Abstract
The human riboflavin (RF) transporter-3 (hRFVT-3; product of the SLC52A3 gene) plays an essential role in the intestinal RF absorption process and is expressed exclusively at the apical membrane domain of polarized enterocytes. Previous studies have characterized different physiological/biological aspects of this transporter, but nothing is known about the glycosylation status of the hRFVT-3 protein and role of this modification in its physiology/biology. Additionally, little is known about the residues in the hRFVT-3 protein that interact with the ligand, RF. We addressed these issues using appropriate biochemical/molecular approaches, a protein-docking model, and established intestinal/renal epithelial cells. Our results showed that the hRFVT-3 protein is glycosylated and that glycosylation is important for its function. Mutating the predicted N-glycosylation sites at Asn94 and Asn168 led to a significant decrease in RF uptake; it also led to a marked intracellular (in the endoplasmic reticulum, ER) retention of the mutated proteins as shown by live-cell confocal imaging studies. The protein-docking model used in this study has identified a number of putative substrate-interacting sites: Ser16, Ile20, Trp24, Phe142, Thr314, and Asn315. Mutating these potential interacting sites was indeed found to lead to a significant inhibition in RF uptake and to intracellular (ER) retention of the mutated proteins (except for the Phe142 mutant). These results demonstrate that the hRFVT-3 protein is glycosylated and this glycosylation is important for its function and cell surface expression. This study also identified a number of residues in the hRFVT-3 polypeptide that are important for its function/cell surface expression.
Keywords: RFVT-3, riboflavin transport, glycosylation, structure-function activity
in its biologically active forms, i.e., flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD), the water-soluble vitamin B2 (riboflavin; RF) plays critical roles in a variety of cellular metabolic reactions and functions. This includes oxidation-reduction reactions and protein folding; it also possesses antioxidant (4, 13, 33, 48) as well as anti-inflammatory (21) properties. In addition, important roles for RF in normal immune function (23, 38) and in the maintenance of normal intestinal homeostasis (28) have been reported in recent years. Thus, it is not surprising that disturbances in normal RF body homeostasis lead to a variety of clinical abnormalities such as degenerative changes in the nervous system, anemia, and growth retardation (30, 36). RF deficiency and suboptimal levels are observed in a variety of clinical conditions including chronic alcoholism (1, 32), inflammatory bowel disease (6, 18), inborn errors of RF metabolism (2, 9, 11, 14), and diabetes mellitus (16). In addition, it has been suggested that low maternal RF levels may increase the risk for development of fetal anomalies including transverse and longitudinal limb deficiency, as well as congenital heart defects (29, 31, 39). In contrast to the negative effects of RF deficiency, optimizing RF body homeostasis is effective in the treatment of patients with Fazio Londe syndrome, Brown-Vialetto Van Laere syndrome (BVVLS) (2, 9, 11, 14), and those with RF-responsive multiple acyl-CoA dehydrogenase deficiency (20, 56).
Humans and other mammals have lost the capability for de novo synthesis of RF and therefore must acquire this micronutrient from exogenous sources via intestinal absorption. Two sources of RF are available to the intestine: a dietary source (which is absorbed in the small intestine) and a microbiota source (which is absorbed in the large intestine) (12, 15, 34, 40). Absorption of both sources of RF occurs via an efficient and specific carrier-mediated process (24, 34, 35, 37). The molecular identity of the transport systems involved in the absorption of RF has been recently delineated following the cloning of three mammalian RF transporters, i.e., RFVT-1, -2, and -3 (products of the SLC52A1, SLC52A2, and SLC52A3 genes) (51–53). All these transporters are expressed in the human intestine with expression of hRFVT-3 being the highest (45, 52, 54). Other studies have utilized live-cell confocal imaging to show that the hRFVT-3 is expressed exclusively at the apical membrane domain of polarized epithelia, the hRFVT-1 is mainly expressed at the basolateral membrane (BLM) domain of these cells, while expression of hRFVT-2 appears to be mostly intracellular (with some at the BLM domain) (41, 45). An essential role for RFVT-3 in intestinal RF uptake has also been established in studies by our group utilizing in vitro gene-specific silencing approach (siRNA) and an intestinal-specific conditional RFVT-3 (SLC53A3) knockout mouse model (45, 46).
The human RFVT-3 is a 469 amino acid protein, and is predicted to have 11 transmembrane domains (TMDs), with the NH2-terminal tail being oriented toward the cell interior while the COOH-terminal tail is oriented toward the cell exterior (41, 51). Few structural features of the hRFVT-3 protein that are important for its cell biology and function have been identified. These include the determination that the COOH-terminal sequence of the hRFVT-3 protein is essential for its targeting to the apical membrane domain of polarized epithelia, and the identification of several clinical mutations in the SLC52A3 gene in patients with BVVLS (3, 41). Further studies, however, are needed to delineate the structure-function activity of this important vitamin transporter. Thus, our aim in this study was to address this issue with a specific focus on determining the glycosylation status of the hRFVT-3 protein, and on identifying residues that are important for the function and cell biology of the membrane transporter. Our results showed that the hRFVT-3 protein is glycosylated and that glycosylation is important for its function. Also a number of residues (those located at positions 16, 20, 24, 142, 314, and 315) were identified as being important for the function and cell biology of the hRFVT-3 polypeptide.
MATERIALS AND METHODS
Materials
[3H]riboflavin (specific activity > 21.2 Ci/mmol, radiochemical purity > 98%) was obtained from American Radiolabeled Chemicals (ARC, St. Louis, MO). The human RFVT-3 polyclonal antibodies were raised in rabbits using a commercial vendor (Alpha Diagnostics International, San Antonio, TX), and anti-rabbit IRDye-800 (catalog no. 926-32211) secondary antibody was purchased from LI-COR Bioscience (Lincoln, NE). The mutant oligonucleotide primers were synthesized using Sigma Genosys (Woodlands, TX). The molecular biology grade chemicals and reagents used in these studies were obtained from commercial sources.
Site-Directed Mutagenesis
For site-directed mutagenesis studies, the full-length hRFVT-3 cloned into GFP-C3 vector (Clontech) was used as a template. The Quick Change II site-directed mutagenesis kit from Stratagene (La Jolla, CA) was used for mutating N-glycosylation and potentially interacting with RF residues and utilizing specifically designed primers for each mutation (Table 1). The PCR amplification and transformation procedures were performed following manufacturer’s instructions (Stratagene). Mutated clones were sequenced and verified for the presence of mutations from isolated plasmid DNA (Laragen).
Table 1.
Combination of primers used for PCR
| Forward and Reverse Primers (5′–3′) | |
|---|---|
| S16A |
TTCGGAATGGGCGCCTGGGTGACCATC; GATGGTCACCCAGGCGCCCATTCCGAA |
| I20A |
TCCTGGGTGACCGCCAATGGGCTCTGG; CCAGAGCCCATTGGCGGTCACCCAGGA |
| W24A |
ATCAATGGGCTCGCGGTAGAGCTGCCC; GGGCAGCTCTACCGCGAGCCCATTGAT |
| N94Q |
GCCTTCCTCTGGCAAATGACCTCCTGG; CCAGGAGGTCATTTGCCAGAGGAAGGA |
| F142A |
CTCACCACCTTCGCTGTGGGTGAAGGA; TCCTTCACCCACAGCGAAGGTGGTGAG |
| C120A |
CTGGCCCTGGTGGACGCCACCTCTTCAGTGACC GGTCACTGAAGAGGTGGCGTCCACCAGGGCCAG |
| N168Q |
ACTACCTGCGTCCAAGTCACTGAGATA; TATCTCAGTGACTTGGACGCAGGTAGT |
| T314A |
GTCAACGCGCTCGCCAACGGCATGCTG; CAGCATGCCGTTGGCGAGCGCGTTGAC |
| N315A |
AACGCGCTCACCGCCGGCATGCTGCCC; GGGCAGCATGCCGGCGGTGAGCGCGTT |
The nucleotide changes in forward and reverse primers (boldface) used for generating point mutations.
Cell Culture, Transient and Stable Transfections
Human duodenum epithelial (HuTu-80; adenocarcinoma, male Caucasian, 53 yr old) and Madin-Darby canine kidney (MDCK; normal adult female dog) cell lines were used in these studies. These cells were purchased from ATCC (Manassas, VA) and were cultured in EMEM media at 37°C in 5% CO2-95% air atmosphere. The culture medium was supplemented with 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 µg) for optimal growth. The HuTu-80 cells grown in 12-well cell culture plates (Corning, NY) were used for uptake experiments, and HuTu-80 and MDCK cells grown in sterile glass-bottomed petri dishes (MatTek, Ashland, MA) were used for live-cell confocal imaging. Cells that reached 80–90% confluence were transfected with 3 µg of GFP-hRFVT-3 wild type (WT) or mutated constructs with 3 µl of lipofectamine 2000 (Invitrogen). Cells were used for uptake or imaging studies after 24–48 h posttransfection. For stable transfections, HuTu-80 cells were selected using G418 (0.5 mg/ml) (Invitrogen) for 6–8 wk as described before (41).
Uptake Analysis
Transiently transfected HuTu-80 cells with GFP-hRFVT-3 (WT) or with the mutated constructs as well as the tunicamycin treated stable GFP-hRFVT-3 expressing HuTu-80 cells were grown in 12-well plates, and [3H]RF uptake was performed in Krebs-Ringer (K-R) buffer at 37°C for 3 min (initial rate) as described before (35, 41, 45). Protein contents were estimated using a Bio-Rad protein assay kit.
Tunicamycin and PNGase F Treatment
GFP-hRFVT-3 stably expressing HuTu-80 cells were treated with 2 µg/ml tunicamycin for 24 h and then these cells were used for Western blotting, [3H]RF uptake, and live-cell confocal imaging analysis. Total membrane protein (~60 µg) was isolated from GFP-hRFVT-3 stably expressing HuTu-80 cells and incubated with denaturing buffer at 100°C for 10 min. The denatured sample was then incubated with peptide-N-glycosidase F (PNGase F) (10 µg protein/µl of PNGase F; NEB, Ipswich, MA) for 1 h at 37°C and subjected to Western blot analysis.
Western Blot Analysis
Protein (60 µg) samples prepared from GFP-hRFVT-3-expressing HuTu-80 cells were separated in NuPAGE 4–12% Bis-Tris gradient minigel (Invitrogen). The separated proteins were transferred onto immobilon polyvinylidene difluoride membrane (PVDF) (Fisher Scientific). The membrane was hybridized with anti-hRFVT-3 (1:200) and the immunodetection was performed utilizing anti-rabbit IRDye-800 antibody (1:30,000). The immunoreactive bands (which were verified with the use of antigenic peptide; data not shown) were visualized and quantified using the Odyssey infrared imaging system (LI-COR Bioscience).
Live-Cell Confocal Imaging
HuTu-80 or MDCK cell monolayers grown on petri dishes were imaged for construct expression using an inverted Nikon C-1 confocal microscope (Nikon Instruments, Melville, NY). The green fluorescent protein (GFP) was excited with a 488 nm line from an argon ion laser and the red fluorescent protein (DsRed) was excited with a 543 nm line from a HeNe ion laser. Emitted fluorescence was monitored at 515 ± 30 nm short-pass and 570 ± 50 nm long-pass filters, respectively. Images were captured and processed using Nikon C-1 software and Photoshop.
Comparative Protein Structure Modeling
To obtain homology models, we subjected the hRFVT-3 protein sequence to “Phyre2” fold recognition program (http://www.sbg.bio.ic.ac.uk/phyre2) (17). We then validated the models using “ProSA-Web” (https://prosa.services.came.sbg.ac.at/prosa.php) (50). The model we selected was the one that uses the structure of the glucose transporter of Staphylococcus epidermidis (PDB ID: c4LDSB) as the most suitable template (5), as it gave a 93% confidence. The generated three-dimensional (3D) structure of hRFVT-3 was visualized by “RasMol” (http://www.rasmol.org). This generated model was then subjected to “DockingServer” (https://www.dockingserver.com) (10) to identify the putative interacting residues with RF.
Data Presentation and Statistical Analysis
Uptake data are means ± SE of least three independent experiments and are expressed in fmoles per mg of protein per 3 min; data are presented as percentage relative to simultaneously performed controls. Significance level was calculated by Student’s t-test, and a P value of < 0.05 was considered as being significant. Western blotting analysis and confocal imaging studies were all performed on at least three separate experiments.
RESULTS
Glycosylation Status of the hRFVT-3 Protein, and Role of Glycosylation in Transport Function
The hRFVT-3 protein is glycosylated and glycosylation is important for its function.
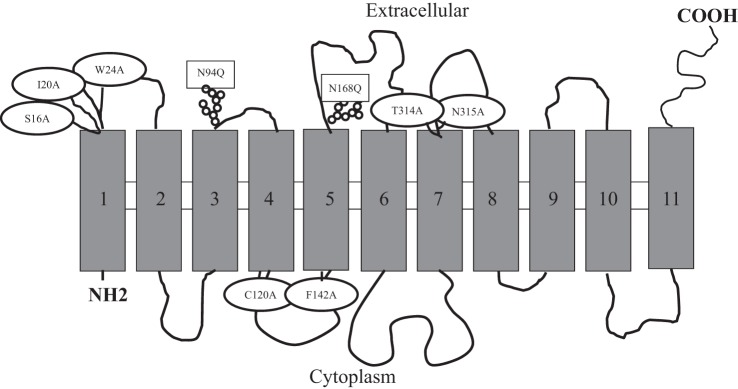
The hydrophobicity analysis of the hRFVT-3 protein (469 amino acids) predicts the polypeptide as having 11 transmembrane domains (TMDs), with the intracellular NH2- being oriented toward the cell interior while the COOH-terminal tail is oriented toward the cell exterior (41, 51) (Fig. 1). This analysis also predicts the hRFVT-3 protein as having two putative N-glycosylation sites (http://www.cbs.dtu.dk/services/NetNGlyc), Asn94 (N-M-T) and Asn168 (N-V-T), located within the extracellular loops between the predicted TMDs 3 and 4 and 5 and 6, respectively. Both of the predicted glycosylation sites conform to the established human and mouse species consensus (N-X-S/T) (44, 49).
Fig. 1.
The predicted membrane topology of human riboflavin transporter-3 (hRFVT-3). hRFVT-3 is predicted to have 11 transmembrane domains (TMDs) with intracellular NH2- and extracellular COOH-terminal tails. The predicted N-glycosylation and substrate interaction residue sites are depicted in rectangular and oval shapes, respectively.
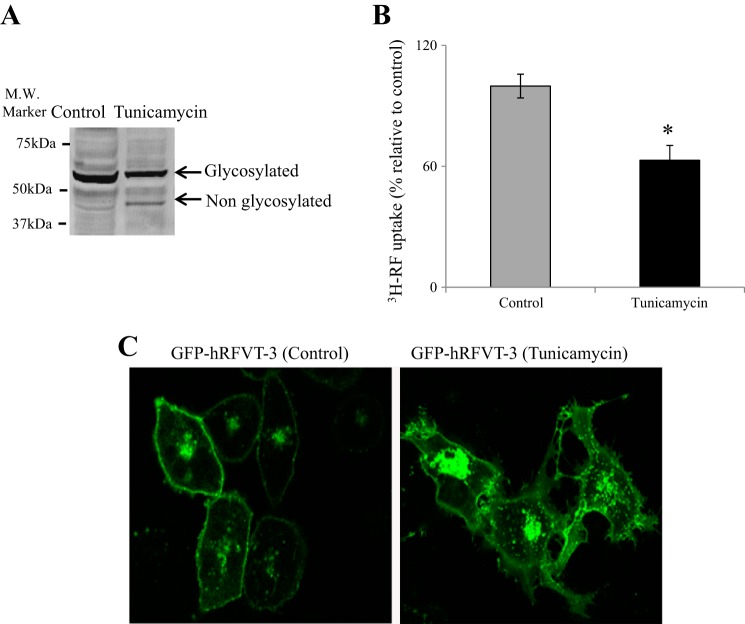
Since N-glycosylation of a variety of membrane transporters (including those involved in the transport of water-soluble vitamins) plays a role in their function, membrane targeting, and stability (8, 19, 22, 25, 26, 44, 47, 49, 55), we aimed in this study to determine whether the hRFVT-3 protein is glycosylated, and if so, the effect of this glycosylation on its transport function. For this we pretreated GFP-hRFVT-3-expressing intestinal epithelial HuTu-80 cells (a previously established intestinal cell model in such investigations; 41) with the glycosylation inhibitor tunicamycin (2 µg/ml for 24 h; 8, 25, 26), then subjected the protein extract of these cells to Western blot analysis using specific hRFVT-3 antibodies. The results (Fig. 2A) showed that tunicamycin treatment led to a clear shift in the molecular mass of the endogenous hRFVT-3 from ~52 kDa to ~48 kDa. A similar shift in the endogenous hRFVT-3 immunoreactive band was observed when cells were treated with PNGase F (data not shown). To determine the effect of N-linked glycosylation on functionality of the intestinal hRFVT-3, we examined the effect of tunicamycin treatment on initial rate of carrier-mediated [3H]RF (14 nM) uptake in HuTu-80 cells expressing the GFP-hRFVT-3. The results showed that tunicamycin treatment causes a significant (P < 0.01) inhibition in [3H]RF uptake compared with untreated control cells (Fig. 2B). In other studies, we performed live-cell confocal imaging of HuTu-80 cells expressing GFP-hRFVT-3 and treated with tunicamycin and observed numerous GFP-hRFVT-3-containing intracellular vesicles in the tunicamycin treated compared with the untreated cells (Fig. 2C). The latter suggests that N-glycosylation is important for membrane targeting of the hRFVT-3 protein. Taken together, these results suggest that the hRFVT-3 polypeptide is glycosylated and this glycosylation is important for its function.
Fig. 2.
Effect of tunicamycin treatment on hRFVT-3 properties in HuTu-80 cells. A: Western blot analysis after tunicamycin treatment of green fluorescent protein (GFP)-hRFVT-3-expressing HuTu-80 cells. Western blotting was performed as described in materials and methods. The images are representative of three independent experiments with similar results. B: effect of tunicamycin treatment on riboflavin (RF) uptake. Uptake of [3H]RF (14 nM) was performed in Krebs-Ringer (K-R) buffer (pH 7.4) at 37°C for 3 min on GFP-hRFVT-3-expressing HuTu-80 cells with or without tunicamycin treatment (2 µg/ml, 24 h). Data are means ± SE of multiple determinations from at least four independent experiments. *P < 0.01. C: lateral (xy) images of GFP-hRFVT-3-expressing HuTu-80 cells: untreated control and treated with tunicamycin (2 µg/ml, 24 h).
Effect of mutating the predicted N-glycosylation sites of the hRFVT-3 protein (i.e., Asn94 and Asn168) on function and membrane expression.
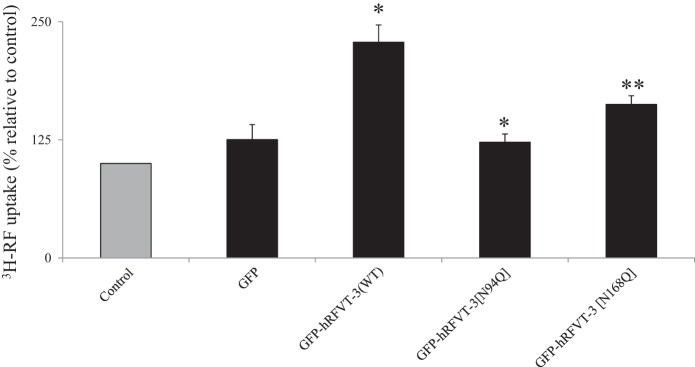
As mentioned earlier, the hRFVT-3 protein is predicted to have two potential N-glycosylation sites. In this study, we examined the effect of mutating these sites (asparagine to glutamine) on functionality of the transport protein. For this, HuTu-80 cells were transiently transfected with hRFVT-3 (WT) or with the N-glycosylation mutants followed by examination of the [3H]RF uptake. As expected (41), cells expressing the WT hRFVT-3 showed significant (P < 0.01) induction in RF uptake compared with (untransfected) control cells (Fig. 3). On the other hand, cells expressing the Asn94 and Asn168 hRFVT-3 mutants showed a significantly (P < 0.01 for both) lower induction in RF uptake compared with those expressing the WT hRFVT-3 (Fig. 3).
Fig. 3.
Effect of mutating the putative N-glycosylation sites of hRFVT-3 on transport function. HuTu-80 cells transiently expressing GFP (vector alone), wild type (WT), or N-glycosylation mutants were used for RF uptake determinations. [3H]RF (14 nM) uptake was performed in K-R buffer (pH 7.4) at 37°C for 3 min. Data are means ± SE of multiple determinations from at least four independent uptake experiments. *P < 0.01, **P < 0.05.
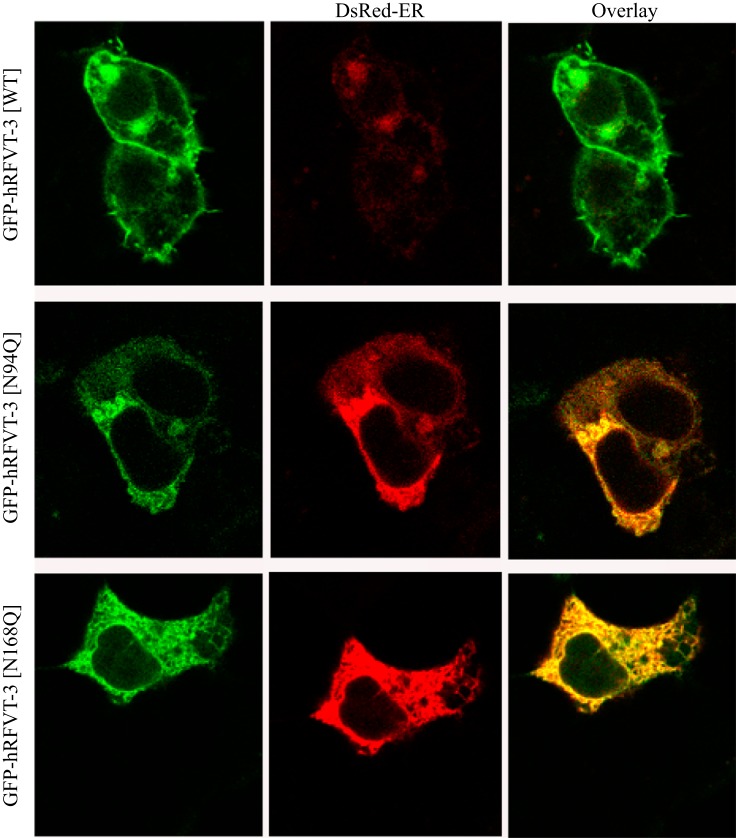
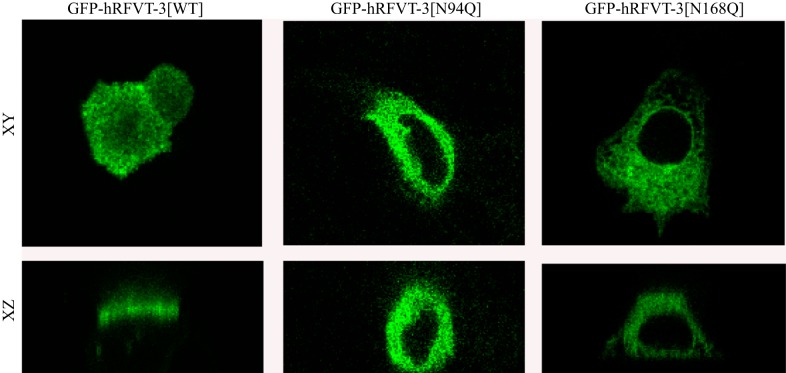
To determine whether the decrease in RF uptake by cells expressing the Asn94 and Asn168 hRFVT-3 mutants is due to impairment in membrane expression of the transporter, we transiently expressed each of the N-glycosylation mutants, i.e., GFP-hRFVT-3[N94Q] and GFP-hRFVT-3[N168Q], in intestinal epithelial HuTu-80 cells and performed live-cell confocal imaging; expression was compared with that of WT GFP-hRFVT-3. Results with HuTu-80 cells showed the known pattern of expression of the WT GFP-hRFVT-3 at the cell membrane (41). On the other hand, cells expressing the GFP-hRFVT-3[N94Q] mutant showed the protein to be retained intracellularly; expression of the GFP-hRFVT-3[N168Q] mutant, however, showed a mixed phenotype with some cells showing expression at the cell surface while others retained the protein intracellularly (Fig. 4). The intracellular retention of the mutated hRFVT-3 proteins appeared to be in the endoplasmic reticulum (ER) as shown by colocalization studies with the ER marker (DsRed-ER) (Fig. 4). We obtained similar findings when the polarized renal epithelial MDCK cells were used for imaging purposes, in that expression of the WT GFP-hRFVT-3 was found (as before; 41, 45) to be at the apical membrane domain (41, 45), that of the GFP-hRFVT-3 [N94Q] mutant to be intracellular, and that of the GFP-hRFVT-3 [N168Q] mutant to be of mixed phenotype with some cells expressing the protein at the apical membrane domain (data not shown) while others expressing it intracellularly (Fig. 5). These observations suggest that the impairment in [3H]RF uptake by cells expressing the Asn94 and Asn168 hRFVT-3 mutants is due to impairment in membrane expression of the RF transporter.
Fig. 4.
Effect of mutating the putative N-glycosylation sites of hRFVT-3 on cell surface expression of hRFVT-3 in HuTu-80 cells. Coexpression of GFP-hRFVT-3 (WT) or N-glycosylation mutants (left) along with DsRed-ER (middle), displayed overlaid images (right). The transiently coexpressing HuTu-80 cells were imaged 48 h posttransfection.
Fig. 5.
Effect of mutating the putative N-glycosylation sites of hRFVT-3 on the apical membrane expression of hRFVT-3 in polarized Madin-Darby canine kidney (MDCK) cells. Lateral (xy) and axial (xz) sections of MDCK cells transiently expressing GFP-hRFVT-3 (WT) and N-glycosylation mutants. The transfected MDCK cells were imaged after 48 h of transient transfection.
Structure-Function Characterization of the hRFVT-3 Protein
Generation of a comparative protein structure modeling for hRFVT-3 and prediction of the RF interacting residues.
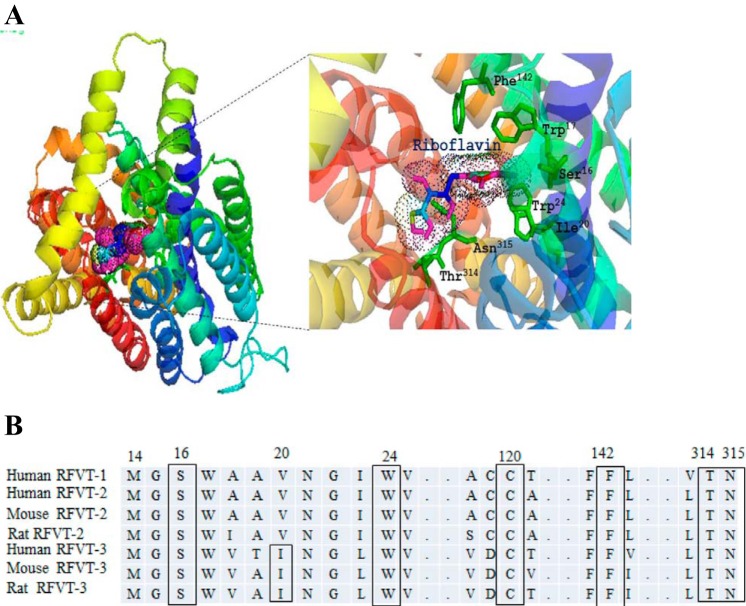
In these investigations, we subjected the hRFVT-3 polypeptide to “Phyre2” fold recognition program to obtain a homology model as described in materials and methods. We then validated the models using “ProSA” computer program. The model we selected was the one that uses the structure of the glucose transporter of Staphylococcus epidermidis (PDB ID: c4LDSB) as the template (5), as it gave a 93% confidence. This model was then subjected to the “DockingServer” to identify putative interacting sites with RF. This docking model predicted that amino acids Ser16, Trp17, Ile20, Trp24, Phe142, Thr314, and Asn315 interact with the RF molecule (Fig. 6A). As predicted by “TMpred program” the amino acids Ser16, Trp17, Ile20, and Trp24 are located in the 1st TMD; Phe142 is located in TMD5, and both Thr314 and Asn315 are located in TMD7 (41, 51). Since the amino acid Trp17 was previously characterized, it was not considered for further analysis (27). All of the predicted amino acids were found to be conserved among RFVTs in different species (human, rat, and mouse) except Ile20, which is conserved only in human, rat, and mouse RFVT-3 (Fig. 6B). The schematic representation of the hRFVT-3 secondary structure showing the predicted site of potential interacting mutations locations in its 11 TMDs is shown in Fig. 1.
Fig. 6.
Homology modeling of the hRFVT-3 protein showing location of the putative substrate interaction residues. A: the docking model of hRFVT-3 with RF was generated using the docking server. The protein (hRFVT-3) is shown as a multicolor ribbon representation and RF as wire mesh pattern. The homology modeling of hRFVT-3 was generated based on the crystal structure of the glucose transporter (PDB ID:c4LDSB) (5). B: RFVT amino acid sequences in humans, mouse, and rat. The locations of the predicted substrate interaction residues are shown in box.
Effect of mutating the predicted potential interacting residues on hRFVT-3 function.
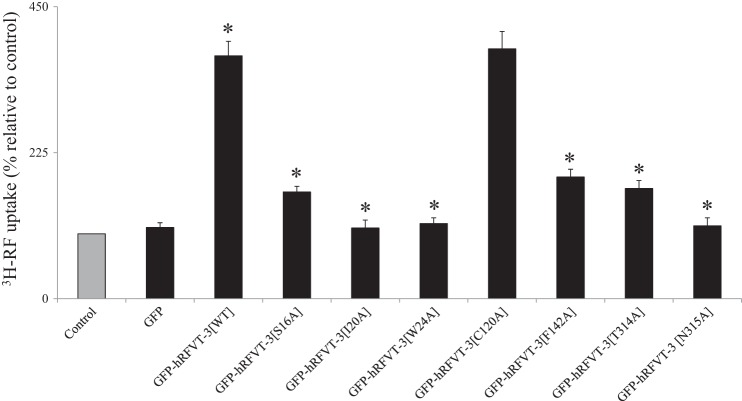
In these investigations, we mutated amino acids Ser16, Ile20, Trp24, Phe142, Thr314, and Asn315 (to alanine) then examined the effect of such mutations on functionality of the hRFVT-3. We also mutated (to alanine) a randomly chosen residue (Cys120) that was not predicted by “Docking Server,” to serve as a control. We then transiently transfected hRFVT-3 (WT) and the different mutants into HuTu-80 cells followed by examination of the [3H]RF uptake. The results showed that mutating Ser16, Ile20, Trp24, Phe142, Thr314, and Asn315 led to a significant (P < 0.01) decrease in the RF uptake compared with uptake by cells expressing hRFVT-3 (WT) (Fig. 7) or mutant Cys120, which did not affect RF uptake.
Fig. 7.
Effect of mutating the predicted substrate interaction residues in the of hRFVT-3 protein on RF uptake. Uptake of [3H]RF (14 nM) by HuTu-80 cells transiently expressing GFP (vector alone), GFP-hRFVT-3 (WT), and the substrate interaction residue mutants was performed in K-R buffer (pH 7.4) at 37°C for 3 min. Data are means ± SE of multiple determinations from at least four independent experiments. *P < 0.01.
Cellular distribution of hRFVT-3 mutant residues that were predicted as substrate-interacting sites in HuTu-80 and MDCK cells: confocal imaging analysis.
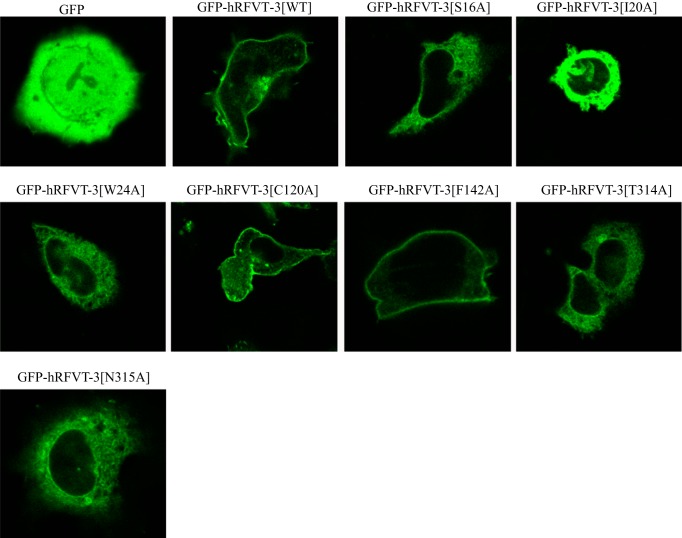
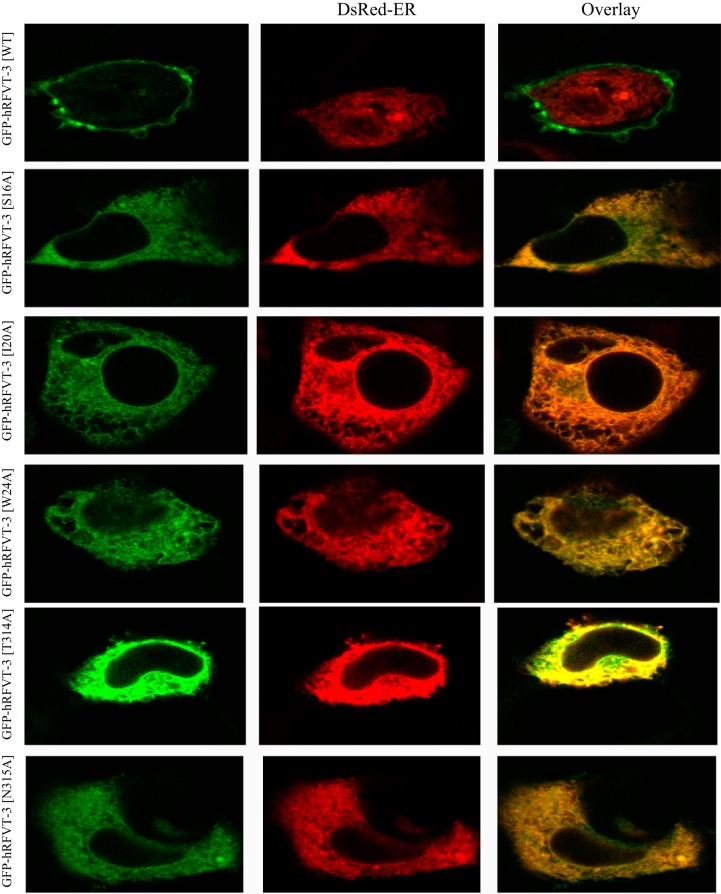
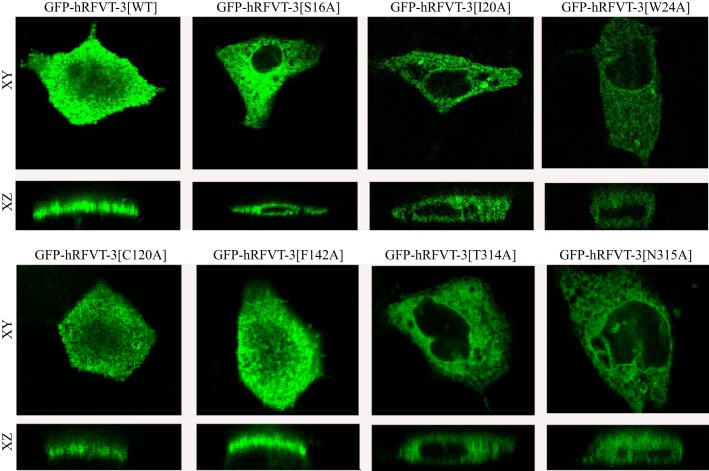
To determine whether the decrease in the rate of [3H]RF uptake observed with the different hRFVT-3 mutants (sites predicted to interact with the RF molecule) is due to defect in the targeting of the mutated protein to the cell membrane, we transiently expressed the GFP-hRFVT-3 (WT) and the individual mutant constructs in HuTu-80 and MDCK cells and performed live-cell confocal imaging. The results with the HuTu-80 cells showed that mutants GFP-hRFVT-3[C120A] and GFP-hRFVT-3[F142A], like hRFVT-3(WT), are expressed at the cell membrane (Fig. 8). On the other hand, expression of GFP-hRFVT-3[S16A], GFP-hRFVT-3[T314A], and GFP-hRFVT-3[N315A] showed a mixed phenotype, with some cells showing expression at the cell surface, while others showing the protein being retained intracellularly (most likely in the ER as shown by the significant overlap with the ER marker DsRed-ER) (Fig. 9). In contrast, mutants GFP-hRFVT-3[I20A] and GFP-hRFVT-3[W24A] were uniformly retained intracellularly (Fig. 8), and colocalized with the ER marker DsRed-ER (Fig. 9). In MDCK cells, the confocal axial (xz) sections showed that mutants (GFP-hRFVT-3[C120A] and GFP-hRFVT-3[F142A]) were similar to hRFVT-3 (WT) in that they are expressed at the apical membrane domain (Fig. 10). On the other hand, expression of mutants (GFP-hRFVT-3[S16A], GFP-hRFVT-3[T314A], and GFP-hRFVT-3[N315A]) were of mixed pattern, with some cells showing expression at the apical membrane (data not shown), while others retained intracellularly (Fig. 10). Expression of mutants (GFP-hRFVT-3[I20A] and GFP-hRFVT-3[W24A]), on the other hand, resulted in complete intracellular localization of the protein (Fig. 10). These findings provide explanation for the impairment in [3H]RF uptake observed with all the mutants in the predicted interacting site of the hRFVT-3 protein, i.e., the impairment is due to partial or complete retention of the mutant protein intracellularly. The exception was GFP-hRFVT-3[F142A], where the mutant was found to be expressed at the cell surface, but showed impaired function. This raises the possibility that mutation at the 142 site of the hRFVT-3 leads to conformational change in the transport protein which affects its interaction with its substrate; however, this suggestion remains to be confirmed.
Fig. 8.
Cellular distribution of the GFP-hRFVT-3 (WT) and the substrate interaction residue mutants in HuTu-80 cells. Lateral (xy) sections of HuTu-80 cells transiently expressing hRFVT-3 (WT) and substrate interaction residue mutants. The transfected HuTu-80 cells were imaged after 48 h.
Fig. 9.
Coexpression of GFP-hRFVT-3 (WT) or substrate interaction residue mutants with DsRed-ER in HuTu-80 cells. Coexpression of GFP-hRFVT-3 (WT) and mutants (left) along with DsRed-ER (middle), displayed overlaid images (right). The transiently coexpressing HuTu-80 cells were imaged after 48 h of transfection.
Fig. 10.
Polarized expression of the GFP-hRFVT-3 (WT) and substrate interaction residue mutants in MDCK cells. Lateral (xy) and axial (xz) sections of MDCK cells transiently expressing hRFVT-3 (WT) and substrate interaction residue mutants. The transfected MDCK cells were imaged after 48 h.
DISCUSSION
The hRFVT-3 is a physiologically and clinically relevant transporter as it is responsible for intestinal uptake of an essential micronutrient, RF, and its mutation leads to significant clinical implications as seen in patients with BVVLS (2, 3). Recent studies from our laboratory and those of others have characterized different physiological and biological aspects of the hRFVT-3 system (7, 27, 41–43, 45, 51). However, nothing is currently known about the N-glycosylation status of the hRFVT-3 protein and the effect of that modification on the physiology/cell biology of the membrane transporter; also little is known about the structure-function relationship of this important carrier protein. Our aim in this study was to address these issues and for that we employed hydrophobicity/NetNglyc programs to predict the potential N-glycosylation sites, a comparative protein-docking modeling approach to predict putative substrate-interacting residues in the hRFVT-3 protein that may influence its function/cell biology, and live-cell confocal imaging of appropriate intestinal/renal epithelial cell models.
The hydrophobicity/NetNglyc programs predicted the hRFVT-3 polypeptide to have two putative N-glycosylation sites, one at Asn94 and the other at Asn168. Thus we first determined the N-glycosylation status of hRFVT-3 in intestinal epithelial cells. For that we used tunicamycin (a nucleoside antibiotic that inhibits N-glycosylation) to inhibit N-glycosylation de novo in intestinal HuTu-80 cells and observed a clear shift of the hRFVT-3 band from ~52 kDa to ~48 kDa. A similar shift in the molecular mass of the hRFVT-3 band was seen with cellular protein of HuTu-80 cells treated with PNGase F. The N-glycosylation of the hRFVT-3 protein appeared to be important for the function of this transporter as significant inhibition in RF uptake in HuTu-80 cells pretreated with tunicamycin was observed. This is not unique to RFVT-3 as glycosylation has been shown to affect the function of many membrane transporters including those of other water-soluble vitamin transporters (8, 19, 22, 25, 26, 44, 47, 49, 55). Moreover, glycosylation of hRFVT-3 could also play a role in its interaction with potential luminal pathogens, but further investigations are needed to test this possibility. To determine which of the two putative N-glycosylation sites in the hRFVT-3 polypeptide is/are important for hRFVT-3 function, we mutated these sites individually and then examined the effect of that mutation on functionality of the hRFVT-3. Our results showed that both of the N-glycosylation sites (i.e., Asn94 and Asn168) are important for hRFVT-3 function and their mutation leads to impairment in RF uptake. The latter appears to be due to retention (partial or complete) of the hRFVT-3-mutated protein intracellularly (most likely in ER) as evidenced by live-cell imaging analysis.
In examining the amino acid residues in the hRFVT-3 protein that are predicted by the protein-docking model to interact with the RF molecule (Ser16, Ile20, Trp24, Phe142, Thr314, and Asn315), we have mutated these sites individually then examined the effect of the particular mutation on RF uptake by intestinal epithelial HuTu-80 cells. We found that all the predicted residues to be important for hRFVT-3 function, as evidenced by the marked decrease in RF uptake in cells transfected with these mutants compared with those transfected with the WT hRFVT-3. Mutating a randomly selected residue (Cys120), on the other hand, did not affect RFVT-3 function. This impairment in RF uptake upon mutating the substrate interacting residues appears to be, at least in part, due to partial or complete retention of the mutant protein intracellularly as evidenced by live-cell imaging analysis. The only exception was the Phe142 hRFVT-3 mutant where expression of this mutant was found at the cell surface, suggesting that mutation at this site may lead to impairment in function via possible conformational change in the membrane transporter. Future studies are needed to confirm the latter suggestion. It is of interest to note here that the predicted interacting residues in the distal part of the 1st TMD (Ser16, Ile20, and Trp24) and in the 7th TMD (Thr314 and Asn315) of the hRFVT-3 polypeptide are located in regions where clusters of hRFVT-3 clinical mutations have been identified in BVVLS patients and appear to be functionally impaired (2, 3).
In summary, results of the present study provide important information regarding the N-glycosylation status and structure-function relationship of hRFVT-3, a transporter that plays a key role in the intestinal RF uptake process.
GRANTS
This study was supported by the Department of Veterans Affairs and National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases Grants DK58057 and DK56057 (H. M. Said), and DK107474 (V. S. Subramanian).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
V.S.S., S.S., and H.M.S. conceived and designed research; V.S.S., S.S., and T.T. performed experiments; V.S.S., S.S., J.A.B., and H.M.S. analyzed data; V.S.S., S.S., and H.M.S. interpreted results of experiments; V.S.S. and S.S. prepared figures; V.S.S. and H.M.S. drafted manuscript; V.S.S., S.S., T.T., and H.M.S. edited and revised manuscript; V.S.S., S.S., T.T., J.A.B., and H.M.S. approved final version of manuscript.
REFERENCES
- 1.Bonjour JP. Vitamins and alcoholism. V. Riboflavin, VI. Niacin, VII. Pantothenic acid, and VIII. Biotin. Int J Vitam Nutr Res 50: 425–440, 1980. [PubMed] [Google Scholar]
- 2.Bosch AM, Abeling NG, Ijlst L, Knoester H, van der Pol WL, Stroomer AE, Wanders RJ, Visser G, Wijburg FA, Duran M, Waterham HR. Brown-Vialetto-Van Laere and Fazio Londe syndrome is associated with a riboflavin transporter defect mimicking mild MADD: a new inborn error of metabolism with potential treatment. J Inherit Metab Dis 34: 159–164, 2011. doi: 10.1007/s10545-010-9242-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosch AM, Stroek K, Abeling NG, Waterham HR, Ijlst L, Wanders RJ. The Brown-Vialetto-Van Laere and Fazio Londe syndrome revisited: natural history, genetics, treatment and future perspectives. Orphanet J Rare Dis 7: 83, 2012. doi: 10.1186/1750-1172-7-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooperman JM, Lopez R. Riboflavin, in Handbook of Vitamins: Nutritional, Biochemical and Clinical Aspects, edited by Machlin LJ. New York: Dekker, 1984, p. 299–327. [Google Scholar]
- 5.Dang S, Sun L, Huang Y, Lu F, Liu Y, Gong H, Wang J, Yan N. Structure of a fucose transporter in an outward-open conformation. Nature 467: 734–738, 2010. doi: 10.1038/nature09406. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez-Banares F, Abad-Lacruz A, Xiol X, Gine JJ, Dolz C, Cabre E, Esteve M, Gonzalez-Huix F, Gassull MA. Vitamin status in patients with inflammatory bowel disease. Am J Gastroenterol 84: 744–748, 1989. [PubMed] [Google Scholar]
- 7.Fujimura M, Yamamoto S, Murata T, Yasujima T, Inoue K, Ohta KY, Yuasa H. Functional characteristics of the human ortholog of riboflavin transporter 2 and riboflavin-responsive expression of its rat ortholog in the small intestine indicate its involvement in riboflavin absorption. J Nutr 140: 1722–1727, 2010. doi: 10.3945/jn.110.128330. [DOI] [PubMed] [Google Scholar]
- 8.Ghosal A, Subramanian VS, Said HM. Role of the putative N-glycosylation and PKC-phosphorylation sites of the human sodium-dependent multivitamin transporter (hSMVT) in function and regulation. Biochim Biophys Acta 1808: 2073–2080, 2011. doi: 10.1016/j.bbamem.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green P, Wiseman M, Crow YJ, Houlden H, Riphagen S, Lin JP, Raymond FL, Childs AM, Sheridan E, Edwards S, Josifova DJ. Brown-Vialetto-Van Laere syndrome, a ponto-bulbar palsy with deafness, is caused by mutations in c20orf54. Am J Hum Genet 86: 485–489, 2010. doi: 10.1016/j.ajhg.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazai E, Kovacs S, Demko L, Bikadi Z. DockingServer (https://www.dockingserver.com). Budapest, Hungary:Virtual Drug, 2009. [Google Scholar]
- 11.Ho G, Yonezawa A, Masuda S, Inui K, Sim KG, Carpenter K, Olsen RK, Mitchell JJ, Rhead WJ, Peters G, Christodoulou J. Maternal riboflavin deficiency, resulting in transient neonatal-onset glutaric aciduria Type 2, is caused by a microdeletion in the riboflavin transporter gene GPR172B. Hum Mutat 32: E1976–E1984, 2011. doi: 10.1002/humu.21399. [DOI] [PubMed] [Google Scholar]
- 12.Iinuma S. Synthesis of riboflavin by intestinal bacteria. J Vitaminol (Kyoto) 1: 6–13, 1955. doi: 10.5925/jnsv1954.1.2_6. [DOI] [PubMed] [Google Scholar]
- 13.Iwanaga K, Hasegawa T, Hultquist DE, Harada H, Yoshikawa Y, Yanamadala S, Liao H, Visovatti SH, Pinsky DJ. Riboflavin-mediated reduction of oxidant injury, rejection, and vasculopathy after cardiac allotransplantation. Transplantation 83: 747–753, 2007. doi: 10.1097/01.tp.0000256283.06469.d4. [DOI] [PubMed] [Google Scholar]
- 14.Johnson JO, Gibbs JR, Megarbane A, Urtizberea JA, Hernandez DG, Foley AR, Arepalli S, Pandraud A, Simón-Sánchez J, Clayton P, Reilly MM, Muntoni F, Abramzon Y, Houlden H, Singleton AB. Exome sequencing reveals riboflavin transporter mutations as a cause of motor neuron disease. Brain 135: 2875–2882, 2012. doi: 10.1093/brain/aws161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasper H. Vitamin absorption in the colon. Am J Proctol 21: 341–345, 1970. [PubMed] [Google Scholar]
- 16.Kodentsova VM, Vrzhesinskaia OA, Sokol’nikov AA, Kharitonchik LA, Spirichev VB. [Metabolism of B group vitamins in patients with insulin-dependent and non-insulin dependent forms of diabetes mellitus]. Vopr Med Khim 39: 26–29, 1993. [PubMed] [Google Scholar]
- 17.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858, 2015. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuroki F, Iida M, Tominaga M, Matsumoto T, Hirakawa K, Sugiyama S, Fujishima M. Multiple vitamin status in Crohn’s disease. Correlation with disease activity. Dig Dis Sci 38: 1614–1618, 1993. doi: 10.1007/BF01303168. [DOI] [PubMed] [Google Scholar]
- 19.Kuze K, Graves P, Leahy A, Wilson P, Stuhlmann H, You G. Heterologous expression and functional characterization of a mouse renal organic anion transporter in mammalian cells. J Biol Chem 274: 1519–1524, 1999. doi: 10.1074/jbc.274.3.1519. [DOI] [PubMed] [Google Scholar]
- 20.Law LK, Tang NL, Hui J, Fung SL, Ruiter J, Wanders RJ, Fok TF, Lam CW. Novel mutations in ETFDH gene in Chinese patients with riboflavin-responsive multiple acyl-CoA dehydrogenase deficiency. Clin Chim Acta 404: 95–99, 2009. doi: 10.1016/j.cca.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 21.Liu D, Zempleni J. Low activity of LSD1 elicits a pro-inflammatory gene expression profile in riboflavin-deficient human T Lymphoma Jurkat cells. Genes Nutr 9: 422, 2014. doi: 10.1007/s12263-014-0422-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martínez-Maza R, Poyatos I, López-Corcuera B, Núñez E, Giménez C, Zafra F, Aragón C. The role of N-glycosylation in transport to the plasma membrane and sorting of the neuronal glycine transporter GLYT2. J Biol Chem 276: 2168–2173, 2001. doi: 10.1074/jbc.M006774200. [DOI] [PubMed] [Google Scholar]
- 23.Mazur-Bialy AI, Buchala B, Plytycz B. Riboflavin deprivation inhibits macrophage viability and activity - a study on the RAW 264.7 cell line. Br J Nutr 110: 509–514, 2013. doi: 10.1017/S0007114512005351. [DOI] [PubMed] [Google Scholar]
- 24.Middleton HM., III Uptake of riboflavin by rat intestinal mucosa in vitro. J Nutr 120: 588–593, 1990. [DOI] [PubMed] [Google Scholar]
- 25.Muthusamy S, Malhotra P, Hosameddin M, Dudeja AK, Borthakur S, Saksena S, Gill RK, Dudeja PK, Alrefai WA. N-glycosylation is essential for ileal ASBT function and protection against proteases. Am J Physiol Cell Physiol 308: C964–C971, 2015. doi: 10.1152/ajpcell.00023.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nabokina SM, Subramanian VS, Said HM. The human colonic thiamine pyrophosphate transporter (hTPPT) is a glycoprotein and N-linked glycosylation is important for its function. Biochim Biophys Acta 1858: 866–871, 2016. doi: 10.1016/j.bbamem.2016.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nabokina SM, Subramanian VS, Said HM. Effect of clinical mutations on functionality of the human riboflavin transporter-2 (hRFT-2). Mol Genet Metab 105: 652–657, 2012. doi: 10.1016/j.ymgme.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakano E, Mushtaq S, Heath PR, Lee ES, Bury JP, Riley SA, Powers HJ, Corfe BM. Riboflavin depletion impairs cell proliferation in adult human duodenum: identification of potential effectors. Dig Dis Sci 56: 1007–1019, 2011. doi: 10.1007/s10620-010-1374-3. [DOI] [PubMed] [Google Scholar]
- 29.Powers HJ. Riboflavin (vitamin B-2) and health. Am J Clin Nutr 77: 1352–1360, 2003. [DOI] [PubMed] [Google Scholar]
- 30.Rivilin RS. Riboflavin. In: Encyclopedia of Dietary Supplements, edited by Coates PM, Betz JM, Blackman MR, Cragg GM, Levin M, Moss J, White JD. Boca Raton, FL: CRC Press, 2010. doi: 10.1201/b14669-80. [DOI] [Google Scholar]
- 31.Robitaille J, Carmichael SL, Shaw GM, Olney RS; National Birth Defects Prevention Study . Maternal nutrient intake and risks for transverse and longitudinal limb deficiencies: data from the National Birth Defects Prevention Study, 1997–2003. Birth Defects Res A Clin Mol Teratol 85: 773–779, 2009. doi: 10.1002/bdra.20587. [DOI] [PubMed] [Google Scholar]
- 32.Rosenthal WS, Adham NF, Lopez R, Cooperman JM. Riboflavin deficiency in complicated chronic alcoholism. Am J Clin Nutr 26: 858–860, 1973. [DOI] [PubMed] [Google Scholar]
- 33.Sanches SC, Ramalho LN, Mendes-Braz M, Terra VA, Cecchini R, Augusto MJ, Ramalho FS. Riboflavin (vitamin B-2) reduces hepatocellular injury following liver ischaemia and reperfusion in mice. Food Chem Toxicol 67: 65–71, 2014. doi: 10.1016/j.fct.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 34.Said HM, Arianas P. Transport of riboflavin in human intestinal brush border membrane vesicles. Gastroenterology 100: 82–88, 1991. doi: 10.1016/0016-5085(91)90586-A. [DOI] [PubMed] [Google Scholar]
- 35.Said HM, Ma TY. Mechanism of riboflavine uptake by Caco-2 human intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol 266: G15–G21, 1994. [DOI] [PubMed] [Google Scholar]
- 36.Said HM, Ross C. Riboflavin. In: Encyclopedia of Dietary Supplements (11th ed.), edited by Ross AC, Caballero B, Cousins RJ, Tucker KL, Ziegler TR. Baltimore, MD: Lippincott Williams & Wilkins, 2014, p. 325–330. [Google Scholar]
- 37.Said HM, Mohammadkhani R. Uptake of riboflavin across the brush border membrane of rat intestine: regulation by dietary vitamin levels. Gastroenterology 105: 1294–1298, 1993. doi: 10.1016/0016-5085(93)90131-U. [DOI] [PubMed] [Google Scholar]
- 38.Schramm M, Wiegmann K, Schramm S, Gluschko A, Herb M, Utermöhlen O, Krönke M. Riboflavin (vitamin B2) deficiency impairs NADPH oxidase 2 (Nox2) priming and defense against Listeria monocytogenes. Eur J Immunol 44: 728–741, 2014. doi: 10.1002/eji.201343940. [DOI] [PubMed] [Google Scholar]
- 39.Smedts HP, Rakhshandehroo M, Verkleij-Hagoort AC, de Vries JH, Ottenkamp J, Steegers EA, Steegers-Theunissen RP. Maternal intake of fat, riboflavin and nicotinamide and the risk of having offspring with congenital heart defects. Eur J Nutr 47: 357–365, 2008. doi: 10.1007/s00394-008-0735-6. [DOI] [PubMed] [Google Scholar]
- 40.Sorrell MF, Frank O, Thompson AD, Aquino A, Baker H. Absorption of vitamins from the large intestine. Nutr Rep Int 3: 143–148, 1971. [Google Scholar]
- 41.Subramanian VS, Rapp L, Marchant JS, Said HM. Role of cysteine residues in cell surface expression of the human riboflavin transporter-2 (hRFT2) in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol 301: G100–G109, 2011. doi: 10.1152/ajpgi.00120.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subramanian VS, Ghosal A, Subramanya SB, Lytle C, Said HM. Differentiation-dependent regulation of intestinal vitamin B2 uptake: studies utilizing human-derived intestinal epithelial Caco-2 cells and native rat intestine. Am J Physiol Gastrointest Liver Physiol 304: G741–G748, 2013. doi: 10.1152/ajpgi.00018.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramanian VS, Ghosal A, Kapadia R, Nabokina SM, Said HM. Molecular mechanisms mediating the adaptive regulation of intestinal riboflavin uptake process. PLoS One 10: e0131698, 2015. doi: 10.1371/journal.pone.0131698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subramanian VS, Marchant JS, Reidling JC, Said HM. N-glycosylation is required for Na+-dependent vitamin C transporter functionality. Biochem Biophys Res Commun 374: 123–127, 2008. doi: 10.1016/j.bbrc.2008.06.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subramanian VS, Subramanya SB, Rapp L, Marchant JS, Ma TY, Said HM. Differential expression of human riboflavin transporters -1, -2, and -3 in polarized epithelia: a key role for hRFT-2 in intestinal riboflavin uptake. Biochim Biophys Acta 1808: 3016–3021, 2011. doi: 10.1016/j.bbamem.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subramanian VS, Lambrecht N, Lytle C, Said HM. Conditional (intestinal-specific) knockout of the riboflavin transporter-3 (RFVT-3) impairs riboflavin absorption. Am J Physiol Gastrointest Liver Physiol 310: G285–G293, 2016. doi: 10.1152/ajpgi.00340.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanaka K, Xu W, Zhou F, You G. Role of glycosylation in the organic anion transporter OAT1. J Biol Chem 279: 14961–14966, 2004. doi: 10.1074/jbc.M400197200. [DOI] [PubMed] [Google Scholar]
- 48.Tu BP, Ho-Schleyer SC, Travers KJ, Weissman JS. Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science 290: 1571–1574, 2000. doi: 10.1126/science.290.5496.1571. [DOI] [PubMed] [Google Scholar]
- 49.Unal ES, Zhao R, Qiu A, Goldman ID. N-linked glycosylation and its impact on the electrophoretic mobility and function of the human proton-coupled folate transporter (HsPCFT). Biochim Biophys Acta 1778: 1407–1414, 2008. doi: 10.1016/j.bbamem.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35: W407–W410, 2007. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamamoto S, Inoue K, Ohta KY, Fukatsu R, Maeda JY, Yoshida Y, Yuasa H. Identification and functional characterization of rat riboflavin transporter 2. J Biochem 145: 437–443, 2009. doi: 10.1093/jb/mvn181. [DOI] [PubMed] [Google Scholar]
- 52.Yao Y, Yonezawa A, Yoshimatsu H, Masuda S, Katsura T, Inui K. Identification and comparative functional characterization of a new human riboflavin transporter hRFT3 expressed in the brain. J Nutr 140: 1220–1226, 2010. doi: 10.3945/jn.110.122911. [DOI] [PubMed] [Google Scholar]
- 53.Yonezawa A, Masuda S, Katsura T, Inui K. Identification and functional characterization of a novel human and rat riboflavin transporter, RFT1. Am J Physiol Cell Physiol 295: C632–C641, 2008. doi: 10.1152/ajpcell.00019.2008. [DOI] [PubMed] [Google Scholar]
- 54.Yonezawa A, Inui K. Novel riboflavin transporter family RFVT/SLC52: identification, nomenclature, functional characterization and genetic diseases of RFVT/SLC52. Mol Aspects Med 34: 693–701, 2013. doi: 10.1016/j.mam.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 55.Zhou F, Xu W, Hong M, Pan Z, Sinko PJ, Ma J, You G. The role of N-linked glycosylation in protein folding, membrane targeting, and substrate binding of human organic anion transporter hOAT4. Mol Pharmacol 67: 868–876, 2005. doi: 10.1124/mol.104.007583. [DOI] [PubMed] [Google Scholar]
- 56.Zhu M, Zhu X, Qi X, Weijiang D, Yu Y, Wan H, Hong D. Riboflavin-responsive multiple Acyl-CoA dehydrogenation deficiency in 13 cases, and a literature review in mainland Chinese patients. J Hum Genet 59: 256–261, 2014. doi: 10.1038/jhg.2014.10. [DOI] [PubMed] [Google Scholar]