Abstract
In both multiple sclerosis and experimental autoimmune encephalomyelitis (EAE), the C-C chemokine receptor 6 (CCR6) is critical for pathogenic T helper 17 (Th17) cell migration to the central nervous system (CNS). Whereas many cytokines and their receptors are potently regulated via post-transcriptional mechanisms in response to various stimuli, how CCR6 expression is post-transcriptionally regulated in Th17 cells is unknown. Here, using RNA-binding protein HuR conditional knock-out (KO) and wild-type (WT) mice, we present evidence that HuR post-transcriptionally regulates CCR6 expression by binding to and stabilizing Ccr6 mRNA and by promoting CCR6 translation. We also found that HuR down-regulates several microRNA expressions, which could target the 3′-UTR of Ccr6 mRNA for decay. Accordingly, knock-out of HuR reduced CCR6 expression on Th17 cells and impaired their migration to CNS compared with the response of WT Th17 cells and thereby ameliorated EAE. Together, these findings highlight how HuR contributes to Th17 cell-mediated autoimmune neuroinflammation and support the notion that targeting HuR might be a potential therapeutic intervention for managing autoimmune disorders of the CNS.
Keywords: gene expression, mouse, mRNA decay, neuroinflammation, post-transcriptional regulation, RNA binding protein, T helper cells, CCR6, Th17 cells, experimental autoimmune encephalomyelitis
Introduction
T helper 17 (Th17)4 cells play critical roles in inflammatory disorders, including multiple sclerosis and experimental autoimmune encephalomyelitis (EAE) (1–5). The signature cytokine produced by Th17 cells, IL-17 (2, 6), has been found previously to be involved in host defense against bacterial infection (7, 8). Expression of the C-C chemokine receptor 6 (CCR6) on Th17 cells is required for migration of pathogenic Th17 cells to sites of inflammation (9–12). In the CNS of mice with EAE, Th17 cells expressing CCR6 are attracted by the chemokine ligand 20 (CCL20), which is constitutively secreted by choroid plexus epithelial cells (12–14); accordingly, anti-CCL20 antibody treatment in vivo significantly suppressed pathogenic CD4+ T cell accumulation and the development of EAE (15, 16). Signaling by CCL20 through CCR6 allows Th17 cells to cross the epithelial barrier of the choroid plexus and enter the cerebrospinal fluid. Thus, the initial trigger of inflammation is induced by CCR6-dependent autoreactive Th17 cell infiltration of the uninflamed CNS (12–14). In agreement with this notion, CCR6−/− mice are resistant to development of EAE (12). Moreover, CCR6-expressing Th17 cells are enriched in the cerebrospinal fluid of patients with early clinical symptoms of multiple sclerosis (12, 17, 18). Therefore, further understanding the mechanisms that underlie CCR6 expression in Th17 cells may uncover novel therapeutic targets for treatment of Th17 cell-mediated autoimmune diseases.
Differentiation of Th17 cells is induced by activation of naive CD4+ T cells in the presence of a milieu of inflammatory cytokines. TGF-β and IL-6 potently induce naive CD4+ T cells to differentiate into Th17 cells, which are reinforced by IL-23 (6, 19, 20). During cytokine-mediated Th17 cell differentiation, the transcription factor STAT3 and two orphan nuclear receptors, RORγt and RORα, function to regulate Th17 cell differentiation (21, 22). The transcriptional regulation of CCR6 gene expression on Th17 cells is known to be regulated by TGF-β and to require RORγt and RORα (11).
Despite recent progress in understanding the regulation and function of CCR6 on Th17 cells, it is still unclear how CCR6 expression is post-transcriptionally regulated. Given the importance of post-transcriptional gene regulation in eliciting quick responses to stimuli, defining the mechanisms mediating post-transcriptional regulation is a very active area of research (23, 24). Mammalian HuR is the homolog of the ELAV (embryonic lethal abnormal vision)-like protein in Drosophila melanogaster (25) and is ubiquitously expressed in all tissues. HuR binds to target mRNAs bearing specific sequence elements, often U- and AU-rich and generally found in the mRNA 3′-UTRs, and plays a critical role in their post-transcriptional regulation (26). HuR stabilizes many target mRNAs encoding proteins with roles in cell proliferation, survival, immune responses, and differentiation (27). Although HuR is known to stabilize many of these mRNAs and/or modulate their translation, the molecular mechanisms by which HuR affects the fate of target mRNA remain unknown. In addition, recent studies indicate that HuR may mediate some of its effects through interplay with microRNAs (miRNAs) associated with the same target mRNAs (27).
In this study, we focused on investigating the role of HuR in mediating expression of CCR6 on pathogenic Th17 cells in EAE. HuR binds to Ccr6 mRNA to prolong its half-life and moderately increases its translation, leading to increased CCR6 expression. In addition, our data indicate that HuR negatively regulates the expression of some miRNAs, and thus HuR may prevent these miRNAs from binding to and degrading Ccr6 mRNA, further enhancing CCR6 expression. Knock-out of HuR decreases CCR6 expression on Th17 cells and impairs their migration in response to its ligand, CCL20, thereby ameliorating EAE. These results further support the notion that targeting HuR might be a novel therapeutic intervention for autoimmune encephalomyelitis (28).
Results
Th17 cells express high levels of HuR and CCR6
In previous studies, we demonstrated that in comparison with WT Th17 cells, HuR conditional knock-out (HuR KO) Th17 cells induced less severe EAE after transfer into naive C57BL/6 mice (28), which indicated that HuR plays a role in the initiation of EAE. When using Rag1−/− mice as recipients, the difference in induction of EAE between WT and HuR KO Th17 cells was much more significant, and the difference in EAE severity between the two groups lasted for 3 weeks, at which time the experiment was ended (data not shown). Because recipients that received HuR KO Th17 cells had fewer CD4+ T cell infiltrations in the CNS (28), we speculated that HuR KO Th17 cells may exhibit impaired migration into the inflamed CNS. Because CCR6 is crucial for the pathogenic Th17 cell migration in rheumatoid arthritis and EAE (10, 12, 29), we sought to investigate whether HuR regulates expression of CCR6 on Th17 cells.
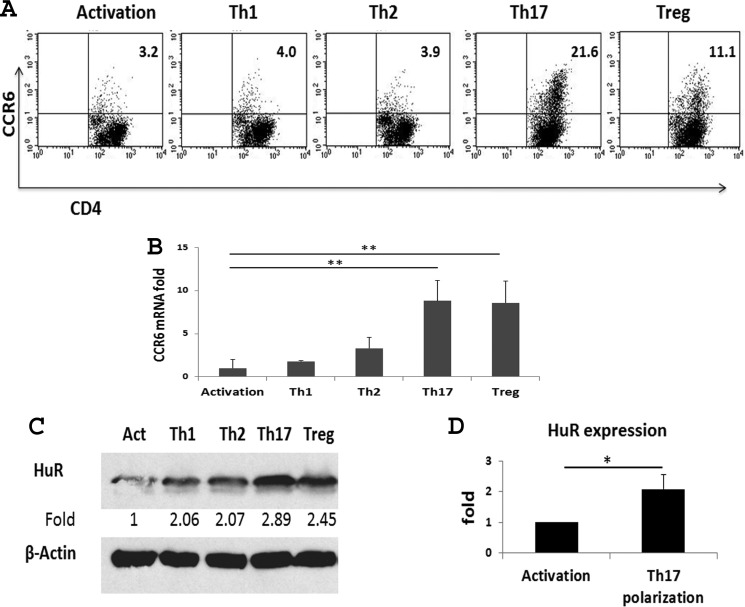
Our previous studies showed that naive CD4+ T cells expressed low levels of HuR protein, and activation of CD4+ T cells by anti-CD3 plus anti-CD28 increased expression of HuR (28). Further analysis showed that anti-CD3 alone induced an intermediate level of HuR protein expression, and anti-CD28 or IL-2 synergized with anti-CD3 to induce a high level of HuR protein expression (data not shown). These findings supported the idea that HuR may play an important role in T cell activation, in agreement with our previous studies (30). To determine whether there is a relationship between expression of CCR6 and HuR protein in T helper cells, naive CD4+ T cells from WT mice were isolated and cultured under different T helper cell or T regulatory (Treg) cell polarization conditions. The expression levels of CCR6 and HuR on Th1, Th2, Th17, and Treg cells were measured by qRT-PCR, flow cytometry, and Western blot analysis, respectively. As shown, CCR6 was highly expressed on Th17 cells, modestly expressed on Treg cells, and expressed at low levels on Th1 and Th2 cells, and the level of Ccr6 mRNA expression correlates well with level of CCR6 protein expression in these different Th subsets (Fig. 1, A and B). Western blot analysis demonstrated a similar pattern of HuR expression in these T subset cells, with the highest expression of HuR in Th17 cells, moderate HuR levels in Treg cells, and lowest expression in Th1 cells (Fig. 1C). Furthermore, differentiation of Th17 cells resulted in an average 2-fold increase in HuR protein expression levels in comparison with CD4+ T cells activated with anti-CD3 plus anti-CD28 and without Th17 cell polarization cytokines (Fig. 1, C and D). Together, these findings suggest that the expression of CCR6 correlates with the level of HuR protein in Th17 cells, supporting our hypothesis that HuR positively promotes expression of CCR6 in Th17 cells.
Figure 1.
Expression of CCR6 and HuR on T helper cells and Treg cells. Naive CD4+ T cells were isolated from spleen of WT mice and activated or polarized as described under “Experimental procedures.” Activation (Act), Th1, Th2, Th17, and Treg cells were analyzed after 5 days of cell culture. Expressions of CCR6 on T helper cells and Treg cells were analyzed by flow cytometry (A) and qRT-PCR (B). C, HuR protein levels in CD4+ T cells were assessed by Western blot analysis. D, summary of HuR protein level in Th17 cells compared with that of activated CD4+ T cells. One of three independent experiments is shown in A and C. Data in B and D are derived from at least three independent experiments and presented as mean ± S.E. (error bars). *, p < 0.05; **, p < 0.01.
HuR deficiency reduces the expression of Ccr6 mRNA and protein levels in Th17 cells
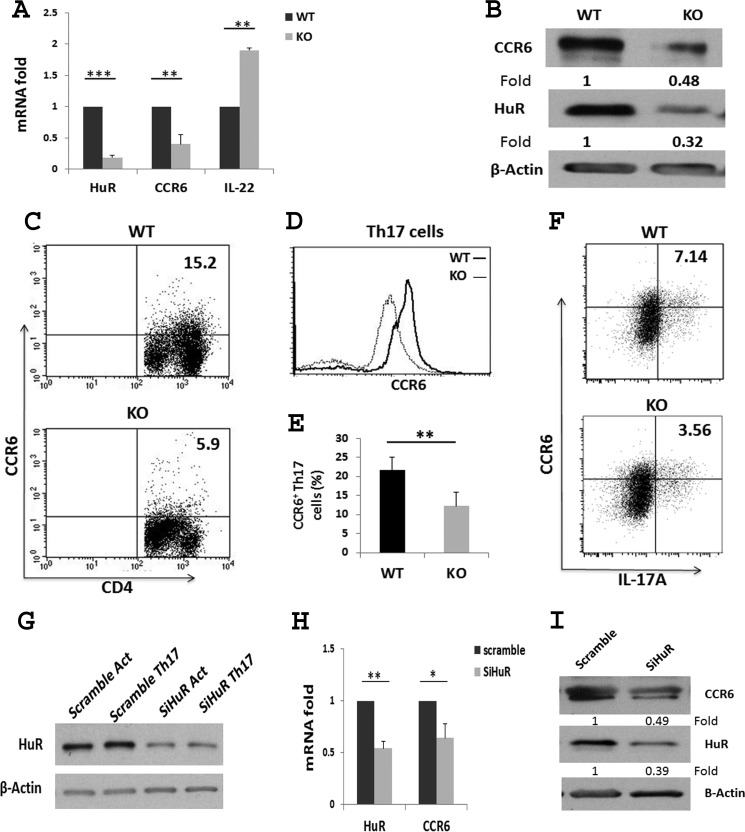
We utilized HuR conditional knock-out mice to more definitively evaluate the relationship between HuR and CCR6 expression in Th17 cells. We isolated naive CD4+ T cells from spleens of WT and HuR KO mice and cultured the cells under Th17 cell polarization conditions (28). Reverse transcription followed by qRT-PCR revealed that expression of Ccr6 mRNA, but not il22 mRNA, was significantly reduced in HuR KO Th17 cells in comparison with WT Th17 cells (Fig. 2A). Consistent with the qRT-PCR data, Western blot and flow cytometric analyses revealed that the level of CCR6 protein was also decreased in HuR KO Th17 cells compared with WT Th17 cells (Fig. 2, B–F). These results further supported the notion that HuR positively regulates CCR6 expression on Th17 cells.
Figure 2.
Knock-out of HuR reduces CCR6 expression on Th17 cells. Naive CD4+ T cells were isolated from spleen of WT and HuR KO mice and cultured under Th17 cell–polarizing conditions for 5 days. A, Ccr6 mRNA levels were measured by qRT-PCR analysis. B, Western blot analysis of CCR6 expression on WT and HuR KO Th17 cells. C and D, flow cytometric analysis of CCR6 expression on Th17 cells. E, summary of flow cytometric analysis of three independent experiments. F, the percentage of CCR6+IL-17+ cells was reduced in HuR KO Th17 cells compared with that of WT Th17 cells. G, human peripheral blood CD4+ T cells were isolated and transfected with scrambled and HuR siRNA and cultured in the presence (Th17) or absence (Act) of Th17 cell–polarizing cytokines. HuR protein expression levels were then assessed by Western blot analysis. H, qRT-PCR analysis of Ccr6 mRNA levels following transfection of human Th17 cells with scramble and HuR siRNAs (SiHuR). I, the level of CCR6 protein was also reduced in HuR siRNA-transfected human Th17 cells compared with that with scramble siRNA by Western blots. Data in B–D, F, and I represent one of three independent experiments. Data in G represent one of three independent experiments. Data in A, E, and H are summarized at least three independent experiments and are presented as mean ± S.E. (error bars). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Computational analysis showed that mouse Ccr6 mRNA with short 3′-UTR contains one potential HuR binding site. However, the human CCR6 mRNA contains several HuR potential binding sites in the ORF and the 3′-UTR (data not shown). Thus, we hypothesized that HuR may also regulate CCR6 expression on human CD4+ T cells, which might have clinically relevant consequences, given the identification of CCR6-positive Th17 cells in multiple sclerosis (17). To test this possibility, naive CD4+ T cells were isolated from human peripheral blood, and HuR expression was reduced by transfection with siRNA directed at the HuR mRNA. The cells were differentiated using Th17 cell culture conditions (28). The results revealed that CCR6 expression was reduced following knockdown of HuR in human CD4+ T cells compared with cells treated with scrambled control siRNA (Fig. 2, G–I), suggesting that HuR may play a similar role in controlling CCR6 expression on human CD4+ T cells.
Because macrophages treated with LPS express high levels of CCR6 (31, 32), we evaluated whether knockdown of HuR altered CCR6 expression on macrophages. The results shows that silencing HuR significantly reduced the expression of HuR in RAW 264.7 macrophages (data not shown). Flow cytometric analysis revealed that HuR silencing also selectively decreased CCR6 expression on macrophages (data not shown). These data further support the notion that HuR positively regulates CCR6 expression.
HuR modulates CCR6 expression by binding to and stabilizing Ccr6 mRNA
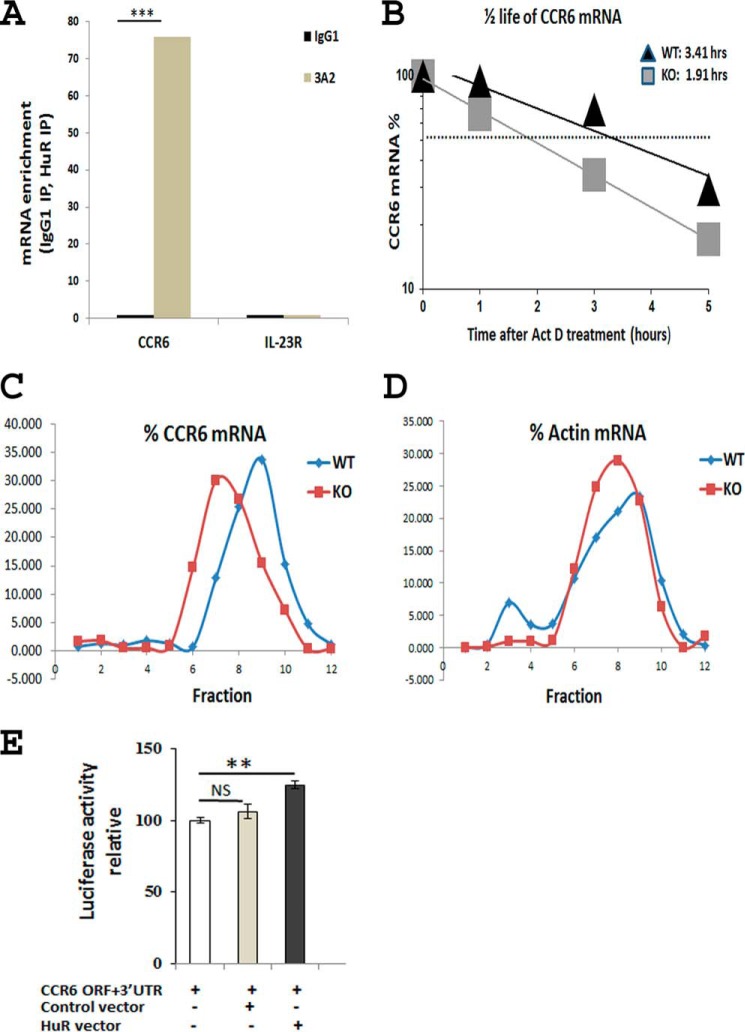
HuR has three RNA recognition motifs through which it interacts with target mRNAs (26). To explore the mechanism underlying the regulation of CCR6 expression by HuR, we performed an RNA immunoprecipitation (RIP) assay to examine whether HuR proteins physically associate with Ccr6 mRNA. Following RIP using anti-HuR antibody (3A2) beads in Th17 cell lysates, the RNA was isolated from the immunoprecipitated material, reverse transcribed, and quantitated by qRT-PCR analysis. Control immunoprecipitation (IP) reactions were carried out in parallel using an IgG. As shown in Fig. 3A, Ccr6 mRNA was significantly enriched in anti-HuR IP samples compared with isotype-matched IgG controls, after normalization of RNA levels in all IP samples by amplifying a Gapdh mRNA, which encodes a housekeeping protein and is not a target of HuR. These findings indicate that HuR forms a complex with Ccr6 mRNA.
Figure 3.
HuR binds to and stabilizes Ccr6 mRNA. Naive CD4+ T cells were isolated from spleens of WT mice and stimulated under Th17 cell–polarizing conditions. A, RIP analysis was performed to detect Ccr6 mRNA enrichment after IP using anti-HuR (3A2) or isotype-matched antibody (IgG1) from Th17 cell cytoplasmic extracts. Il23r mRNA was assayed as negative control. B, WT and HuR KO Th17 polarized cells were either left untreated or treated with actinomycin D (Act D; 3 μg/ml) and harvested 1, 3, and 5 h later. Ccr6 mRNA levels were determined by qRT-PCR analysis. C, WT and HuR KO Th17 cell lysates were size-fractionated through sucrose gradients, and RNA was isolated from each of 12 fractions, reverse-transcribed, and measured by qRT-PCR analysis. The relative distribution of Ccr6 mRNA along the gradient was calculated and plotted as a percentage of the total Ccr6 mRNA in the gradient. D, the distribution of actin mRNA, encoding a housekeeping protein, was included as a negative control. E, overexpression of HuR by HuR plasmid (0.2 μg) transfection increased the luciferase activity of vector containing Ccr6 mRNA ORF and 3′-UTR (0.2 μg) in HeLa cells in comparison with empty vector control groups. One of three independent experiments is shown in A–D. Data in E represent one of two individual experiments. **, p < 0.01; ***, p < 0.001; NS, not significant. Error bars, S.E.
HuR regulates many target mRNAs by modulating their stability and/or translation rate (27). To determine whether HuR functions to protect Ccr6 mRNA from degradation, we isolated naive CD4+ T cells from WT and KO mice and cultured them under Th17 polarization conditions. The polarized Th17 cells were treated with the RNA polymerase II inhibitor actinomycin D to inhibit de novo transcription, and the Ccr6 mRNA decay rates were measured by qRT-PCR analysis. As indicated in Fig. 3B, the half-life of Ccr6 mRNA in HuR KO Th17 cells was much shorter than that in WT Th17 cells (1.91 h versus 3.41 h), suggesting that HuR binds to and stabilizes Ccr6 mRNA to prolong its half-life.
We then asked whether HuR functions to modulate Ccr6 mRNA translation. Polysome fractionation analysis is useful to analyze the RNA-binding protein regulation of target mRNA translation (33). We compared the relative sizes of Ccr6 mRNA associated with polyribosomes in WT and HuR KO Th17 polarized cells. Following fractionation through sucrose gradients, Ccr6 mRNA polyribosomes in HuR KO Th17 cells showed moderately smaller size (shifted toward polysomes of lower molecular weight) compared with those in WT Th17 cells (Fig. 3C). By contrast, actin mRNA, encoding a housekeeping protein, did not show this shift in distribution toward smaller molecular weight (Fig. 3D). These findings suggested that in addition to promoting the stability of Ccr6 mRNA, HuR also moderately promoted Ccr6 mRNA translation. Indeed, overexpression of HuR by HuR plasmid transfection increases the luciferase activity of vector containing Ccr6 mRNA ORF and 3′-UTR in HeLa cells compared with the control group (Fig. 3E).
miRNA-mediated Ccr6 mRNA decay
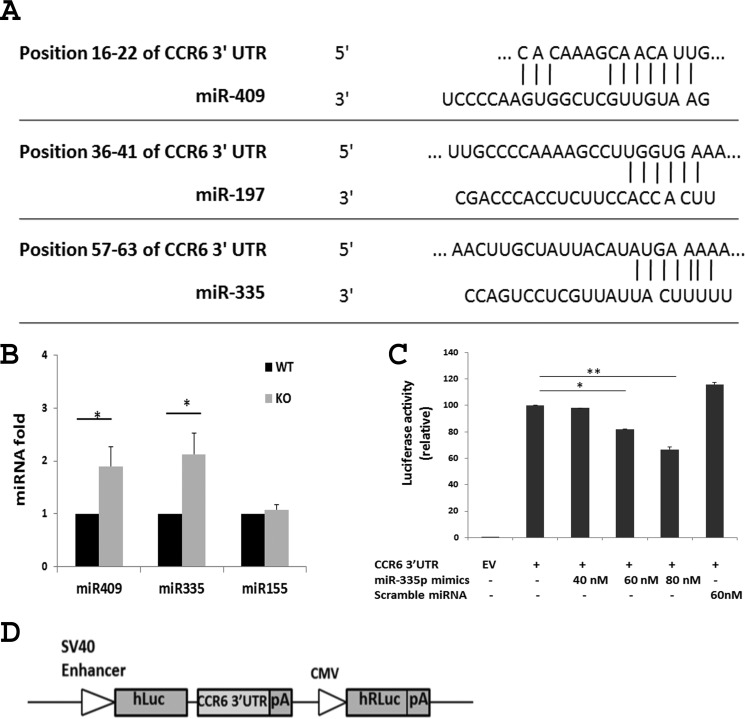
Besides RNA-binding proteins, microRNAs also regulate gene expression in a sequence-specific manner, often through interplay with RNA-binding proteins that associate with the same mRNAs (34). The physical and functional interaction of HuR with miRNAs has been documented (27). To investigate the possibility that HuR affects the fate of Ccr6 mRNA via miRNAs, computational analyses were performed using the TargetScan and miRanda algorithms to identify potential miRNA binding sites in the Ccr6 3′-UTR. Three sites were identified: miR-409, miR-197, and miR-335 (Fig. 4A). To determine whether there was any functional link between HuR and miRNAs predicted to bind the Ccr6 3′-UTR, qRT-PCR analysis was performed to detect these miRNAs in WT and HuR KO Th17 cells. Interestingly, whereas miR-155 levels were unchanged in the two Th17 populations, miR-409 and miR-335 levels were increased 2–3-fold (Fig. 4B), and miR-197 level increased 1.5-fold (data not shown) in HuR KO Th17 cells compared with WT Th17 cells, suggesting that HuR may negatively impact the levels of certain miRNAs.
Figure 4.
MiR-335 negatively regulates Ccr6 mRNA. A, TargetScan analysis prediction that miR-335, miR-197, and miR-409 have potential binding sites on the CCR6 3′-UTR. B, qRT-PCR analysis shows that knock-out of HuR increased the level of miR-335 and miR-409 in Th17 cells. C, overexpression of miR-335 reduced the activity of luciferase reporter containing the CCR6 3′-UTR. HeLa cells were transfected with a luciferase reporter construct containing CCR6 3′-UTR, in which overexpression of miR-335 decreased the luciferase activity. Data in B and C were derived from at least three independent experiments. D, sketch diagram of a luciferase reporter construct containing CCR6 3′-UTR. *, p < 0.05; **, p < 0.01. Error bars, S.E.
Because miRNAs robustly regulate gene expression by degrading and/or repressing the translation of target mRNAs (34), we investigated whether miR-409 and miR-335 bind to their predicted sites of Ccr6 mRNA 3′-UTR and regulate its expression. A reporter vector expressing firefly luciferase from a transcript that contained the Ccr6 3′-UTR (Fig. 4D) was cotransfected with miRNA mimics into HeLa cells. Analysis of luciferase activity in HeLa cells revealed that overexpression of miR-335 reduced the luciferase activity of the reporter construct, whereas overexpression of control scrambled miRNAs did not (Fig. 4C), suggesting that miR-335 was capable of selectively degrading Ccr6 mRNA. Similarly, overexpression of miR-409 in HeLa cells elicited a similar reduction in reporter luciferase activity (data not shown).
Genetic ablation of HuR impairs Th17 cell migration in response to CCL20 in vitro and in vivo
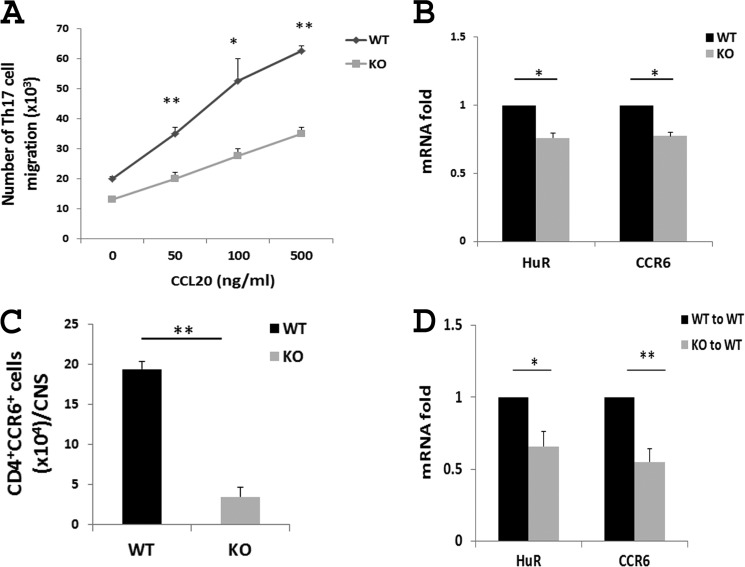
CCR6 is critical for Th17 cell migration to sites of inflammation (10, 12, 29, 35, 36). The lack of CCR6 on Th17 cells in mice decreases their susceptibility to autoimmune diseases, including EAE (12, 29, 35, 36). To evaluate the function of HuR in Th17 cell migration, we used a 24-well plate carrying a transwell-permeable membrane to compare the capacity of WT with HuR KO Th17 cells to migrate toward the chamber containing CCL20, as previously described (11). WT and HuR KO Th17 cells were cultured in the upper well in transmembrane plates, and CCL20 was added to the lower chamber. As shown in Fig. 5A, a large number of WT Th17 cells migrated into the lower chamber in response to CCL20. In contrast, HuR KO Th17 cell migration was significantly decreased, indicating that knock-out of HuR in Th17 cells, where CCR6 expression was impaired, resulted in decreased Th17 cell migration, which is consistent with the in vivo findings that fewer CD4+ T cells were present in the CNS of recipients of HuR KO Th17 cells (28).
Figure 5.
Knock-out of HuR impairs Th17 cell migration in vitro and in vivo. A, Transwell chemotaxis assay of WT and HuR KO Th17 cells was performed in the presence of CCL20 (CCR6 ligand). The migrated cells were analyzed at 3 h of culture. HuR deficiency significantly reduced Th17 cell migration to CCL20. B, CCR6 mRNA level in splenic CD4+ T cells from WT and HuR KO and mice with EAE. C, number of CD4+CCR6+ cells in the CNS of WT and HuR KO mice with EAE measured by flow cytometric analysis. D, CCR6 expression on CNS monocytes from recipients of WT and HuR KO Th17 cells. The results shown in A–D are a summary of at least three independent experiments. All data are expressed as the mean ± S.E. (error bars). *, p < 0.05; **, p < 0.01.
Consistent with this experiment in cultured cells, ex vivo experiments revealed that expression of Ccr6 mRNA in splenic CD4+ T cells was significantly reduced in HuR KO mice compared with that of WT mice with EAE (Fig. 5B), suggesting that HuR may also promote expression of CCR6 by inflammatory cells in vivo. Indeed, flow cytometric analysis revealed that the number of inflammatory CD4+CCR6+ cells was significantly reduced in the CNS of HuR KO mice relative to WT mice with EAE (Fig. 5C). Furthermore, Ccr6 mRNA was much lower in CNS monocytes from mice that received HuR KO Th17 cells than mice receiving WT Th17 cells (Fig. 5D).
HuR regulates CCR6 expression on Th17 cells for promoting the initiation of EAE
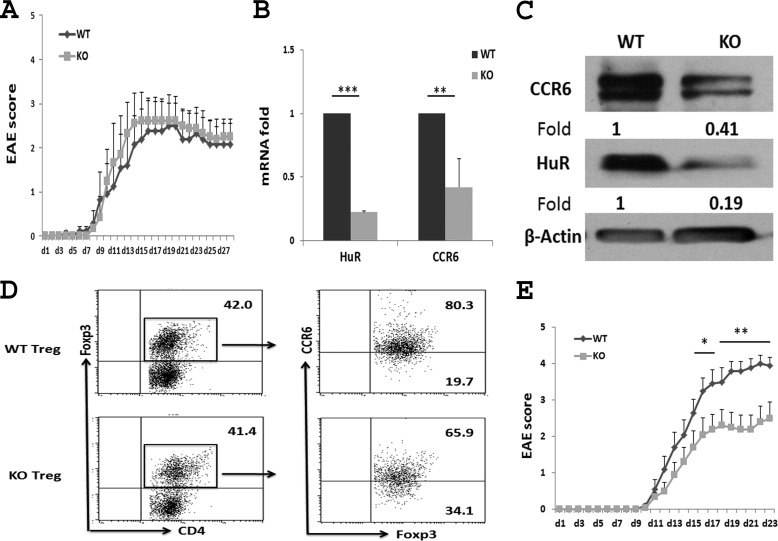
We previously demonstrated that transfer of HuR−/− Th17 cells induced less severe EAE in naive C57BL/6 recipients compared with that of WT Th17 cells (28). However, in active EAE, the clinical score in HuR KO mice is only slightly less severe than that of WT mice at early stages of immunization (days 5 to 10), and the disease severity in both groups is similar at the late stages of induction (Fig. 6A). It is well known that Treg cells play an important role in the resolution of EAE (37–39). Because CCR6 is also expressed on Treg cells, we hypothesized that knock-out of HuR reduces the expression of CCR6 on Treg cells, resulting in fewer Treg cells migrating into the CNS and less suppression of EAE pathogenesis. To test this hypothesis, WT and HuR KO Treg cells were generated from naive CD4+ T cells under Treg cell culture conditions. The levels of Ccr6 mRNA (Fig. 6B) and protein (Fig. 6, C and D) in HuR KO Treg cells were drastically lower than those in WT Treg cells. To exclude the effect of HuR on Treg cells and to better understand the function of HuR in Th17 cells, we depleted Treg cells by injection of anti-CD25 (PC61) 10 days before induction of EAE in WT and HuR KO mice. This treatment regimen was chosen given our previous findings that one injection of anti-CD25 was able to transiently deplete most Treg cells in the spleen and lymph nodes (40, 41). In addition, anti-CD25 antibody was degraded 10–14 days later, which would not deplete the effector T cells when mice are immunized for induction of EAE. The results showed that when Treg cells were transiently depleted, HuR KO mice developed significantly less severe EAE than did WT mice (Fig. 6E), which further demonstrated that knock-out of HuR reduces CCR6 expression on Th17 cells, leading to impaired initiation of EAE.
Figure 6.
After Treg cell depletion, the severity of EAE is lower in HuR KO than in WT mice. A, EAE in WT and HuR KO mice without Treg cell depletion. Each group contains six mice. CCR6 levels on WT and HuR KO Treg cells were analyzed by qRT-PCR (B), Western blots (C), and flow cytometry (D). E, EAE in WT and HuR KO mice in which Treg cells were depleted by anti-CD25 10 days earlier. Each group contains 11 mice. One of two representative experiments is shown in A, C, D, and E. Data in B are derived from at least three independent experiments and presented as mean ± S.E. (error bars). *, p < 0.05; **, p < 0.01.
Discussion
In this study, we have demonstrated that HuR post-transcriptionally regulates CCR6 expression on Th17 cells. Mechanistically, HuR binds to Ccr6 mRNA to promote its stability and prolong its half-life. HuR deficiency reduces Ccr6 mRNA and protein levels, leading to reduced Th17 cell migration, in turn impairing the initiation of EAE.
Animal model studies showed that CCR6 is essential for development of autoimmune diseases, and Ccr6−/− mice are resistant to EAE, rheumatoid arthritis, and atherosclerosis (10, 12, 29, 35, 36). Clinical studies showed that CCR6 is expressed at significantly higher levels in lesions of psoriatic skin than in normal skin (42). CCR6 was identified as a risk factor for rheumatoid arthritis. Moreover, a nucleotide polymorphism within the CCR6 locus was shown to be associated with susceptibility to rheumatoid arthritis, Graves' disease, and Crohn's disease (43, 44). Mechanistic studies further demonstrated that CCR6 is required for pathogenic Th17 cell migration to inflamed tissues (9, 10, 29). More specifically, CCR6-positive Th17 cells cross the blood–brain barrier through the interaction between CCR6 and its ligand CCL20 in choroid plexus to induce EAE and multiple sclerosis (12). Therefore, further understanding the regulation of CCR6 expression may provide a novel venue for drug design to treat autoimmune inflammation.
Previous studies indicated that TGF-β promotes CCR6 expression on Th17 cells through transcription factors RORγt and RORα (11). Further studies identified a noncoding region of the human CCR6 gene with methylation-sensitive transcriptional activity in CCR6+ T cells that controls stable CCR6 expression via epigenetic mechanisms (43). However, the post-transcriptional modulation of CCR6 expression is unknown. Understanding this process is important because post-transcriptional gene regulation permits 1) synthesis of protein by translation from mRNA stored in cytoplasmic granules more rapidly than from newly synthesized transcripts, 2) metabolic economy, because translational repression allows the rapid turning off of protein production while preserving the mRNA, which allows reinitiation at a later time if conditions warrant it, and 3) sensitivity to environmental stimuli via receptors on the cell surface linked to signaling pathways that regulate the translation and decay of mRNA (24). Thus, we studied how CCR6 expression is post-transcriptionally regulated by RNA-binding protein HuR. Interestingly, compared with Th1 and Th2 cells, Th17 cells express the highest level of HuR, and Treg cells express moderate levels of HuR (Fig. 1), consistent with previous reports that SMAD transcriptionally activates HuR expression (45) and that TGF-β required for Th17 and Treg cell differentiation is able to activate SMAD (6, 46, 47).
We previously showed that knock-out of HuR in Th17 cells reduces IL-17 mRNA stability (28). We now provide evidence that Ccr6 mRNA is enriched in anti-HuR IP complexes, indicating that HuR protein interacts with Ccr6 mRNA. In addition, we demonstrate that HuR−/− CD4+ T cells display reduced Ccr6 mRNA half-life when cells are polarized under Th17 cell culture conditions (Fig. 3). Although HuR binds to and stabilizes Ccr6 mRNA to promote its expression, we could not exclude the possibility of HuR modulating transcription factors to modulate CCR6 expression in Th17 cells. Work is in progress to address this possibility.
RNA-binding proteins can coordinate with miRNAs to regulate target gene expression post-transcriptionally (34). Several miRNAs like miR-335 and miR-409 are expressed in Th17 cells. Reporter analyses revealed that miR-335 and miR-409 reduced the activity of vectors containing the Ccr6 3′-UTR. Interestingly, knock-out of HuR in CD4+ T cells increased the expression of these miRNAs, indicating that HuR may negatively regulate their expression. Additional studies are needed to define the underlying mechanism by which HuR inhibits the expression of certain miRNAs. One possible explanation is that HuR cooperates with the miRNA let-7 in down-regulating the expression of c-Myc (48, 49), a transcription factor that promotes microRNA biogenesis (50), and thus knock-out of HuR may increase c-Myc expression, which, in turn, promotes expression of microRNAs. This possibility remains to be tested experimentally.
Our previous work showed that HuR KO Th17 cells are less pathogenic than WT Th17 cells for inducing EAE (28). Our current study revealed that HuR deficiency reduces CCR6 expression in Th17 cells and impairs their migration in response to its ligand CCL20. This finding is consistent with our previous report that fewer CD4+ T cells migrated into the CNS in mice receiving HuR KO Th17 cells compared with those transferred with WT Th17 cells (28). In addition, early reports showed that CCR6-deficient autoreactive Th17 cells failed to migrate to the CNS, which resulted in the development of less severe EAE (12).
Because CCR6 is also expressed on Treg cells (11), knock-out of HuR also reduced CCR6 expression in Treg cells (Fig. 6), resulting in reduced Treg cell migration to the CNS, which might account for the moderate difference in severity at early stages between WT and HuR KO mice in actively induced EAE. To test this possibility, we depleted Treg cells by anti-CD25 in WT and HuR KO mice before the induction of the disease. As expected, Treg cell-depleted HuR KO mice develop significantly less severe EAE than do WT mice (Fig. 6E), supporting the idea that knock-out of HuR dampens the pathogenicity of Th17 cells for induction of EAE. Taken together, our results suggest that HuR modulates CCR6 expression on Th17 cells to contribute to autoimmune neuroinflammation.
Experimental procedures
Animals
HuRflox/flox mice were generated as described previously (28, 51). 8–10-week-old control (HuRflox/flox) mice and HuR KO mice (OX40-cre HuRflox/flox) were used. All mice were on the C57BL/6 background and were bred at the animal facility of Arkansas Bioscience Institute at Arkansas State University and/or the Thomas Jefferson University. Animal experiments were approved by the institutional animal care and use committee and performed according to federal and institutional guidelines.
Isolation and differentiation of CD4+ T cells in vitro
Naive CD4+ T cells were purified from splenocytes using CD4 negative selection kits (catalog no. 19852A, StemCell Technologies, Vancouver, Canada) following the manufacturer's protocol. Single splenocytes were prepared by using a 70-μm nylon mesh cell strainer (catalog no. 22363548, Fisher) and incubated with mouse CD4+ T cell isolation mixture (catalog no. 19852c.1, StemCell Technologies) followed by adding Straptavidin RapidSpheres (catalog no. 50001, StemCell Technologies), placing the tube containing the splenocytes into a magnet for 3 min and pouring the unlabeled CD4+ T cells into a new tube. The isolated CD4+ T cells were cultured as described previously (28, 52). Purified naive CD4+ T Cells were activated with plate-bound anti-CD3 (10 μg/ml) (14-0032, eBioscience (San Diego, CA)) and anti-CD28 (3 μg/ml) (14-0281, eBioscience). For Th1 polarization, IL-12 (10 ng/ml) (419-ML-010, R&D Systems (Minneapolis, MN)) and anti-IL-4 (10 μg/ml) (16-7041, eBioscience) were additionally used. For Th17 polarization, TGF-β (3 ng/ml) (rH100-21, PeproTech), IL-6 (20 ng/ml) (9216-6, PeproTech), IL-23 (20 ng/ml) (14-8231, eBioscience), anti-IFN-γ (10 μg/ml) (16-7312, eBioscience), and anti-IL-4 (10 μg/ml) were additionally used. For Th2 cell polarization, IL-4 (10 μg/ml), IL-2 (10 ng/ml) (212-12, PeproTech), and anti-IFN-γ (10 μg/ml) were added. For Treg cell polarization, TGF-β (5 ng/ml), IL-2 (10 ng/ml), anti-IFN-γ (10 μg/ml), and anti-IL-4 (10 μg/ml) were additionally used. All cytokines were purchased from R&D Systems and PeproTech (Rocky Hill, NJ), and antibodies were purchased from eBioscience.
RNA isolation and qRT-PCR
Cells were collected, and total RNA was extracted using TRIzol (Invitrogen). 500 ng of RNA was reverse-transcribed into cDNA using the SuperScript III kit (Invitrogen) according to the manufacturer's protocols. The cDNA was subjected to quantitative real-time PCR analysis using the CFX96 real-time PCR detection system (Bio-Rad) with SYBR Green reagent kit (Invitrogen) according to the manufacturer's protocols. The Cq data of test mRNAs were normalized to the data of Gapdh mRNA for each sample, and the gene expression of WT was designated as 1. The gene expression in the KO group was relative to those of WT (28). Forward and reverse primers for specific murine target genes are listed in Table 1. Sequences of HuR siRNA and scrambled siRNA are listed in Table 2. Forward and reverse primers for human target genes are listed in Table 3.
Table 1.
Sequences of mouse quantitative RT-PCR primers
| Primers | Sequences |
|---|---|
| CCR6 forward | 5′-CCTCACATTCTTAGGACTGGAGC-3′ |
| CCR6 reverse | 5′-GGCAATCAGAGCTCTCGGA-3′ |
| HuR forward | 5′-ACTGCAGGGATGACATTGGGAGAA-3′ |
| HuR reverse | 5′-AAGCTTTGCAGATTCAACCTCGCC-3′ |
| IL-23R forward | 5′-TTCAGATGGGCATGAATGTTTCT-3′ |
| IL-23R reverse | 5′-CCAAATCCGAGCTGTTGTTCTAT-3′ |
| IL-17A forward | 5′-TGTGTCTCTGATGCTGTTGCTGCT-3′ |
| IL-17A reverse | 5′-AGGAAGTCCTTGGCCTCAGTGTTT-3′ |
| GAPDH forward | 5′-TCAACAGCAACTCCCACTCTTCCA-3′ |
| GAPDH reverse | 5′-ACCCTGTTGCTGTAGCCGTATTCA-3′ |
| IL-22 forward | 5′-TTGAGGTGTCCAACTTCCAGCA-3′ |
| IL-22 reverse | 5′-AGCCGGACGTCTGTGTTGTTA-3′ |
| β-Actin forward | 5′-GGCTGTATTCCCCTCCATCG-3′ |
| β-Actin reverse | 5′-CCAGTTGGTAACAATGCCATGT-3′ |
Table 2.
Sequences of siRNA for human T cell transfection
| siRNAs | Sequences |
|---|---|
| HuR siRNA | 5 ′-GAGUGAAGGAGUUGAAACUTT-3′ |
| Scramble siRNA | 5′-GCCAAUUCAUCAGCAAUGGTT-3′ |
Table 3.
Sequences of human quantitative RT-PCR primers
| Primers | Sequences |
|---|---|
| CCR6 forward | 5′-TGAGCGGGGAATCAATGAATT-3′ |
| CCR6 reverse | 5′-TCCTGCAAGGAGCACAGTAACA-3′ |
| HuR forward | 5′-AACTACGTGACCGCGAAGG-3′ |
| HuR reverse | 5′-CGCCCAAACCGAGAGAACA-3′ |
| IL-23R forward | 5′-ACATGCTTCTATGTACTGCACTG-3′ |
| IL-23R reverse | 5′-TGTGTCTATGTAGGTGAGCTTCC-3′ |
| GAPDH forward | 5′-GGAGCGAGATCCCTCCAAAAT-3′ |
| GAPDH reverse | 5′-GGCTGTTGTCATACTTCTCATGG-3′ |
In vitro migration assay
The migration assay was performed in 24-well plates (Costar, Corning, NY) carrying Transwell-permeable supports with a 5-μm polycarbonate membrane for T cells. Briefly, 5 × 105 polarized Th17 cells were added to the upper wells in 200 μl of culture medium, and the recombinant CCL20 (250-27, PeproTech) was added to the lower chambers in 600 μl of culture medium. Cells were cultured for 3 h, and cell migration was determined by counting the number of T cells in the lower chambers.
Transient depletion of Treg cells and EAE induction
To deplete Treg cells, anti-CD25 antibody (0.5 mg/mouse) (PC61, BioXcell) was injected i.p. 10 days before EAE induction. For induction of active EAE, 8–10-week-old female WT mice and HuR conditional KO mice were immunized with MOG(35–55) and complete Freund's adjuvant followed by injection of pertussis toxin. EAE was clinically assessed by daily assignment of scores on a scale of 0–5 as described previously (28).
Isolation of mononuclear cells from CNS
Mononuclear cells were extracted from inflamed CNS tissue of EAE mice as described previously (28, 52). Briefly, mice were perfused with cold PBS to remove blood from internal organs. The spinal cord was flushed out by hydrostatic pressure. Brain and spinal cords were digested in a solution with 0.2 units/ml Liberase DL (Roche Applied Science) and 1 mg/ml DNase I (Roche Applied Science) in DMEM at 37 °C for 45 min. A single-cell suspension was prepared by passing through a 70-μm cell strainer, followed by centrifugation through a Percoll gradient (37%/70%). Mononuclear cells in the interphase layer were transferred into a fresh tube and used in subsequent experiments for RNA isolation and for flow cytometry analysis.
Western blotting
Whole-cell lysates were prepared in RIP buffer (catalog no. 89900, Thermo Scientific), and Western blot analysis was performed as described previously (28, 52). The concentration of protein was determined with BCA protein assay reagent (Thermo Scientific). Protein samples were separated by 10% SDS-PAGE and transferred to a nitrocellulose membrane (Bio-Rad). The membranes were incubated with primary antibodies recognizing HuR (Sc-5261, Santa Cruz Biotechnology, Inc., Dallas, TX), β-actin (Sc-130657, Santa Cruz Biotechnology), or CCR6 (NBP2-25220, Novus Biologicals) followed by incubation with goat anti-mouse or goat anti-rat secondary Ig conjugated with HRP (Jackson ImmunoResearch Laboratories Inc., West Grove, PA). Membranes were developed by SuperSignal West Pico chemiluminescent substrate (Thermo Scientific).
Flow cytometry
Cells obtained from in vitro and ex vivo culture were stained for surface markers with allophycocyanin-conjugated anti-CD4 and phycoerythrin-conjugated anti-CCR6 (129804, Biolegend, San Diego, CA). Acquisitions were made with a BD FACSCalibur (BD Biosciences). Cell Quest software was used for data analysis.
Immunoprecipitation of endogenous messenger ribonucleoprotein complexes
RIP was performed according to the established protocols (28, 53). Briefly, Th17 polarized cells were lysed using polysome lysis buffer (28). The lysates were precleared for 30 min at 4 °C by adding 30 μg of IgG1 (BD Biosciences) and 50 μl of Protein A–Sepharose beads (P3391, Sigma-Aldrich) swollen in NT2 buffer with 5% BSA (27, 30). Beads (100 μl) were coated by adding 30 μg of either IgG1 (BD Biosciences) as control or anti-HuR antibody 3A2 and incubated overnight at 4 °C. After extensive washes of precoated Protein A–Sepharose beads, 100 μl of precleared lysate was added and incubated for 4 h at 4 °C, and then 30 μg of proteinase K was added and incubated for 30 min at 55 °C to digest protein. RNA was extracted and reverse-transcribed, and the presence of specific target mRNAs was quantified by quantitative PCR analysis.
Transfection and luciferase assay
HeLa cells (CCL-2, ATCC) were cultured in DMEM containing 10% FBS, penicillin (100 units/ml), streptomycin (100 μg/ml), 2 mm l-glutamine, and non-essential amino acids (Invitrogen). A total of 2 × 105 cells in 1 ml of cell culture medium were seeded into each well of 24-well plates and incubated overnight. Transient transfections were performed using Lipofectamine 2000 (Invitrogen) with 0.3 μg of firefly luciferase plasmid DNA (GeneCopoeia, Rockville, MD) and 0.15 μg of pRL-CMV-renilla-luciferase plasmid. Cells were co-transfected with 40–80 nmol of miRNA (Life Sciences). 24 h later, cells were collected, and luciferase activities were measured using a Dual-Luciferase reporter assay system according to the manufacturer's instructions (Promega, Madison, WI). Firefly luciferase activity was normalized to Renilla luciferase in the same transfection groups.
Statistical analysis
Student's t test was used to analyze the differences between two groups for the experiments. The data are expressed as the mean ± S.E. A p value < 0.05 was considered statistically significant.
Author contributions
J. C. and S. Y. designed and performed most of the experiments; J. L. M. did the polysome assay; C. C., M. G., U. A., P. D. D., and S. Y. analyzed the data; J. C., P. D. D., M. G., and S. Y. wrote the manuscript. All authors reviewed the results and approved the final version of manuscript.
This work was supported by National Institutes of Health Grant p20GM103429 from the IDeA Networks of Biomedical Research Excellence (INBRE) Program of the National Center for Research Resources and National Institutes of Health Grant R01AI119135. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- Th17
- T helper 17
- Treg
- T regulatory
- EAE
- experimental autoimmune encephalomyelitis
- HuR KO
- HuR conditional knockout
- CCR6
- C-C chemokine receptor 6
- RIP
- RNA immunoprecipitation
- CCL20
- chemokine ligand 20
- miRNA
- microRNA
- qRT-PCR
- quantitative RT-PCR
- IP
- immunoprecipitation.
References
- 1. Korn T., Bettelli E., Oukka M., and Kuchroo V. K. (2009) IL-17 and Th17 Cells. Annu. Rev. Immunol. 27, 485–517 [DOI] [PubMed] [Google Scholar]
- 2. Park H., Li Z., Yang X. O., Chang S. H., Nurieva R., Wang Y. H., Wang Y., Hood L., Zhu Z., Tian Q., and Dong C. (2005) A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 6, 1133–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harrington L. E., Hatton R. D., Mangan P. R., Turner H., Murphy T. L., Murphy K. M., and Weaver C. T. (2005) Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 6, 1123–1132 [DOI] [PubMed] [Google Scholar]
- 4. Veldhoen M., Hocking R. J., Flavell R. A., and Stockinger B. (2006) Signals mediated by transforming growth factor-β initiate autoimmune encephalomyelitis, but chronic inflammation is needed to sustain disease. Nat. Immunol. 7, 1151–1156 [DOI] [PubMed] [Google Scholar]
- 5. Kebir H., Kreymborg K., Ifergan I., Dodelet-Devillers A., Cayrol R., Bernard M., Giuliani F., Arbour N., Becher B., and Prat A. (2007) Human TH17 lymphocytes promote blood–brain barrier disruption and central nervous system inflammation. Nat. Med. 13, 1173–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mangan P. R., Harrington L. E., O'Quinn D. B., Helms W. S., Bullard D. C., Elson C. O., Hatton R. D., Wahl S. M., Schoeb T. R., and Weaver C. T. (2006) Transforming growth factor-β induces development of the T(H)17 lineage. Nature 441, 231–234 [DOI] [PubMed] [Google Scholar]
- 7. Ye P., Rodriguez F. H., Kanaly S., Stocking K. L., Schurr J., Schwarzenberger P., Oliver P., Huang W., Zhang P., Zhang J., Shellito J. E., Bagby G. J., Nelson S., Charrier K., Peschon J. J., and Kolls J. K. (2001) Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J. Exp. Med. 194, 519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Conti H. R., Shen F., Nayyar N., Stocum E., Sun J. N., Lindemann M. J., Ho A. W., Hai J. H., Yu J. J., Jung J. W., Filler S. G., Masso-Welch P., Edgerton M., and Gaffen S. L. (2009) Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J. Exp. Med. 206, 299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hedrick M. N., Lonsdorf A. S., Shirakawa A. K., Richard Lee C. C., Liao F., Singh S. P., Zhang H. H., Grinberg A., Love P. E., Hwang S. T., and Farber J. M. (2009) CCR6 is required for IL-23-induced psoriasis-like inflammation in mice. J. Clin. Invest. 119, 2317–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hirota K., Yoshitomi H., Hashimoto M., Maeda S., Teradaira S., Sugimoto N., Yamaguchi T., Nomura T., Ito H., Nakamura T., Sakaguchi N., and Sakaguchi S. (2007) Preferential recruitment of CCR6-expressing Th17 cells to inflamed joints via CCL20 in rheumatoid arthritis and its animal model. J. Exp. Med. 204, 2803–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yamazaki T., Yang X. O., Chung Y., Fukunaga A., Nurieva R., Pappu B., Martin-Orozco N., Kang H. S., Ma L., Panopoulos A. D., Craig S., Watowich S. S., Jetten A. M., Tian Q., and Dong C. (2008) CCR6 regulates the migration of inflammatory and regulatory T cells. J. Immunol. 181, 8391–8401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reboldi A., Coisne C., Baumjohann D., Benvenuto F., Bottinelli D., Lira S., Uccelli A., Lanzavecchia A., Engelhardt B., and Sallusto F. (2009) C-C chemokine receptor 6-regulated entry of TH-17 cells into the CNS through the choroid plexus is required for the initiation of EAE. Nat. Immunol. 10, 514–523 [DOI] [PubMed] [Google Scholar]
- 13. Ransohoff R. M. (2009) Immunology: in the beginning. Nature 462, 41–42 [DOI] [PubMed] [Google Scholar]
- 14. Axtell R. C., and Steinman L. (2009) Gaining entry to an uninflamed brain. Nat. Immunol. 10, 453–455 [DOI] [PubMed] [Google Scholar]
- 15. Arima Y., Harada M., Kamimura D., Park J. H., Kawano F., Yull F. E., Kawamoto T., Iwakura Y., Betz U. A., Márquez G., Blackwell T. S., Ohira Y., Hirano T., and Murakami M. (2012) Regional neural activation defines a gateway for autoreactive T cells to cross the blood–brain barrier. Cell 148, 447–457 [DOI] [PubMed] [Google Scholar]
- 16. Tracey K. J. (2012) Immune cells exploit a neural circuit to enter the CNS. Cell 148, 392–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Acosta-Rodriguez E. V., Rivino L., Geginat J., Jarrossay D., Gattorno M., Lanzavecchia A., Sallusto F., and Napolitani G. (2007) Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat. Immunol. 8, 639–646 [DOI] [PubMed] [Google Scholar]
- 18. Palm N. W., and Medzhitov R. (2007) Antifungal defense turns 17. Nat. Immunol. 8, 549–551 [DOI] [PubMed] [Google Scholar]
- 19. Zhou L., Ivanov I. I., Spolski R., Min R., Shenderov K., Egawa T., Levy D. E., Leonard W. J., and Littman D. R. (2007) IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 8, 967–974 [DOI] [PubMed] [Google Scholar]
- 20. Yang X. O., Panopoulos A. D., Nurieva R., Chang S. H., Wang D., Watowich S. S., and Dong C. (2007) STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J. Biol. Chem. 282, 9358–9363 [DOI] [PubMed] [Google Scholar]
- 21. Ivanov I. I., McKenzie B. S., Zhou L., Tadokoro C. E., Lepelley A., Lafaille J. J., Cua D. J., and Littman D. R. (2006) The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 [DOI] [PubMed] [Google Scholar]
- 22. Yang X. O., Pappu B. P., Nurieva R., Akimzhanov A., Kang H. S., Chung Y., Ma L., Shah B., Panopoulos A. D., Schluns K. S., Watowich S. S., Tian Q., Jetten A. M., and Dong C. (2008) T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR α and ROR γ. Immunity 28, 29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kafasla P., Skliris A., and Kontoyiannis D. L. (2014) Post-transcriptional coordination of immunological responses by RNA-binding proteins. Nat. Immunol. 15, 492–502 [DOI] [PubMed] [Google Scholar]
- 24. Anderson P. (2008) Post-transcriptional control of cytokine production. Nat. Immunol. 9, 353–359 [DOI] [PubMed] [Google Scholar]
- 25. Ma W. J., Cheng S., Campbell C., Wright A., and Furneaux H. (1996) Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 271, 8144–8151 [DOI] [PubMed] [Google Scholar]
- 26. López de Silanes I., Zhan M., Lal A., Yang X., and Gorospe M. (2004) Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. U.S.A. 101, 2987–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Srikantan S., Tominaga K., and Gorospe M. (2012) Functional interplay between RNA-binding protein HuR and microRNAs. Curr. Protein Pept. Sci. 13, 372–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen J., Cascio J., Magee J. D., Techasintana P., Gubin M. M., Dahm G. M., Calaluce R., Yu S., and Atasoy U. (2013) Posttranscriptional gene regulation of IL-17 by the RNA-binding protein HuR is required for initiation of experimental autoimmune encephalomyelitis. J. Immunol. 191, 5441–5450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liston A., Kohler R. E., Townley S., Haylock-Jacobs S., Comerford I., Caon A. C., Webster J., Harrison J. M., Swann J., Clark-Lewis I., Korner H., and McColl S. R. (2009) Inhibition of CCR6 function reduces the severity of experimental autoimmune encephalomyelitis via effects on the priming phase of the immune response. J. Immunol. 182, 3121–3130 [DOI] [PubMed] [Google Scholar]
- 30. Atasoy U., Watson J., Patel D., and Keene J. D. (1998) ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J. Cell Sci. 111, 3145–3156 [DOI] [PubMed] [Google Scholar]
- 31. Fujiie S., Hieshima K., Izawa D., Nakayama T., Fujisawa R., Ohyanagi H., and Yoshie O. (2001) Proinflammatory cytokines induce liver and activation-regulated chemokine/macrophage inflammatory protein-3α/CCL20 in mucosal epithelial cells through NF-κB [correction of NK-κB]. Int. Immunol. 13, 1255–1263 [DOI] [PubMed] [Google Scholar]
- 32. Ito T., Carson W. F. 4th, Cavassani K. A., Connett J. M., and Kunkel S. L. (2011) CCR6 as a mediator of immunity in the lung and gut. Exp. Cell Res. 317, 613–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lu L., Zheng L., Si Y., Luo W., Dujardin G., Kwan T., Potochick N. R., Thompson S. R., Schneider D. A., and King P. H. (2014) Hu antigen R (HuR) is a positive regulator of the RNA-binding proteins TDP-43 and FUS/TLS: implications for amyotrophic lateral sclerosis. J. Biol. Chem. 289, 31792–31804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xiao C., and Rajewsky K. (2009) MicroRNA control in the immune system: basic principles. Cell 136, 26–36 [DOI] [PubMed] [Google Scholar]
- 35. Varona R., Villares R., Carramolino L., Goya I., Zaballos A., Gutiérrez J., Torres M., Martínez-A C., and Márquez G. (2001) CCR6-deficient mice have impaired leukocyte homeostasis and altered contact hypersensitivity and delayed-type hypersensitivity responses. J. Clin. Invest. 107, R37–R45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wan W., Lim J. K., Lionakis M. S., Rivollier A., McDermott D. H., Kelsall B. L., Farber J. M., and Murphy P. M. (2011) Genetic deletion of chemokine receptor Ccr6 decreases atherogenesis in ApoE-deficient mice. Circ. Res. 109, 374–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu Y., Teige I., Birnir B., and Issazadeh-Navikas S. (2006) Neuron-mediated generation of regulatory T cells from encephalitogenic T cells suppresses EAE. Nat. Med. 12, 518–525 [DOI] [PubMed] [Google Scholar]
- 38. O'Connor R. A., Malpass K. H., and Anderton S. M. (2007) The inflamed central nervous system drives the activation and rapid proliferation of Foxp3+ regulatory T cells. J. Immunol. 179, 958–966 [DOI] [PubMed] [Google Scholar]
- 39. Villares R., Cadenas V., Lozano M., Almonacid L., Zaballos A., Martínez-A C., and Varona R. (2009) CCR6 regulates EAE pathogenesis by controlling regulatory CD4+ T-cell recruitment to target tissues. Eur. J. Immunol. 39, 1671–1681 [DOI] [PubMed] [Google Scholar]
- 40. Yu S., Ellis J. S., Dunn R., Kehry M. R., and Braley-Mullen H. (2012) Transient depletion of B cells in young mice results in activation of regulatory T cells that inhibit development of autoimmune disease in adults. Int. Immunol. 24, 233–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu S., Maiti P. K., Dyson M., Jain R., and Braley-Mullen H. (2006) B cell-deficient NOD.H-2h4 mice have CD4+CD25+ T regulatory cells that inhibit the development of spontaneous autoimmune thyroiditis. J. Exp. Med. 203, 349–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Homey B., Dieu-Nosjean M. C., Wiesenborn A., Massacrier C., Pin J. J., Oldham E., Catron D., Buchanan M. E., Müller A., deWaal Malefyt R., Deng G., Orozco R., Ruzicka T., Lehmann P., Lebecque S., Caux C., and Zlotnik A. (2000) Up-regulation of macrophage inflammatory protein-3 α/CCL20 and CC chemokine receptor 6 in psoriasis. J. Immunol. 164, 6621–6632 [DOI] [PubMed] [Google Scholar]
- 43. Steinfelder S., Floess S., Engelbert D., Haeringer B., Baron U., Rivino L., Steckel B., Gruetzkau A., Olek S., Geginat J., Huehn J., and Hamann A. (2011) Epigenetic modification of the human CCR6 gene is associated with stable CCR6 expression in T cells. Blood 117, 2839–2846 [DOI] [PubMed] [Google Scholar]
- 44. Stahl E. A., Raychaudhuri S., Remmers E. F., Xie G., Eyre S., Thomson B. P., Li Y., Kurreeman F. A., Zhernakova A., Hinks A., Guiducci C., Chen R., Alfredsson L., Amos C. I., Ardlie K. G., et al. (2010) Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 42, 508–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jeyaraj S. C., Singh M., Ayupova D. A., Govindaraju S., and Lee B. S. (2010) Transcriptional control of human antigen R by bone morphogenetic protein. J. Biol. Chem. 285, 4432–4440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen W., Jin W., Hardegen N., Lei K. J., Li L., Marinos N., McGrady G., and Wahl S. M. (2003) Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J. Exp. Med. 198, 1875–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen W., and Konkel J. E. (2015) Development of thymic Foxp3+ regulatory T cells: TGF-β matters. Eur. J. Immunol. 45, 958–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gunzburg M. J., Sivakumaran A., Pendini N. R., Yoon J. H., Gorospe M., Wilce M. C., and Wilce J. A. (2015) Cooperative interplay of let-7 mimic and HuR with MYC RNA. Cell Cycle 14, 2729–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim H. H., Kuwano Y., Srikantan S., Lee E. K., Martindale J. L., and Gorospe M. (2009) HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 23, 1743–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. O'Donnell K. A., Wentzel E. A., Zeller K. I., Dang C. V., and Mendell J. T. (2005) c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435, 839–843 [DOI] [PubMed] [Google Scholar]
- 51. Gubin M. M., Techasintana P., Magee J. D., Dahm G. M., Calaluce R., Martindale J. L., Whitney M. S., Franklin C. L., Besch-Williford C., Hollingsworth J. W., Abdelmohsen K., Gorospe M., and Atasoy U. (2014) Conditional knockout of the RNA-binding protein HuR in CD4+ T cells reveals a gene dosage effect on cytokine production. Mol. Med. 20, 93–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xie L., Chen J., McMickle A., Awar N., Nady S., Sredni B., Drew P. D., and Yu S. (2014) The immunomodulator AS101 suppresses production of inflammatory cytokines and ameliorates the pathogenesis of experimental autoimmune encephalomyelitis. J. Neuroimmunol. 273, 31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stellato C., Gubin M. M., Magee J. D., Fang X., Fan J., Tartar D. M., Chen J., Dahm G. M., Calaluce R., Mori F., Jackson G. A., Casolaro V., Franklin C. L., and Atasoy U. (2011) Coordinate regulation of GATA-3 and Th2 cytokine gene expression by the RNA-binding protein HuR. J. Immunol. 187, 441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]