) at 1,024 × 1,024 pixels using a ×100 (NA 1.4, oil immersion; Leica) objective for boutons staining. All images were initially acquired through LAS X software (Leica). All images were treated with Huygens Professional software.
Immunoprecipitation
The immunoprecipitation experiments were performed as previously described (56). Briefly, fly heads of the desired genotypes were homogenized using a glass homogenizer in ice-cold lysis buffer containing 20 mm Tris (pH 7.5), 150 mm NaCl, 1% Triton X-100, and sodium pyrophosphate, β-glycerophosphate, EDTA, Na3VO4, and leupeptin (Beyotime Biotechnology) with protease inhibitors. The lysates were kept on a shaker at 4 °C for 30 min and centrifuged twice at 12,000 rpm at 4 °C for 10 min. The supernatant was incubated with beads and antibody on a shaker at 4 °C overnight. Then centrifuged at 2,500 rpm at 4 °C for 2 min and the supernatant was discarded. The beads were subsequently washed three times with lysis buffer, and proteins were eluted by boiling the beads in 2× SDS loading buffer. Then centrifuged at 12,000 rpm at room temperature for 2–3 min and the beads were discarded, the remnant buffer was used for the immunoblot analysis.
GST pulldown assay
The methods for the GST pulldown assay has been described previously (57). Briefly, GST fusion proteins were expressed in BL21 Escherichia coli cells with pGEX-5X-1 vector, and then purified and immobilized with glutathione-Sepharose 4B beads (GE Healthcare). Plasmids coding the four Scribble PDZ domains of pCDNA3.1 were transfected into HEK293T cells. After inducing and expressing, transfected cells were lysed in the lysis buffer (Roche Applied Science) and incubated with beads that contained GST-fused protein. The beads were subsequently washed three times with lysis buffer, and proteins were eluted by boiling the beads in 2× SDS loading buffer. The beads were then centrifuged at 12,000 rpm at room temperature for 2–3 min and the beads were discarded, the remnant buffer was used for immunoblot analysis.
Cell culture, transfection, and treatments
DMEM (Sigma) was used to culture HEK293 and supplemented with 10% FBS (Invitrogen). Cells were transfected with X-tremeGENE 9 DNA Transfection Reagent (Roche Applied Science) according to the manufacturer's protocols and grown in a single Petri dish (90 × 90 mm). After incubating and maintaining the cells for 48 h at 37 °C, they were harvested for biochemical analyses.
Electrophysiology
Third instar larvae were dissected as described previously (58). Briefly, third instar larvae were rapidly dissected in Ca2+-free HL3.1 saline, fat and gut in the body were removed, brain and VNC were cut, and the body wall was carefully spread out to expose muscle to avoid damage to the muscle architecture. The free segmental nerve end was drawn into a microelectrode using a injector and stimulated with a Grass S48 stimulator (Astro-Grass) at 0.3 Hz with a suprathreshold stimulating pulse, and we selected NMJ 6 in the A3 segment for recording using recording electrodes (20–50 MΩ) filled with 3 m KCl. mEJPs were recorded, the start was from 8 s after EJP recordings for 20 s. All recordings were conducted in a modified HL3.1 solution containing 70 mm NaCl, 5 mm KCl, 4 mm MgCl2, 10 mm NaHCO3, 0.6 mm CaCl2, 115 mm sucrose, 5 mm trehalose, and 5 mm Hepes (pH 7.2). Recordings were performed at room temperature with an Axoclamp 900A amplifier (Molecular Devices, Sunnyvale, CA) in bridge mode. Recording data were digitized with a Digitizer 1322A (Molecular Devices). Data were analyzed in the Clampfit 10.2 software.
Rac1 activity assay
The heads of ∼500 adult flies were isolated and ground to gain sample. For detecting the relative levels of Rac1-GTP, a Pak-PBD pulldown assay (no. 16118; Thermo Scientific) was used according to the manufacturer's procedures as described previously (36). The anti-human Rac1 monoclonal antibody (1:1,000 dilution) was used to detect total and activated Rac1 levels.
Statistical analysis
Statistical significance was determined using Student's t tests for comparisons of two groups and one-way analysis of variance followed by the appropriate post hoc test for comparisons of multiple group means (GraphPad Prism 5). All asterisks above a line indicate comparisons between the two groups indicated. Data are expressed as the mean ± S.E. p values <0.05 were considered to be statistically significant.
Author contributions
W. X. conceived the project, W. X. and M. L. R. designed the experiments and wrote the manuscript, W. X., M. L. R., J. H. H., and Z. P. J. analyzed data. M. L. R. conducted the experiments. J. J. Q. performed the qRT-PCR experiment. L. J. L., Y. H. C., and H. H. L. helped in recoding and animal analysis.
Supplementary Material
Acknowledgments
We thank Dr. Manzoor A. Bhat for dnrx mutants; the Bloomington Stock Center for providing Drosophila stocks; Tsinghua Stock center for providing RNAi flies, and N. Perrimon for providing scribble mutant flies; and DSHB for antibodies. We thank Dr. Fugen Wu and Dr. Hongyin Wang for valuable assistance in STED imaging. We thank Dr. Renjie Chai for critical reading the manuscript and members of the Xie laboratory for comments on the manuscript and experiment design.
This work was supported by National Natural Science Foundation of China Grants 31430035 and 30930051 (to W. X.) and 31600824 (to L. L.) and China National Basic Research Program 973 Program Grant 2012CB517903 (to W. X. and Z. J.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S7.
- SV
- synaptic vesicle
- ASDs
- autism spectrum disorders
- AZ
- active zone
- BRP
- bruchpilot
- DLG
- Discs large
- DNRX
- Drosophila Neurexin
- DPix
- Drosophila Pix
- EJP
- excitatory junction potential
- GEF
- guanine nucleotide exchange factor
- HRP
- neuronal plasma membrane
- mEJP
- miniature excitatory junction potentials
- NMJ
- neuromuscular junction
- SYN
- synapsin
- SYT
- synaptotagmin
- VNC
- ventral nerve cord
- STED
- stimulated emission depletion
- qRT
- quantitative RT.
References
- 1. Alabi A. A., and Tsien R. W. (2012) Synaptic vesicle pools and dynamics. Cold Spring Harb. Perspect. Biol. 4, a013680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rizzoli S. O., and Betz W. J. (2005) Synaptic vesicle pools. Nat. Rev. Neurosci 6, 57–69 [DOI] [PubMed] [Google Scholar]
- 3. Nelson J. C., Stavoe A. K., and Colon-Ramos D. A. (2013) The actin cytoskeleton in presynaptic assembly. Cell Adh. Migr. 9, 379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sankaranarayanan S., Atluri P. P., and Ryan T. A. (2003) Actin has a molecular scaffolding, not propulsive, role in presynaptic function. Nat. Neurosci. 6, 127–135 [DOI] [PubMed] [Google Scholar]
- 5. Lee J. S., Ho W. K., and Lee S. H. (2012) Actin-dependent rapid recruitment of reluctant synaptic vesicles into a fast-releasing vesicle pool. Proc. Natl. Acad. Sci. U.S.A. 109, E765–E774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shupliakov O., Bloom O., Gustafsson J. S., Kjaerulff O., Low P., Tomilin N., Pieribone V. A., Greengard P., and Brodin L. (2002) Impaired recycling of synaptic vesicles after acute perturbation of the presynaptic actin cytoskeleton. Proc. Natl. Acad. Sci. U.S.A. 99, 14476–14481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Scheiffele P., Fan J., Choih J., Fetter R., and Serafini T. (2000) Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell 101, 657–669 [DOI] [PubMed] [Google Scholar]
- 8. Ushkaryov Y. A., Petrenko A. G., Geppert M., and Südhof T. C. (1992) Neurexins: synaptic cell-surface proteins related to the α-latrotoxin receptor and laminin. Science 257, 50–56 [DOI] [PubMed] [Google Scholar]
- 9. Zeng X., Sun M., Liu L., Chen F., Wei L., and Xie W. (2007) Neurexin-1 is required for synapse formation and larvae associative learning in Drosophila. FEBS Lett. 581, 2509–2516 [DOI] [PubMed] [Google Scholar]
- 10. Kim H. G., Kishikawa S., Higgins A. W., Seong I. S., Donovan D. J., Shen Y., Lally E., Weiss L. A., Najm J., Kutsche K., Descartes M., Holt L., Braddock S., Troxell R., Kaplan L., et al. (2008) Disruption of neurexin 1 associated with autism spectrum disorder. Am. J. Hum. Genet. 82, 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yan J., Noltner K., Feng J., Li W., Schroer R., Skinner C., Zeng W., Schwartz C. E., and Sommer S. S. (2008) Neurexin 1α structural variants associated with autism. Neurosci. Lett. 438, 368–370 [DOI] [PubMed] [Google Scholar]
- 12. Biederer T., and Südhof T. C. (2000) Mints as adaptors: direct binding to neurexins and recruitment of Munc18. J. Biol. Chem. 275, 39803–39806 [DOI] [PubMed] [Google Scholar]
- 13. Hata Y., Butz S., and Südhof T. C. (1996) CASK: a novel dlg/PSD95 homolog with an N-terminal calmodulin-dependent protein kinase domain identified by interaction with neurexins. J. Neurosci. 16, 2488–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun M., Liu L., Zeng X., Xu M., Liu L., Fang M., and Xie W. (2009) Genetic interaction between Neurexin and CAKI/CMG is important for synaptic function in Drosophila neuromuscular junction. Neurosci. Res. 64, 362–371 [DOI] [PubMed] [Google Scholar]
- 15. de Wit J., Sylwestrak E., O'Sullivan M. L., Otto S., Tiglio K., Savas J. N., Yates J. R. 3rd, Comoletti D., Taylor P., and Ghosh A. (2009) LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation. Neuron 64, 799–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ko J., Fuccillo M. V., Malenka R. C., and Südhof T. C. (2009) LRRTM2 functions as a Neurexin ligand in promoting excitatory synapse formation. Neuron 64, 791–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Muhammad K., Reddy-Alla S., Driller J. H., Schreiner D., Rey U., Böhme M. A., Hollmann C., Ramesh N., Depner H., Lützkendorf J., Matkovic T., Götz T., Bergeron D. D., Schmoranzer J., Goettfert F., et al. (2015) Presynaptic spinophilin tunes neurexin signalling to control active zone architecture and function. Nat. Commun. 6, 8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li T., Tian Y., Li Q., Chen H., Lv H., Xie W., and Han J. (2015) The neurexin/N-ethylmaleimide-sensitive factor (NSF) interaction regulates short term synaptic depression. J. Biol. Chem. 290, 17656–17667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Knight D., Xie W., and Boulianne G. L. (2011) Neurexins and neuroligins: recent insights from invertebrates. Mol. Neurobiol. 44, 426–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Südhof T. C. (2008) Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455, 903–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li J., Ashley J., Budnik V., and Bhat M. A. (2007) Crucial role of Drosophila neurexin in proper active zone apposition to postsynaptic densities, synaptic growth, and synaptic transmission. Neuron 55, 741–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Missler M., Zhang W., Rohlmann A., Kattenstroth G., Hammer R. E., Gottmann K., and Südhof T. C. (2003) α-Neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature 423, 939–948 [DOI] [PubMed] [Google Scholar]
- 23. Littleton J. T., Bellen H. J., and Perin M. S. (1993) Expression of synaptotagmin in Drosophila reveals transport and localization of synaptic vesicles to the synapse. Development 118, 1077–1088 [DOI] [PubMed] [Google Scholar]
- 24. Merriam E. B., and Hu X. (2012) Cell adhesion complexes localize presynaptic vesicles by regulating local actin polymerization. J. Neurosci. 32, 3955–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wolf M., Zimmermann A. M., Görlich A., Gurniak C. B., Sassoè-Pognetto M., Friauf E., Witke W., and Rust M. B. (2015) ADF/cofilin controls synaptic actin dynamics and regulates synaptic vesicle mobilization and exocytosis. Cereb. Cortex 25, 2863–2875 [DOI] [PubMed] [Google Scholar]
- 26. Morales M., Colicos M. A., and Goda Y. (2000) Actin-dependent regulation of neurotransmitter release at central synapses. Neuron 27, 539–550 [DOI] [PubMed] [Google Scholar]
- 27. Vitale M. L., Seward E. P., and Trifaró J. M. (1995) Chromaffin cell cortical actin network dynamics control the size of the release-ready vesicle pool and the initial rate of exocytosis. Neuron 14, 353–363 [DOI] [PubMed] [Google Scholar]
- 28. Nunes P., Haines N., Kuppuswamy V., Fleet D. J., and Stewart B. A. (2006) Synaptic vesicle mobility and presynaptic F-actin are disrupted in a N-ethylmaleimide-sensitive factor allele of Drosophila. Mol. Biol. Cell 17, 4709–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cosen-Binker L. I., and Kapus A. (2006) Cortactin: the gray eminence of the cytoskeleton. Physiology 21, 352–361 [DOI] [PubMed] [Google Scholar]
- 30. Ridley A. J. (2006) Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 16, 522–529 [DOI] [PubMed] [Google Scholar]
- 31. Qin Y., Capaldo C., Gumbiner B. M., and Macara I. G. (2005) The mammalian Scribble polarity protein regulates epithelial cell adhesion and migration through E-cadherin. J. Cell Biol. 171, 1061–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bilder D., and Perrimon N. (2000) Localization of apical epithelial determinants by the basolateral PDZ protein Scribble. Nature 403, 676–680 [DOI] [PubMed] [Google Scholar]
- 33. Moreau M. M., Piguel N., Papouin T., Koehl M., Durand C. M., Rubio M. E., Loll F., Richard E. M., Mazzocco C., Racca C., Oliet S. H., Abrous D. N., Montcouquiol M., and Sans N. (2010) The planar polarity protein Scribble1 is essential for neuronal plasticity and brain function. J. Neurosci. 30, 9738–9752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roche J. P., Packard M. C., Moeckel-Cole S., and Budnik V. (2002) Regulation of synaptic plasticity and synaptic vesicle dynamics by the PDZ protein scribble. J. Neurosci. 22, 6471–6479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cervantes-Sandoval I., Chakraborty M., MacMullen C., and Davis R. L. (2016) Scribble scaffolds a signalosome for active forgetting. Neuron 90, 1230–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dong T., He J., Wang S., Wang L., Cheng Y., and Zhong Y. (2016) Inability to activate Rac1-dependent forgetting contributes to behavioral inflexibility in mutants of multiple autism-risk genes. Proc. Natl. Acad. Sci. U.S.A. 113, 7644–7649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen K., Gracheva E. O., Yu S. C., Sheng Q., Richmond J., and Featherstone D. E. (2010) Neurexin in embryonic Drosophila neuromuscular junctions. PloS One 5, e11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sun M., Zeng X., and Xie W. (2016) Temporal and spatial expression of Drosophila neurexin during the life cycle visualized using a DNRX-Gal4/UAS-reporter. Sci. China Life Sci. 59, 68–77 [DOI] [PubMed] [Google Scholar]
- 39. Sun Y., Aiga M., Yoshida E., Humbert P. O., and Bamji S. X. (2009) Scribble interacts with β-catenin to localize synaptic vesicles to synapses. Mol. Biol. Cell 20, 3390–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Audebert S., Navarro C., Nourry C., Chasserot-Golaz S., Lécine P., Bellaiche Y., Dupont J. L., Premont R. T., Sempéré C., Strub J. M., Van Dorsselaer A., Vitale N., and Borg J. P. (2004) Mammalian Scribble forms a tight complex with the βPIX exchange factor. Curr. Biol. 14, 987–995 [DOI] [PubMed] [Google Scholar]
- 41. Richier L., Williton K., Clattenburg L., Colwill K., O'Brien M., Tsang C., Kolar A., Zinck N., Metalnikov P., Trimble W. S., Krueger S. R., Pawson T., and Fawcett J. P. (2010) NOS1AP associates with Scribble and regulates dendritic spine development. J. Neurosci. 30, 4796–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sun Y., and Bamji S. X. (2011) β-Pix modulates actin-mediated recruitment of synaptic vesicles to synapses. J. Neurosci. 31, 17123–17133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Frank S. R., and Hansen S. H. (2008) The PIX-GIT complex: a G protein signaling cassette in control of cell shape. Semin. Cell Dev. Biol. 19, 234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Attias O., Jiang R., Aoudjit L., Kawachi H., and Takano T. (2010) Rac1 contributes to actin organization in glomerular podocytes. Nephron Exp. Nephrol. 114, e93–e106 [DOI] [PubMed] [Google Scholar]
- 45. Tejada-Simon M. V. (2015) Modulation of actin dynamics by Rac1 to target cognitive function. J. Neurochem. 133, 767–779 [DOI] [PubMed] [Google Scholar]
- 46. Zhang H., Webb D. J., Asmussen H., Niu S., and Horwitz A. F. (2005) A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J. Neurosci. 25, 3379–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Erickson J. W., Cerione R. A., and Hart M. J. (1997) Identification of an actin cytoskeletal complex that includes IQGAP and the Cdc42 GTPase. J. Biol. Chem. 272, 24443–24447 [DOI] [PubMed] [Google Scholar]
- 48. Tang D. D., and Gunst S. J. (2004) The small GTPase Cdc42 regulates actin polymerization and tension development during contractile stimulation of smooth muscle. J. Biol. Chem. 279, 51722–51728 [DOI] [PubMed] [Google Scholar]
- 49. Parnas D., Haghighi A. P., Fetter R. D., Kim S. W., and Goodman C. S. (2001) Regulation of postsynaptic structure and protein localization by the Rho-type guanine nucleotide exchange factor dPix. Neuron 32, 415–424 [DOI] [PubMed] [Google Scholar]
- 50. Harris J. C. (2016) The origin and natural history of autism spectrum disorders. Nat. Neurosci. 19, 1390–1391 [DOI] [PubMed] [Google Scholar]
- 51. Zhang H. F., Dai Y. C., Wu J., Jia M. X., Zhang J. S., Shou X. J., Han S. P., Zhang R., and Han J. S. (2016) Plasma oxytocin and arginine-vasopressin levels in children with autism spectrum disorder in China: associations with symptoms. Neurosci. Bull. 32, 423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ganguly I., Mackay T. F., and Anholt R. R. (2003) Scribble is essential for olfactory behavior in Drosophila melanogaster. Genetics 164, 1447–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Delgado R., Maureira C., Oliva C., Kidokoro Y., and Labarca P. (2000) Size of vesicle pools, rates of mobilization, and recycling at neuromuscular synapses of a Drosophila mutant, shibire. Neuron 28, 941–953 [DOI] [PubMed] [Google Scholar]
- 54. Tran D. T., Masedunskas A., Weigert R., and Ten Hagen K. G. (2015) Arp2/3-mediated F-actin formation controls regulated exocytosis in vivo. Nat. Commun. 6, 10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tian Y., Li T., Sun M., Wan D., Li Q., Li P., Zhang Z. C., Han J., and Xie W. (2013) Neurexin regulates visual function via mediating retinoid transport to promote rhodopsin maturation. Neuron 77, 311–322 [DOI] [PubMed] [Google Scholar]
- 56. Chen Y. C., Lin Y. Q., Banerjee S., Venken K., Li J., Ismat A., Chen K., Duraine L., Bellen H. J., and Bhat M. A. (2012) Drosophila neuroligin 2 is required presynaptically and postsynaptically for proper synaptic differentiation and synaptic transmission. J. Neurosci. 32, 16018–16030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nakao S., Platek A., Hirano S., and Takeichi M. (2008) Contact-dependent promotion of cell migration by the OL-protocadherin-Nap1 interaction. J. Cell Biol. 182, 395–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jan L. Y., and Jan Y. N. (1976) Properties of the larval neuromuscular junction in Drosophila melanogaster. J. Physiol. 262, 189–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.