Abstract
Urease is a ubiquitous nickel metalloenzyme. In plants, its activation requires three urease accessory proteins (UAPs), UreD, UreF, and UreG. In bacteria, the UAPs interact with urease and facilitate activation, which involves the channeling of two nickel ions into the active site. So far this process has not been investigated in eukaryotes. Using affinity pulldowns of Strep-tagged UAPs from Arabidopsis and rice transiently expressed in planta, we demonstrate that a urease–UreD–UreF–UreG complex exists in plants and show its stepwise assembly. UreG is crucial for nickel delivery because UreG-dependent urease activation in vitro was observed only with UreG obtained from nickel-sufficient plants. This activation competence could not be generated in vitro by incubation of UreG with nickel, bicarbonate, and GTP. Compared with their bacterial orthologs, plant UreGs possess an N-terminal extension containing a His- and Asp/Glu-rich hypervariable region followed by a highly conserved sequence comprising two potential HXH metal-binding sites. Complementing the ureG-1 mutant of Arabidopsis with N-terminal deletion variants of UreG demonstrated that the hypervariable region has a minor impact on activation efficiency, whereas the conserved region up to the first HXH motif is highly beneficial and up to the second HXH motif strictly required for activation. We also show that urease reaches its full activity several days after nickel becomes available in the leaves, indicating that urease activation is limited by nickel accessibility in vivo. Our data uncover the crucial role of UreG for nickel delivery during eukaryotic urease activation, inciting further investigations of the details of this process.
Keywords: chaperone, enzyme processing, metal ion-protein interaction, metalloenzyme, nickel, protein complex
Introduction
Nitrogen is the mineral nutrient that plants require in the highest amounts and that often limits plant growth. Although urea accounts today for >50% of the N-fertilizer used worldwide (International Fertilizer Industry Association), only little is known about the direct utilization of fertilizer urea by plants. Generally it is assumed that urea is fully degraded in the soil by microbes and that plants mainly take up ammonium and nitrate. However, urea can be actively transported and assimilated by plants (1–4), indicating its significance for plant nutrition. Apart from external sources, urea also originates within plants as a product of arginine catabolism (5). The hydrolysis of urea to ammonia and carbonic acid is facilitated by urease, a nickel metalloenzyme found not only ubiquitously in plants (6) but also in many fungi and bacteria (7). The urease activation process, which is best characterized in Klebsiella aerogenes and Helicobacter pylori (8), requires the carbamylation of a lysine residue in the active site followed by incorporation of two nickel ions. In vivo, the three urease accessory proteins (UAPs)2 UreD, UreF, and UreG are necessary for metallocenter assembly (9), and a series of complexes of UAPs with apo-urease were shown to exist (10). A fourth UAP, UreE, facilitates the activation process by acting as a nickel metallochaperone (8, 11). Crystal structure data of a UreD–UreF–UreG complex from H. pylori showed at atomic resolution how two UreD (called UreH in this organism) interact with dimeric UreF to form a docking site for a UreG dimer (12). UreE delivers nickel to UreG (13, 11), which is channeled via UreG, UreF, and finally UreD to urease (14). UreG acts as GTPase during this process but it is still not entirely clear why GTP hydrolysis is necessary.
In plants, orthologs of UreD, UreF and UreG were identified and shown to be required and sufficient for urease activation (15, 16). A UreE homolog was not found in plants, but it was suggested that plant UreGs might additionally possess UreE function (17–19). This hypothesis was based on the observation that plant UreGs possess a His- and Asp/Glu-rich N terminus not present in the bacterial orthologs that might be involved in the binding and delivery of Ni2+ ions. A similar role has been proposed for bacterial HypB proteins involved in maturation of Ni-Fe-hydrogenases. They possess GTPase activity and contain a His-rich region at the N terminus, which binds nickel ions (20).
In this study we analyzed the interactions between plant urease and its UAPs in detail and present a model for the formation of the activation complex. Urease activation was investigated in vitro using Strep-tagged urease–UAP complexes as well as in vivo expressing a series of N-terminal deletion variants of UreG in a ureG mutant background. Our data demonstrate that UreG is crucial for nickel delivery during the urease activation process, which requires several days in vivo.
Results
Interaction of plant urease and its accessory proteins
To obtain a complete interaction map of a plant urease and its accessory proteins (UAPs), the respective Arabidopsis cDNAs were cloned into binary vectors for transient expression in Nicotiana benthamiana. For each UAP, two constructs were prepared allowing the expression of either untagged or N-terminally Strep-tagged proteins for affinity purification using StrepTactin Macroprep beads. A urease clone in pXCS-HAStrep (V13; Ref. 21) was constructed for the expression of the untagged enzyme. Employing this construct, only small amounts of protein could be generated. To optimize urease expression, bases immediately upstream of the AUG start site were modified to better match the start-site consensus of dicot plants (22). However, expression was not improved by this modification (supplemental Fig. S1). A significant boost of protein quantity could be achieved by expressing urease from a new vector termed pXCScpmv-HAStrep (V69) containing modified 5′- and 3′-UTRs of RNA-2 from the cowpea mosaic virus that act as translational enhancers (23; supplemental Fig. S1).
Urease and UAPs were co-expressed in N. benthamiana in different combinations with generally one of the UAPs Strep-tagged at the N terminus for affinity purification. Successful complementation of UAP mutants with the respective Strep-tagged UAPs in transgenic Arabidopsis plants showed that the tagged versions of these proteins are functional (supplemental Fig. S2). Purified and co-purified UAPs as well as co-purified urease were visualized in Western blots with anti-UAP antisera, newly developed by us, and a commercial anti-urease antibody. The new antisera specifically detected the respective UAPs (supplemental Figs. S2 and S3).
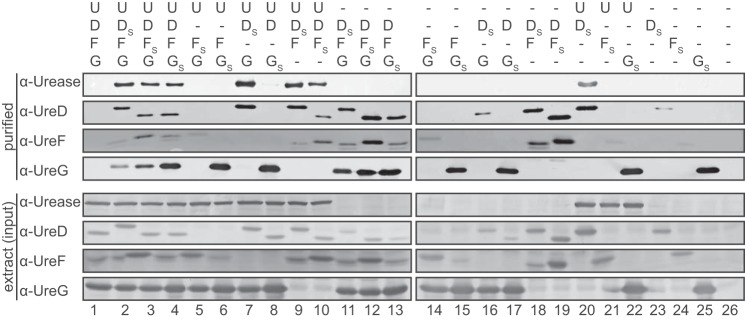
In the absence of urease, direct binding of UreD to UreF was observed (Fig. 1, lanes 18 and 19), demonstrating the formation of a UreD–UreF complex. UreG on the other hand was binding neither UreD (lanes 16 and 17) nor UreF (lanes 14 and 15) alone but was co-purified when expressed with UreD and UreF together (lanes 11–13). These results show that UreG can interact with a UreD–UreF complex forming a UreD–UreF–UreG complex.
Figure 1.
Protein interactions of A. thaliana UAPs and urease. Urease (U) and the accessory proteins UreD, UreF, and UreG (D, F, and G) were co-expressed in leaves of N. benthamiana as indicated. N-terminally Strep-tagged variants of the UAPs (the respective tagged protein is labeled with S) and its interaction partners were affinity-purified from extracts and detected on Western blots. Lower panel, clarified leaf extracts (input). Upper panel, after affinity purification. The experiment was repeated three times, and a representative repeat is shown.
When studying the binding of urease to the UAPs, a direct interaction with urease was only observed for UreD but not for UreF or UreG (lanes 20–22). A direct interaction between urease and UreD had already been shown for the rice proteins (16). Co-purification of urease with tagged UreF was only possible in the presence of UreD (lane 10) and with tagged UreG only in the presence of UreD and UreF (lane 4).
These findings in dicotyledonous Arabidopsis prompted us to investigate the urease activation complex in rice as an important representative of the monocots. From leaves of N. benthamiana transiently expressing rice urease and the three rice UAPs (with UreD tagged), a urease–UreD–UreF–UreG complex could be purified (supplemental Fig. S4). Because orthologs of urease and of the UAPs are encoded by the genomes of all plants sequenced to date (16), it appears likely that the urease–UreD–UreF–UreG activation complex is universal in plants.
An assembly model derived from the co-purification data (Fig. 2) suggests that there are two main routes toward the urease–UreD–UreF–UreG complex: (i) binding of UreD to urease followed by UreF and finally UreG or (ii) the formation of a UreD–UreF–UreG complex by binding of UreG to preformed UreD–UreF, which then can interact with urease. The formation of a urease–UreD–UreF complex by urease and UreD–UreF represents a third intermediate assembly route.
Figure 2.
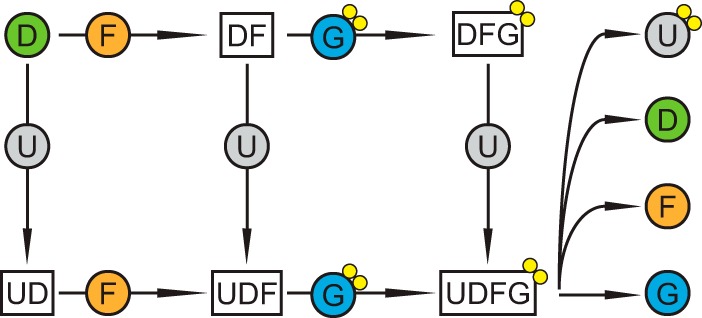
Assembly model for the plant urease activation complex. Urease (U) either binds the accessory proteins UreD (D), UreF (F), and UreG (G) sequentially or the UreD–UreF–UreG complex as a whole. In both cases a urease–UreD–UreF–UreG complex is formed. Nickel ions are shown as yellow circles.
To assess if a preformed UreD–UreF–UreG complex interacts with urease as proposed in this model, such a complex was purified from N. benthamiana as described before but left bound on the StrepTactin matrix and then incubated with a leaf extract expressing only urease (Fig. 3). The matrix was washed, and the proteins were eluted with biotin. Urease could be recovered after incubation with a matrix loaded with UreD–UreF–UreG (Fig. 3, lane 5) but not in a control experiment with empty matrix (Fig. 3, lane 6) demonstrating that urease can bind to a preformed UreD–UreF–UreG complex.
Figure 3.
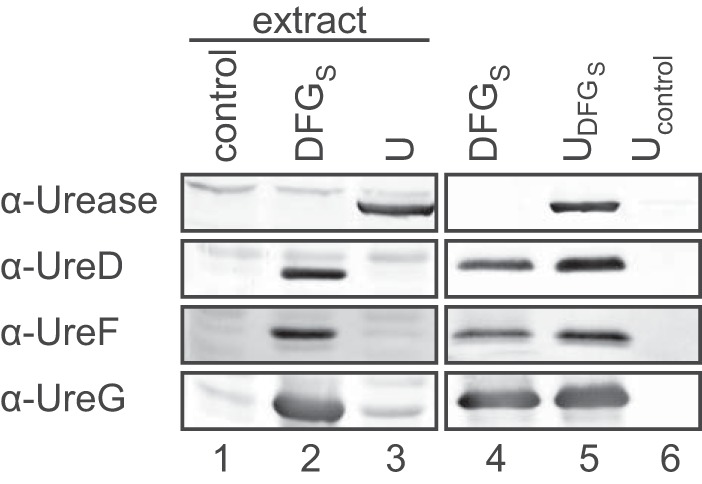
Interaction of the UreD–UreF–UreG complex with urease. The following proteins were transiently expressed in N. benthamiana: lane 1, none (control); lane 2, ureD, ureF, and Strep-tagged ureG (DFGS); lane 3, urease (U). The UreD–UreF–UreGS complex was affinity-purified without eluting the proteins from the matrix, and an aliquot was analyzed (lane 4). The clarified extract from urease-expressing leaves was incubated either with purified UreD–UreF–UreGS complex still attached to Streptactin beads or with fresh Streptactin beads (control). After washing, the respective affinity-bound proteins were eluted and detected (lanes 5 and 6) by Western blot analysis. The experiment was repeated two times, and one of the repeats is shown.
Urease activation in vitro
The peat substrate used for growing N. benthamiana employed for transient expression usually did not supply sufficient nickel for urease activation in planta. Therefore, it was possible to purify inactive urease–UAP complexes from these plants. To investigate whether the urease–UreD–UreF–UreG complex is competent to activate urease, it was purified, and several in vitro activation assays were performed. In a first approach, the complex was incubated in a time course with high concentrations of nickel, bicarbonate, and GTP known to be required for bacterial urease activation (24), and then the resulting urease activity was assessed. The activation of urease was rapid, and the activity did not further increase during the time course (supplemental Fig. S5). Inclusion of EDTA in the urease assay showed that initially the activity was sensitive to EDTA and that an EDTA-insensitive urease activity was formed gradually over several hours. The kinetics of this urease activation was not accelerated, and the total activity was suppressed by the addition of GTP (supplemental Fig. S5B).
A similar GTP-independent activation was also described for the in vitro activation of urease from K. aerogenes but was considered to be physiologically irrelevant because of the high nickel and bicarbonate concentrations required (24). We hypothesized that a positive effect of GTP on the activation might only occur at lower concentrations of NiCl2 and NaHCO3, which by itself (without GTP) would not be sufficient for full activation. To first identify concentrations leading to intermediate activation, a range of bicarbonate and nickel concentration was tested for activation of the purified urease–UreD–UreF–UreG complex. Mid-range activation was found with 2 mm NaHCO3 and 5 μm NiCl2 (Fig. 4, upper and middle panels). With these concentrations set, the activation assay was carried out in the presence of either 0.15 mm or 1.5 mm GTP, respectively. Surprisingly, the addition of GTP to the assay mix did not accelerate the activation of urease but, rather, inhibited it in a dosage-dependent manner (Fig. 4, lower panel). Because magnesium ions were provided in a 2-fold excess of GTP, nickel chelation by the nucleotide as an explanation for the inhibition appears unlikely. Magnesium alone without GTP had no effect on the activation kinetics.
Figure 4.
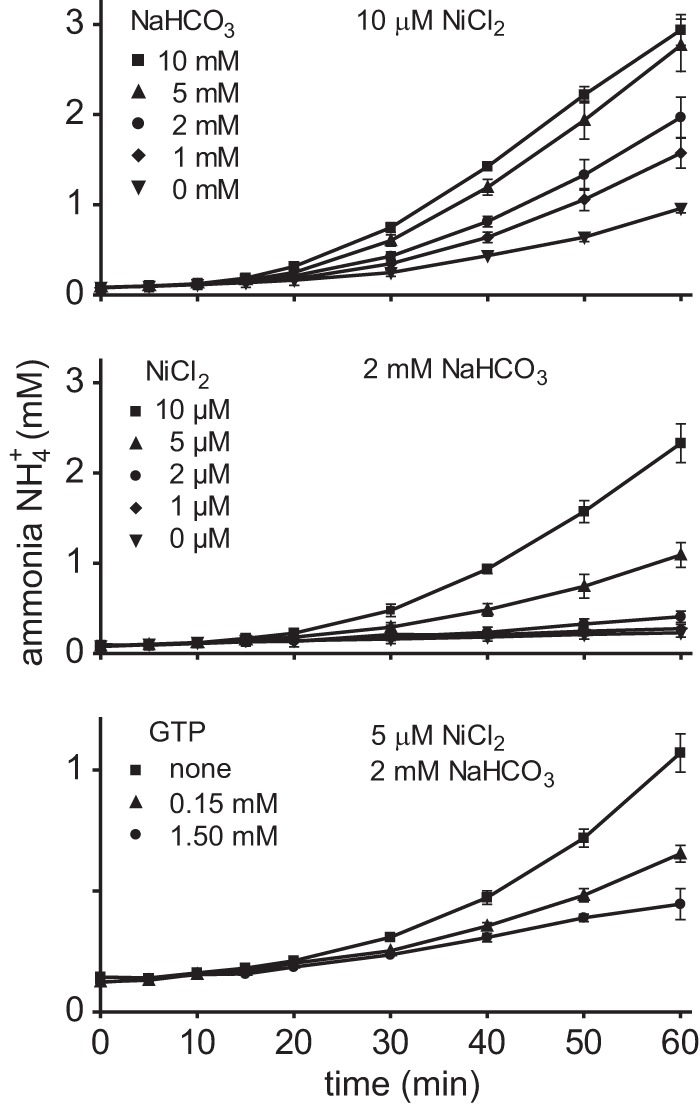
The influence of NiCl2, NaHCO3, and GTP on urease activation in vitro. Purified urease–UreDS–UreF–UreG complex was incubated with 10 μm NiCl2 and different concentrations of NaHCO3 and 5 mm urea, and ammonia release was monitored (upper panel). Identifying 2 mm NaHCO3 as the concentration for moderate urease activation, the experiment was repeated with this concentration of NaHCO3 and different concentrations of NiCl2 (middle panel). Under these conditions, the presence of 5 μm NiCl2 resulted in an intermediate rate of ammonia release. With 2 mm NaHCO3 and 5 μm NiCl2 set, the influence of GTP on the activation was tested (lower panel). The experiments were repeated three times (n = 3, error bars = S.D.).
Next it was tested whether the observed activation was at all dependent on the presence of UreF and UreG. Urease in complex with either UreD, UreF, and UreG or only UreD and UreF or with UreD alone was purified and incubated with 5 μm NiCl2, 2 mm NaHCO3, and 5 mm urea. The urease activities resulting from urease activation were normalized by the relative amount of purified urease protein present in each assay (Fig. 5). Urease from the urease–UreD and urease–UreD–UreF complexes was activated to a higher extent than urease from the urease–UreD–UreF–UreG complex. When these samples were subjected to in-gel urease activity-staining, all three setups resulted in an active urease with identical electrophoretic mobility as found in the control, which was an extract from leaves of the Arabidopsis wild type (Fig. 5, inset). However, active urease detectable by in-gel assays was obtained only after prolonged incubation with NiCl2 and NaHCO3 (>16 h) indicating that urease folding into the native form, which maintains its activity during electrophoresis, required several hours.
Figure 5.
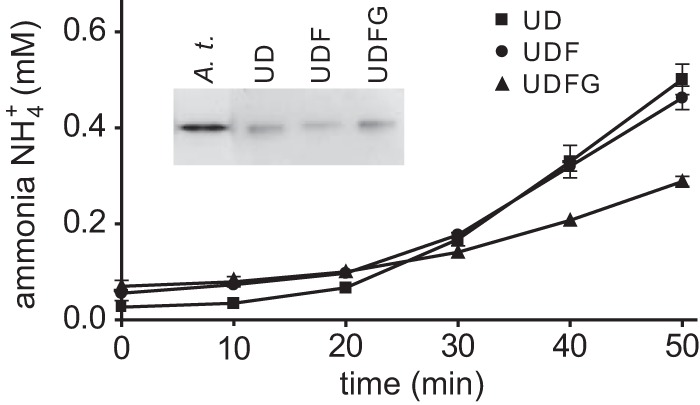
In vitro activation of different urease–UAP complexes. Urease–UreDS (UD)- and urease–UreDSUreF (UDF)- and urease–UreDS–UreF–UreG (UDFG) complexes, transiently expressed in N. benthamiana and purified using N-terminally Strep-tagged UreD, were incubated with 5 μm NiCl2, 2 mm NaHCO3, and 5 mm urea. Ammonia release by urease was measured and normalized to the relative amount of purified urease protein present in each assay, which was quantified by Western blot. The experiment was repeated three times (n = 3, error bars = S.D.). After 16 h incubation, the samples and a clarified extract of Arabidopsis wild-type leaves (A.t.) as positive control were separated by native gel electrophoresis and tested for in-gel urease activity (inset).
Although prolonged in vitro activation generated an EDTA-resistant urease enzyme (supplemental Fig. S5), showing the same electrophoretic mobility as the urease activated in planta (Fig. 5), this activation did not resemble the activation process in vivo where all three UAPs were shown to be required (15). Despite substantial efforts, we were unable to generate conditions where UreF and UreG promoted the activation in the presence of GTP and limiting amounts of nickel and bicarbonate. UreG even had an inhibitory effect (Fig. 5), maybe because it partially blocked the diffusion of nickel and bicarbonate toward the active site of urease.
UreG can acquire in vitro activation competence in vivo
Because GTP and UreG were not required in our in vitro activation trials, we speculated that we did not purify an activation-competent species of UreG and were unable to generate such a species in vitro. This prompted us to further investigate UreG and its proposed role in nickel delivery during activation (18, 17), especially since it was recently suggested that UreG from soybean might not be involved in nickel delivery (19). UreG was expressed alone in N. benthamiana plants, which were either left unfertilized or were fertilized with NiCl2. The peat substrate employed in our experiments usually did not supply sufficient nickel for urease activation, making nickel fertilization mandatory for obtaining active urease in planta. Desalted extracts of UreG-expressing plants were mixed with desalted extracts of plants expressing the urease–UreD–UreF complex (again with or without Ni2+ fertilization), incubated for the indicated times, and then urease activity was monitored in these mixtures (Fig. 6A). Urease activation was only observed when UreG from nickel-fertilized plants was present in the mixtures. UreG from unfertilized plants was not activation-competent, and it was irrelevant whether extracts containing the urease–UreD–UreF complex came from fertilized or unfertilized plants. This indicated that UreG must come into contact with nickel in vivo to be converted into the activation-competent form. Alternatively, a component of the desalted extract from Ni2+-fertilized plants expressing UreG is required for activation. To distinguish these two possibilities, Strep-tagged UreG was purified and mixed either with a urease–UreD–UreF-containing extract or with a purified urease–UreD–UreF complex (purified via Strep-tagged UreD). Urease activation occurred in all cases where UreG was exposed to nickel in vivo, even when purified proteins were combined (Fig. 6B), showing for the first time a UreG-dependent activation of a eukaryotic urease with purified proteins. Note that neither nickel nor bicarbonate nor GTP was added to the activation mixture. These data demonstrate that extract components were not required but that activation competence lay within the UreG protein itself after coming into contact with nickel in vivo.
Figure 6.
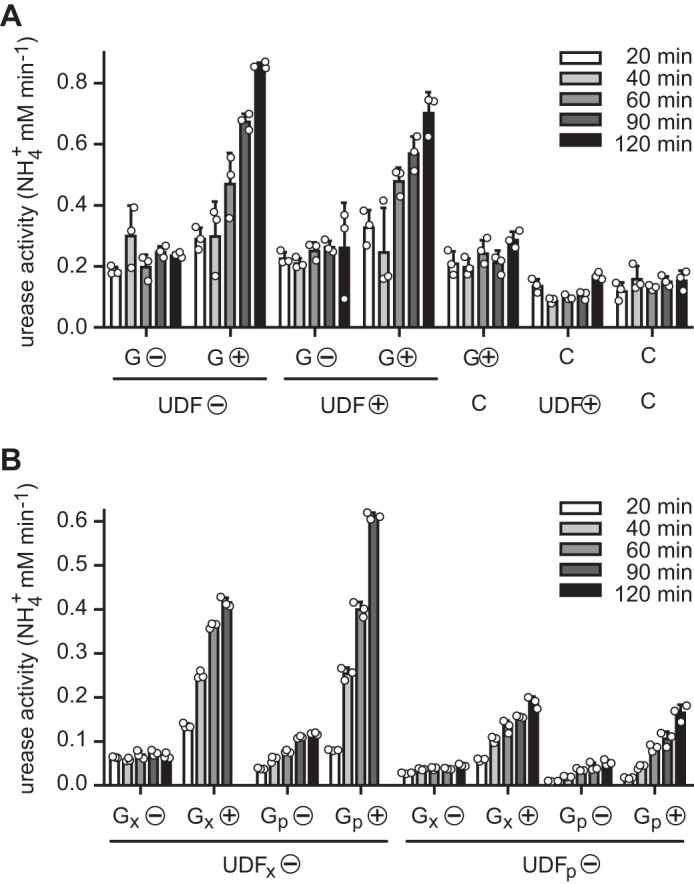
UreG-dependent in vitro activation of urease. A, nickel-fertilized and non-fertilized N. benthamiana plants (⊕ or ⊝) were used to transiently express Arabidopsis UreG or to co-express urease together with UreD and UreF (UreG and UreD were Strep-tagged). Clarified and desalted extracts of these plants (G and UDF) were mixed with each other or with the extract of a control plant (C) not expressing any proteins and incubated for the indicated times. After incubation urea was added, and the urease activity was determined. B, the experiment was in part repeated and expanded mixing clarified and desalted extracts (index: x) and purified proteins (index: p). For purification, N-terminally Strep-tagged UreG (for Gp) and UreD (for UDFp) were employed. Activation assays were carried out in triplicate (individual data points shown as open circles) in both experiments (error bars = S.D.).
Plant UreG plays a role in nickel delivery to urease
It was noted before that plant UreG proteins possess an N-terminal extension in comparison to their bacterial orthologs (17, 18). This extension contains a His- and Asp/Glu-rich stretch immediately downstream of the start codon, which is hypervariable in sequence and length (16), followed by ∼40 amino acids that are highly conserved in plant UreGs comprising two similar HXH motifs: HSHD and H(S/T)H(E/D) beginning at amino acid 38 and 46 in the Arabidopsis sequence, respectively (Fig. 7A).
Figure 7.
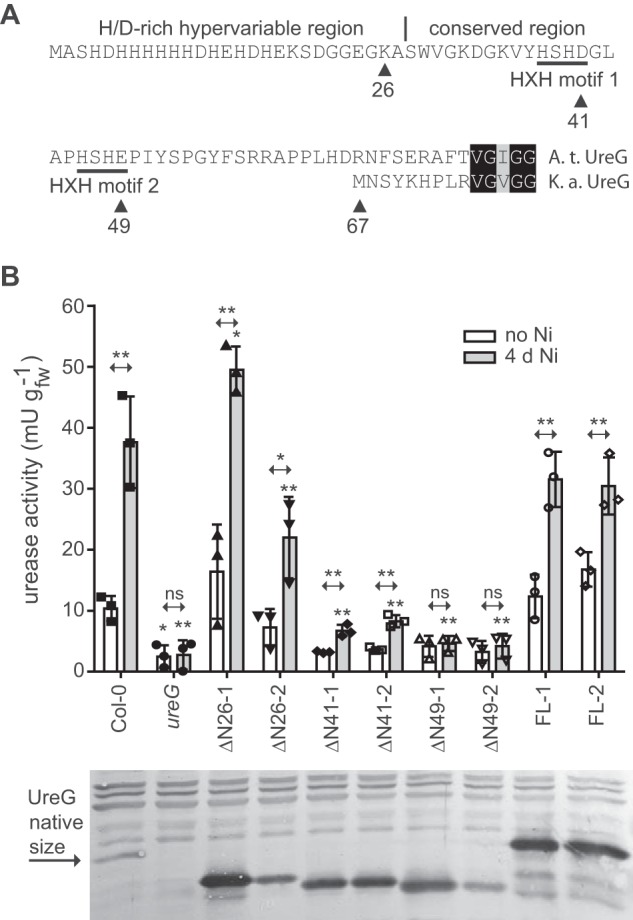
The plant-specific N terminus of UreG is important for urease activation. A, the plant-specific N terminus of UreG (from Arabidopsis (A.t.)) in comparison to the N terminus of UreG from K. aerogenes (K.a.). The HD/E-rich hypervariable region, the conserved region, and the HXH motifs are indicated. Triangles with numbers mark the amino acid up to which the N terminus was deleted in several N-terminal truncation mutants used to transform the Arabidopsis ureG-1 knock-out mutant. B, urease activity was measured in rosette extracts of the wild type (Col-0), the ureG-1 mutant (ureG), and several transformants that over-expressed full-length (FLx) and truncated variants of UreG (ΔNx) in the ureG-1 background. Two independent transgenic lines for each UreG variant (if possible varying in expression) were employed. Three biological replicates (each containing three plants) were assessed per line, and the individual data points were plotted as squares, triangles, circles, or diamonds. The experiment was conducted twice on plants grown in parallel, once with unfertilized and once with NiCl2-fertilized plants (4 days after nickel supplementation). For both experiments a separate analysis of variance analysis was conducted and coupled with a Dunnett's post test comparing the wild type to the other genotypes. Probabilities are indicated by asterisks (*, p < 0.05; **, p < 0.01). Error bars are S.D. t tests were conducted for each genotype comparing activities before and after the nickel addition. The results are indicated at the double arrows over the bars (*, p < 0.05; **, p < 0.01; ns = non significant). Lower panel, Western blot developed with anti-UreG antiserum. Samples from the three biological replicates used in the activity assay were pooled for the blot (each lane represents the average of nine plants). ns, not significant.
A sequence of 15 amino acids rich in His is also found at the C terminus of the K. aerogenes UreE, which shows no further sequence resemblance to plant UreGs. UreE dimers of K. aerogenes can bind up to six Ni2+, four of which are bound by the C terminus. However, the C terminus is not required for urease activation in vivo even at low nickel availability (25). An internal Ni2+-binding site at the dimer interface involving two His, one from each monomer, is conserved in bacterial UreEs (26) and is required for UreE activity (27). More His residues, in some cases organized in an HXH motif, at the C terminus of UreE proteins from different origins are also involved in Ni2+ coordination (28). If plant UreG comprises UreE functionality, we hypothesized that the variable His- and Asp/Glu-rich sequence may be involved in this function as well as the two HXH motifs. These contain all invariant His residues in the conserved region of the plant-specific N-terminal UreG extension. Four N-terminal deletion mutants were generated, (i) eliminating only the hypervariable region of 26 amino acids (mutant ΔN26), (ii) expanding the deletion into the first HXH motif (mutant ΔN41), (iii) including both HXH motives (mutant ΔN49), and (iv) eliminating the entire plant-specific N-terminal extension (mutant ΔN67). Constructs containing a full length UreG as well as the four deletion variants, all driven by a strong 35S promoter, were transformed into the ureG-1 knock-out line (15). To assess dose-dependent effects, lines with stronger and weaker expression of the corresponding variants were selected. UreG-ΔN67 was undetectable in the transgenic lines probably because the protein was unstable; therefore, these lines were not included in further experiments.
To assess the ability of the UreG variants to activate urease in a nickel-dependent manner, the urease activity of 4-week-old plants (just before bolting) grown on peat soil was assessed before and 4 days after nickel fertilization (Fig. 7B). Low-level urease activity was observed in the wild type even before the nickel treatment. As in the ureG mutant, the activity was near to zero (at background level) without nickel addition in the complementation lines containing the ΔN41- and ΔN49-UreG variants. However, a statistically significant (at p < 0.05 level) difference in the wild type was only obtained for the ureG mutant. Nickel treatment led to a significant rise in urease activity in all genotypes except the ureG mutant and the two ΔN49-UreG-expressing lines. With the amount of nickel added, the wild type reached ∼30% of the maximal possible activity (15) showing that activation was not complete. The UreG variants were intentionally assessed at sub-optimal nickel availability to reveal a potential role of the plant-specific N terminus of UreG in nickel efficiency. The ΔN26-UreG variant still activated urease but appeared to be less efficient when expressed at near wild-type level (ΔN26–2), indicating that the hypervariable His- and Asp/Glu-rich N terminus boosts the efficiency of UreG but is not strictly required. The variant lacking the hypervariable region and the first HXH motif (ΔN41) still retains activation competence albeit at strongly reduced efficiency, whereas the ΔN49-UreG variant loses the capacity to activate urease even when strongly overexpressed. These data demonstrate that the conserved part of the plant-specific N-terminal UreG extension plays a major role for UreG functionality.
Urease activation in vivo
Activation of K. aerogenes urease in vitro at near physiological conditions with regard to nickel and bicarbonate concentrations is a slow process requiring up to 10 h (29). Similar results were obtained for in vivo activation, which even after 4 h of nickel supplementation is not completed (30). For plant ureases nothing is known about the kinetics of the in vivo activation. To study this process in a time-resolved manner, Arabidopsis plants were grown hydroponically for 5½ weeks without nickel. After supplementing the nutrient solution with 10 μm NiCl2 (no nickel in the control), plants were harvested in a time course, and urease activity and nickel concentration were assessed. Both remained unchanged in the unfertilized controls (supplemental Fig. 6, gray lines). In fertilized plants, the initial nickel concentration was only ∼10% of the final concentration of ∼0.02 nmol mg−1 fresh weight reached after 4 days (Fig. 8). The initial nickel concentration was sufficient to activate ∼40% of the maximum urease activity of ∼17 milliunits mg−1 protein reached at the end of the experiment (Fig. 8), which is similar to literature values of full urease activation in Arabidopsis Col-0 (15). Within the first 4 h, the leaf nickel concentration doubled and was increased 6 times after 24 h, whereas the urease activity remained unchanged during the first 12 h and only slightly increased after 24 h (Fig. 8). Even after 4 days only 80% of the maximum activity was reached. The amount of urease protein was not affected by nickel availability (supplemental Fig. 4). These data show that in vivo the process of urease activation is rather a matter of days than of hours.
Figure 8.
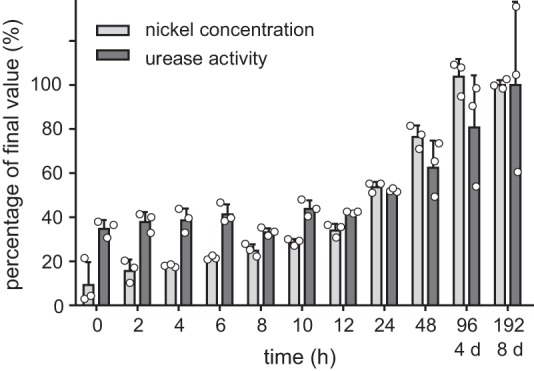
Time course of nickel-dependent activation of urease in vivo. Arabidopsis wild type plants were hydroponically grown without nickel for 5½ weeks. After supplementation of NiCl2 (final concentration: 10 μm), the urease activity and the Ni2+ concentration in the plants were repeatedly monitored. Both are shown relative to the final values reached after 8 days, which were set to 100%. Three biological replicates (three plants per replicate) were analyzed (individual data points shown as open circles; error bars = S.D.). The absolute data is shown in supplemental Fig. S6A.
Discussion
The interaction of eukaryotic UAPs with urease have so far not been investigated in detail. Only for UreD and urease from rice has an interaction been reported (16). The crystal structure of a UreD–UreF–UreG complex from H. pylori was recently solved revealing that UreD (called UreH in this organism) interacts with UreF, which forms dimers. Upon interaction of UreH with UreF, a binding site for a dimer of UreG is formed on the UreF surface (12). Our data for Arabidopsis are consistent with such a model of UAP interactions. That plant UreGs can form dimers and are structurally similar to UreGs from bacteria has been shown for the protein from soybean (19). The amino acid sequences of the UreD and UreF proteins are only weakly conserved between plants and bacteria. Nonetheless, amino acids Arg-179 and Tyr-183 of H. pylori UreF, which are important for interaction with UreH (12), are conserved as Arg/Lys and Tyr/Phe, respectively, in UreF proteins from plants (16). The residues in UreG and UreF of H. pylori and K. aerogenes involved in the interaction between both proteins (31, 12) are as well mostly conserved in plant sequences, although UreF is only 19% identical between H. pylori and Arabidopsis (UreG: 40% identity). For example, for the amino acids Tyr-48 and Arg-250 in UreF of H. pylori, which are absolutely required for the interaction with UreG, the residues Phe and Arg are found in all plant UreF sequences (Phe-35 and Arg-236 in UreF of Arabidopsis; Ref. 16). Our co-purification data together with the particular conservation of residues important for protein-protein interactions suggest that the plant UAPs likely form a (UreD–UreF–UreG)2 complex as found in H. pylori and K. aerogenes (32). In line with this are computational analyses indicating that plant and bacterial UAPs are structurally conserved (19, 16). Although different models exist on how the UAPs interact with ureases of different quaternary structure for activation (32, 12, 8), there also appears to be a significant structural conservation of the urease–UAP activation complex across kingdoms. This is supported by the ability of soybean UreF to complement a UreF mutant of Schizosaccharomyces pombe (33) and by partial complementation of the K. aerogenes urease activation machinery lacking UreG by UreG from potato (18).
Interestingly in H. pylori, UreG dissociates from (UreD–UreF–UreG)2 in the presence of GTP and nickel. However by binding nickel and GTP, a UreG dimer is formed that acquires the capability to interact with the urease–UreH–UreF complex leading to urease activation in the absence of nickel and GTP in solution (12). Similarly, Arabidopsis UreG activated in vivo in nickel-fertilized plants is capable of transferring Ni2+ to a urease–UreD–UreF complex in vitro leading to urease activation without the addition of any nickel and bicarbonate and in the presence of EDTA (Fig. 6). Although the preformed UreD–UreF–UreG complex can interact with urease (Fig. 3) as was also shown in K. aerogenes (34, 32), the activation of urease at least in H. pylori may rather occur via the interaction of a urease–UreD–UreF complex with an activation-competent UreG dimer. UreGs from different sources bind at least one nickel ion per monomer (35, 36, 19, 31) consistent with the idea that a UreG dimer would be capable of delivering the two Ni2+ ions required in each active center of urease. It is also possible that a nickel- and GTP-charged UreG dimer delivers only one nickel then dissociates into monomeric UreG, which is followed by a second round of charged UreG dimer-binding and nickel delivery. This is an attractive model because there is a single metal-binding site at the interface between the UreG dimers (12) that is conserved in bacterial and plant UreGs (16).
Bacterial UreG can be made activation-competent in vitro (24, 12). In contrast, we were unable to generate activation-competent UreG in vitro by the incubation with nickel, bicarbonate, and GTP (also ATP was tested). That plant UreG hydrolyzes GTP in principle was shown for the soybean enzyme (19). However, Arabidopsis UreG isolated from nickel-sufficient plant cells (Fig. 6) or from Escherichia coli (15) was competent for activation. This may be explained by the requirement for a further protein factor needed to deliver the nickel to plant UreG, which was not present in vitro. In H. pylori, HypA and HypB, which are associated with metallocenter biosynthesis of Ni-Fe hydrogenases, also play a role in urease activation. It was shown that HypA binds to a UreE dimer to deliver nickel. In the presence of Mg-GTP, a UreG dimer interacts with (UreE)2, displacing HypA and acquiring nickel from UreE (11, 37). It is possible that plant UreGs, which may unite bacterial UreG and UreE functions in one protein, obtain activation competence only if the nickel is delivered from a protein factor. Alternatively, a protein factor may be required to assist UreG folding into the activation-competent state because UreG was shown to be an intrinsically disordered protein (38, 36).
UreE of K. aerogenes contains a His-rich C terminus, which is not required for urease activation in vivo (25), and K. aerogenes urease can be partially activated even in the absence of UreE (30, 25). The deletion of the His-Asp/Glu-rich part of the N terminus of Arabidopsis UreG also had no strong effect on the functionality of the protein (Fig. 7). However, at a lower level of overexpression (line ΔN26–2), urease activity was significantly lower than in the wild type when limiting nickel had been supplied. The data indicate that the hypervariable N terminus of the protein may serve to enrich metals (nickel). A functional UreE requires a conserved His involved in Ni2+-binding at the dimer interface (26, 27). The deletion of both HXH motifs comprising all conserved His in the plant-specific N terminus of Arabidopsis UreG (mutant ΔN49) abolished urease activation in vivo despite strong overexpression of the variant protein (Fig. 7). We conclude that the plant-specific N terminus of UreG not only boosts urease activation, as does UreE, but is absolutely required, at least at the limiting availability of nickel in our experiment. When only the first HXH motif was removed (mutant ΔN41), the protein remained partially functional after nickel addition, indicating that the N terminus of UreG up to this point may be involved in efficient nickel delivery to the core function of UreG.
Full urease activation in vivo requires several days after nickel addition (Fig. 8) but not because the urease protein is unstable if it is not activated (supplemental Fig. S6). This is similar in K. aerogenes where urease is produced at normal levels when UAPs are missing (9). Also in bacteria, urease activation is not immediate but requires several hours after nickel becomes available (30). An explanation may be that the kinetics of nickel delivery to the urease–UAP complex might be governed by the diffusion rate of nickel-carrying proteins within the cell. Due to the greater size of eukaryotic cells, this process may take longer there than in prokaryotes.
Experimental procedures
Plant material and cultivation
Arabidopsis thaliana, ecotype Col-0, and the ureG-1 mutant line (GK294B06; 15) were used. Arabidopsis and N. benthamiana plants were cultivated as described before (21, 15). Transient expression in N. benthamiana was performed as described by Werner et al. (39) but adjusting the optical densities of the helper Agrobacterium strain containing the p19 silencing inhibitor construct to 0.1 and of all other agrobacteria strains to 0.25 each. In co-purification experiments, equal agrobacteria load was ensured by adding agrobacteria expressing urate oxidase (40), which is unrelated to urease activation, when agrobacteria for the expression of distinct components of the urease–UAP complex were omitted.
Cloning
The viral 5′- and 3′-UTRs from pEAQ-HT (41) were amplified using the primer pairs N2 + N3 and N4 + N5 (supplemental Table S1), respectively, and cloned into pXCS-HA-Strep (V13; Ref. 21) generating pXCScpmv-HA-Strep (V69). A urease cDNA including the stop codon was amplified from pET30-CTH-urease (U25; Ref. 15) with the primer pair N1 + 569 introducing an EcoRI and XmaI site used for cloning into V13 and V69, generating U148 and U149, respectively. For UreD binary constructs, the cDNA was released from pAlter-ureD (U14; Ref. 15) by NcoI and HindIII and cloned into pXS2pat (V51) and pXNS2pat-Strep (V42; Ref. 16). This generated AtureD-pXS2pat (U101) and AtureD-pXNS2pat-Strep (U97). The UreG gene was excised from pCDF-ureG-ureF (U10; Ref. 15) using NcoI and PstI and cloned into the same vectors used for UreD generating AtureG-pXS2pat (U103) and AtureG-pXNS2pat-Strep (U99). UreF was released from U10 by NdeI digest and cloned into pXS1pat (V50) and pXNS1pat-Strep (V41), resulting in AtureF-pXS1pat (U102) and AtureF-pXNS1pat-Strep (U98). For protein expression in E. coli, cDNAs of UreD and UreF from Arabidopsis were cloned with the same restriction sites as used above into pMALc5x (New England BioLabs) generating AtUreD-pMALc5x (U131) and AtUreF-pMALc5x (U132). The cDNA of rice UreD was excised from U104 using NcoI and HindIII and ligated into pMALc5x generating OsUreD-pMALc5x (U133). OsUreF-pMALc5x (U134) was generated by cloning the respective cDNA from U121 (16) with NdeI and NotI. Four N-terminally truncated variants of AtUreG lacking the first 26, 41, 49, and 67 amino acids were generated by amplification of the corresponding cDNA with the forward primers 1760 (ΔN26), 1761 (ΔN41), 1762 (ΔN49), or 1763 (ΔN67) and the reverse primer 1759. Amplicons were cloned via NcoI and PstI into pXS2pat (V51), generating AtUreG-ΔN26-pXS2pat (U110), AtUreG-ΔN41-pXS2pat (U111), AtUreG-ΔN49-pXS2pat (U112), and AtUreG-ΔN67-pXS2pat (U113). For the generation of an anti-AtUreG antibody, first an NcoI site was generated in pET30-CTH (V2; Ref. 42), which was cut with XbaI and EcoRI and ligated with the oligonucleotides 1779 and 1780, resulting in pET30nco-CTH (V48). The cDNA coding for AtUreG-ΔN67 was cloned into pET30nco-CTH via NcoI and PstI, generating AtUreG-ΔN67-pET30nco-CTH (U65). The clones OsUre-pXCS-HA-Strep (U42), OsUreD-pXS2pat (U104), OsUreG-pXS2pat (U106), OsUreD- pXNS2pat-Strep (U107), and OsUreF-pXS1pat (U121) were used for expression of rice urease and accessory proteins (all described in Refs. 16).
Affinity purification
For investigating the urease activation complex, proteins were affinity-purified according to Werner et al. (39), with slight changes in the washing procedure. The affinity matrix was washed only once but with 1.5 ml of wash buffer.
For the binding assay of urease to the UreD–UreF–UreG complex, 1.5 g and 0.75 g of leaf material expressing urease and the complex were extracted, respectively (1:2 ratio with extraction buffer) and clarified (20,000 × g, 4 °C). 1.5 ml of supernatant containing the complex was incubated for 20 min (rotation wheel, 4 °C) with 60 μl Strep-Tactin Macroprep (50% suspension). The beads were washed twice with 1 ml and 0.5 ml of wash buffer, respectively. The supernatant of 3 ml-containing urease was split into two halves to which either 60 μl of fresh Strep-Tactin Macroprep or the beads with bound complex were added. The slurries were incubated for 20 min, and the matrices were washed once with 1.5 ml of wash buffer and then exposed to 150 μl of elution buffer.
In vitro activation of urease
Affinity-purified urease–UAP complexes were incubated in 25 mm HEPES, pH 8.0, 0–10 mm NaHCO3, 0–10 μm NiCl2, 0–1.5 mm GTP, and 5 mm urea. The reaction components were incubated at 50 °C, and the reaction (volume 300 μl) was started with the addition of purified complex. For urease assay, samples of 20 μl were placed into 80 μl of water in a time course, and 25 μl of phenol nitroprusside followed by 50 μl of hypochloride solution were added. The resulting color was quantified colorimetrically at 639 nm (6).
For urease activation experiments in mixed N. benthamiana leaf extracts, plants expressing Strep-tagged UreG or co-expressing urease, UreD, and UreF (with UreD Strep-tagged) or non-infiltrated control plants were employed. Conditionally, plants were fertilized with 1 mm NiCl2 (6 ml per pot) 1 day before infiltration for transient expression. Clarified extracts were desalted on 5-ml HiTrap G25 Sephadex columns (GE Healthcare) equilibrated with 25 mm HEPES, pH 8.0, 75 mm NaCl, 2.5 mm EDTA, and 2 mm DTT. Extracts were mixed and incubated at 50 °C for the indicated times followed by the quantification of urease activities. Each incubation was repeated three times.
Generation of polyclonal antisera
AtUreD, AtUreF, OsUreD, and OsUreF were expressed as maltose-binding protein fusion proteins in E. coli BL21 (DE3) cells from the pMAL-c5X vector (New England BioLabs). Cells were grown in 100 ml of LB medium and induced by isopropyl-β-d-thiogalactoside after reaching an optical density of 0.5. Cells were harvested 3 h after induction, and cell pellets were dissolved in column buffer (20 mm Tris-HCl, pH 7.5; 200 mm NaCl; 1 mm EDTA) at a ratio of 2.5 ml per 100 ml cell culture. A spatula tip of lysozyme powder and 1.5 μl of DNase I (10 mg ml−1) were added. Cells were incubated at room temperature for 30 min, sonicated four times (Branson S150-D; level 3; 15 s each), and centrifuged (20 min, 20,000 × g, 4 °C). AtUreD, AtUreF, and OsUreD formed inclusion bodies, whereas OsUreF was soluble.
The supernatant-containing OsUreF was loaded on a 1-ml amylose matrix (New England BioLabs) pre-equilibrated with 5 ml of column buffer and washed 5 times with 2 ml of column buffer per wash. OsUreF was eluted by the repeated addition (10 times) of 0.75 ml of elution buffer (column buffer + 10 mm maltose). After examining purity and concentration of the protein on SDS gels, the two fractions with the highest protein content were pooled and desalted using a 5-ml HiTrap column (GE Healthcare).
The inclusion body pellets of AtUreD, AtUreF, and OsUreD were washed by resuspending them in 1.25 ml of column buffer each and incubating in an ice-cold ultrasonic bath (5 min) followed by the addition of 12.5 μl of Triton X-100 per ml and centrifugation (20 min; 20,000 × g; 4 °C). The washing step was repeated, and inclusion body pellets were resuspended in 300 μl of SDS-PAGE loading buffer. Proteins were separated on porous SDS gels and processed for antisera production as described by Dahncke and Witte (43).
The untagged and truncated AtUreG variant ΔN67 was expressed in E. coli from pET30nco-CTH by isopropyl-β-d-thiogalactoside induction as described above. The soluble protein was purified using a 13% SDS gel column on a Model 491 PrepCell (Bio-Rad) system. Fractions containing the purified protein were precipitated with methanol chloroform.
N-terminally truncated AtUreG as well as AtUreD, AtUreF, OsUreD, and OsUreF fusion proteins were used for commercial antisera production (Biogenes GmbH, Berlin).
Electrophoresis, in-gel activity staining, and Western blotting
Native SDS gel electrophoresis and in-gel activity stains were performed as described by Witte and Medina-Escobar (6). Denaturating SDS gel electrophoresis and Western blotting were performed according to Witte et al. (21). Urease was detected using a polyclonal rabbit anti-jack bean urease antibody (Rockland 100-4182; dilution 1:1,000) and a mouse anti-rabbit IgG alkaline phosphatase conjugate (Sigma A2306; dilution 1:10,000). For detection of the accessory proteins, generated polyclonal rabbit antisera (diluted 1:1,000) were used. For urease quantification on Western blots, the polyclonal rabbit anti-jack bean urease antibody (dilution 1:20,000), a goat anti-rabbit IgG HRP- conjugate (Thermo 32460; dilution 1:10,000), and a commercial enhanced chemiluminescence (ECL) kit (SuperSignal West Femto Chemiluminescent Substrate; Thermo Scientific) were employed.
Urease activation in complementation lines expressing UreG variants
The following lines were used: Col-0, ureG-1 mutant (294B06, Ref. 15) and ureG-1 complementation lines expressing untagged full-length as well as N-terminal truncation variants of UreG: FL (1, line 1; 2, line 7); ΔN26 (1, line 5; 2, line 18), ΔN41 (1, line 2; 2, line 7), ΔN49 (1, line 1; 1, line 4). The seeds were germinated on soil, and after 1 week the plantlets from segregating transgenic lines were sprayed with Basta (Bayer) to select for the transgene. After reaching the 2–4 leaf stage, plants were transferred to pots (d = 8 cm, containing Steckmedium from Klasmann-Deilmann, Germany, 3 plants per pot, 6 pots per genotype) and grown randomized for 4 weeks in a growth chamber (16 h of light, 100 μmol m−2 s−1, 22 °C day, 20 °C night, 70% relative humidity) to rosettes just before bolting. For each replicate, 3 rosettes from 1 pot were ground with 3 volumes of extraction buffer (50 mm HEPES, pH 7.5, 150 mm NaCl, 5 mm EDTA, 0.2 mm 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF), 10 mm DTT) in a mortar. The extract was clarified (20,000 × g, 15 min, 4 °C) and 100-μl desalted using spin columns equilibrated with half-strength extraction buffer without AEBSF and DTT. Urease activity from three replicates per line was determined with colorimetric detection (6). The experiment was repeated with plants that had been fertilized with 1 ml of 100 μm NiCl2 applied to the soil 4 days before harvest near the center of each rosette (3 ml of Ni2+ solution per pot). The expression of the UreG variants was monitored by Western blot using the antibody raised against ΔN67-UreG. For each line the sample for the blot consisted of a pool of extracts of all replicates.
Urease activation and nickel content in hydroponically grown plants
Col-0 was grown in a hydroponic growth system (Araponics) for 12 h in the light (22 °C) and 12 h in the dark (20 °C) in modified Hoagland medium containing 1.5 mm KNO3, 0.75 mm Ca(NO3)2, 0.5 mm (NH4)2SO4, 0.75 mm MgSO4, 0.5 mm K2HPO4, 0.5 mm KCl, 0.1 mm FeEDTA, 50 μm H3BO4, 10 μm MnSO4, 20 μm ZnSO4, 1 μm CuSO4, 0.1 μm Na2MoO4, and 1 μm Na2O3Si, adjusted to pH 5.5 with HCl. Five-and-a-half weeks after germination, the growth medium was supplemented with 10 μm NiCl2. Three plants from one tank were pooled and homogenized with 4 volumes extraction buffer per weight (50 mm HEPES, pH 7.5, 150 mm NaCl, 5 mm EDTA). After centrifugation (20,000 × g, 15 min, 4 °C), 100 μl of the supernatant were desalted as described in Cao et al. (16), and urease activity was determined, whereas the rest was stored for Western blotting and nickel analysis. To determine the nickel content, 100 μl of plant extract were mixed with 900 μl of double-distilled H2O. After the addition of 1 ml of diluted nitric acid (2.3%), samples were analyzed with an inductively coupled plasma mass spectrometer (ICP-MS; 7500CX, Agilent).
Data analysis
Statistical analyses were performed with the GraphPad Prism software package.
Author contributions
C.-P. W. and T. M. designed the experiments, which were performed by T. M. with the exception of the experiment shown in Fig. 7, which was performed by C.-P. W. and A. Z. The manuscript was written by C.-P. W. and T. M. with revision by A. Z. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank André Specht for technical support quantifying nickel contents and Feng Qiu Cao for the preparation of the anti-UreG antibody.
This work was supported by the German Academic Exchange Service from funds of the German Federal Ministry for Education and Research, program Modern Applications of Biotechnology–Binational Junior Research Groups (Grant ID 57041752). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S6 and Tables S1.
- UAP
- urease accessory protein
- AEBSF
- 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride.
References
- 1. Liu G.-W., Sun A.-L., Li D.-Q., Athman A., Gilliham M., and Liu L.-H. (2015) Molecular identification and functional analysis of a maize (Zea mays) DUR3 homolog that transports urea with high affinity. Planta 241, 861–874 [DOI] [PubMed] [Google Scholar]
- 2. Liu L.-H., Ludewig U., Frommer W. B., and von Wirén N. (2003) AtDUR3 encodes a new type of high-affinity urea/H+ symporter in Arabidopsis. Plant Cell 15, 790–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang W.-H., Köhler B., Cao F.-Q., Liu G.-W., Gong Y.-Y., Sheng S., Song Q.-C., Cheng X.-Y., Garnett T., Okamoto M., Qin R., Mueller-Roeber B., Tester M., and Liu L.-H. (2012) Rice DUR3 mediates high-affinity urea transport and plays an effective role in improvement of urea acquisition and utilization when expressed in Arabidopsis. New Phytol. 193, 432–444 [DOI] [PubMed] [Google Scholar]
- 4. Zanin L., Tomasi N., Wirdnam C., Meier S., Komarova N. Y., Mimmo T., Cesco S., Rentsch D., and Pinton R. (2014) Isolation and functional characterization of a high affinity urea transporter from roots of Zea mays. BMC Plant Biol. 14, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goldraij A., and Polacco J. C. (1999) Arginase is inoperative in developing soybean embryos. Plant Physiol. 119, 297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Witte C. P., and Medina-Escobar N. (2001) In-gel detection of urease with nitroblue tetrazolium and quantification of the enzyme from different crop plants using the indophenol reaction. Anal. Biochem. 290, 102–107 [DOI] [PubMed] [Google Scholar]
- 7. Mobley H. L., and Hausinger R. P. (1989) Microbial ureases: significance, regulation, and molecular characterization. Microbiol. Rev. 53, 85–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Farrugia M. A., Macomber L., and Hausinger R. P. (2013) Biosynthesis of the urease metallocenter. J. Biol. Chem. 288, 13178–13185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee M. H., Mulrooney S. B., Renner M. J., Markowicz Y., and Hausinger R. P. (1992) Klebsiella aerogenes urease gene cluster: sequence of ureD and demonstration that four accessory genes (ureD, ureE, ureF, and ureG) are involved in nickel metallocenter biosynthesis. J. Bacteriol. 174, 4324–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Park I. S., and Hausinger R. P. (1995) Evidence for the presence of urease apoprotein complexes containing UreD, UreF, and UreG in cells that are competent for in vivo enzyme activation. J. Bacteriol. 177, 1947–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang X., Li H., Lai T.-P., and Sun H. (2015) UreE-UreG complex facilitates nickel transfer and preactivates GTPase of UreG in Helicobacter pylori. J. Biol. Chem. 290, 12474–12485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fong Y. H., Wong H. C., Yuen M. H., Lau P. H., Chen Y. W., and Wong K.-B. (2013) Structure of UreG/UreF/UreH complex reveals how urease accessory proteins facilitate maturation of Helicobacter pylori urease. PLos Biol. 11, e1001678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boer J. L., Quiroz-Valenzuela S., Anderson K. L., and Hausinger R. P. (2010) Mutagenesis of Klebsiella aerogenes UreG to probe nickel binding and interactions with other urease-related proteins. Biochemistry 49, 5859–5869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Farrugia M. A., Wang B., Feig M., and Hausinger R. P. (2015) Mutational and computational evidence that a nickel-transfer tunnel in UreD is used for activation of Klebsiella aerogenes urease. Biochemistry 54, 6392–6401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Witte C.-P., Rosso M. G., and Romeis T. (2005) Identification of three urease accessory proteins that are required for urease activation in Arabidopsis. Plant Physiol. 139, 1155–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cao F.-Q., Werner A. K., Dahncke K., Romeis T., Liu L.-H., and Witte C.-P. (2010) Identification and characterization of proteins involved in rice urea and arginine catabolism. Plant Physiol. 154, 98–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Freyermuth S. K., Bacanamwo M., and Polacco J. C. (2000) The soybean Eu3 gene encodes an Ni2+-binding protein necessary for urease activity. Plant J. 21, 53–60 [DOI] [PubMed] [Google Scholar]
- 18. Witte C. P., Isidore E., Tiller S. A., Davies H. V., and Taylor M. A. (2001) Functional characterisation of urease accessory protein G (ureG) from potato. Plant Mol. Biol. 45, 169–179 [DOI] [PubMed] [Google Scholar]
- 19. Real-Guerra R., Staniscuaski F., Zambelli B., Musiani F., Ciurli S., and Carlini C. R. (2012) Biochemical and structural studies on native and recombinant glycine max UreG: a detailed characterization of a plant urease accessory protein. Plant Mol. Biol. 78, 461–475 [DOI] [PubMed] [Google Scholar]
- 20. Olson J. W., and Maier R. J. (2000) Dual roles of Bradyrhizobium japonicum nickelin protein in nickel storage and GTP-dependent Ni2+ mobilization. J. Bacteriol. 182, 1702–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Witte C.-P., Noël L. D., Gielbert J., Parker J. E., and Romeis T. (2004) Rapid one-step protein purification from plant material using the eight-amino acid StrepII epitope. Plant Mol. Biol. 55, 135–147 [DOI] [PubMed] [Google Scholar]
- 22. Nakagawa S., Niimura Y., Gojobori T., Tanaka H., and Miura K. (2008) Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Nucleic Acids Res. 36, 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sainsbury F., and Lomonossoff G. P. (2008) Extremely high-level and rapid transient protein production in plants without the use of viral replication. Plant Physiol. 148, 1212–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Soriano A., and Hausinger R. P. (1999) GTP-dependent activation of urease apoprotein in complex with the UreD, UreF, and UreG accessory proteins. Proc. Natl. Acad. Sci. U.S.A. 96, 11140–11144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brayman T. G., and Hausinger R. P. (1996) Purification, characterization, and functional analysis of a truncated Klebsiella aerogenes UreE urease accessory protein lacking the histidine-rich carboxyl terminus. J. Bacteriol. 178, 5410–5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bellucci M., Zambelli B., Musiani F., Turano P., and Ciurli S. (2009) Helicobacter pylori UreE, a urease accessory protein: specific Ni2+- and Zn2+-binding properties and interaction with its cognate UreG. Biochem. J. 422, 91–100 [DOI] [PubMed] [Google Scholar]
- 27. Colpas G. J., Brayman T. G., Ming L. J., and Hausinger R. P. (1999) Identification of metal-binding residues in the Klebsiella aerogenes urease nickel metallochaperone, UreE. Biochemistry 38, 4078–4088 [DOI] [PubMed] [Google Scholar]
- 28. Zambelli B., Banaszak K., Merloni A., Kiliszek A., Rypniewski W., and Ciurli S. (2013) Selectivity of Ni(II) and Zn(II) binding to Sporosarcina pasteurii UreE, a metallochaperone in the urease assembly: a calorimetric and crystallographic study. J. Biol. Inorg. Chem. 18, 1005–1017 [DOI] [PubMed] [Google Scholar]
- 29. Soriano A., Colpas G. J., and Hausinger R. P. (2000) UreE stimulation of GTP-dependent urease activation in the UreD-UreF-UreG-urease apoprotein complex. Biochemistry 39, 12435–12440 [DOI] [PubMed] [Google Scholar]
- 30. Colpas G. J., and Hausinger R. P. (2000) In vivo and in vitro kinetics of metal transfer by the Klebsiella aerogenes urease nickel metallochaperone, UreE. J. Biol. Chem. 275, 10731–10737 [DOI] [PubMed] [Google Scholar]
- 31. Boer J. L., and Hausinger R. P. (2012) Klebsiella aerogenes UreF: identification of the UreG binding site and role in enhancing the fidelity of urease activation. Biochemistry 51, 2298–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Farrugia M. A., Han L., Zhong Y., Boer J. L., Ruotolo B. T., and Hausinger R. P. (2013) Analysis of a soluble (UreD–UreF–UreG)2 accessory protein complex and its interactions with Klebsiella aerogenes urease by mass spectrometry. J. Am. Soc. Mass Spectrom. 24, 1328–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bacanamwo M., Witte C.-P., Lubbers M. W., and Polacco J. C. (2002) Activation of the urease of Schizosaccharomyces pombe by the UreF accessory protein from soybean. Mol. Genet. Genomics 268, 525–534 [DOI] [PubMed] [Google Scholar]
- 34. Carter E. L., Boer J. L., Farrugia M. A., Flugga N., Towns C. L., and Hausinger R. P. (2011) Function of UreB in Klebsiella aerogenes urease. Biochemistry 50, 9296–9308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zambelli B., Stola M., Musiani F., De Vriendt K., Samyn B., Devreese B., Van Beeumen J., Turano P., Dikiy A., Bryant D. A., and Ciurli S. (2005) UreG, a chaperone in the urease assembly process, is an intrinsically unstructured GTPase that specifically binds Zn2+. J. Biol. Chem. 280, 4684–4695 [DOI] [PubMed] [Google Scholar]
- 36. Zambelli B., Turano P., Musiani F., Neyroz P., and Ciurli S. (2009) Zn2+-linked dimerization of UreG from Helicobacter pylori, a chaperone involved in nickel trafficking and urease activation. Proteins 74, 222–239 [DOI] [PubMed] [Google Scholar]
- 37. Yang X., Li H., Cheng T., Xia W., Lai Y.-T., and Sun H. (2014) Nickel translocation between metallochaperones HypA and UreE in Helicobacter pylori. Metallomics 6, 1731–1736 [DOI] [PubMed] [Google Scholar]
- 38. D'Urzo A., Santambrogio C., Grandori R., Ciurli S., and Zambelli B. (2014) The conformational response to Zn(II) and Ni(II) binding of Sporosarcina pasteurii UreG, an intrinsically disordered GTPase. J. Biol. Inorg. Chem. 19, 1341–1354 [DOI] [PubMed] [Google Scholar]
- 39. Werner A. K., Sparkes I. A., Romeis T., and Witte C.-P. (2008) Identification, biochemical characterization, and subcellular localization of allantoate amidohydrolases from Arabidopsis and soybean. Plant Physiol. 146, 418–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hauck O. K., Scharnberg J., Escobar N. M., Wanner G., Giavalisco P., and Witte C.-P. (2014) Uric acid accumulation in an Arabidopsis urate oxidase mutant impairs seedling establishment by blocking peroxisome maintenance. Plant Cell 26, 3090–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sainsbury F., Thuenemann E. C., and Lomonossoff G. P. (2009) pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7, 682–693 [DOI] [PubMed] [Google Scholar]
- 42. Glinski M., Romeis T., Witte C.-P., Wienkoop S., and Weckwerth W. (2003) Stable isotope labeling of phosphopeptides for multiparallel kinase target analysis and identification of phosphorylation sites. Rapid Commun. Mass Spectrom. 17, 1579–1584 [DOI] [PubMed] [Google Scholar]
- 43. Dahncke K., and Witte C.-P. (2013) Plant purine nucleoside catabolism employs a guanosine deaminase required for the generation of xanthosine in Arabidopsis. Plant Cell 25, 4101–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.