Abstract
Electron bifurcation has recently gained acceptance as the third mechanism of energy conservation in which energy is conserved through the coupling of exergonic and endergonic reactions. A structure-based mechanism of bifurcation has been elucidated recently for the flavin-based enzyme NADH-dependent ferredoxin NADP+ oxidoreductase I (NfnI) from the hyperthermophillic archaeon Pyrococcus furiosus. NfnI is thought to be involved in maintaining the cellular redox balance, producing NADPH for biosynthesis by recycling the two other primary redox carriers, NADH and ferredoxin. The P. furiosus genome encodes an NfnI paralog termed NfnII, and the two are differentially expressed, depending on the growth conditions. In this study, we show that deletion of the genes encoding either NfnI or NfnII affects the cellular concentrations of NAD(P)H and particularly NADPH. This results in a moderate to severe growth phenotype in deletion mutants, demonstrating a key role for each enzyme in maintaining redox homeostasis. Despite their similarity in primary sequence and cofactor content, crystallographic, kinetic, and mass spectrometry analyses reveal that there are fundamental structural differences between the two enzymes, and NfnII does not catalyze the NfnI bifurcating reaction. Instead, it exhibits non-bifurcating ferredoxin NADP oxidoreductase-type activity. NfnII is therefore proposed to be a bifunctional enzyme and also to catalyze a bifurcating reaction, although its third substrate, in addition to ferredoxin and NADP(H), is as yet unknown.
Keywords: archaea, crystal structure, extreme thermophile, metabolism, nicotinamide, Nfn, electron bifurcation, elemental sulfur
Introduction
Pyrococcus furiosus is a hyperthermophillic archaeon that grows optimally near 100 °C and is able to utilize a wide range of simple and complex carbohydrates and peptides as carbon sources to produce acetate, CO2, and H2 or, in the presence of elemental sulfur, H2S. The carbohydrate metabolism of P. furiosus proceeds through a modified Embden–Meyerhof pathway that only utilizes ferredoxin (Fd)2 as an electron acceptor, and NAD+ is not required (1). NADPH for biosynthesis is thought to be generated from evolved H2 recycled by soluble hydrogenases (SHI and SHII) (2), as well as by a soluble Fd-dependent NADP+ oxidoreductase (FNOR) (3, 4). Fd generated from glycolysis is in turn oxidized by an energy-conserving membrane-bound hydrogenase, resulting in the production of H2 (5). Upon the addition of elemental sulfur, P. furiosus metabolism is shifted rapidly to shut down the hydrogenases and H2 production while initiating the production of H2S, probably via the membrane-bound oxidoreductase complex, together with the NADPH-dependent sulfur reductase (NSR) (6). The presence of sulfur causes a dramatic decrease in the expression of genes encoding the three hydrogenases and an increase in the expression of membrane-bound oxidoreductase and other so-called sulfur response genes (7). Specifically, the regulatory transcription factor SurR (sulfur response regulator) orchestrates the metabolic switch from H2 to H2S formation and also functions as a global regulator of electron flow pathways in P. furiosus (8–10). P. furiosus metabolism also changes dramatically, depending on whether the carbon source is carbohydrates or peptides (11). During growth on sugar (maltose), Fd serves as the main cellular redox carrier, whereas during growth on peptides (in the presence of sulfur), both Fd (generated from keto-acid oxidoreductases) and NADPH (generated from amino acid deamination) are thought to serve as primary reductants (11).
Recently, a new type of enzyme system was described to regulate the redox pools of Fd, NAD(H), and NADP(H) in anaerobes, which lack the canonical transhydrogenases. The redox pools in some anaerobes are balanced by a bifurcating enzyme called NADH-dependent ferredoxin NADP+ oxidoreductase (Nfn). This was first reported in Clostridium kluyveri and catalyzes the production of NADPH coupled to the simultaneous oxidation of NADH and reduced Fd. This enzyme carried out flavin-based electron bifurcation, a fundamental mechanism of biological energy conservation (12, 13). In anaerobes, Nfn is directly involved in shuttling electrons between the three main redox carriers in anaerobes: Fd, NADH, and NADPH.
Nfn effectively couples the endergonic reduction of NADP by NADH and the exergonic reduction of NADP+ by reduced Fd (Fdred), thereby maintaining a high ratio of NADPH/NADP to drive biosynthesis (13, 14). The genome of P. furiosus contains two Nfn homologs. One (PF1327-28) was initially purified as a sulfide dehydrogenase (4) and was subsequently found to have FNOR (3) activity.
This enzyme was recently renamed NfnI because it was shown to catalyze Reaction 1. NfnI was functionally and structurally characterized, and the mechanism of flavin-based electron bifurcation was elucidated (14). NfnI contains a large (L) subunit of 53 kDa harboring two [4Fe4S] clusters and one FAD and a small (S) subunit of 31 kDa containing one [2Fe2S] cluster and one FAD. Nfn homologs are found mainly in anaerobic microorganisms, both bacteria and archaea, and the subunit and cofactor composition are highly conserved (14).
The second Nfn homolog of P. furiosus, termed NfnII (PF1910-11), has yet to be structurally and functionally characterized and is the subject of this study. Interestingly, the expression of NfnI and NfnII are dependent upon sulfur availability and carbon source in a reciprocal fashion (10, 11). Specifically, expression of nfnI is up-regulated under H2-producing conditions (sugar fermentation), whereas nfnII is up-regulated under S0-reducing conditions (with sugars or peptides as the carbon source). It was assumed that NfnI and NfnII have similar functions in shuttling electrons between NAD(P)H and Fd, adapting to the cells needs during growth, but why the cell needs two such enzymes and how they differ from each other was not at all clear, especially given their high sequence similarity.
Our current understanding of the physiological functions of Nfn in redox metabolism is very limited. Deletion of the two genes encoding Nfn in the fermentative bacterium Clostridium thermocellum was reported to have no significant effect on its metabolism (15). In Thermoanaerobacterium saccharolyticum, Nfn was proposed to play a role in the NADPH-dependent ethanol production pathway because its deletion decreased ethanol yield in the NADPH-dependent strain (16). In this study, we assess the distribution and evolution of Nfn in the order Thermococcales and determine the importance of the Nfn enzymes in P. furiosus through characterization of Nfn deletion mutants. In contrast to what was reported for C. thermocellum, both NfnI and NfnII play key roles in the metabolism of P. furiosus. In addition, we provide the first biochemical and structural characterization of P. furiosus NfnII.
Results and discussion
NfnSL taxonomic distribution
Genes encoding homologs of the S and L subunits of P. furiosus NfnI were found in 72 archaeal genomes (or 31% of the total). Within these genomes, a total of 92 archaeal NfnSL homologs were identified, with 17 genomes encoding multiple copies. (supplemental Fig. 1). Of the total 72 archaeal genomes, 66 were from the phylum Euryarchaeota, one from the phylum Korarchaeota, and five from the phylum Crenarchaeota. Of the 66 Euryarchaeota genomes that contained NfnSL, 41 genomes belonged to the order Methanomicrobia, 23 to the Thermococci, five to the Thermoprotei, and two to an unclassified order. Furthermore, all 17 genomes that encoded two or three isoforms of NfnSL were in the order Thermococcales (Fig. 1). Interestingly, genes encoding for homologs of P. furiosus NfnII are always present in the genomes of the order Thermococcales together with either NfnI or a third type of isoform, NfnIII. The Thermococcales strains Thermococcus onnurineus, Pyrococcus sp. ST04, and Pyrococcus sp. NCB100 encode for homologs of all three isoforms of Nfn.
Figure 1.
Phylogenetic reconstruction of a subset of Nfn homologs encoded by Archaea within the Thermococci and Thermoprotei classes (n = 45). NfnSL were concatenated prior to phylogenetic reconstruction. *, genomes that comprise multiple copies of Nfn.
Phylogenetic analysis of a concatenation of NfnSL homologs reveals monophyly of Nfn isoforms at the taxonomic level (e.g. evidence for vertical inheritance). To provide insight into the physiological role of Nfn in archaeal genomes, in particular enzymatic processes or pathways that are NADP(H)-, NAD(H)-, or Fd-specific, we investigated the proteins encoded in the gene neighborhood of those encoding for NfnSL in the Thermococci and Methanomicrobia. In Thermococci, neighboring genes included those encoding [NiFe]-hydrogenases and transferases (e.g. nicotinate phosphoribosyltranserase-like and carboxyl methyltransferase-like proteins.). In contrast, in Methanomicrobia, genes encoding isomerases (e.g. ketol-acid reductoisomerase-like and mannose phosphate isomerase-like proteins) and nitroreductases, which are NADP(H)-dependent enzymes (17–19), were enriched in the neighborhood of nfn (supplemental Fig. 2).
Phylogenetic reconstruction of archaeal NfnSL also revealed that the multiple isoforms were the result of at least two independent gene duplication events, yielding monophyletic lineages that we have termed NfnI, NfnII, and NfnIII. To provide more insight into specific NADP(H)-, NAD(H)-, or Fd-requiring processes that might lead to selective pressure to retain multiple copies of Nfn, we analyzed the proteins encoded in the gene neighborhood of all three Nfn groups. This analysis revealed unique genes that were abundant in the flanking regions of each enzyme complex (supplemental Fig. 3). Genes encoding for [NiFe]-hydrogenases were abundant in the neighborhood of the NfnI group. Likewise, genes encoding for enzymes involved in glycine cleavage as well as biotin synthase-like enzymes that are NADP(H)- or NAD(H)-dependent (20, 21) were abundant in the flanking region in the NfnII group. Genes encoding for nicotinate phosphoribosyltransferase, which is one of the primary enzymes involved in NAD+ synthesis, was clearly enriched in the flanking regions of the NfnIII group (22).
Effects of deleting genes encoding NfnI and NfnII in P. furiosus
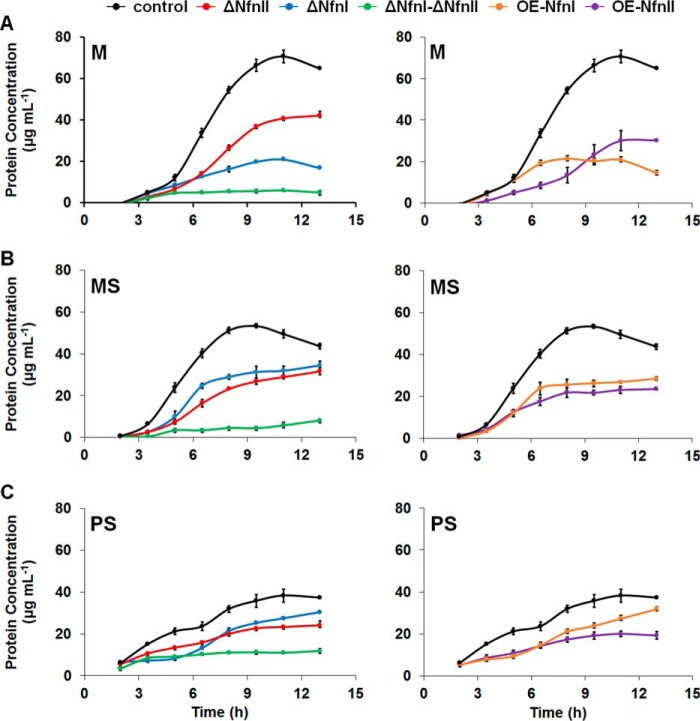
To analyze the roles of NfnI and NfnII under different carbon (sugar versus peptides) and redox (H+ versus S0 reduction) metabolisms, strains harboring deletions of the nfnI and nfnII genes were constructed for phenotypic characterization. Multiple attempts at constructing a strain containing deletions of both NfnI and NfnII were made; however, this strain could not be completely purified to prevent the reemergence of the wild-type nfnI allele during subsequent growth. All cultures used as inocula in this comparative growth study were subcultured once from the revived glycerol stocks, to maintain a higher fraction of the NfnI and NfnII double deletions in the ΔNfnI-ΔNfnII strain. The single and double deletions of NfnI and NfnII were directly compared with the control strain COM1c under three different growth conditions: maltose (M), maltose and sulfur (MS), or peptides and sulfur (PS). Growth yields, represented by the total protein concentration, were compared, and the doubling time (TD) was calculated for each strain.
During growth with maltose (M), the single deletion strains lacking either NfnI or NfnII exhibited moderate to severe growth phenotypes (Fig. 2A and supplemental Table S2). The growth phenotype was less prominent in the ΔNfnII strain, with similar TD and final protein concentration of ∼60% that of the control strain (42 ± 2 versus 71 ± 3 μg ml−1). Deletion of NfnI caused a more severe growth phenotype, with a 2-fold increase in TD and a final protein concentration approximately 4 times lower than the control strain (17 ± 1 versus 71 ± 3 μg ml−1). However, the phenotype observed with the ΔNfnI strain under sulfur-reducing conditions was less severe, with the final protein concentration similar to that of ΔNfnII and having about 75% of the growth of the control (34 ± 2 and 32 ± 2 versus 44 ± 1 μg ml−1) (Fig. 2B). Furthermore, when peptides were used as the carbon source (PS), deletion of NfnII caused a more pronounced phenotype than NfnI (Fig. 2C). The ΔNfnI strain grew more slowly but reached a final protein concentration close to that of the control strain (30 ± 1 versus 37 ± 1 μg ml−1). There was no significant difference in growth of the ΔNfnII strain compared with that grown in MS medium. To rule out any polar effects caused by deleting nfnI and nfnII, RT-qPCR analysis were performed to determine whether there were any changes in expression of the neighboring genes, PF1326, PF1329, PF1909, and PF1912. Although there was a small increase in the expression of PF1326, PF1329, and PF1912, these changes are not thought to be significant (supplemental Fig. 4).
Figure 2.
Growth analysis of COM1c (black; control strain), ΔNfnI (blue), ΔNfnII (red), ΔNfnI–ΔNfnII (green), OE-NfnI (orange), and OE-NfnII (purple) strains in M (A), MS (B), and PS media (C) at 90 °C. The S.D. (error bars) analyses were taken from three independent biological samples.
Deletion of both NfnI and NfnII caused the most severe growth phenotype in all three growth conditions, suggesting that NfnI and NfnII are important and required for robust and healthy growth of P. furiosus. Taken altogether, the phenotype caused by deletion of NfnI was the most severe in M medium lacking S0 and less severe in MS and PS, whereas the ΔNfnII strain maintained a similar phenotype in the different media used for growth. The result of this phenotypic study was in agreement with the expression analysis of NfnI and NfnII genes in all three growth media. Specifically, the expression of NfnI was decreased in the presence of S0, and more so when peptides were used as a carbon source, whereas the expression of NfnII followed an opposite trend, increasing in the presence of S0 and peptides (supplemental Fig. 5). This suggests that NfnI and NfnII are differentially expressed, depending on the availability of carbon sources and whether or not S0 is present as the terminal electron acceptor. NfnI appears to be more necessary for P. furiosus during carbohydrate metabolism, whereas NfnII is utilized more during sulfur and peptide metabolism. Also, at least one of the two Nfn paralogs appears to be required to sustain observable growth because attempts at deleting both NfnI and NfnII resulted in generation of an impure strain retaining ∼50% of the wild-type NfnI allele. Furthermore, growth of this strain in all three medium formulations tested was minimal (Fig. 2).
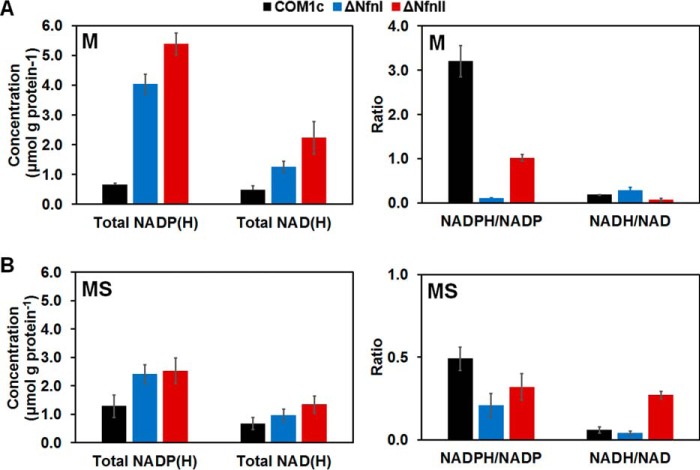
To determine whether the growth defects observed in the ΔNfnI and ΔNfnII strains were linked to changes in the internal redox pools, total NADP(H) and NAD(H) concentrations and their reduced/oxidized ratios were determined under the various growth conditions. In general, deletion of either NfnI or NfnII caused an increase in total NADP(H) and NAD(H) pools and decreased the ratio of NADPH/NADP+ (Fig. 3 and supplemental Fig. 6). During growth with maltose, deletion of NfnI caused a 30-fold decrease in the NADPH/NADP+ ratio compared with the control strain COM1c (0.10 ± 0.01 versus 3.2 ± 0.4), and deletion of NfnII also affected this ratio, but only by 3-fold (1.0 ± 0.1 versus 3.2 ± 0.4). Importantly, no significant changes were observed in the NADH/NAD+ ratio in these Nfn deletion strains (Fig. 3A). These results imply that the physiological functions of both NfnI and NfnII are indeed to generate NADPH, presumably for biosynthesis. In the presence of S0 (MS medium), the effects on the total amounts and the ratio of reduced to oxidized nicotinamide nucleotides were not as dramatic as those seen in the absence of S0 (Fig. 3B). The total NADP(H) and NAD(H) only doubled in the Nfn deletion strains, and the redox ratio was only reduced by half in the ΔNfnI strain and even less than that in the ΔNfnII strain. Another noticeable increase was for the NADH/NAD+ ratio in the ΔNfnII strain (0.30 ± 0.02 versus 0.10 ± 0.02), whereas this ratio measured in the ΔNfnI strain was comparable with that in the control COM1c strain. Hence, under peptide growth conditions, deletion of NfnI and NfnII mainly affects NADPH formation, reflected in an increase in total NADP(H) and the redox ratio, but NADH/NAD+ values remain unchanged, although the total amount of NAD(H) slightly increased (supplemental Fig. 6). Contrary to the differential expression of NfnI and NfnII regulated by S0 (supplemental Fig. 5), our redox pool analysis does not support the hypothesis that NfnII is more important for growth in the presence of S0. However, these results show that the growth phenotypes observed when either NfnI or NfnII is deleted are due at least in part to internal changes of the nucleotide ratios and suggest that NfnI and NfnII are the two major enzymes responsible for NADPH production in P. furiosus.
Figure 3.
Redox nucleotide pool analysis: total concentration of NADP(H), NAD(H), NADPH/NADP+ and NADH/NAD+ ratios of COM1c (black; control), ΔNfnI (blue), and ΔNfnII (red) strains in M (A) and MS media (B) at 90 °C. The S.D. (error bars) analyses were taken from three independent biological samples.
To determine whether deletion of either NfnI or NfnII had any effect on expression of other related genes, RT-qPCR analysis was performed. There was no significant change of expression in either NfnI or NfnII genes in the respective ΔNfnII or ΔNfnI strains, under sulfur or non-sulfur reducing conditions, indicating that there is no transcriptional compensation between the paralogs when the other Nfn is deleted (supplemental Fig. 7). Expression of key genes in the NAD+ salvage pathway was also tested because of the roles the Nfn paralogs are expected to play in balancing the redox pools. The genes encoding NAD+ kinase (PF1103), (NH3)-dependent NAD+ synthase (PF0098), and NAD+ diphosphorylase (PF0458) did not change significantly in expression; however, a significant change occurred in the gene encoding l-aspartate oxidase (NadB; PF1976) in the ΔNfnI strain. NadB catalyzes the conversion of l-aspartate to iminoaspartate, the very first step in the NAD+ salvage pathway. When NfnI was deleted, NadB expression decreased by nearly 8-fold in the presence of sulfur compared with the control strain (supplemental Fig. 7). If NfnI is the major enzyme producing NADPH for biosynthesis from NADH and reduced ferredoxin and is responsible for regulating the favorable concentration of NADH and NADPH in P. furiosus, deletion of NfnI would most likely cause an accumulation of NADH internally. Thus, NadB expression was decreased as a result of feedback inhibition, thereby turning off or slowing down the NAD+ salvage pathway to avoid excess NADH from accumulating.
Unlike NfnI, deletion of NfnII had little to no effect on NadB and other NAD+ synthesis enzymes, despite the growth phenotype observed under sulfur-reducing conditions. In addition, deletion of NfnI or NfnII did not change the expression of Fd significantly compared with that of the control strain. However, it is possible that the reduced/oxidized Fd ratio is altered in the Nfn deletion strains. Because there are many other Fd-utilizing enzymes that have a relatively high transcription level (e.g. pyruvate oxidoreductase, membrane-bound hydrogenase, etc.), it is challenging to determine the effect of an Nfn deletion on the ratio of reduced/oxidized Fd in the cell.
Expression of other major NAD(P)(H)-utilizing enzymes was also analyzed in the Nfn deletion strains under sulfur and non-sulfur growth conditions, including genes encoding subunits of SHI and SHII and the gene encoding NSR. Overall, deletion of NfnI or NfnII did not affect expression of the SHI and SHII operons (supplemental Fig. 7), because the transcription levels are comparable with those of the control strain. However, expression of NSR decreased significantly in the ΔNfnI strain but not in the ΔNfnII strain. It is not clear whether there is a strong connection between NfnI and NSR because these two enzymes are differentially expressed under sulfur-reducing conditions. NSR has a preference for NADPH over NADH; therefore, it is possible that the expression of NSR decreased in response to the drop of NADPH level in the ΔNfnI strain.
Effect of NfnI and NfnII overexpression in P. furiosus
To analyze the phenotypic effects of overexpressing NfnI and NfnII, strains that contained either NfnI or NfnII placed under the control of the strong, constitutive slp (S-layer protein) promoter were constructed in P. furiosus. These are referred to as OE-NfnI and OE-NfnII, respectively. The gene encoding the S subunit for each also contained a His tag at the N terminus for purification purposes (see below). To screen for possible growth phenotypes due to expressing these redox enzymes at high levels, the OE-NfnI and OE-NfnII strains were grown in M, MS, and PS media. When grown using different carbon sources (maltose or peptides) and terminal electron acceptors (H+ or S0), both OE-NfnI and OE-NfnII strains showed growth defects at different levels of severity. OE-NfnI had most severe phenotype in M medium, similar to that observed with the ΔNfnI strain (Fig. 2A). The phenotype is less prominent in the MS medium and insignificant in PS medium. This observation, together with the decrease in transcription of NfnI during sulfur and peptide metabolism, suggests that NfnI function is more important in carbohydrate metabolism. On the other hand, the OE-NfnII strain displayed similar phenotypes when grown in all three media, reaching ∼50% of the growth compared with the control strain. It appears that the NfnI and NfnII overexpression strains have similar and sometimes more severe growth phenotypes compared with the ΔNfnI and ΔNfnII strains. It is likely that these growth defects are due to the increased NfnI and NfnII activities changing the composition of the three main redox pools (Fd, NAD(H), and NADP(H)), thereby affecting the activities of other redox enzymes. Taken together, these data suggest that the native expression levels of the NfnI and NfnII operons are finely tuned to regulate redox balance in P. furiosus metabolism.
The differential expression of NfnI and NfnII together with the different phenotypes observed in the overexpression and deletion strains suggest that NfnI and NfnII play pivotal roles in carbohydrate and peptides metabolism, whereas S0 metabolism adds an additional layer of complexity. In short, NfnI is mainly responsible for the transfer of electrons from NADH and reduced Fd to NADP+, which is used for biosynthesis (14) under normal carbohydrate metabolism in P. furiosus. However, whether NfnII has the same function as NfnI is still unclear because during growth with peptides, where NfnII expression is highest, expression of glutamate dehydrogenase, another NAD(P)H-forming enzyme, is also up-regulated (23). Judging from sequence homology, NfnI and NfnII could be catalyzing very similar reactions but with different kinetics. However, to better understand the roles of each Nfn, characterization of NfnII was undertaken.
Purification of NfnII in P. furiosus
To obtain protein for structural and biochemical characterization, His-tagged NfnII was purified from the P. furiosus OE-NfnII strain. Approximately 43 mg of purified NfnII was obtained from 60 g of wet P. furiosus cells. All peak fractions from the nickel–nitrilotriacetic acid column that were brownish to yellow in color (from flavin and iron-sulfur clusters) and had NADPH-dependent BV reduction activity were pooled, concentrated, and further purified via anion-exchange chromatography. A similar two-step purification protocol was used previously for the purification of NfnI (14). The total protein recovery after the two-column purification was 62% (Table 1). The NfnII protein was purified as the expected heterodimer containing L and S subunits, as shown by gel electrophoresis (supplemental Fig. 8).
Table 1.
Purification of NfnII
| Steps | Volume | Total protein | Total activity | Specific activity | Purification | Yield |
|---|---|---|---|---|---|---|
| ml | mg | units | units mg−1 | -fold | % | |
| S100 | 450 | 6480 | 96 | 17 | 1.0 | 100 |
| His-trap | 28 | 165 | 62 | 74 | 4.1 | 65 |
| QHP | 17 | 42 | 59 | 275 | 15 | 62 |
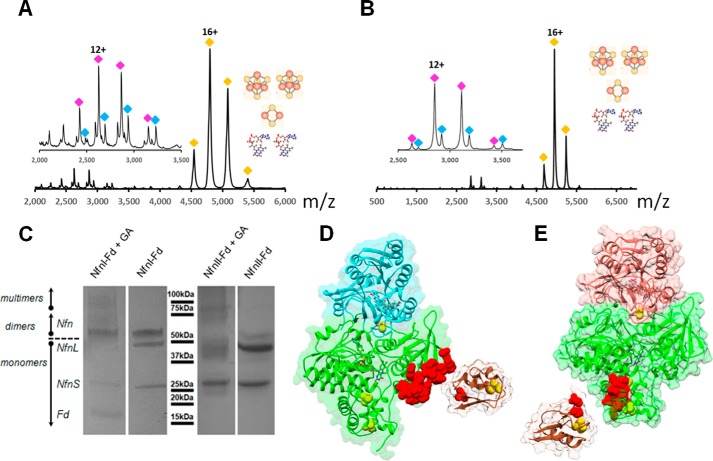
In-gel digestion and LC-MS analysis confirmed the identity of the L and S subunits of both NfnI and NfnII. We suspected that the lower-molecular weight protein band seen on the electrophoresis gel was Fd, as described for NfnI (14). LC-MS analysis of both samples after digestion with the low-specificity protease pepsin confirmed the presence of Fd in both samples. Note that Fd is a small protein (7.5 kDa) and does not stain well on an electrophoresis gel (supplemental Fig. 8). These results inspired us to pursue a more in-depth analysis of the as-purified complexes using native mass spectrometry. Analysis of the purified complexes confirmed the heterodimeric state and were consistent with the presence of two FADs, two [4Fe4S] clusters, and one [2Fe–2S] cluster in each NfnI and II dimer in solution (Fig. 4), based on the crystallography data presented below and by Lubner et al. (14). By activating the complex with low collision energy, the dimer was dissociated, revealing a small subunit with and without one FAD. At this point, the two complexes appeared highly similar. However, further collision-induced disassociation at higher energies showed that the NfnI complex was much more tightly associated than NfnII. Even at high collision energy (200 V) or in the presence of acetonitrile, a fraction of NfnI remained dimeric. Fd appears to have a lower affinity for NfnII, because the trimeric complex was not observed in the gas phase.
Figure 4.
Native and chemical cross-linking mass spectrometry of NfnI and NfnII. Native mass spectrum of NfnI complex (A) and NfnII complex (B) in the gas phase. Yellow diamonds, charge states of the intact complex centered around charge 16+. After deconvolution, the intact NfnI complex is 86,407 Da (expected: 86,425 Da) and NfnII is 88,951 Da (expected 88,901 Da), consistent with complexes containing one small and large subunit, two FAD molecules, two [4Fe–4S] clusters, and one [2Fe–2S] cluster. Under conditions of low collision energy (80 V for NfnI and 60 V for NfnII), the subunits dissociated (insets in A and B). Blue and magenta diamonds, charge envelopes of small subunits with and without one FAD cofactor. C, SDS-PAGE of glutaraldehyde (GA)–cross-linked NfnI–Fd and NfnII–Fd complexes. LC-MS analysis revealed that ferredoxin (brown) interacts with the large subunits (green) of NfnI (D) and NfnII (E). Red space-filling regions show cross-linked peptides of NfnI and NfnII. Small subunits are colored in cyan and red, respectively. In all experiments, Nfn enzymes were used “as purified.” The NfnI structure is PDB entry 5JFC; NfnII structure is PDB entry 5VJ7.
To capture the trimeric complex, chemical cross-linking using glutaraldehyde was employed. Cross-linked complexes were separated on SDS-PAGE, and then cross-linked species where subjected to in-gel proteolysis and LC-MS analysis (Fig. 4C). NfnI (Lys371–Phe392) interacted with the N terminus of Fd, whereas NfnII (residues Gly55–Lys68) cross-linked around Lys32 on Fd (Fig. 4, D and E). Thus, whereas the NfnI and NfnII have the same stoichiometry and cofactor composition, the complexes have specific differences in stability and potentially in the Fd docking site.
Structure determination of NfnII
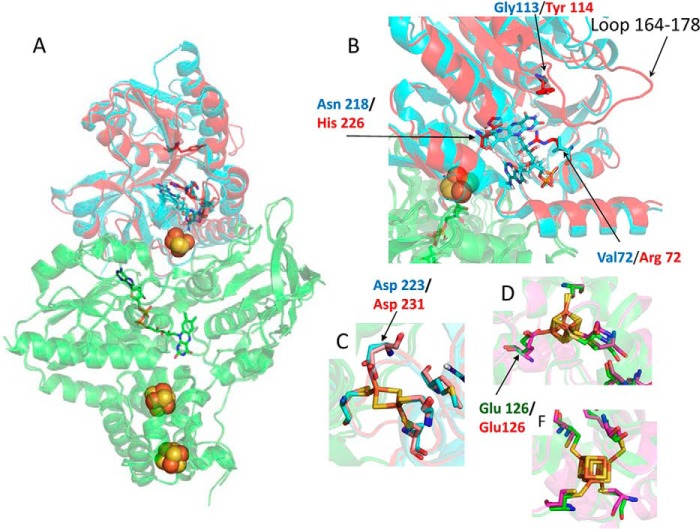
The NfnII enzyme was crystallized as a heterodimer with a large (NfnII-L) and a small (NfnII-S) subunit. The structure was determined by molecular replacement using the known structure of NfnI as a search model and refined to 2.6 Å resolution. NfnII-L contains one FAD (L-FAD) and two [4Fe–4S] clusters, whereas NfnII-S contains one FAD (S-FAD) and one [2Fe–2S] cluster (Fig. 5A and supplemental Figs. 9 and 10). The [2Fe–2S] and the [4Fe–4S] clusters proximal to L-FAD are coordinated by aspartate and glutamate ligands, respectively (Fig. 5, C and D). The NfnII-L is very similar to NfnI-L, with a root mean square deviation (RMSD) of 0.70 Å. The main differences between the two enzymes are in the small subunit, having an RMSD of 1.06 Å. The Fe–S clusters in NfnII have similar coordination environments as in NfnI (Fig. 5, C, D, and F). The most notable difference in the structure is the presence of an extended loop in the S subunit, composed of residues 164–178, which appear to occlude the assumed NAD(H) binding site, as determined by comparison with the structure of NfnI. In addition, within this FAD binding site in the S subunit, there are substitutions in NfnII of key residues that help to coordinate NADH binding in the NfnI structure. Specifically, Asn218 (in NfnI) is replaced by His226 (in NfnII), Gly113 replaces Tyr114, and Val72 replaces Arg72 (numbering according to NfnI structure; PDB entry 5JFC). Hence, given that the NAD(H) binding site is blocked and key binding residues are absent, the primary conclusion from the crystal structure of NfnII is that this enzyme is unlikely to use NAD(H) as a substrate. Accordingly, as described below, NfnII did not exhibit NADH-dependent dye reduction, in contrast to NfnI.
Figure 5.
Structure of NfnII. A, superposition of NfnII and NfnI (PDB entry 5JFC) structures shows the similarity in the large NfnII and NfnI (green) subunits and differences in the small NfnII (red) and NfnI (cyan) subunits. Protein structures are presented as schematics, FeS clusters as balls, and FAD molecules as sticks. B, difference between the NfnII (red) and NfnI (cyan) small subunits at the S-FAD binding site. The pairs of amino acids, which potentially result in the blockage of NAD(H) binding to the active site, are shown in sticks and are colored according to the color of the subunits. Similarity between NfnII and NfnI in the coordination of the 2Fe–2S cluster (C), proximal 4Fe–4S cluster (D), and distal 4Fe–4S cluster (F) is shown in sticks. Iron atoms are shown in brown, and sulfur atoms are shown in yellow. Amino acids are colored according to the color of the subunits.
Comparison of NfnII and NfnI in vitro activities
A comprehensive list of the catalytic activities of NfnI and NfnII measured in this study are given in Table 2. Consistent with the structural data, NfnII did not exhibit significant NADH-dependent reduction of the dye BV, which was ∼60 times lower than that observed with NfnI (1.1 ± 0.3 versus 61 ± 7 units mg−1). However, the NADPH-dependent reduction of BV activities were similar between NfnI and NfnII (1277 ± 165 and 408 ± 60 units mg−1). In light of the NADH-dependent data, it was not surprising to find that the Nfn bifurcating activity of NfnII (using NADH, NADP+, and Fdox) was insignificant (<0.05 units mg−1), compared with that measured using NfnI (21 ± 3 units mg−1, measured by adding NfnI into the same assay cuvette used to measure NfnII activity). To rule out the possibility that NfnII was inactive after purification and/or lacked some active cofactor, the bifurcating Nfn activity was measured using the cytosolic extracts of the following strains under non-sulfur and sulfur-reducing conditions: COM1c (as the positive control), ΔNfnI (to separate NfnII activity from NfnI), and ΔNfnII (as the negative control). There was no bifurcating activity detected in the cytosolic extract of the ΔNfnI strain under both conditions, whereas comparable activities were measured in the extract of the COM1c and ΔNfnII strains (supplemental Table S3). These data suggest that NfnI is the sole enzyme responsible for the Nfn bifurcating activity in P. furiosus and that “NfnII” has FNOR-type (measured by the NADPH-dependent reduction of BV) rather than true Nfn bifurcating activity.
Table 2.
Activities of NfnI and NfnII
S.D. values were derived from three technical measurements.
| Electron donors | Electron acceptors | NfnI activity | NfnII activity |
|---|---|---|---|
| units mg−1 | units mg−1 | ||
| NADPH | NAD+ + Fdox | 21 ± 3 | <0.01a |
| NADPH | BV | 1277 ± 165 | 408 ± 60 |
| NADH | BV | 61 ± 7 | 1.1 ± 0.3 |
| NADPH | Fdox | 0.7 ± 0.01 | <0.01a |
| Fd70redb | NADP+ | 0.4 ± 0.2 | 5.0 ± 0.8 |
| Fd70redb | NAD+ | 0.7 ± 0.2 | 0.6 ± 0.2 |
a Below detection limit of 0.01 units mg−1.
b Chemically reduced by ∼70% with titanium citrate.
The apparent FNOR-like activity of NfnII was confirmed using the physiological substrate Fd in place of the artificial electron carrier BV. By using Fd, both the oxidative and reductive directions of the FNOR reaction could be measured. No significant NADPH-dependent Fd reduction activity was detected with NfnII, although NfnI had measureable activity (0.70 ± 0.01 units mg−1; Table 2). To measure the NAD(P)+ reduction activity, titanium citrate was used to obtain ∼70% reduced Fd to serve as the electron donor (to mimic the cellular redox ratio of Fd). The Fdred-dependent NAD+ and NADP+reduction activities of NfnI were low because electron transfer was gated in this “tight” bifurcating enzyme. There was little activity unless all three substrates (NAD(H), NADP(H), and Fdred/ox) were present, which prevented Fd from directly reducing NAD+ or NADP+ in the absence of the other cofactor (14). In contrast, this was not the case with NfnII, which exhibited significant Fd-dependent NADP+ reduction activity compared with NfnI (5.0 ± 0.8 versus 0.4 ± 0.2 units mg−1; Table 2).
These results show that NfnII does not have Nfn bifurcating activity and does not utilize NAD(H) as a substrate. It exhibits one related activity, the Fd-dependent reduction of NADP+ (FNOR activity). The lack of NAD(H)-linked activity is consistent with the structure of the enzyme discussed above (Fig. 5B). Interestingly, this additional loop in the NfnII structure that blocks NAD(H) binding is found in the sequences of all of the NfnII homologs in the order Thermoccocales (supplemental Fig. 11) but is not found in the sequences of NfnI or NfnIII. Additionally, Fd was found to bind differently in NfnI and NfnII (Fig. 4, D and E). Investigation is under way to determine whether this considerable difference in Fd mode of binding affects NfnII activity. However, as expected from thermodynamic considerations, the FNOR activity exhibited by NfnII is catalytically biased toward NADP+ reduction (supplemental Table S4), in agreement with the proposed physiological role of NfnII, which is to generate NADPH from reduced Fd, independent of NAD(H).
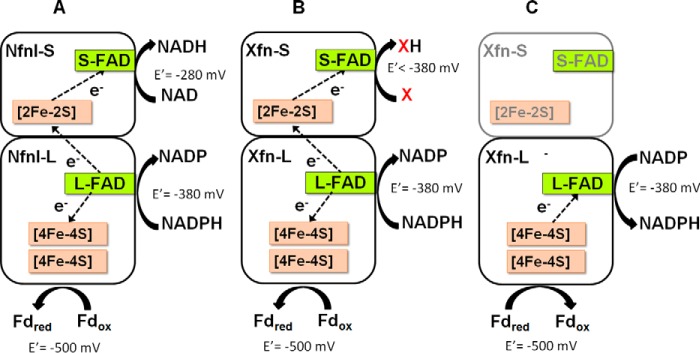
Hence, unlike NfnI, NfnII is not a bifurcating Nfn. However, whether it is another type of bifurcating enzyme utilizing another substrate (in place of NAD(H)) or a new type of non-bifurcating FNOR enzyme is unclear at this point. There are two scenarios to be considered (Fig. 6). First, if NfnII functions as a non-bifurcating FNOR, then structurally, only the NfnII-L subunit contributes to the catalytic activity of the holoenzyme, begging the question of the role of its S-subunit and why NfnII retains a structure and cofactor composition similar to NfnI. Alternatively, given that the NfnII-L subunit contains a bifurcating flavin (by analogy with NfnI), NfnII may be a “bifunctional” enzyme in that, as well as FNOR activity, it also bifurcates using a third substrate that is not NAD(H). We hypothesize that, if this is the case, then the third substrate of NfnII is somehow involved in peptide and sulfur metabolism by P. furiosus. However, NfnII is clearly not a bifurcating NADH-dependent ferredoxin NADP+ oxidoreductase, and the term Nfn is not appropriate. At present, we favor the notion that NfnII is a bifunctional enzyme with both non-bifurcating (FNOR) and bifurcating activity. Henceforth, it will be referred to as Xfn, with X representing the unknown third substrate used in bifurcation. We are currently using spectroscopic tools to determine whether the FAD in the L-subunit of Xfn has the expected and characteristic signature of a bifurcating flavin and are attempting to identify substrate X. Substrate X could be another redox protein, an oxidoreductase, or a small molecule. Understanding the properties of Xfn will add another layer to our limited knowledge of both bifurcating enzymes and the primary redox metabolism of P. furiosus.
Figure 6.
Schematic models depicting the cofactor contents and the reactions catalyzed by NfnI (A) and Xfn in two possible scenarios, where Xfn is a bifurcating enzyme with substrate X that replaces NAD(H) (B) or Xfn is a non-bifurcating FNOR type enzyme where only the large subunit (Xfn-L) contributes to the catalytic activity (C).
In conclusion, in the order Thermococcales, NfnI is an important bifurcating enzyme that functions in primary redox metabolism by balancing three separate pools of redox cofactors, Fd, NADPH, and NADH, during carbohydrate metabolism. NfnII, a homolog of NfnI, which we now term Xfn, appears to be a bifunctional enzyme and plays an important role in maintaining the primary redox pool during sulfur and peptide metabolism. In P. furiosus, the physiological functions of NfnI and Xfn are overlapping and crucial, and no other enzyme can compensate for them.
Experimental procedures
Characterization of nfnS gene neighborhood
Homologs of NfnS and NfnL were identified in publicly available complete archaeal genomes (n = 230) using previously characterized NfnS (TM1639) and NfnL (TM1640) from Thermotoga maritima (24) as BLASTp queries. Only NfnSL that were co-localized in the genome were retained for further phylogenetic and gene neighborhood analyses. A custom Python (version 2.7.6) script was used to extract gene sequences (10 upstream and 10 downstream) that flanked nfnS (see supplemental material). The 20 inferred protein sequences were subjected to pairwise alignment and were clustered using the cd-hit program (25). Inferred protein sequences were clustered using identity thresholds of 90, 60, and 30% while holding the pairwise sequence coverage threshold constant at >60%. The clusters generated by the three-step clustering method were later combined to obtain a final “averaged” cluster identity. Protein sequence clusters were then used to generate a binary matrix describing the presence or absence of clusters for use in statistical analyses.
Network analysis
The binary matrix describing the distribution of protein bins in the gene neighborhood of nfnS was organized based on the taxonomic rank of the host genome. To predict the potential functional role of Nfn without bias, only the archaeal phyla that contained >10 nfnS encoding genomes were considered further, and these were ultimately analyzed at the class taxonomic level or at the level of genes that flank individual Nfn isoforms. Proteins that were in high abundance in the flanking region of nfnS (>20%) were visualized using Cytoscape, specifying the force-directed organic layout (26).
Phylogenetic analysis
Identified NfnS and NfnL proteins were aligned individually with ClustalW, specifying default settings (27). NfnS and NfnL alignment blocks were concatenated using a custom Python (version 2.7.6) script. Phylogenetic reconstruction was performed with PhyML (version 3.1) (28), specifying the LG substitution matrix, and Chi2 to approximate the likelihood ratios. Trees were projected using Itol (29).
PCR product construction
For the overexpressed (OE-) strain, a linear PCR product was assembled to overexpress NfnII by targeted replacement of the native promoter with the promoter (slp) of the gene encoding the highly expressed S-layer protein, similar to the overexpression cassette for NfnI (15). The full linear ∼2.5-kb PCR construct was assembled via splicing by overlap extension and PCR (30). The NfnI and NfnII deletion cassettes were also designed to knock out the NfnI (PF1327-1328) and NfnII (PF1910-1911) genes in the P. furiosus genome. These deletion cassettes containing the pyrF pop-out marker cassette, including the 65-bp identical flanking region, were inserted between the homologous up- and downstream flanking regions of the NfnI and NfnII genes. The full ∼2.3-kb PCR constructs were obtained using a PCR technique similar to that described above. All primers designed and used in this study are reported in supplemental Table S1.
Strain constructions
Strains constructed for this study are listed in Table 3. The linear PCR constructs were designed to insert an NfnII overexpression cassette into the P. furiosus genome at the native NfnII gene location, as described above. The overexpression PCR construct was transformed into P. furiosus COM1, as described previously (31). The transformants were cultured and purified three times on liquid and solid defined cellobiose-containing medium (31). Genomic DNA was isolated using the ZymoBeadTM Genomic DNA Kit (Zymo Research), and isolates were screened by PCR, using primers that target outside of the homologous flanking regions. A strain containing the NfnII overexpression constructs was sequence-verified and designated as MW333 or OE-NfnII, respectively.
Table 3.
Strains used and constructed in this study
| Strain number | Alias | Genotype | Source/Reference |
|---|---|---|---|
| MW002 | COM1/parent | ΔpyrF | Ref. 31 |
| MW004 | COM1c/control | ΔpyrF::pyrF | Ref. 31 |
| MW187 | ΔNfnII | ΔpyrF ΔnfnII | This study |
| MW190 | ΔNfnI | ΔpyrF ΔnfnI::Ppep pyrF | This study |
| MW379 | ΔNfnII | ΔpyrF::pyrF ΔnfnII | This study |
| MW367 | OE-NfnI | ΔpyrF PslpHis9-Gly-nfnI::Ppep pyrF | Ref. 14 |
| MW333 | OE-NfnII | ΔpyrF Pslp His9-Gly-nfnII::Ppep pyrF | This study |
| MW383 | ΔNfnI-ΔNfnII | ΔpyrF ΔnfnII ΔnfnI::Ppep pyrF | This study |
The single deletion mutant of NfnI (PF1327-28) was also constructed using methods similar to those described above, and the sequence-verified strain was termed MW190 and will be referred to herein as the ΔNfnI strain. The single-deletion mutant of NfnII mutant was also constructed, but the genes encoding it (PF1910-11) are located downstream and transcribed divergently from the gene encoding ferredoxin, PF1909. Therefore, to avoid any polar effect, the genetic marker pyrF was immediately spliced out by counterselection for the loss of the marker cassette, as described previously (32, 33). The markerless strain, after sequence verification, was termed MW187. This strain was subsequently transformed with linear DNA that targeted restoration of the pyrF gene at its native location. This new strain was named MW379 and is referred to as the ΔNfnII strain.
To generate the double deletion mutant lacking both NfnI and NfnII, the NfnI deletion construct was transformed into the markerless MW187 strain, as described above. This strain was named MW383 and is referred to herein as the ΔNfnI-ΔNfnII strain. It should be noted that attempts to construct this double deletion strain were unsuccessful as judged by DNA-gel electrophoresis (data not shown), with roughly 50% of the cells still containing genes encoding NfnI.
Growth of P. furiosus
Strains (Table 3) were cultured in artificial seawater medium containing the following per liter: 1× base salts (34), 1× trace minerals (34), 10 μm sodium tungstate (34), 0.25 μg of resazurin, 10 μm riboflavin, 10 μm cobalamin, 0.5 g of cysteine, 1 g of sodium bicarbonate, and 1 mm potassium phosphate buffer, with the pH adjusted to 6.8. 50 or 75 ml of medium were aliquoted into 100- or 150-ml serum bottles. The medium bottles were then capped, and the headspace was replaced with argon after three cycles of vacuum. For the growth experiments, three types of medium were used: the minimal maltose medium (M), which was supplemented with 5 g of maltose and 0.5 g of yeast extract per liter (35); the maltose sulfur (MS) medium, which is the M medium containing 2 g of elemental sulfur per liter; and the minimal peptide sulfur medium (PS), which contained 5 g of casein, 0.5 g of yeast extract, and 2 g of elemental sulfur per liter (11). P. furiosus cells were inoculated to ∼3 × 106 cells ml−1, and cultures were incubated at 90 °C with shaking at ∼200 rpm. Growth was monitored by cell counting using a Petroff–Hausser counting chamber. Cell protein was also quantified from 1 ml culture samples using the Bradford protein assay kit (Bio-Rad). Cells were harvested by centrifugation and lysed by osmotic shock in an equal volume of water with vortexing. Lysate was centrifuged at 10,000 × g for 1 min to pellet insoluble cell debris before quantitation of soluble cell protein. The overexpressing recombinant strains, OE-NfnI and OE-NfnII, were each grown on a 20-liter scale as described previously (14). The growth medium was supplemented with 5 g of maltose and 5 g of yeast extract per liter. After reaching the optimal cell density, cells were collected by centrifuging at 6000 × g for 10 min and were stored at −80 °C until use.
Measurements of NAD(H) and NADP(H) in P. furiosus
The COM1c, ΔNfnI, and ΔNfnI strains were grown in 50-ml medium bottles of M, MS, and PS media until mid-log phase at 90 °C with shaking at ∼200 rpm. Cells were collected via centrifugation at 6000 × g for 10 min at 4 °C and lysed with 100 μl of 50 mm ammonium acetate anaerobically inside a Coy chamber. Lysates were filtered through 10-kDa cut-off filters (Merck Millipore) to remove proteins. The flow-through samples were immediately used for NAD(H) and NADP(H) analysis by high-performance liquid chromatography (HPLC). HPLC measurements were performed according to a method described previously (36) with some modification. A Hydrosphere C18 column (5 μm, 150 × 4.6-mm inner diameter, 12 nm (YMC Co., Ltd., Kyoto, Japan) was used connected to a YMC Guard Cartridge column and run on an Agilent 1260 HPLC (Hewlett-Packard, Wilmington, DE). Filtered lysates were kept anaerobically, and 20-μl aliquots were injected for analysis. NAD(P)(H) (5 μm) was added to samples as an internal control. NADPH, NADH, NADP+, and NAD+ were quantified based on absorption at 340 and 260 nm, respectively. Calculated concentrations of NAD(P)(H) were normalized based on the protein concentrations of the lysates. One unit represents 1 μmol of nicotinamide nucleotide per g of protein.
RNA extraction
RNA was extracted using a phenol/chloroform method as described previously (37). Contaminating genomic DNA was digested using TURBO DNase (Ambion). RNA quality was assessed by A260/A280 ratios and qPCR.
Quantitative RT-PCR
Synthesis of cDNA was performed with 1 μg of purified RNA using the Affinity Script QPCR cDNA synthesis kit (Agilent). The Brilliant III SYBR® Green QPCR Master Mix (Agilent) was used for quantitative RT-PCR experiments with primers designed to amplify a ∼150-bp product within the target gene. The constitutively expressed PF0983 gene encoding the sliding clamp subunit of the DNA polymerase was used as a reference.
Purification of the recombinant His-tagged NfnI and NfnII
Approximately 60 g of frozen cells were lysed by resuspending them in 300 ml of 50 mm phosphate buffer, pH 7.0, under strict anaerobic conditions inside a Coy chamber (CoyLab, Grass Lake, MI). The S100 cell-free extract fraction was obtained by ultracentrifugation at 100,000 × g for 1 h at 12 °C to remove the membrane remnants and cell debris. Anaerobic conditions were maintained during Nfn (I and II) purification by adding 1 mm cysteine to all buffers. The S100 was loaded onto a 5-ml HisTrapTM FF crude column (GE Healthcare), and the His-tagged protein was eluted by applying a gradient of imidazole in 50 mm phosphate buffer, pH 7.0, per the manufacturer's instructions. All fractions that contained Nfn activity, measured by the NADPH-dependent reduction of benzyl viologen, were pooled and concentrated using an Amicon Ultra-4 ultrafiltration centrifugal filter (10-kDa cut-off; Merck Millipore). The concentrated fractions were loaded onto a 5-ml HiTrapTM Q HP column (GE Healthcare) equilibrated with 25 mm Tris/HCl, pH 8.0, and Nfn was eluted with a gradient from 0 to 500 mm NaCl. The purity of active fractions was judged by gel electrophoresis before pooling and storing at −80 °C until use. Approximately 0.5 mg of protein was purified from 1 g of wet cell paste for both NfnI and NfnII.
Protein identification
Digestion of gel bands and proteins in solution was performed according to standard protocols recommended by the manufacturers using a trypsin (Promega) protease/complex ratio of 1:50–1:100 overnight and pepsin (Sigma) protease/complex ratio of 1:10 for 60 s. Proteins were identified as described (38), using a maXis Impact UHR-QTOF instrument (Bruker Daltonics, Billerica, MA) interfaced with a Dionex 3000 nano-uHPLC (Thermo Fisher Scientific), followed by data analysis in a peptide shaker (39). Intact protein analysis was performed as described previously using a Bruker Micro-TOF mass spectrometer (Bruker Daltonics) (40).
Chemical cross-linking
Protein cross-linking was performed using 10 mm glutaraldehyde (Sigma) and 20 μg of the NfnI and NfnII at 14 μm, complexed with Fd in a 1:1 ratio (Fd, 14 μm). The reaction was carried out in 50 mm HEPES, pH 7.2, 150 mm NaCl at room temperature. The reaction was quenched after 10 min by the addition of 1.7 m Tris buffer, pH 8, to a final concentration of 100 mm. The resulting mixtures were separated by SDS-PAGE (4–20% linear gradient gel; Bio-Rad) and stained with Coomassie Brilliant Blue (Thermo Fisher Scientific). Protein bands of interest were excised from the gel, digested with trypsin, and analyzed by LC-MS as described above.
Native mass spectrometry
Non-covalent mass spectrometry under native conditions was conducted on a SYNAPT G2-Si instrument (Waters) in a fashion similar to what was previously described (41). Briefly, the NfnI and NfnII complex samples were buffer-exchanged with 200 mm ammonium acetate, pH 7 (Sigma), using Mr 3000 cut-off spin filters (Pall Corp.) and infused from in-house prepared gold-coated borosilicate glass capillaries to electrospray source at a protein concentration of 5 μm and a rate of ∼90 nl min−1. The instrument was tuned to enhance performance in the high mass-to-charge range. Settings were as follows: source temperature 30 °C, capillary voltage 1.7 kV, trap bias voltage 16 V, and argon flow in collision cell (trap) 7 ml min−1. The transfer collision energy was held at 10 V, whereas trap energy varied between 10 and 200 V. To determine an accurate mass of individual protein components, complexes were denatured by dilution in a 50:50 solution of 1% formic acid (Sigma) and acetonitrile (Thermo Fisher Scientific). Data analysis was performed in MassLynx software version 4.1 (Waters). Molecular graphics were created using the UCSF Chimera package (42). Intact protein analysis showed that all proteins, except NfnII-S, have missing N-terminal Met residues. NfnII-S 34,029.1 Da (calculated 34,028.71 Da), NfnII-L 52,368.37 Da (calculated 52,497.8 Da, missing N-terminal Met residue); NfnI-S 31,376.7 Da (calculated: 31,507.9 Da, missing N-terminal Met residue), NfnI-L 52,597.8 Da (calculated 52,729 Da, missing N-terminal Met residue); Fd 7,167.0 Da (calculated: 7,298.2 Da, missing N-terminal Met residue).
Structure determination and refinement
NfnII crystals were obtained by the vapor diffusion method under anaerobic conditions in a Coy anaerobic chamber using 0.22 m magnesium sulfate, 27% (w/v) polyethylene glycol 3350, and 0.4% (v/v) ethyl acetate in the presence of 1 mm sodium dithionite. In addition, before flash freezing in liquid nitrogen, NfnII crystals under argon flow were dragged through crystallization solution containing 10% (v/v) glycerol as a cryoprotectant. The data were collected from flash-cooled crystals with a continuous flow of liquid nitrogen at 100 K on BL12-2 (SLAC National Accelerator Laboratory). The diffraction images were indexed, integrated, and scaled using HKL2000 (43). The structure was solved to 2.6 Å by molecular replacement using the structure of NfnI (sequence identity 48.3%, PDB code 5JFC), with phenix.phaser (44). The solutions were refined and improved by phenix.refine (45) with final R/Rfree to 20.9%/27.4% (Table 4). Model building was subsequently completed manually using COOT (46). Figures were prepared using PyMOL (47). The RMSD was calculated using SUPERPOSE (48). Composition of the crystal was confirmed by dissolving the protein and running on SDS-PAGE. Protein bands were digested in gel with trypsin and identified using LC-MS as described under “Chemical cross-linking.” The NfnII structure was deposited in the PBD with code 5VJ7.
Table 4.
Data collection and refinement statistics for NfnII
| Parameters | Values |
|---|---|
| Data collection | |
| Wavelength (Å) | 0.9369 |
| Unit cell parameters | |
| a, b, c (Å) | 55.74, 73.14, 99.96 |
| α, β, γ (degrees) | 90.0, 96.7, 90.0 |
| Space group | P 21 |
| Resolution range (Å) | 39–2.6 |
| Total reflections | 108,586 |
| Unique reflections | 25,877 |
| Rmerge (%) | 10.0 (36.2)a |
| Rpim (%) | 6.1 (21.5) |
| CC1/2 | 0.991 (0.934) |
| I/σ(I) | 7.7 (2.6) |
| Completeness (%) | 98.8 (99.1) |
| Redundancy | 4.2 (4.2) |
| Refinement | |
| Resolution limits (Å) | 39–2.6 |
| No. of reflections | 25,825 |
| No. of atoms | 6196 |
| Rfactor (%) | 20.9 |
| Rfree (%) | 27.4 |
| Wilson B-factor (Å2) | 35.3 |
| Ramachandran plot | |
| Favorable region (%) | 95.0 |
| Allowed regions (%) | 5.0 |
| Disallowed regions (%) | 0 |
| RMSD from ideality | |
| Bond distance (Å) | 0.010 |
| Angles (degrees) | 1.19 |
a Values in parentheses are for the highest-resolution shell.
Nfn and FNOR dye-linked activity assay
NADH- and NADPH-dependent dye-linked activity assays were performed in 50 mm MOPS, pH 7.5, and 2 mm benzyl viologen (BV) was used as the electron acceptor. BV reduction was monitored at 600 nm, ϵ = 7.4 mm−1 cm−1. The assay contained 1 mm NAD(P)H and 2 mm BV. One unit is defined as 1 μmol of BV reduced per min.
Nfn bifurcating activity assay
The bifurcating activity was measured based on the following reaction: 2 NADPH + 2 Fdox + NAD+ →2 NADP+ + H+ + NADH + 2 Fdred. Fd was purified based on the protocol described previously (49). Fd reduction was monitored at 425 nm (ϵ = 13 mm−1 cm−1) using 1 mm NADPH, 2 mm NAD+, and 25 μm Fd in 50 mm MOPS, pH 7.5, at 80 °C. One unit of bifurcating activity is defined as 1 μmol of Fd reduced per min.
FNOR activity assay
The FNOR activity assay was measured according to the following reaction: 2 NAD(P)+ + 2 Fdred ↔ 2 NAD(P)H + 2 Fdox. Fd reduction or oxidation was monitored at 425 nm using 1 mm NADP(H) and 50 μm Fd (to an absorbance of ∼0.9) in 50 mm MOPS, pH 7.5, at 80 °C. Where reduced Fd was used, the protein was chemically reduced with freshly prepared 7 mm titanium citrate at the beginning of each assay. One unit of FNOR activity is defined as 1 μmol of Fd reduced or oxidized per min.
Author contributions
D. M. N. N., G. J. S., G. L. L., and M. W. W. A. conceived and coordinated the study and wrote the paper. S. P. and E. S. B. carried out the phylogenetic analyses, O. A. Z. and J. W. P. performed the crystallographic study, and M. T.-L. and B. B. carried out the MS analyses. D. M. N. N., G. L. L., and G. J. S. designed and performed all of the genetic, enzymatic, and physiological studies with technical assistance from L. A. A., J. T. D., and W. J. N. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank the Montana State University Microfabrication Facility for help in preparation of gold-coated borosilica capillaries for non-covalent mass spectrometry and Dr. Ravi Kant for assistance with protein identification. The Mass Spectrometry Facility at Montana State University is supported in part by the Murdock Charitable Trust and National Institutes of Health IDEA Program Grant P20GM103474. The use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the Department of Energy Office of Biological and Environmental Research, and by NIGMS, National Institutes of Health (including Grant P41GM103393).
This work was supported as part of the Biological Electron Transfer and Catalysis Energy Frontier Research Center funded by the U. S. Department of Energy, Office of Science, Basic Energy Sciences under Award DE-SC0012518. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Tables S1–S4 and Figs. S1–S11.
The atomic coordinates and structure factors (code 5VJ7) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- Fd
- ferredoxin
- Fdred
- reduced ferredoxin
- Fdox
- oxidized ferredoxin
- Nfn
- NADH-dependent ferredoxin NADP+ oxidoreductase
- SH
- soluble hydrogenase
- FNOR
- ferredoxin-dependent NAD(P)+ oxidoreductase
- NSR
- NADPH-dependent sulfur reductase
- M
- maltose
- MS
- maltose sulfur
- PS
- peptide sulfur
- NadB
- l-aspartate oxidase
- BV
- benzyl viologen
- OE
- overexpression
- qPCR
- quantitative PCR
- RMSD
- root mean square deviation
- PDB
- Protein Data Bank.
References
- 1. Verhees C. H., Kengen S. W., Tuininga J. E., Schut G. J., Adams M. W., De Vos W. M., and Van Der Oost J. (2003) The unique features of glycolytic pathways in Archaea. Biochem. J. 375, 231–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schut G. J., Nixon W. J., Lipscomb G. L., Scott R. A., and Adams M. W. W. (2012) Mutational analyses of the enzymes involved in the metabolism of hydrogen by the hyperthermophilic archaeon Pyrococcus furiosus. Front. Microbiol. 3, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ma K., and Adams M. W. W. (2001) Ferredoxin:NADP oxidoreductase from Pyrococcus furiosus. Methods Enzymol. 334, 40–45 [DOI] [PubMed] [Google Scholar]
- 4. Ma K., and Adams M. W. W. (1994) Sulfide dehydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus: a new multifunctional enzyme involved in the reduction of elemental sulfur. J. Bacteriol. 176, 6509–6517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sapra R., Bagramyan K., and Adams M. W. W. (2003) A simple energy-conserving system: proton reduction coupled to proton translocation. Proc. Natl. Acad. Sci. U.S.A. 100, 7545–7550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bridger S. L., Clarkson S. M., Stirrett K., DeBarry M. B., Lipscomb G. L., Schut G. J., Westpheling J., Scott R. A., and Adams M. W. W. (2011) Deletion strains reveal metabolic roles for key elemental sulfur-responsive proteins in Pyrococcus furiosus. J. Bacteriol. 193, 6498–6504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schut G. J., Bridger S. L., and Adams M. W. W. (2007) Insights into the metabolism of elemental sulfur by the hyperthermophilic archaeon Pyrococcus furiosus: characterization of a coenzyme A- dependent NAD(P)H sulfur oxidoreductase. J. Bacteriol. 189, 4431–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lipscomb G. L., Keese A. M., Cowart D. M., Schut G. J., Thomm M., Adams M. W. W., and Scott R. A. (2009) SurR: a transcriptional activator and repressor controlling hydrogen and elemental sulfur metabolism in Pyrococcus furiosus. Mol. Microbiol. 71, 332–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang H., Lipscomb G. L., Keese A. M., Schut G. J., Thomm M., Adams M. W. W., Wang B. C., and Scott R. A. (2010) SurR regulates hydrogen production in Pyrococcus furiosus by a sulfur-dependent redox switch. Mol. Microbiol. 77, 1111–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lipscomb G. L., Schut G. J., Scott R. A., and Adams M. W. W. (2017) SurR is a master regulator of the primary electron flow pathways in the order Thermococcales. Mol. Microbiol. 104, 869–881 [DOI] [PubMed] [Google Scholar]
- 11. Schut G. J., Brehm S. D., Datta S., and Adams M. W. W. (2003) Whole-genome DNA microarray analysis of a hyperthermophile and an archaeon: Pyrococcus furiosus grown on carbohydrates or peptides. J. Bacteriol. 185, 3935–3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang S., Huang H., Moll J., and Thauer R. K. (2010) NADP+ reduction with reduced ferredoxin and NADP+ reduction with NADH are coupled via an electron-bifurcating enzyme complex in Clostridium kluyveri. J. Bacteriol. 192, 5115–5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Buckel W., and Thauer R. K. (2013) Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim. Biophys. Acta 1827, 94–113 [DOI] [PubMed] [Google Scholar]
- 14. Lubner C. E., Jennings D. P., Mulder D. W., Schut G. J., Zadvornyy O. A., Hoben J. P., Tokmina-Lukaszewska M., Berry L., Nguyen D. M., Lipscomb G. L., Bothner B., Jones A. K., Miller A. F., King P. W., Adams M. W. W., and Peters J. W. (2017) Mechanistic insights into energy conservation by flavin-based electron bifurcation. Nat. Chem. Biol. 13, 655–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lo J., Olson D. G., Murphy S. J., Tian L., Hon S., Lanahan A., Guss A. M., and Lynd L. R. (2017) Engineering electron metabolism to increase ethanol production in Clostridium thermocellum. Metab. Eng. 39, 71–79 [DOI] [PubMed] [Google Scholar]
- 16. Lo J., Zheng T., Olson D. G., Ruppertsberger N., Tripathi S. A., Tian L., Guss A. M., and Lynd L. R. (2015) Deletion of nfnAB in Thermoanaerobacterium saccharolyticum and its effect on metabolism. J. Bacteriol. 197, 2920–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kanai T., Ito S., and Imanaka T. (2003) Characterization of a cytosolic NiFe-hydrogenase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185, 1705–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nixon J. E., Wang A., Field J., Morrison H. G., McArthur A. G., Sogin M. L., Loftus B. J., and Samuelson J. (2002) Evidence for lateral transfer of genes encoding ferredoxins, nitroreductases, NADH oxidase, and alcohol dehydrogenase 3 from anaerobic prokaryotes to Giardia lamblia and Entamoeba histolytica. Eukaryot. Cell 1, 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bastian S., Liu X., Meyerowitz J. T., Snow C. D., Chen M. M., and Arnold F. H. (2011) Engineered ketol-acid reductoisomerase and alcohol dehydrogenase enable anaerobic 2-methylpropan-1-ol production at theoretical yield in Escherichia coli. Metab. Eng. 13, 345–352 [DOI] [PubMed] [Google Scholar]
- 20. Maddocks O. D., Labuschagne C. F., and Vousden K. H. (2014) Localization of NADPH production: a wheel within a wheel. Mol. Cell 55, 158–160 [DOI] [PubMed] [Google Scholar]
- 21. Kikuchi G. (1973) The glycine cleavage system: composition, reaction mechanism, and physiological significance. Mol. Cell Biochem. 1, 169–187 [DOI] [PubMed] [Google Scholar]
- 22. Berger F., Ramírez-Hernández H. H., and Ziegler M. (2004) The new life of a centenarian: signalling functions of NAD(P). Trends Biochem. Sci. 29, 111–118 [DOI] [PubMed] [Google Scholar]
- 23. Robb F. T., Park J. B., and Adams M. W. W. (1992) Characterization of an extremely thermostable glutamate dehydrogenase: a key enzyme in the primary metabolism of the hyperthermophilic archaebacterium, Pyrococcus furiosus. Biochim. Biophys. Acta 1120, 267–272 [DOI] [PubMed] [Google Scholar]
- 24. Demmer J. K., Huang H., Wang S., Demmer U., Thauer R. K., and Ermler U. (2015) Insights into flavin-based electron bifurcation via the NADH-dependent reduced ferredoxin:NADP oxidoreductase structure. J. Biol. Chem. 290, 21985–21995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li W., and Godzik A. (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 [DOI] [PubMed] [Google Scholar]
- 26. Smoot M. E., Ono K., Ruscheinski J., Wang P. L., and Ideker T. (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thompson J. D., Gibson T. J., and Higgins D. G. (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics 10.1002/0471250953.bi0203s00 [DOI] [PubMed] [Google Scholar]
- 28. Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., and Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 [DOI] [PubMed] [Google Scholar]
- 29. Letunic I., and Bork P. (2007) Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128 [DOI] [PubMed] [Google Scholar]
- 30. Horton R. M., Hunt H. D., Ho S. N., Pullen J. K., and Pease L. R. (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77, 61–68 [DOI] [PubMed] [Google Scholar]
- 31. Lipscomb G. L., Stirrett K., Schut G. J., Yang F., Jenney F. E. Jr., Scott R. A., Adams M. W. W., and Westpheling J. (2011) Natural competence in the hyperthermophilic archaeon Pyrococcus furiosus facilitates genetic manipulation: construction of markerless deletions of genes encoding the two cytoplasmic hydrogenases. Appl. Environ. Microbiol. 77, 2232–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Farkas J., Stirrett K., Lipscomb G. L., Nixon W., Scott R. A., Adams M. W. W., and Westpheling J. (2012) Recombinogenic properties of Pyrococcus furiosus strain COM1 enable rapid selection of targeted mutants. Appl. Environ. Microbiol. 78, 4669–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thorgersen M. P., Stirrett K., Scott R. A., and Adams M. W. W. (2012) Mechanism of oxygen detoxification by the surprisingly oxygen-tolerant hyperthermophilic archaeon, Pyrococcus furiosus. Proc. Natl. Acad. Sci. U.S.A. 109, 18547–18552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Keller M. W., Lipscomb G. L., Loder A. J., Schut G. J., Kelly R. M., and Adams M. W. (2015) A hybrid synthetic pathway for butanol production by a hyperthermophilic microbe. Metab. Eng. 27, 101–106 [DOI] [PubMed] [Google Scholar]
- 35. Keller M. W., Schut G. J., Lipscomb G. L., Menon A. L., Iwuchukwu I. J., Leuko T. T., Thorgersen M. P., Nixon W. J., Hawkins A. S., Kelly R. M., and Adams M. W. W. (2013) Exploiting microbial hyperthermophilicity to produce an industrial chemical, using hydrogen and carbon dioxide. Proc. Natl. Acad. Sci. U.S.A. 110, 5840–5845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sporty J. L., Kabir M. M., Turteltaub K. W., Ognibene T., Lin S. J., and Bench G. (2008) Single sample extraction protocol for the quantification of NAD and NADH redox states in Saccharomyces cerevisiae. J. Sep. Sci. 31, 3202–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Weinberg M. V., Schut G. J., Brehm S., Datta S., and Adams M. W. W. (2005) Cold shock of a hyperthermophilic archaeon: Pyrococcus furiosus exhibits multiple responses to a suboptimal growth temperature with a key role for membrane-bound glycoproteins. J. Bacteriol. 187, 336–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang Z. Y., Ledbetter R., Shaw S., Pence N., Tokmina-Lukaszewska M., Eilers B., Guo Q., Pokhrel N., Cash V. L., Dean D. R., Antony E., Bothner B., Peters J. W., and Seefeldt L. C. (2016) Evidence that the Pi release event is the rate-limiting step in the nitrogenase catalytic cycle. Biochemistry 55, 3625–3635 [DOI] [PubMed] [Google Scholar]
- 39. Vaudel M., Burkhart J. M., Zahedi R. P., Oveland E., Berven F. S., Sickmann A., Martens L., and Barsnes H. (2015) PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat. Biotechnol. 33, 22–24 [DOI] [PubMed] [Google Scholar]
- 40. Poudel S., Tokmina-Lukaszewska M., Colman D. R., Refai M., Schut G. J., King P. W., Maness P.-C., Adams M. W. W., Peters J. W., Bothner B., and Boyd E. S. (2016) Unification of [FeFe]-hydrogenases into three structural and functional groups. Biochim. Biophys. Acta 1860, 1910–1921 [DOI] [PubMed] [Google Scholar]
- 41. Luo M. L., Jackson R. N., Denny S. R., Tokmina-Lukaszewska M., Maksimchuk K. R., Lin W., Bothner B., Wiedenheft B., and Beisel C. L. (2016) The CRISPR RNA-guided surveillance complex in Escherichia coli accommodates extended RNA spacers. Nucleic Acids Res. 44, 7385–7394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 43. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 44. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 47. DeLano W. L. (2015) The PyMOL Molecular Graphics System, version 1.8, Schrödinger, LLC, New York [Google Scholar]
- 48. Krissinel E., and Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 49. Aono S., Bryant F. O., and Adams M. W. W. (1989) A novel and remarkably thermostable ferredoxin from the hyperthermophilic archaebacterium Pyrococcus furiosus. J. Bacteriol. 171, 3433–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.