Abstract
The discovery of multiple RNA modifications in the past few years has broadened our views of the structures and potential functions of RNA species, but deciphering which modifications are made where and how remains a challenge. A new study by Xu et al. applies a combination of mass spectrometry, biochemistry, genetics, and cellular biology tools to reveal the two mammalian methyltransferases that are responsible for m3C installation in tRNA and a third that mediates the previously unknown installation of m3C in mammalian mRNA.
Introduction
Unlike genomic DNA, which tends to have a limited number of chemical modifications, RNA species can have many more types of modifications: To date, >100 different RNA modifications have been identified that encompass a wide variety of chemical diversity. Early studies on the abundant RNAs such as rRNA, snRNA, and tRNA demonstrated that this diversity of modifications leads to additional cellular functions for different RNA species (1). For example, rRNA modifications affect translation accuracy and efficiency and likely facilitate ribosome biogenesis. The studies of these modifications, particularly in the context of mRNA and lncRNA, embody the new concept of “epitranscriptomics,” in which the functional significance of chemical alterations is controlled by three groups of proteins: “writers” to install, “erasers” to remove, and “readers” to recognize modifications and thus determine the cellular fate of the modified RNA species. Xu et al. (2) now report the characterization of three “writers” in the form of mammalian methyltransferases anticipated to introduce N3-methylcytosine (m3C)2 modifications. Their data show that two of the enzymes act on tRNA as suspected, whereas the third surprisingly uses mRNA as a substrate, defining a new modification for this RNA species in mammals.
Efforts in epitranscriptomics are aided by the fact that many RNA modifications are conserved across most eukaryotes. For example, the conserved m3C modification in tRNA (3–6), the most heavily modified type of RNA, is installed in yeast by the methyltransferase Trm140 or the complex of Trm140 and Trm141 (7–9). m3C has been identified in tRNA and plant mRNA (10) but has not been reported in mammalian mRNA. However, N6-methyladenosine (m6A), pseudouridine (Ψ), 5-methylcytosine (m5C), N1-methyladenosine (m1A), and 2′O-methylation were previously shown to be present in mRNA (1) and may play versatile roles in mRNA processing and impact mRNA fates. For instance, m6A, the most abundant mRNA modification, appears to affect almost every phase of mRNA metabolism and function, thereby impacting diverse biological processes. Known modification enzymes that install Ψ and m5C in mRNA, PUS1/PUS7 and NSUN2, can also install the same modifications on tRNAs, suggesting that certain tRNA/rRNA modification enzymes could also act on mRNA.
The study by Fu and colleagues (2) begins with three mammalian methyltransferases: the two homologs of yeast Trm140 and Trm141 (METTL2 and METTL6) and another enzyme that also possesses a methyltransferase domain (METTL8). The authors knocked out Mettl2, Mettl6, and Mettl8 in mice and in human cell lines using CRISPR/Cas9. Using liquid chromatography triple quadrupole mass spectrometry (LC-MS/MS), they quantified m3C in tRNA fractions in brain and liver tissues from wild-type mice and Mettl2, Mettl6, and Mettl8 knockout (KO) mutants. The results showed that Mettl2 and Mettl6 KO led to a 35 and 12% reduction in m3C levels in tRNA, respectively. These observations were further confirmed in Mettl2A and Mettl2B KO HEK293T cells. Furthermore, the authors defined the location of the METTL2-dependent m3C in specific tRNAs by using primer extension assays: tRNAThr(UGU) and tRNAArg(CCU) extracted from wild-type mice generated RNA bands consistent with blockage of the reverse transcriptase at the expected position 32 of the tRNA. The blockage was significantly reduced in the tRNA extracted from Mettl2 KO mice. Reconstitution of METTL2B restored the polymerase-blocking modification in Mettl2 KO cells. The authors further identified two serine-specific tRNAs as substrates for METTL6, and followed up on a recent discovery in yeast suggesting that an interaction with the seryl-tRNA synthase might stimulate methyltransferase activity. In human cell lines, the authors observed that METTL6 interacts with seryl-tRNA synthase in an RNA-dependent manner, with the association blocked by mutation of the SAM-binding domain of METTL6 or upon RNase treatment.
In contrast to these results, METTL8 deficiency did not produce a significant change in tRNA m3C levels. Instead, Xu et al. (2) provide definitive proof through stringent purification of mRNA and quantification by LC-MS/MS that METTL8 acts on mRNA, functionally annotating this enzyme and defining m3C as a modification in mRNA for the first time in mammalian systems. Specifically, tRNA was first removed from the total RNA sample by size-exclusion chromatography. The rest of the RNA fraction was subsequently subjected to poly(A) enrichment and rRNA depletion. m1A, known to be more abundant in 28S rRNA than mRNA, and N6-threonylcarbamoyladenosine, present in tRNA but not mRNA, were utilized as standards to confirm the purity of mRNA extracted. The results validated the presence of the known m1A, m6A, and m5C modifications in addition to showing the presence of m3C in purified mRNA. Quantification of their results further demonstrated that m1A, m3C, and m5C are at approximately similar levels, whereas m6A is more abundant. mRNA extracted from Mettl8 KO mice showed lower levels of m3C in comparison with the control, with no noticeable changes observed for the m3C levels in tRNA. At a functional level, the authors did not observe developmental defects in Mettl2, Mettl6, and Mettl8-null-mutant mice or changes in the growth rate of HEK293T cells for Mettl2, Mettl6, or Mettl8 KO cells. Interestingly, the Mettl8 KO in the HCT116 cell line decreased the ratio of polysomes over monosomes, suggesting that Mettl8 could have an influence on cellular translation.
The work by Fu and colleagues (6) provides several immediate angles to explore. Initial primer extension assays suggested METTL6 acts near position 32 of the two serine-specific tRNAs, but further data will be needed to determine the specific substrate nucleotide. Similarly, the locations of the METTL8-installed modifications remain unknown. Furthermore, the functional roles of these enzymes and the m3C modifications and whether these vary under different stress conditions or during cellular signaling remain to be elucidated.
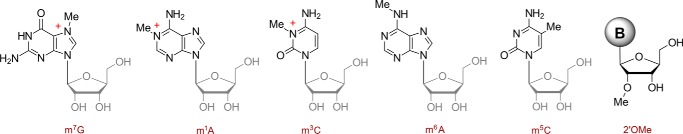
Identifying and deciphering the roles of RNA modifications is more than just a biochemical treasure hunt: Defects of certain RNA-modifying enzymes are known to be associated with human diseases. Moving beyond the abundant RNAs to RNA species such as mRNA and long noncoding RNA, coupled with the discoveries of chemical modifications such as m6A, m1A, m5C, Ψ, 2′O, and now m3C methylations, is opening new directions in understanding RNA modification-mediated RNA processing and gene expression regulation. For example, the m3C, m7G, and m1A modifications introduce a positive charge to their RNA strand (Fig. 1), the presence of which could lead to enhanced protein-RNA interactions, secondary structure changes, and impacts on translation. The explosive discoveries of regulatory RNAs in the past decade have changed our views on the diverse functions possible for RNA species; overlaying our burgeoning knowledge of RNA modifications makes this a very exciting time indeed for the RNA field.
Figure 1.
The known chemical modifications in mammalian mRNA, now including m3C due to the work of Xu et al. (2).
The authors declare that they have no conflicts of interest with the contents of this article.
- m3C
- N3-methylcytosine
- m6A
- N6-methyladenosine
- m5C
- 5-methylcytosine
- m1A
- N1-methyladenosine
- Ψ
- pseudouridine.
References
- 1. Roundtree I. A., Evans M. E., Pan T., and He C. (2017) Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xu L., Liu X., Sheng N., Oo K. S., Liang J., Chionh Y. H., Xu J., Ye F., Gao Y. G., Dedon P. C., and Fu X. Y. (2017) Three distinct 3-methylcytidine (m3C) methyltransferases modify tRNA and mRNA in mice and humans. J. Biol. Chem. 292, 14695–14703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cozen A. E., Quartley E., Holmes A. D., Hrabeta-Robinson E., Phizicky E. M., and Lowe T. M. (2015) ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods 12, 879–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Olson M. V., Page G. S., Sentenac A., Piper P. W., Worthington M., Weiss R. B., and Hall B. D. (1981) Only one of two closely related yeast suppressor tRNA genes contains an intervening sequence. Nature 291, 464–469 [DOI] [PubMed] [Google Scholar]
- 5. Iwanami Y., and Brown G. M. (1968) Methylated bases of ribosomal ribonucleic acid from HeLa cells. Arch. Biochem. Biophys. 126, 8–15 [DOI] [PubMed] [Google Scholar]
- 6. Clark W. C., Evans M. E., Dominissini D., Zheng G., and Pan T. (2016) tRNA base methylation identification and quantification via high-throughput sequencing. RNA 22, 1771–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. D'Silva S., Haider S. J., and Phizicky E. M. (2011) A domain of the actin binding protein Abp140 is the yeast methyltransferase responsible for 3-methylcytidine modification in the tRNA anti-codon loop. RNA 17, 1100–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Han L., Marcus E., D'Silva S., and Phizicky E. M. (2017) S. cerevisiae Trm140 has two recognition modes for 3-methylcytidine modification of the anticodon loop of tRNA substrates. RNA 23, 406–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Noma A., Yi S., Katoh T., Takai Y., Suzuki T., and Suzuki T. (2011) Actin-binding protein ABP140 is a methyltransferase for 3-methylcytidine at position 32 of tRNAs in Saccharomyces cerevisiae. RNA 17, 1111–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vandivier L. E., Campos R., Kuksa P. P., Silverman I. M., Wang L. S., and Gregory B. D. (2015) Chemical modifications mark alternatively spliced and uncapped messenger RNAs in Arabidopsis. Plant Cell 27, 3024–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]