Recent research has shown that acetylation of mitochondrial fatty acid oxidation enzymes has greatly contrasting effects on their activity in different tissues. Here, we provide new evidence that acetylation of cardiac mitochondrial fatty acid oxidation enzymes by GCN5L1 significantly upregulates their activity in diet-induced obese mice.
Keywords: mitochondria, acetylation, GCN5L1, sirtuin 3, fatty acid oxidation, high-fat diet, heart
Abstract
Lysine acetylation is a reversible posttranslational modification and is particularly important in the regulation of mitochondrial metabolic enzymes. Acetylation uses acetyl-CoA derived from fuel metabolism as a cofactor, thereby linking nutrition to metabolic activity. In the present study, we investigated how mitochondrial acetylation status in the heart is controlled by food intake and how these changes affect mitochondrial metabolism. We found that there was a significant increase in cardiac mitochondrial protein acetylation in mice fed a long-term high-fat diet and that this change correlated with an increase in the abundance of the mitochondrial acetyltransferase-related protein GCN5L1. We showed that the acetylation status of several mitochondrial fatty acid oxidation enzymes (long-chain acyl-CoA dehydrogenase, short-chain acyl-CoA dehydrogenase, and hydroxyacyl-CoA dehydrogenase) and a pyruvate oxidation enzyme (pyruvate dehydrogenase) was significantly upregulated in high-fat diet-fed mice and that the increase in long-chain and short-chain acyl-CoA dehydrogenase acetylation correlated with increased enzymatic activity. Finally, we demonstrated that the acetylation of mitochondrial fatty acid oxidation proteins was decreased after GCN5L1 knockdown and that the reduced acetylation led to diminished fatty acid oxidation in cultured H9C2 cells. These data indicate that lysine acetylation promotes fatty acid oxidation in the heart and that this modification is regulated in part by the activity of GCN5L1.
NEW & NOTEWORTHY Recent research has shown that acetylation of mitochondrial fatty acid oxidation enzymes has greatly contrasting effects on their activity in different tissues. Here, we provide new evidence that acetylation of cardiac mitochondrial fatty acid oxidation enzymes by GCN5L1 significantly upregulates their activity in diet-induced obese mice.
high energetic demand requires that the heart continuously generates ATP to sustain cardiac contractile function (23, 25). The heart uses several fuel substrates for energy metabolism (22, 24), with fatty acid oxidation (FAO) accounting for 50–70% of the total ATP generated (6, 20, 23). Whereas healthy hearts have the flexibility to use various substrates, a shift toward increased fatty acid utilization is a key feature of obesity and diabetes (21). Elevated levels of circulating free fatty acids and triacylglycerol, combined with alterations in FAO enzyme abundance or activity, promote cardiomyocyte lipid accumulation, cardiac dysfunction, and heart failure (7, 9, 16, 40). As the prevalence of obesity and diabetes continues to increase, it is imperative that we examine the processes that regulate cardiac energy metabolism under these conditions.
Lysine acetylation, a reversible posttranslational modification, has recently been identified as a novel regulator of mitochondrial bioenergetic function. A growing body of literature has found that the majority of mitochondrial metabolic enzymes are acetylated (37, 41), and the regulation of FAO enzyme activity by acetylation appears to be particularly important (2, 26, 29, 39, 41). Mitochondrial protein acetylation is significantly increased in various tissues during high-fat diet (HFD) feeding (14, 17), and fatty acids are the main source of the acetyl-CoA used as a cofactor for lysine acetylation (28). These data imply that there is an intrinsic link between nutritional inputs, lysine acetylation, and metabolic activity. Taken together, changes in the acetylation status of cardiac FAO enzymes may greatly impact their activity and could be a key cellular mechanism that drives metabolic dysfunction in the hearts of obese individuals.
The regulation of mitochondrial lysine acetylation has predominantly focused on the activity of sirtuin 3 (SIRT3), the major mitochondrial deacetylase enzyme (13, 14, 33). SIRT3 has been shown to target a number of different regulators of energy metabolism, including the FAO enzyme long-chain acyl-CoA dehydrogenase (LCAD) (13, 14), and the catalytic α-subunit of pyruvate dehydrogenase (PDH) (15). In many tissues, the deacetylation of mitochondrial enzymes promotes their enzymatic function (4, 5, 14); however, there are contrasting reports in the heart (1, 8), which highlights the need for further study. Although there have been numerous reports on the regulation of mitochondrial lysine deacetylation, few studies have addressed the counteracting process of lysine acetylation. In particular, the function of the recently discovered mitochondrial acetyltransferase protein GCN5L1 (35) in cardiac energy metabolism has yet to be intensively explored.
GCN5L1 is localized in the mitochondrial matrix and has been shown to counter the activity of the mitochondrial deacetylase protein SIRT3 (35). Deletion of GCN5L1 blunts mitochondrial protein acetylation and bioenergetics, which promotes mitochondrial dysfunction and cellular energy depletion in mouse embryonic fibroblasts (34). In the present study, we examined the effect of a long-term HFD on the regulation of GCN5L1 and lysine acetylation in the heart. We showed that nutrient excess promotes increased mitochondrial lysine acetylation, which correlates with upregulated expression of GCN5L1 at the expense of SIRT3. HFD feeding led to the hyperacetylation of mitochondrial FAO proteins, which was associated with increased enzymatic activity in in vitro assays. Finally, we found that knockdown of GCN5L1 reduced both the acetylation and activity of FAO enzymes, which culminated in a reduction in mitochondrial bioenergetic output in cardiac-derived H9C2 cells.
MATERIALS AND METHODS
Animal care and use.
Male C57BL/6J mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and housed in the University of Pittsburgh animal facility. Animals were housed under standard conditions with ad libitum access to water and food and maintained on a constant 12:12-h light-dark cycle. Animals were fed either a standard chow diet (67% carbohydrate, 13% fat, 20% protein, 3.6 kcal/g, no. 01351, Harlan Teklad, Madison, WI) or a high-fat chow (40% carbohydrate, 41% fat, 19% protein, 4.4 kcal/g, no. 96001, Harlan Teklad) for 24 wk. At the end of 24 wk, animals were euthanized and cardiac tissues excised for analysis. Experiments were conducted in compliance with National Institutes of Health guidelines and followed procedures approved by the University of Pittsburgh Institutional Animal Care and Use Committee.
Protein isolation.
For whole cardiac protein lysates, tissues were minced and lysed in CHAPS buffer on ice for ~2 h. Homogenates were spun at 10,000 g, and the supernatants collected for Western blot analysis or coimmunoprecipitation experiments. For mitochondrial assays, tissues were lysed in detergent-free buffer and mitochondrial proteins were isolated using the QProteome Mitochondrial Isolation Kit (Qiagen) using the manufacturer’s protocol. For activity assays, muscle homogenates were prepared in modified Chappell-Perry medium A buffer (120 mM KCl, 20 mM HEPES, 5 mM MgCl2, and 1 mM EGTA, pH ~7.2) supplemented with 5 mg/ml fat-free BSA. After centrifugation at 10,000 g, supernatants were used to assay enzyme activity.
Western blot analysis and coimmunoprecipitation.
Protein lysates were prepared in LDS sample buffer, separated using Bolt SDS-PAGE 4–12% or 12% bis-Tris gels, and transferred to nitrocellulose membranes (all Life Technologies). Protein expression was analyzed using the following primary antibodies: rabbit SIRT3, rabbit acetyl-lysine (Ac-K), rabbit glutamate dehydrogenase (GDH), rabbit PDH, and rabbit cytochrome c oxidase subunit 4 (COX IV) from Cell Signaling Technologies; rabbit phospho-PDH (Ser293) from Novus; rabbit LCAD, rabbit short-chain acyl-CoA dehydrogenase (SCAD), and rabbit hydroxyacyl-CoA dehydrogenase (HADHA) from Proteintech; and GCN5L1 as previously reported (35). Fluorescent anti-mouse or anti-rabbit secondary antibodies (red, 700 nm; green, 800 nm) from LiCor were used to detect expression levels. For coimmunoprecipitation experiments, protein lysates were harvested in CHAPS buffer, and equal amounts of total protein were incubated overnight at 4°C with the relevant antibody or an IgG control. Immunocaptured proteins were isolated using protein G-agarose beads (Cell Signaling Technology), washed multiple times with CHAPS buffer, and then eluted in LDS sample buffer at 95°C. Samples were separated on 12% bis-Tris Bolt gels and probed with the appropriate antibodies. Protein densitometry was measured using ImageJ software (National Institutes of Health, Bethesda, MD). Protein loading was further confirmed using GDH or COX IV loading controls where appropriate.
Cellular assays.
Acyl-CoA dehydrogenase assays were performed following the protocol by Verity et al. (36). Palmitoyl-CoA (60 µM) and butyryl-CoA (60 µM) were used as substrates for LCAD and SCAD activity, respectively. Briefly, 25 µg protein was incubated with 0.1 M potassium phosphate buffer, 50 µM 2,6-dichlorophenolindophenol, 2 mM phenazine ethosulfate, 0.2 mM N-ethylmaleimide, 0.4 mM potassium cyanide, and 0.1% Triton X-100 at 37°C for 4 min. The reaction was initiated with 60 µM palmitoyl-CoA or 60 µM butyryl-CoA, and the rate of absorbance change was measured at 600 nm over 5 min. Activities were converted to nanomoles of substrate oxidized per minute per milligram of protein against a standard curve. LDH release assays (MAK066 kit, Sigma) and crystal violet staining were performed on H9C2 cells after exposure to 50 mM H2O2 for 30 min.
RNA isolation and quantitative RT-PCR.
For quantitative RT-PCR, mRNA was isolated using a total RNA extraction kit (Qiagen), and cDNA was produced using a first-strand synthesis kit (Invitrogen). Transcript levels were measured using validated gene-specific primers (Qiagen). Each experiment was performed at least three times, and representative results are shown. Gapdh was used as the internal control.
GCN5L1 knockdown stable cell lines.
H9C2 cells were purchased from the American Type Culture Collection (Manassas, VA). Cells were transduced (multiplicity of infection: 2) with Mission lentiviral shRNA particles targeting GCN5L1 [2 different shRNAs either used individually (KD1 or KD2) or as a pool (KD3)] or a scrambled control sequence. Transduced cells were selected using 1.25 μg/ml puromycin (determined after a kill-curve experiment), and cultured in this concentration for several passages. Stable cell lines were verified by quantitative RT-PCR and Western blot analysis for gene knockdown efficiency.
Mitochondrial bioenergetics measurements.
The O2 consumption rate (OCR) was measured in GCN5L1 control and knockdown stable H9C2 cell lines using the Seahorse XFe96 system (Seahorse Bioscience). Cells (5 × 103 cells/well) in each condition were plated (n = 8) and allowed to attach overnight in DMEM. On the assay day, media were changed to unbuffered DMEM supplemented with 5 mM glucose, 2 mM glutamine, and 0.5 mM carnitine, and the plate was incubated in a non-CO2 incubator (37°C, 15 min) before being run. The basal OCR in each well was measured followed by serial treatment with palmitate (200 μM), etomoxir (40 µM), FCCP (7.5 µM), and rotenone (2 µM). After completion, viability was assessed by crystal violet staining, and the OCR was normalized to cell number.
Statistics.
Means ± SE were calculated for all data sets. Data were analyzed using two-tailed Student’s t-tests, one-way ANOVA, or two-way ANOVA (with Tukey post hoc tests) as appropriate. P < 0.05 was considered significant.
RESULTS
Mitochondrial fuel metabolism gene expression is modulated by HFD.
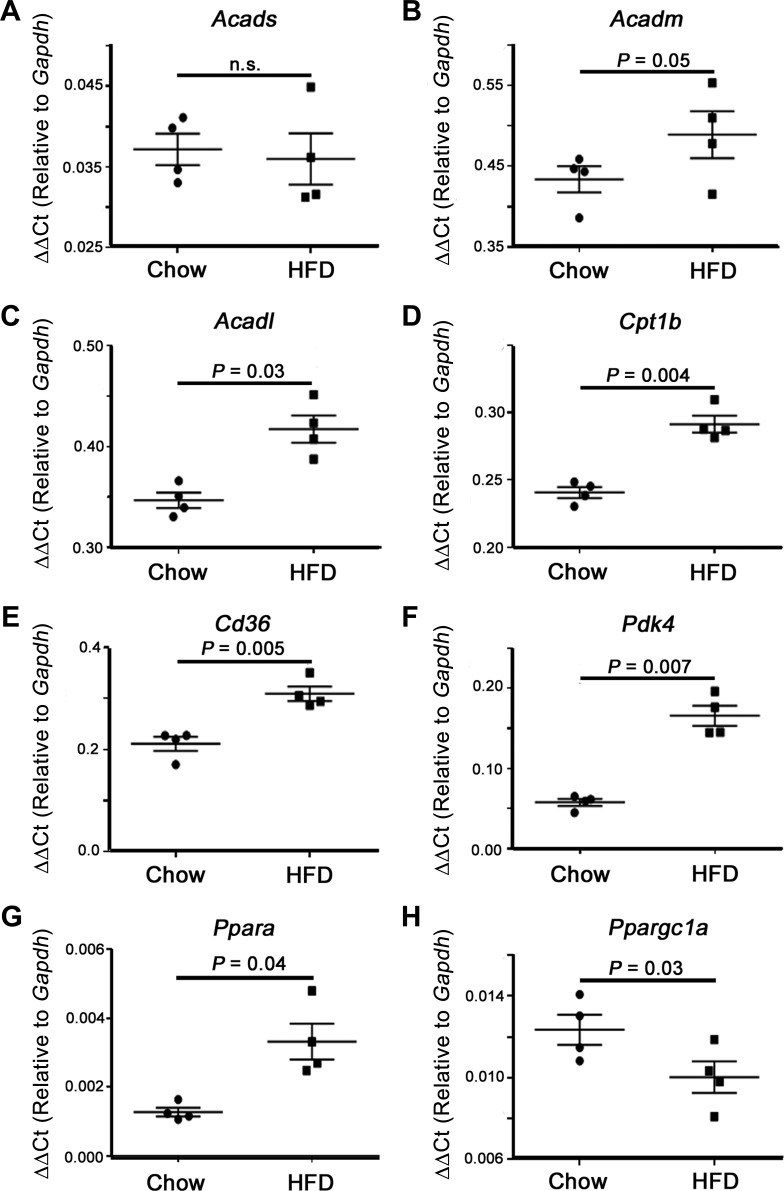
Significant increases in heart mass were observed in animals after 24 wk of HFD (P < 0.05; Table 1), suggesting a negative impact on overall cardiac health. To begin to understand how HFD may regulate cardiac fuel metabolism, we first assessed the expression levels of cardiac fatty acid metabolism genes. Animals fed a HFD showed a general rise in FAO gene expression, with significant increases in long-chain acyl-coenzyme A dehydrogenase (Acadl), carnitine palmitoyltransferase 1b (Cpt1b), and the fatty acid translocase Cd36 (Fig. 1, C–E). The increase in FAO gene expression corresponded with enhanced expression of the PDH negative regulator pyruvate dehydrogenase kinase 4 (Pdk4), indicating a shift from pyruvate oxidation toward FAO in HFD-fed animals (Fig. 1F). Whereas the expression of the transcriptional coactivator peroxisome proliferator-activated receptor (PPAR)-γ coactivator-1α (Ppargc1a) was downregulated in HFD-fed mice, there was a significant increase in the main FAO-related transcription factor PPAR-α (Ppara) (Fig. 1, G and H). In summary, the transcriptional profile of cardiac tissue in HFD-fed mice suggests an increase in FAO at the expense of other fuel substrates, in line with previous reports (1, 7, 10).
Table 1.
Characteristic features of chow- and high-fat diet-fed mice
| Chow | High-Fat Diet | |
|---|---|---|
| Body weight, g | 33.90 ± 0.62 | 48.75 ± 1.05* |
| Heart weight, mg | 124.9 ± 2.23 | 137.7 ± 1.35* |
| Tibial length, mm | 11.63 ± 0.18 | 11.50 ± 0.19 |
| Heart weight/tibia length, mg/mm | 10.75 ± 0.21 | 11.99 ± 0.21* |
Values are means ± SE; n = 8 mice/group. Body weight, heart weight, and heart weight/tibia length were significantly increased in high-fat diet-fed animals.
P < 0.05, chow vs. high-fat diet using a two-way Student’s t-test.
Fig. 1.
Expression of cardiac fuel metabolism genes. High-fat diet (HFD) promotes an enhanced fatty acid oxidation phenotype in the heart. A and B: no significant difference was observed in short-chain acyl-CoA dehydrogenase (Acads, SCAD) or medium-chain acyl-CoA dehydrogenase (Acadm, MCAD) levels. C–G: significant increases in the mRNA content of long-chain acyl-CoA dehydrogenase (Acadl, LCAD), carnitine palmitoyltransferase 1b (Cpt1b), Cd36, pyruvate dehydrogenase kinase 4 (Pdk4), and peroxisome proliferator-activted receptor-α (Ppara) were observed with HFD feeding, whereas peroxisome proliferator-activated receptor-γ coactivator-1α (Ppargc1a; H) was significantly decreased. Values are expressed as means ± SE; n = 4 per group. P value significance is as shown in the graphs using a two-way Student’s t-test.
HFD increases lysine acetylation of cardiac mitochondrial proteins.
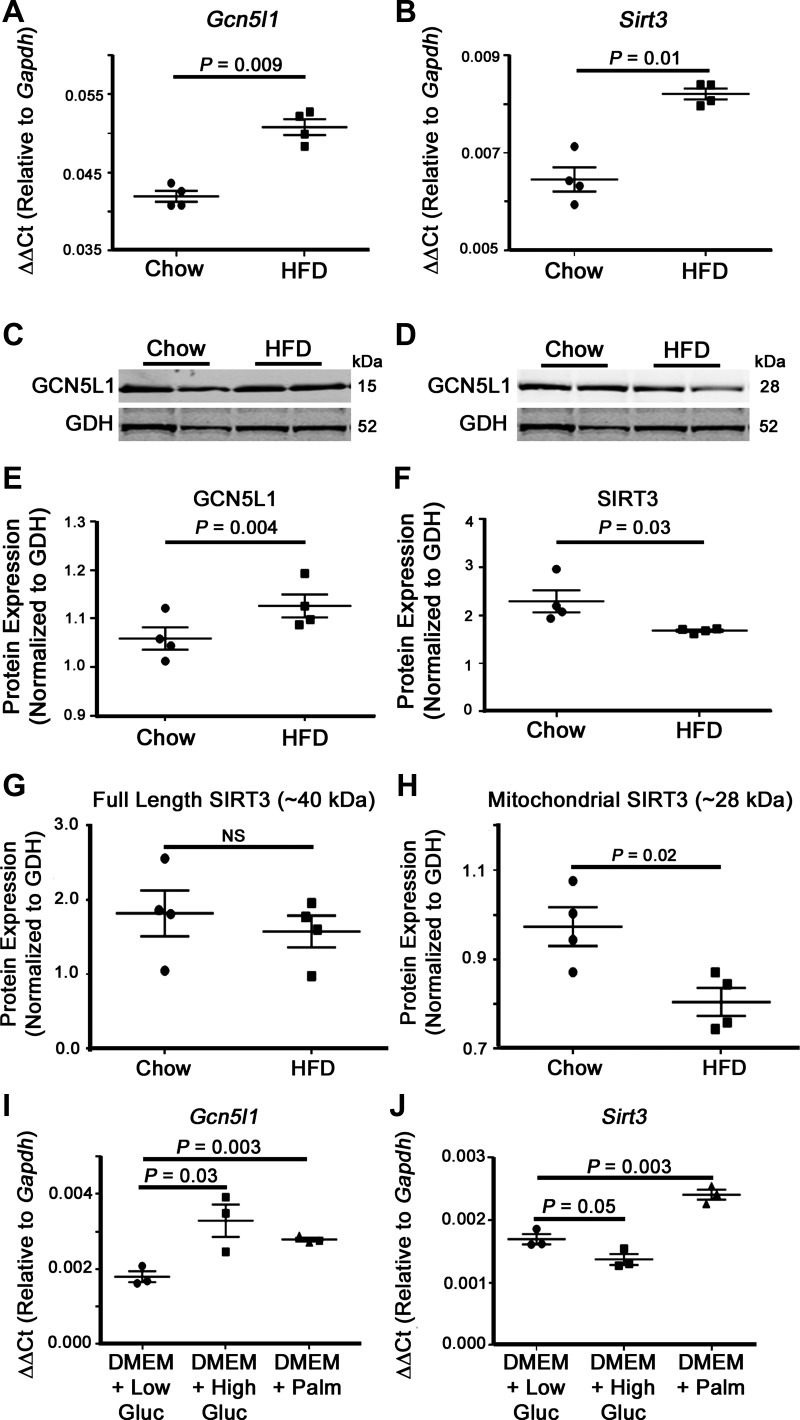
HFD has been linked to increased mitochondrial protein lysine acetylation, a posttranslational modification that has the capacity to modulate the activity of numerous metabolic enzymes (for a review, see Ref. 41). To further investigate how HFD controls mitochondrial acetylation in the heart, we first examined the expression of two mitochondrial acetylation regulators, GCN5L1 and SIRT3. Transcriptional analyses of Gcn5l1, a mitochondrial acetyltransferase-related protein (35), demonstrated a significant increase in gene expression in the hearts of mice after HFD feeding (Fig. 2A). This elevation was matched by a significant increase in Sirt3 mRNA levels [a mitochondria-localized deacetylase protein (32, 33)] under the same conditions (Fig. 2, A and B). Interestingly, whereas GCN5L1 gene and protein expression levels were coordinately upregulated, SIRT3 protein expression was significantly reduced under conditions where its mRNA abundance was elevated (Fig. 2, C–F). SIRT3 is a nuclear-encoded protein that is imported into the mitochondrial matrix after the removal of an NH2-terminal localization sequence (32). We therefore checked to see whether the reduced abundance of the mitochondrial form of SIRT3 (~28 kDa) was caused by a decrease in production of the full-length cytosolic form (~40 kDa). Western blot analysis demonstrated that there was no significant difference in the preimport form of SIRT3 between chow- and HFD-fed animals (Fig. 2, G and H), suggesting that the reduced abundance of mitochondrial SIRT3 in HFD-fed mouse hearts is not caused by a defect in SIRT3 translation.
Fig. 2.
Expression of mitochondrial acetylation regulators in cardiac mitochondria. HFD feeding led to a proacetylation phenotype in cardiac mitochondrial proteins. A and B: mitochondrial acetyltransferase Gcn5l1 and deacetylase sirtuin 3 (Sirt3) mRNA expression were significantly increased in cardiac mitochondria from chronic HFD-fed mice. C–E: whereas the GCN5L1 protein level was significantly increased in HFD-fed animals, SIRT3 protein abundance was significantly decreased. Western blots for GCN5L1 and SIRT3 were obtained from the same gel, and therefore the same GDH loading control was used for each blot. G: there was no significant difference in the abundance of the 40-kDa, preimport, cytosolic form of SIRT3 in chow- and HFD-fed mice. H: in contrast, there was a significant decrease in the 28-kDa mitochondrial form of the enzyme in HFD-fed animals. I and J: Gcn5l1 gene expression in H9C2 cells was significantly increased after 4 h of high glucose (25 mM) and palmitate (200 μM) exposure in basal DMEM relative to low glucose (5 mM) exposure, whereas Sirt3 expression was only elevated in response to palmitate. Values are expressed as means ± SE; n = 4 per group. P value significance is as shown in the graphs using two-way Student’s t-test.
As exposure to a long-term HFD raises both circulating fatty acid and glucose levels, we next aimed to determine which metabolic substrate was responsible for the elevated gene expression of Gcn5l1 and Sirt3. We cultured rat cardiac-derived H9C2 cells in media containing either low glucose (5 mM), high glucose (25 mM), or palmitate (200 μM) for 4 h and then measured the relative expression of each gene by quantitative RT-PCR. Exposure to palmitate led to a significant increase in both Gcn5l1 and Sirt3 gene expression (Fig. 2, I and J), which tallied with the elevated expression seen in HFD-fed mice (Fig. 2, A and B). In contrast, exposure to a high glucose concentration had opposite effects on these genes, with Gcn5l1 expression showing a significant increase (P = 0.03) and Sirt3 trending toward decreased expression (P = 0.05; Fig. 2, I and J). These data suggest that the increase in Gcn5l1 expression is caused by a nonsubstrate-specific increase in available nutrients, whereas Sirt3 expression is controlled by substrate type.
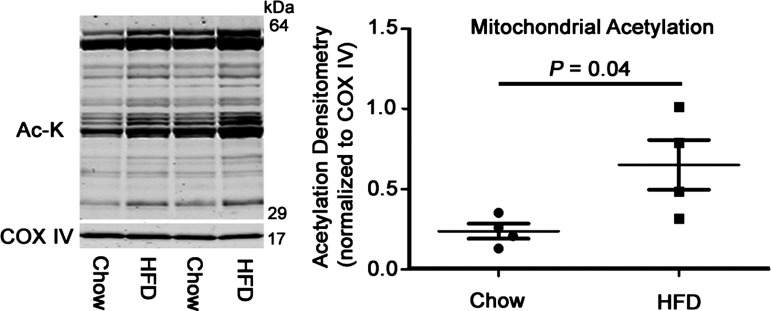
Finally, our overall protein expression data from these acetylation regulators suggested a shift toward a proacetylation phenotype. To test this, we isolated mitochondria from chow- and HFD-fed mice and examined the acetylation status of mitochondrial proteins using a pan-acetylation antibody. This confirmed that there was indeed an increase in this modification in the heart after HFD feeding (Fig. 3).
Fig. 3.
Global mitochondrial lysine acetylation in chow- and HFD-fed hearts. Overall cardiac mitochondrial protein acetylation was significantly increased, with no changes observed in the protein content of cytochrome c oxidase subunit 4 (COX IV). Values are expressed as means ± SE; n = 4 per group. P value significance is as shown in the graphs using two-way Student’s t-test.
Hyperacetylated FAO proteins from HFD mice show increased enzymatic activity.
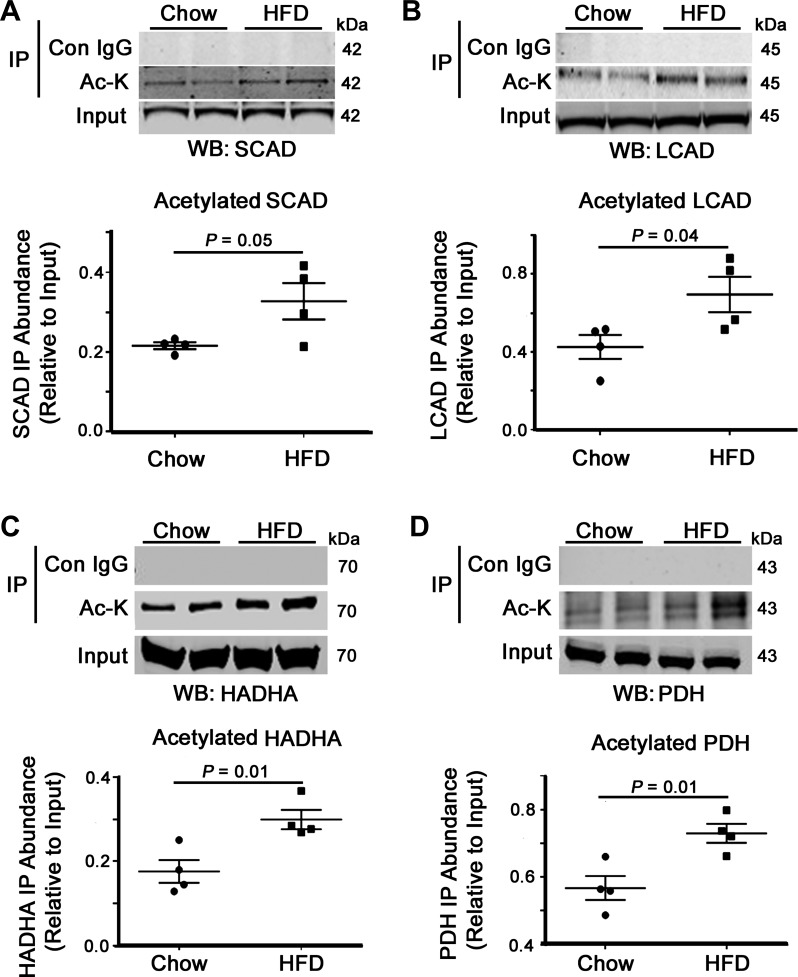
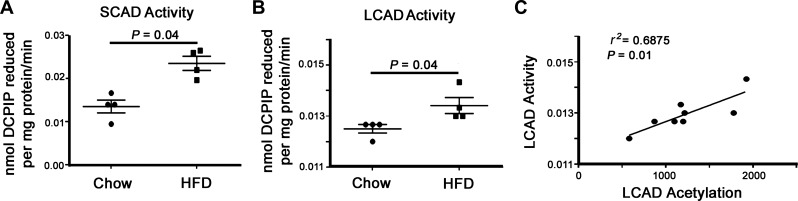
The role of acetylation in regulating metabolic enzyme activity has been extensively studied in several tissues, particularly in the liver and skeletal muscle (13–15, 17, 18, 28, 41). However, the role of this modification in the heart remains to be fully elucidated. In an attempt to understand the effects of increased cardiac lysine acetylation on FAO, we examined the acetylation status and activity of mitochondrial fuel substrate oxidation enzymes. Although there was no significant difference in the cardiac protein abundance of SCAD and LCAD between chow- and HFD-fed animals, there was an increase in the acetylation of these two enzymes (Fig. 4, A and B). Examination of a further FAO enzyme (HADHA) and the α-subunit of PDH showed a similar increase in acetylation (Fig. 4, C and D). In the case of the latter, acetylation of PDH has been shown to inhibit its activity, which could further exacerbate the switch to a pro-FAO metabolic phenotype in the obese heart. We next examined whether HFD had any effect on FAO enzyme activity and found that both SCAD and LCAD oxidation rates were significantly upregulated in samples obtained from overfed mice (Fig. 5, A and B). We performed a regression analysis between LCAD acetylation status and enzyme activity and found that increased acetylation positively correlated with elevated LCAD activity (r2 = 0.6875, P = 0.01; Fig. 5C). These data indicate that a HFD leads to increased FAO enzyme acetylation and that this modification correlates with increased FAO activity in the heart.
Fig. 4.
Impact of HFD on metabolic enzyme acetylation status. HFD feeding led to increased fatty acid oxidation (FAO) enzyme acetylation, which contributes to upregulated fatty acid utilization in diet-induced obese mice. A–C: the acetylated lysine pulldown showed an increase in the acetylation levels of the FAO proteins SCAD, LCAD, and hydroxyacyl-CoA dehydrogenase (HADHA) and the pyruvate oxidation enzyme pyruvate dehydrogenase (PDH; D) from HFD-fed cardiac tissue relative to chow-fed controls. Values are expressed as means ± SE; n = 4 per group. P value significance is as shown in the graphs using two-way Student’s t-test.
Fig. 5.
Impact of HFD-related acetylation on in vitro FAO enzyme activity. A and B: enzymatic activity of SCAD and LCAD was significantly elevated in cardiac tissues from HFD-fed mice. C: regression analysis showed a significant correlation between LCAD acetylation status and enzymatic activity from all tested mice. Values are expressed as means ± SE, n = 4 per group. P value significance is as shown in the graphs using two-way Student’s t-test.
Reductions in GCN5L1 expression limit cellular FAO activity.
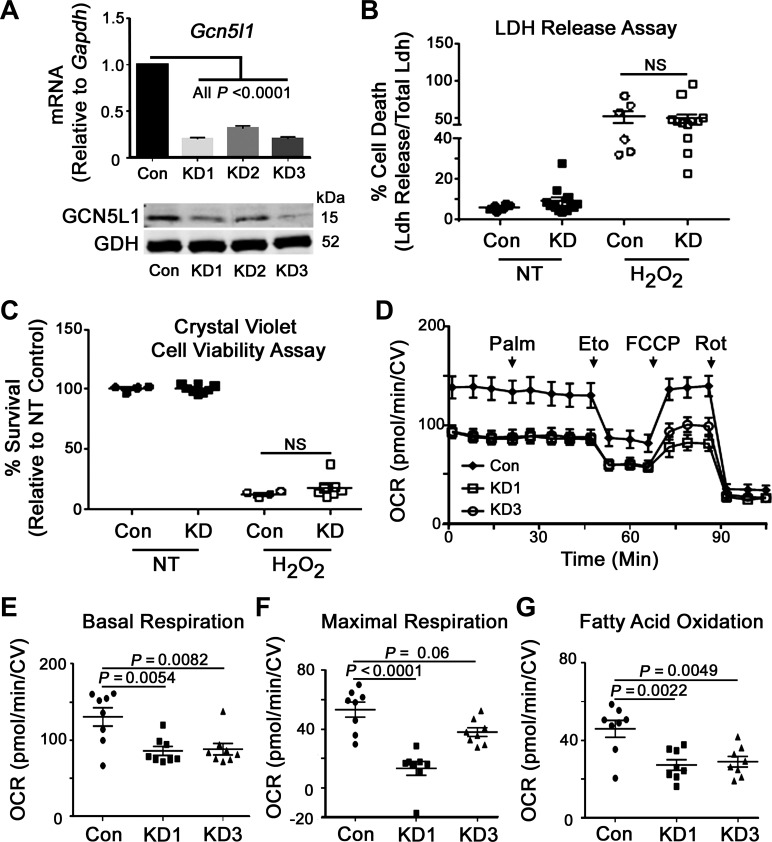
The observed differences in FAO enzymatic activities and mitochondrial protein acetylation, coupled with a significant increase in GCN5L1 protein content in obese mice, led us to further examine the regulatory role of GCN5L1 in cardiac mitochondrial bioenergetics. To aid these experiments, we created three stable GCN5L1 knockdown H9C2 cell lines using different lentivirus-delivered shRNAs. GCN5L1 mRNA levels and protein content were significantly decreased in two lines (KD1 and KD3; Fig. 6A), which were then selected for further characterization. We first tested whether loss of GCN5L1 had any effect on overall cellular fitness and found that GCN5L1 depletion in H9C2 cells had no effect on viability at baseline or after exposure to ROS (Fig. 6, B and C). We next analyzed whether loss of GCN5L1 had an effect on mitochondrial bioenergetic function using the Seahorse XF system (Fig. 6D). Both GCN5L1 knockdown cell lines had significantly impaired baseline respiration, and there was a significant decrease in maximal respiration in KD1, which was matched with a trend toward reduced uncoupled respiration in KD3 (Fig. 6, E and F), which complements similar findings seen previously in GCN5L1 knockout mouse embryonic fibroblast cells (34). In addition, both GCN5L1 knockdown H9C2 cell lines displayed significantly attenuated decreases in FAO in response to treatment with the CPT1 inhibitor etomoxir (Fig. 6G), indicating that these cells predominantly use glucose-based respiratory substrates at the expense of FAO.
Fig. 6.
Regulation of cellular respiration and FAO by GCN5L1. GCN5L1 is a key regulator of FAO in cardiac cells. A: mRNA and protein levels of three stable GCN5L1 knockdown H9C2 cell lines created using different lentivirus-derived shRNAs. Glutamate dehydrogenase (GDH) was used as the loading control. Significant decreases in both mRNA and protein content were observed in two lines (KD1 and KD3). B and C: there was no significant difference in either cell death (LDH release) or cell viability (crystal violet staining) in GCN5L1 knockdown cells relative to the control when exposed to 50 mM H2O2 for 30 min. D: mitochondrial bioenergetic profile of GCN5L1 control, KD1, and KD3 cells after the addition of palmitate (Palm), etomoxir (Eto), carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone (FCCP), and rotenone (Rot). E–G: there was a significant decrease in basal respiration, maximal uncoupled respiration (for KD1), and FAO in GCN5L1 knockdown cells relative to the control. Values are expressed as means ± SE; n = 4 per group for GCN5L1 mRNA and protein levels and n = 8 per group for respiration measurements. P value significance is as shown in the graphs. One-way ANOVA was used for A and E−G; two-way ANOVA was used for B and C; Tukey post hoc testing was performed in each case.
GCN5L1 knockdown decreases acetylation and enzymatic activity of FAO proteins.
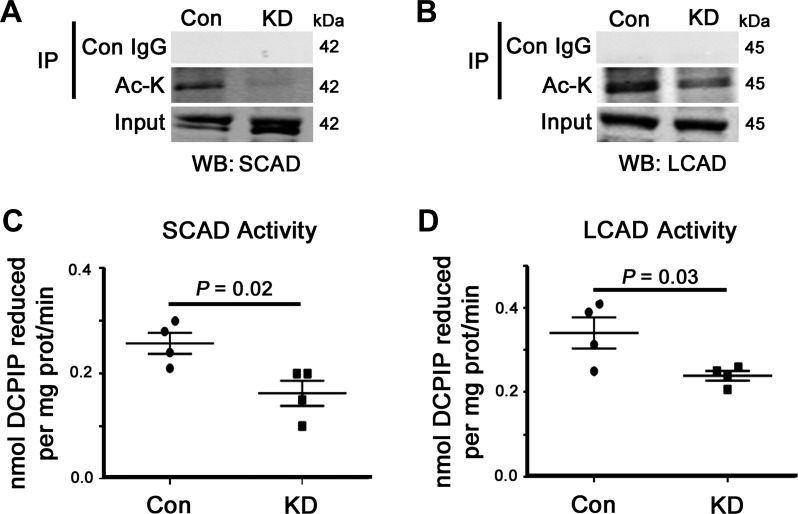
Finally, we attempted to investigate whether there was a mechanistic link between GCN5L1-regulated mitochondrial protein acetylation and FAO rates in H9C2 cells. Using our immunoprecipitation-based acetylation assay, we found a large decrease in the acetylation status of both SCAD and LCAD in GCN5L1 knockdown cells relative to the control (Fig. 7, A and B), although no change in acetylation level was seen in HADHA and PDH using the same methodology (data not shown). We then assessed the activity of SCAD and LCAD in vitro and found that loss of GCN5L1 significantly reduced the oxidation capacity of both dehydrogenases (Fig. 7, C and D). From these experiments, we conclude that GCN5L1 promotes the acetylation of mitochondrial FAO proteins and that this modification is strongly associated with increased FAO activity in cardiac cells.
Fig. 7.
Impact of GCN5L1 knockdown on cardiac FAO enzyme acetylation and activity. GCN5L1 promoted the acetylation and activity of cardiac FAO proteins. A and B: our immunoprecipitation-based acetylation assay showed a large decrease in the acetylation status of SCAD and LCAD proteins in GCN5L1 knockdown cells relative to the control. C and D: this decrease correlated with reduced enzymatic activities of SCAD and LCAD in vitro. Values are expressed as means ± SE; n = 4 per group. P value significance is as shown in the graphs using two-way Student’s t-test.
DISCUSSION
Mitochondrial protein acetylation is controlled by the activity of opposing acetyltransferase and deacetylase enzymes in response to changing nutritional conditions. To further understand the importance of this posttranslational modification in vivo, we examined the regulatory role played by lysine acetylation on cardiac bioenergetics during extended periods of excess dietary fatty acids. Our study demonstrated that increased acetylation status of cardiac mitochondrial proteins after HFD feeding correlated with increased GCN5L1 expression and decreased SIRT3 abundance. Furthermore, we show that prolonged exposure to a HFD led to increased acetylation of mitochondrial FAO enzymes, which was linked to an increase in their enzymatic activity. Finally, we found that knockdown of GCN5L1 in our stable H9C2 cell lines decreased the acetylation and activity of SCAD and LCAD, which resulted in a significant reduction in cellular FAO rates. These findings suggest that GCN5L1 activity may promote cardiac bioenergetic output through the acetylation and activation of multiple mitochondrial FAO enzymes.
In our HFD-induced obesity model, increased expression of Ppara, Acadl, Cpt1b, Cd36, and Pdk4 suggests an increased reliance on FAO, at the expense of pyruvate oxidation. Similar increases in FAO have been observed in human subjects undergoing 7 days of HFD feeding (31) as well as in rats with 3 wk of HFD (10). This shift toward increased utilization of fatty acids for ATP production is commonly observed in obesity and diabetes and may be exacerbated by cardiac insulin resistance and decreased glucose uptake (21). As excess fat conditions persist, changes in cardiac substrate handling may lead to elevated levels of circulating free fatty acids and triacylglycerol, culminating in myocardial lipid accumulation and decreased cardiac output (7, 40). Identification of the mechanisms involved in regulating this metabolic perturbation are therefore of interest in both basic and translational research settings.
Lysine acetylation is a reversible posttranslational modification that controls numerous cellular processes and energetic pathways (2, 26, 29, 41). Proteomic studies have shown that enzymes involved in almost every major metabolic pathway are acetylated (37, 41). Furthermore, studies assessing acetylation dynamics in several tissues, including the liver (17), skeletal muscle (19), and heart (1), have shown that multiple mitochondrial proteins are hyperacetylated. Our study is in agreement with these findings and demonstrates that there is a significant increase in cardiac mitochondrial protein acetylation after extended high-fat exposure (Fig. 3). Although numerous studies have shown that SIRT3 abundance and activity are regulated by prevailing nutrient conditions (13, 17), there have been few reports of the effect of diet on GCN5L1. Livers from fasted mice show a significant decrease in GCN5L1 abundance (38), although there have been conflicting reports of the effect of nutrient excess on GCN5L1 expression in the heart in both nonsurgical (1) and surgical investigations of cardiac metabolism (30). Our present findings are in direct contrast to a previous publication by Alrob et al. (1), which showed that GCN5L1 expression was not significantly changed in the heart after HFD feeding. Here, we showed that GCN5L1 is significantly upregulated at both the gene and protein level in HFD-exposed mice and that high concentrations of both glucose and palmitate can upregulate Gcn5l1 expression in H9C2 cells. The contrasting findings may also explain why we found a significant increase in PDH acetylation not observed previously (1). The reasons behind the diverse study outcomes are currently unclear but may be related to simple differences in diet composition (41% vs. 60% fat) or study length (16–18 wk vs. 24 wk).
In addition to our findings on GCN5L1 regulation, we observed an interesting discrepancy between the Sirt3 gene and SIRT3 protein expression in our HFD-fed animals. Numerous studies have shown that SIRT3 protein abundance is decreased in response to a HFD; however, there is little published information on how Sirt3 gene expression is affected by nutrient excess. We show here that Sirt3 is significantly upregulated in both HFD mouse hearts and in palmitate-treated H9C2 cells, whereas its protein abundance is reduced in vivo under the same conditions (Fig. 2). The mechanism by which the mitochondrial form of SIRT3 is reduced, despite unchanged levels of the preimport cytosolic form, remains to be determined but may be linked to increased SIRT3 degradation under high-fat conditions. Overall, we conclude that regulators of both mitochondrial lysine acetylation and deacetylation are closely controlled by nutrient availability and that this system has evolved to regulate mitochondrial bioenergetics in response to changing nutritional and physiological conditions.
We have previously shown that GCN5L1 modulates the acetylation of electron transport chain proteins (35), and its loss negatively impacts mitochondrial bioenergetic output (34). Here, we focused on the effect of GCN5L1 on FAO enzymes SCAD and LCAD, which oxidize short- and long-chain fatty acids, respectively. LCAD function is particularly important in maintaining cardiac bioenergetic output, and loss of LCAD activity has been shown to result in an elevated reliance on glucose oxidation (3). Conversely, increased activity of LCAD has been associated with increased FAO in the heart (1). We found that SCAD and LCAD are hyperacetylated after a long-term HFD and have increased enzymatic activity in in vitro assays. As these changes correlated with an increase in GCN5L1 abundance, we investigated whether this protein had a functional effect on FAO enzyme activity. Knockdown of GCN5L1 in our stable cardiac cell line resulted in decreased acetylation of SCAD and LCAD and significantly reduced the enzymatic activity of LCAD. Furthermore, we show, for the first time, that reductions in GCN5L1 abundance led to a significant decrease in cellular FAO rates. These findings suggest that GCN5L1 function is central to the regulation of cardiac mitochondrial bioenergetics and is indispensable for the regulation of acetylation and enzymatic activity of FAO proteins. Future studies examining the impact of GCN5L1 loss on fatty acid metabolism and bioenergetics in vivo should help to further elucidate the importance of lysine acetylation in cardiac metabolism.
The outcomes of the present study are in broad agreement with a very recent publication, which showed that reduced GCN5L1 expression led to a significant decrease in the in vitro activity of LCAD and β-hydroxy-acyl-CoA dehydrogenase (11, 12). Importantly, this study also showed that GCN5L1 abundance increased during early development, which closely correlated with the switch from a glycolytic to a FAO metabolic profile in the heart (11, 12). This indicates that an increase in the acetylation of cardiac FAO enzymes plays a functional role in upregulating their activity, and corroborates several recent studies in the heart (1, 30) which have shown clear links between upregulated FAO enzyme acetylation and increased cardiac fatty acid utilization.
However, it must be noted that in the majority of tissues examined (particularly the liver), an increase in the acetylation of FAO enzymes has been linked to a decrease in their activity. For example, hyperacetylation of LCAD, caused by loss of SIRT3, leads to a decrease in enzymatic activity and hepatic lipid accumulation (5, 13). This discrepancy in the impact of acetylation on FAO in different tissues clearly requires further investigation. One potential explanation is that different lysine residues on the same protein (e.g., LCAD) are acetylated under the same HFD conditions in different tissues, which may increase or decrease its enzymatic activity in response to the contrasting metabolic needs of each organ (e.g., liver vs. heart). Proteomic studies have shown differing acetylation patterns in different organs from the same mouse (for a review, see Ref. 27), and it may be that each tissue uses acetylation to regulate metabolic activity differently, even on a protein-by-protein basis. Given that the heart is particularly reliant on FAO (21) and that fatty acids are the main source of acetyl-CoA for acetylation (28), it may be logical that increases in fatty acid availability in the heart would not immediately downregulate FAO via acetylation, as it does in other tissues. Future comparative studies investigating the sites of lysine acetylation on the same protein in different tissues should help to answer some of these questions.
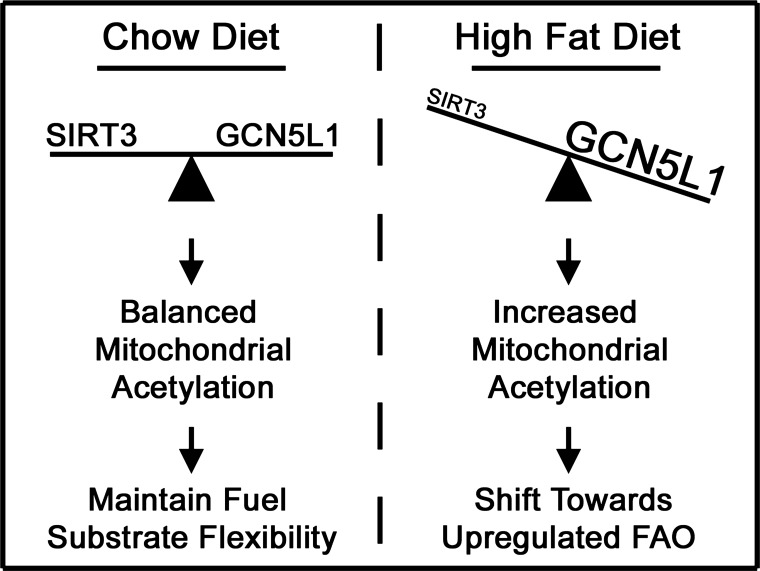
In conclusion, our findings in this study suggest that the regulation of lysine acetylation in mitochondria plays a major role in controlling cardiac bioenergetic output and that the utilization of fatty acids in the heart is enhanced when increased nutrient availability promotes the acetylation of FAO enzymes (Fig. 8). Further work on the function of GCN5L1 and SIRT3 in this context will likely yield important information on cardiac bioenergetics and may in the future uncover new therapeutic targets in cardiac metabolism under various disease states.
Fig. 8.
Model of lysine acetylation and FAO in cardiac metabolism. Under normal conditions, a balance between lysine acetylation and deacetylation allows mitochondria to maintain bioenergetic output under changing physiological conditions. In obese mice, increased acetylation of mitochondrial proteins drives a pro-FAO metabolic phenotype, thereby reducing substrate flexibility in the heart.
GRANTS
This work was funded in part by NHLBI Grants T32-HL-110849 (to J. R. Manning) and K22-HL-116728 and R56-HL-132917 (to I. Scott).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
D.T. and I.S. conceived and designed research; D.T., M.Z., J.R.M., D.G., and M.S. performed experiments; D.T., J.R.M., D.G., and I.S. analyzed data; D.T. and I.S. interpreted results of experiments; D.T. and I.S. prepared figures; D.T. and I.S. drafted manuscript; D.T., M.Z., J.R.M., D.G., M.S., R.M.O., S.S., and I.S. approved final version of manuscript; I.S. edited and revised manuscript.
ACKNOWLEDGMENTS
We thank Dr. Michael Sack [National Heart, Lung, and Blood Institute (NHLBI), Bethesda, MD] for critical reading of the manuscript, and members of the O’Doherty laboratory for help with animal care.
REFERENCES
- 1.Alrob OA, Sankaralingam S, Ma C, Wagg CS, Fillmore N, Jaswal JS, Sack MN, Lehner R, Gupta MP, Michelakis ED, Padwal RS, Johnstone DE, Sharma AM, Lopaschuk GD. Obesity-induced lysine acetylation increases cardiac fatty acid oxidation and impairs insulin signalling. Cardiovasc Res 103: 485–497, 2014. doi: 10.1093/cvr/cvu156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson KA, Hirschey MD. Mitochondrial protein acetylation regulates metabolism. Essays Biochem 52: 23–35, 2012. doi: 10.1042/bse0520023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakermans AJ, Dodd MS, Nicolay K, Prompers JJ, Tyler DJ, Houten SM. Myocardial energy shortage and unmet anaplerotic needs in the fasted long-chain acyl-CoA dehydrogenase knockout mouse. Cardiovasc Res 100: 441–449, 2013. doi: 10.1093/cvr/cvt212. [DOI] [PubMed] [Google Scholar]
- 4.Banks AS, Kon N, Knight C, Matsumoto M, Gutiérrez-Juárez R, Rossetti L, Gu W, Accili D. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab 8: 333–341, 2008. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bharathi SS, Zhang Y, Mohsen AW, Uppala R, Balasubramani M, Schreiber E, Uechi G, Beck ME, Rardin MJ, Vockley J, Verdin E, Gibson BW, Hirschey MD, Goetzman ES. Sirtuin 3 (SIRT3) protein regulates long-chain acyl-CoA dehydrogenase by deacetylating conserved lysines near the active site. J Biol Chem 288: 33837–33847, 2013. doi: 10.1074/jbc.M113.510354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bing RJ, Siegel A, Ungar I, Gilbert M. Metabolism of the human heart. II. Studies on fat, ketone and amino acid metabolism. Am J Med 16: 504–515, 1954. doi: 10.1016/0002-9343(54)90365-4. [DOI] [PubMed] [Google Scholar]
- 7.Carley AN, Atkinson LL, Bonen A, Harper ME, Kunnathu S, Lopaschuk GD, Severson DL. Mechanisms responsible for enhanced fatty acid utilization by perfused hearts from type 2 diabetic db/db mice. Arch Physiol Biochem 113: 65–75, 2007. doi: 10.1080/13813450701422617. [DOI] [PubMed] [Google Scholar]
- 8.Chen T, Liu J, Li N, Wang S, Liu H, Li J, Zhang Y, Bu P. Mouse SIRT3 attenuates hypertrophy-related lipid accumulation in the heart through the deacetylation of LCAD. PLoS One 10: e0118909, 2015. doi: 10.1371/journal.pone.0118909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu HC, Kovacs A, Ford DA, Hsu FF, Garcia R, Herrero P, Saffitz JE, Schaffer JE. A novel mouse model of lipotoxic cardiomyopathy. J Clin Invest 107: 813–822, 2001. doi: 10.1172/JCI10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole MA, Murray AJ, Cochlin LE, Heather LC, McAleese S, Knight NS, Sutton E, Jamil AA, Parassol N, Clarke K. A high fat diet increases mitochondrial fatty acid oxidation and uncoupling to decrease efficiency in rat heart. Basic Res Cardiol 106: 447–457, 2011. doi: 10.1007/s00395-011-0156-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukushima A, Alrob OA, Zhang L, Wagg CS, Altamimi T, Rawat S, Rebeyka IM, Kantor PF, Lopaschuk GD. Acetylation and succinylation contribute to maturational alterations in energy metabolism in the newborn heart. Am J Physiol Heart Circ Physiol 311: H347–H363, 2016. doi: 10.1152/ajpheart.00900.2015. [DOI] [PubMed] [Google Scholar]
- 12.Fukushima A, Lopaschuk GD. Cardiac fatty acid oxidation in heart failure associated with obesity and diabetes. Biochim Biophys Acta 1861: 1525–1534, 2016. doi: 10.1016/j.bbalip.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 13.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV Jr, Alt FW, Kahn CR, Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464: 121–125, 2010. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, Stančáková A, Goetzman E, Lam MM, Schwer B, Stevens RD, Muehlbauer MJ, Kakar S, Bass NM, Kuusisto J, Laakso M, Alt FW, Newgard CB, Farese RV Jr, Kahn CR, Verdin E. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol Cell 44: 177–190, 2011. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jing E, O’Neill BT, Rardin MJ, Kleinridders A, Ilkeyeva OR, Ussar S, Bain JR, Lee KY, Verdin EM, Newgard CB, Gibson BW, Kahn CR. Sirt3 regulates metabolic flexibility of skeletal muscle through reversible enzymatic deacetylation. Diabetes 62: 3404–3417, 2013. doi: 10.2337/db12-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalaivanisailaja J, Manju V, Nalini N. Lipid profile in mice fed a high-fat diet after exogenous leptin administration. Pol J Pharmacol 55: 763–769, 2003. [PubMed] [Google Scholar]
- 17.Kendrick AA, Choudhury M, Rahman SM, McCurdy CE, Friederich M, Van Hove JL, Watson PA, Birdsey N, Bao J, Gius D, Sack MN, Jing E, Kahn CR, Friedman JE, Jonscher KR. Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. Biochem J 433: 505–514, 2011. doi: 10.1042/BJ20100791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell 23: 607–618, 2006. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 19.Lantier L, Williams AS, Williams IM, Yang KK, Bracy DP, Goelzer M, James FD, Gius D, Wasserman DH. SIRT3 Is Crucial for Maintaining Skeletal Muscle Insulin Action and Protects Against Severe Insulin Resistance in High-Fat-Fed Mice. Diabetes 64: 3081–3092, 2015. doi: 10.2337/db14-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopaschuk GD, Belke DD, Gamble J, Itoi T, Schönekess BO. Regulation of fatty acid oxidation in the mammalian heart in health and disease. Biochim Biophys Acta 1213: 263–276, 1994. doi: 10.1016/0005-2760(94)00082-4. [DOI] [PubMed] [Google Scholar]
- 21.Lopaschuk GD, Folmes CD, Stanley WC. Cardiac energy metabolism in obesity. Circ Res 101: 335–347, 2007. doi: 10.1161/CIRCRESAHA.107.150417. [DOI] [PubMed] [Google Scholar]
- 22.Lopaschuk GD, Ussher JR, Folmes CD, Jaswal JS, Stanley WC. Myocardial fatty acid metabolism in health and disease. Physiol Rev 90: 207–258, 2010. doi: 10.1152/physrev.00015.2009. [DOI] [PubMed] [Google Scholar]
- 23.Neely JR, Morgan HE. Relationship between carbohydrate and lipid metabolism and the energy balance of heart muscle. Annu Rev Physiol 36: 413–459, 1974. doi: 10.1146/annurev.ph.36.030174.002213. [DOI] [PubMed] [Google Scholar]
- 24.Neely JR, Rovetto MJ, Oram JF. Myocardial utilization of carbohydrate and lipids. Prog Cardiovasc Dis 15: 289–329, 1972. doi: 10.1016/0033-0620(72)90029-1. [DOI] [PubMed] [Google Scholar]
- 25.Neubauer S. The failing heart–an engine out of fuel. N Engl J Med 356: 1140–1151, 2007. doi: 10.1056/NEJMra063052. [DOI] [PubMed] [Google Scholar]
- 26.Newman JC, He W, Verdin E. Mitochondrial protein acylation and intermediary metabolism: regulation by sirtuins and implications for metabolic disease. J Biol Chem 287: 42436–42443, 2012. doi: 10.1074/jbc.R112.404863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peinado JR, Diaz-Ruiz A, Frühbeck G, Malagon MM. Mitochondria in metabolic disease: getting clues from proteomic studies. Proteomics 14: 452–466, 2014. doi: 10.1002/pmic.201300376. [DOI] [PubMed] [Google Scholar]
- 28.Pougovkina O, te Brinke H, Ofman R, van Cruchten AG, Kulik W, Wanders RJ, Houten SM, de Boer VC. Mitochondrial protein acetylation is driven by acetyl-CoA from fatty acid oxidation. Hum Mol Genet 23: 3513–3522, 2014. doi: 10.1093/hmg/ddu059. [DOI] [PubMed] [Google Scholar]
- 29.Sack MN, Finkel T. Mitochondrial metabolism, sirtuins, and aging. Cold Spring Harb Perspect Biol 4: a013102, 2012. doi: 10.1101/cshperspect.a013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sankaralingam S, Abo Alrob O, Zhang L, Jaswal JS, Wagg CS, Fukushima A, Padwal RS, Johnstone DE, Sharma AM, Lopaschuk GD. Lowering body weight in obese mice with diastolic heart failure improves cardiac insulin sensitivity and function: implications for the obesity paradox. Diabetes 64: 1643–1657, 2015. doi: 10.2337/db14-1050. [DOI] [PubMed] [Google Scholar]
- 31.Schrauwen P, Wagenmakers AJ, van Marken Lichtenbelt WD, Saris WH, Westerterp KR. Increase in fat oxidation on a high-fat diet is accompanied by an increase in triglyceride-derived fatty acid oxidation. Diabetes 49: 640–646, 2000. doi: 10.2337/diabetes.49.4.640. [DOI] [PubMed] [Google Scholar]
- 32.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci USA 103: 10224–10229, 2006. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol 158: 647–657, 2002. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott I, Webster BR, Chan CK, Okonkwo JU, Han K, Sack MN. GCN5-like protein 1 (GCN5L1) controls mitochondrial content through coordinated regulation of mitochondrial biogenesis and mitophagy. J Biol Chem 289: 2864–2872, 2014. doi: 10.1074/jbc.M113.521641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott I, Webster BR, Li JH, Sack MN. Identification of a molecular component of the mitochondrial acetyltransferase programme: a novel role for GCN5L1. Biochem J 443: 655–661, 2012. doi: 10.1042/BJ20120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verity MA, Turnbull DM. Assay of acyl-CoA dehydrogenase activity in frozen muscle biopsies: application to medium-chain acyl-CoA dehydrogenase deficiency. Biochem Med Metab Biol 49: 351–362, 1993. doi: 10.1006/bmmb.1993.1036. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, Ning ZB, Zeng R, Xiong Y, Guan KL, Zhao S, Zhao GP. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327: 1004–1007, 2010. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webster BR, Scott I, Han K, Li JH, Lu Z, Stevens MV, Malide D, Chen Y, Samsel L, Connelly PS, Daniels MP, McCoy JP Jr, Combs CA, Gucek M, Sack MN. Restricted mitochondrial protein acetylation initiates mitochondrial autophagy. J Cell Sci 126: 4843–4849, 2013. doi: 10.1242/jcs.131300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiong Y, Guan KL. Mechanistic insights into the regulation of metabolic enzymes by acetylation. J Cell Biol 198: 155–164, 2012. doi: 10.1083/jcb.201202056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young ME, Guthrie PH, Razeghi P, Leighton B, Abbasi S, Patil S, Youker KA, Taegtmeyer H. Impaired long-chain fatty acid oxidation and contractile dysfunction in the obese Zucker rat heart. Diabetes 51: 2587–2595, 2002. doi: 10.2337/diabetes.51.8.2587. [DOI] [PubMed] [Google Scholar]
- 41.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science 327: 1000–1004, 2010. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]