Abstract
The Saccharomyces cerevisiae α-factor receptor Ste2p has been used extensively as a model to understand the molecular mechanism of signal transduction by G protein-coupled receptors (GPCRs). Single and double cysteine mutants of Ste2p were created and served as surrogates to detect intramolecular interactions and dimerization of Ste2p using disulfide cross-linking methodology. When a mutation was introduced into the phylogenetically conserved tyrosine residue at position 26 (Y26C) in the N-terminus of Ste2p, dimerization was increased greatly. The amount of dimer formed by this Y26C mutant was greatly reduced by ligand binding even though the ligand binding site is far removed from the N-terminus; the lowering of the dimer formation was consistent with a conformational change in the N-terminus of the receptor upon activation. Dimerization was decreased by double mutations Y26C/V109C or Y26C/T114C indicating that Y26 is in close proximity to V109 and T114 of extracellular loop 1 in native Ste2p. Combined with earlier studies, these results indicate previously unrecognized roles for the N-terminus of Ste2p, and perhaps of GPCRs in general, and reveal a specific N-terminus residue or region, that is involved in GPCR signaling, intrareceptor interactions, and receptor dimerization.
Graphical abstract
Introduction
Transmission of extracellular signals across the plasma membrane is one of the most fundamental biological processes. G protein-coupled receptors (GPCRs), the largest family of signaling molecules on the cell surface, play central roles in the signaling processes by responding to a plethora of signals, including photons, hormones, neurotransmitters, ions and lipids [1-4]. Despite the diversity of signals and the pathways they regulate, all GPCRs have a common architecture consisting of seven transmembrane domains. Upon translation of these signals into appropriate responses, cells regulate many crucial physiological and pathophysiological processes [5]. Inappropriate and / or altered GPCR functions cellular are directly or indirectly associated with many diseases including schizophrenia, Alzheimer's, cancer, blindness, obesity, hypertension, and diabetes [2, 6, 7]. Not surprisingly, GPCRs are the therapeutic targets for a large portion of currently prescribed drugs [8-12].
Ligand binding promotes a conformational change in the receptor that triggers the cellular response via intracellular transducers - the heterotrimeric (α, β, γ subunits) guanine nucleotide binding proteins (G proteins) and/or β-arrestin [13, 14]. The conformational changes involve the movement of transmembrane domains [15-21]. However, concomitant changes are also expected to occur in other domains of the receptor, including the loop regions and the N- and C-termini. Despite containing several distinct regions, the majority of GPCR structure and function studies have focused on the transmembrane helices. However, a growing number of studies indicate that the N-terminus also plays an important role in receptor function [22-27]. For example, studies with class B secretin GPCRs indicate that the N-terminus is the ligand binding domain for these receptors [28]. It has been proposed that binding of the cognate ligand to the N-terminus induces a conformational change in the receptor's N-terminus. This enables a segment of the N-terminus to dock near the top of transmembrane domain 6 and this in turn triggers a conformational change in the heptahelical bundle to initiate downstream signaling [29]. In class C glutamate receptors, the conserved N-terminal Venus flytrap module has been reported to regulate ligand binding and receptor activation [22, 24, 25, 30]. The N-termini of protease-activated receptors (PARs) and glycoprotein hormone receptors (GpHRs) have also been associated with receptor activation [31, 32]. The N-terminus of adhesion G protein-coupled receptor GPR56 has been reported to constrain receptor activity [33]. Truncation of the N-terminus of several GPCRs including CB1 cannabinoid [34], α 1D adrenergic [35] and GPR37 [36] has been shown to enhance cell surface expression [37].
A growing number of studies indicate that GPCRs interact with each other to form dimers and / or oligomers with important functional consequences. For example, dimerization is a prerequisite for the expression of a fully functional γ-aminobutyric acid (GABAb) receptor at the plasma membrane [38-40]. Taste T1 receptors also exist as obligate heteromers [41]. Dimerization between D2 and D3 receptors was reported to confer novel functional properties [42]. GPR83, an orphan receptor involved in body weight regulation and the control of circulating adenopectin levels, can form homodimers and heterodimers with other GPCRs. Heterodimerization of this receptor with the ghrelin receptor (GHSR1a) has been shown to impede GHSR activity [43]. The dimerization across the various CXCR4 crystal structures is consistent with reports of CXCR4 homo- and heterodimerization in cells and its possible physiological relevance [44]. These and many other studies have confirmed that dimerization and / or oligomerization play important roles in receptor function.
The Saccharomyces cerevisiae pheromone receptor Ste2p is a GPCR activated upon binding α factor, a 13-residue peptide, which triggers signaling through a cytoplasmic heterotrimeric G protein in MATa haploid cells [45, 46]. Ste2p has been used as a model for understanding structure-function relationships of GPCRs using the power of yeast genetics and analysis of the yeast pheromone response pathway. Although Ste2p lacks strong sequence similarity to mammalian GPCRs, some mammalian GPCRs have been expressed in yeast and are capable of coupling to and activating the yeast mating pathway [47, 48]. Ste2p also exhibits signaling when expressed in mammalian cells [49].
The N-terminus of Ste2p is ∼50 amino acids long and contains two glycosylation sites, neither of which is essential for receptor function [50]. The N-terminus was reported to be involved in forming a domain for Ste2p dimerization [51, 52]. Previous studies of the first extracellular loop using the substituted cysteine accessibility method (SCAM) reported that several residues (L102C, N105C, S108C, Y111C and T114C) in this loop were inaccessible to the sulfhydryl reagent (MTSEA-biotin) used to assess solvent accessibility [53]. It was also reported that mutation of these residues to cysteine affected the glycosylation pattern of the receptor. Because two glycosylation sites of the receptor are located in the N-terminus at N25 and N32 [50] and the mutations in the ECL1 affected the glycosylation pattern, we hypothesized that the N-terminus interacted with ECL1. More recently, several residues in the N-terminus, including Y26C, were also found to be inaccessible to MTSEA-biotin and the Y26C mutant also exhibited markedly increased dimerization [51]. This residue is in the consensus sequence of N-glycosylation N-X-S/T (where X is any amino acid except Pro). The tyrosine in this position is conserved among the α-factor receptors in several fungal species (Fig. 1). These observations stimulated this investigation into whether ECL1 interacts with N-terminus. In this paper we report on an interaction between the N-terminus and ECL1 influenced by ligand binding.
Figure 1.
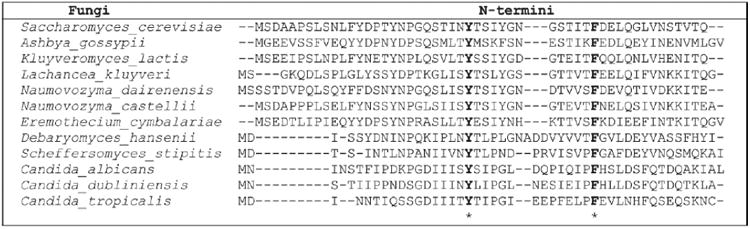
Sequence alignments of yeast α-factor pheromone receptors from different fungi. The N-terminal regions of S. cerevisiae, Ashybya gossypii, Candida albicans, Candida dubliniensis, Candida tropicalis, Debaryomyces hansenii, Eremothecium cymbalariae, Kluyveromyces lactis, Lachancea kluyveri, Naumovozyma castelllii, Naumovozyma dairenesis, and Scheffersomyces stipits α-factor pheromone receptors were compared by amino acid sequence alignment. Invariably conserved residues are shown in bold letters with asterisk (*) at the bottom of the aligned sequences.
Materials and Methods
Media, Reagents, Strains, and Plasmids
S. cerevisiae strain LM102 [MATa ste2 FUS1-lacZ∷URA3 bar1 ura3 leu2 his4 trp1 met1] [54] was used for growth arrest, FUS1-lacZ gene induction and saturation binding assays, and the protease-deficient strain BJS21 [MATa, prc1-407 prb1-1122 pep4-3 leu2 trp1 ura3-52 ste2∷KanR [55] was used for protein isolation, immunoblot analyses, and disulfide cross-linking studies in order to minimize receptor degradation during analysis. To facilitate disulfide cross-linking, plasmid pBEC2-FXa was constructed from plasmid pBEC2 [53] containing a Cys-less Ste2p using primers to introduce tandem Factor Xa protease cleavage sites between residues T78 and P79 in ICL1. The plasmid pBEC2-FXa containing C-terminal FLAG™ and His-tagged STE2 with a tandem Factor Xa cleavage site was transformed by the method of Geitz [56]. Transformants were selected by growth on yeast media [57] lacking tryptophan (designated as MLT) to maintain selection for the plasmid. The cells were cultured in MLT (2% glucose, 1% casamino acids (Research Products International Corp., IL, USA), 0.17% yeast nitrogen base without ammonium sulfate (Research Products International Corp., IL, USA), 0.5% ammonium sulfate (Research Products International Corp., IL, USA), amino acid dropout mix containing (arginine 0.026 g/L, asparagine 0.058 g/L, aspartic acid 0.14 g/L, glutamic acid 0.14 g/L, histidine 0.028 g/L, isoleucine 0.058 g/L, leucine 0.083 g/L, lysine 0.042 g/L, methionine 0.028 g/L, phenylalanine 0.69 g/L, serine 0.52 g/L, threonine 0.28 g/L, tyrosine 0.042 g/L, valine 0.21 g/L, adenine sulfate 0.058 g/L, uracil 0.028 g/L) and grown to mid log phase at 30°C with shaking (200 rpm) for all assays.
Growth Arrest Assays
S. cerevisiae strain LM102 expressing Cys-less Ste2p (ICL1-Xa2) or site-directed Cys mutants was grown at 30°C overnight in MLT, harvested, washed three times with water, and resuspended to a final concentration of 5× 106 cells/mL [58]. Cells (1 mL) were combined with 3.5 mL of agar noble (1.1%) and poured as a top agar lawn onto a MLT medium agar plate. Filter disks (BD, Franklin Lakes, NJ) impregnated with α-factor (4, 2, 1, 0.5, and 0.25 μg/disk) were placed on the top agar. The plates were incubated at 30°C for 24h and then observed for clear halos around the disks. The experiment was repeated at least three times, and reported values represent the mean of these tests.
FUS1-lacZ Gene Induction Assay
Cells expressing Cys-less Ste2p (ICL1-Xa2) and Cys mutants were grown at 30°C in MLT, harvested, washed three times with fresh medium and resuspended to a final concentration of 5 × 107 cells/mL. Cells (500 μl) were combined with α-factor pheromone (final concentration of 1μM) and incubated at 30°C for 90 min. The cells were transferred to a 96-well flat bottom plate (Corning Incorporated, Corning, NY) in triplicate, permeabilized with 0.5% Triton X-100 in 25 mM PIPES buffer (pH 7.2) and then β-galactosidase assays were carried out using fluorescein di-β-galactopyranoside (Molecular Probes, Inc., Eugene, OR) as a substrate as described previously [59]. The reaction mixtures were incubated at 37°C for 60 min. The fluorescence of the samples (excitation of 485 nm and emission of 530 nm) was determined using a 96-well plate reader Synergy2 (BioTek Instruments, Inc., Winooski, VT). The data were analyzed using Prism software (GraphPad Prism version 6.02 for Windows, GraphPad Software, San Diego CA). The experiments were repeated at least three times and reported values represent the mean of these tests.
Immunoblots
Immunoblot analysis of Ste2p was carried out as described previously [60]. Cells (BJS21) expressing various Ste2p constructs grown in MLT were used to prepare total cell membranes as previously described [53, 61]. For studies of disulfide cross-linking, membranes were solubilized in SDS sample buffer (30% glycerol, 3% SDS, 0.01% bromphenol blue, 0.1875 M Tris, pH 6.8) without β-mercaptoethanol (β-ME). Proteins were fractioned by SDS-PAGE (10% acrylamide) along with pre-stained Precision Plus protein standards (BioRad, Hercules, CA) and transferred to an Immobilon™ P membrane (Millipore Corp., Bedford, MA). The blot was probed with anti-FLAG™ M2 antibody (Sigma/Aldrich Chemical, St. Louis, MO), and bands were visualized with the West Pico chemiluminescent detection system (Pierce, Rockford, IL) using ChemiDoc™ XRS+ system (BioRad, Hercules, CA). The intensity of Ste2p signals from blots was measured by densitometry using Image Lab™ software (version 4.1, BioRad, Hercules, CA). Although blots were overexposed to be able to see the mutants, blots with less exposure time were used for accurate quantification of band intensity. Multiple repeats of immunoblot experiments yielded similar results. The constitutively-expressed membrane protein Pma1p was used as a loading control as described previously [62] using Pma1p antibody (Thermo Scientific, Rockford, IL).
Disulfide Cross-Linking with Cu-Phenanthroline
Disulfide cross-linking was carried out as described previously [60]. One hundred micrograms of the membrane preparation was incubated in the absence or presence α-factor (1 μM final concentration) for 30 min prior to Cu-P treatment in experiments performed to examine the influence of ligand on dimerization. To initiate the cross-linking reaction, the membrane proteins were treated with a fresh preparation of Cu(II)-1,10-phenanthroline (Cu-P; final concentration, 2.5 μM CuSO4 and 7.5 μM phenanthroline, in phosphate-buffered saline, pH 7.4). The reaction was carried out at room temperature for 20 min, terminated with 50 mM EDTA, and kept on ice for 20 min followed by adding SDS sample buffer without β-mercaptoethanol.
Factor Xa digestion
The membrane protein preparation (40 μg) was incubated with 0.4 unit of Factor Xa (Novagen) in Factor Xa cleavage buffer (0.1M NaCl, 50 mM Tris-HCl, 5 mM CaCl2, pH 8.0) containing 0.1% Triton X-100 for 16h at 4°C. Each sample was divided into two portions. The reactions were terminated by adding one-third the volume of Laemmli sample buffer (30% glycerol, 3% SDS, 0.01% bromphenol blue, 0.1875 M Tris, pH 6.8). To simulate reducing conditions β-mercaptoethanol (final concentration, 1%, v/v) was added to one of the portions. Samples were analyzed by SDS-PAGE and Western blotting as described above.
Results
Construction of Ste2p Cys mutants
To investigate possible intra-receptor interactions one residue (Y26) in the extracellular N-terminus (NT) and five residues (N105, S108, V109, Y111 and T114) in the extracellular loop 1 (ECL1) of Ste2p were chosen for mutation to Cys. These residues were chosen for several reasons: (i) Y26 was found to be one of the two invariably conserved residues in the N-terminal domains of α-factor pheromone receptors of many fungal species: Saccharomyces cerevisiae, Ashybya gossypii, Candida albicans, Candida dubliniensis, Candida tropicalis, Debaryomyces hansenii, Eremothecium cymbalariae, Kluyveromyces lactis, Lachancea kluyveri, Naumovozyma castelllii, Naumovozyma dairenesis, and Scheffersomyces stipits. The N- terminal regions of the α-factor pheromone receptors of these fungi were predicted using TMHMM 2.0 [63] and aligned (Fig. 1) by Clustal Omega [64]. The twelve receptors analyzed in this study were predicted to have an N-terminal domain of 45-53 residues, (ii) Y26 was determined to be solvent inaccessible by the substituted cysteine accessibility method (SCAM) suggesting a possible interaction between Y26 and extracellular loop of Ste2p [51], (iii) Y26 is located within one of the two known glycosylation sites (N25 and N32) of Ste2p [50], and (iv) mutation of residues N105, S108, V109, Y111 and T114 to Cys led to changes in the glycosylation pattern of the receptors [53].
The template for the introduction of these mutations was a Cys-less receptor to eliminate the possibility of cross-linking between Y26 and endogenous Cys residue in the wild-type receptor [51, 53, 60]. The Cys-less template contained two C-terminal epitope tags (FLAG and 6XHIS) and also tandem Factor Xa cleavage sites (IEGRIEGR) in the first intracellular loop in order to allow detection of interdomain cross-linking (Fig. 2). The Cys-less Ste2p-FLAG-His receptor (referred to herein as “wild-type” and used extensively in previous studies from our lab), and the receptor with the Factor Xa cleavage sites in ICL1 (referred to herein as “ICL1-Xa2”), demonstrated similar expression levels as well as almost identical biological activities in growth arrest and FUS1-lacZ assays to that of wild-type Ste2p indicating that incorporating the protease cleavage sites and other modifications of Ste2p did not alter receptor function (Fig. 3).
Figure 2.
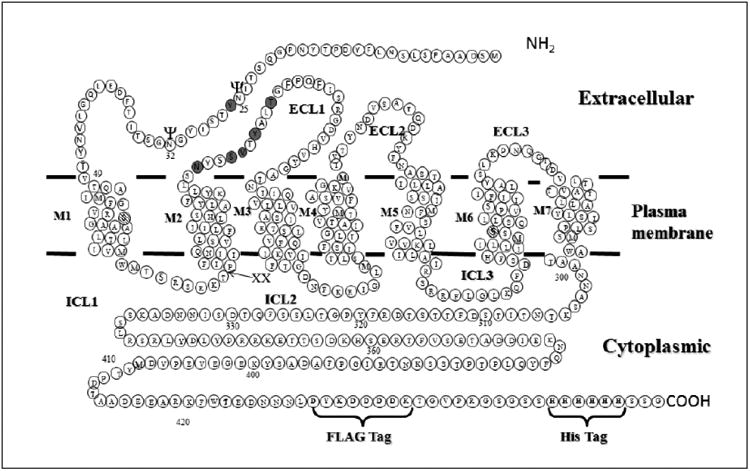
Diagram of Ste2p showing positions of Cys mutations and modifications introduced to facilitate disulfide cross-linking and intramolecular interactions. “XX” marks the site of the tandem Factor Xa protease cleavage (IEGRIEGR) engineered into ICL1. The FLAG and HIS epitope tags engineered into the C-terminus are also shown. The two endogenous Cys residues (C59 and C252 – hatched circles) were mutated to Ser to generate Cys-less Ste2p. The sites of Cys mutation engineered into the NT and ECL1 regions for disulfide cross-linking (Y26C, N105C, S108C, V109C, Y111C and T114C) are shown in grey circles. The two known glycosylation sites are shown with “ψ” symbol.
Figure 3.
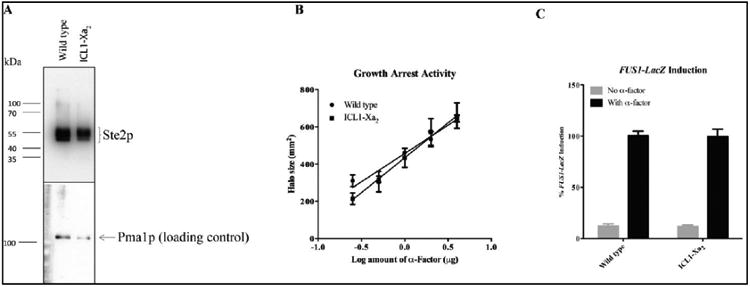
Comparison of the expression levels and signaling activities of the WT and ICL1-Xa2 receptor used in this study. (A) total membranes prepared from the cells expressing wild-type and ICL1-Xa2 constructs were immunoblotted using anti-FLAG antibody. The bottom panel shows the same immunoblot re-probed using antibody against Pma1p, a constitutively expressed plasma membrane protein used as a loading control. (B) The zone of growth inhibition of strains carrying the indicated receptors was measured in response to various concentrations of α-factor. (C) Signaling activities of the constructs determined by pheromone-induced FUS1-lacZ activity. The grey bars represent the constitutive signaling and the black bars represent the α-factor induced signaling activity. The signaling was normalized to that of the wild-type construct.
The signaling activities of the cysteine mutants
Signaling activities of the Cys mutants were examined by two independent assays: pheromone-induced growth arrest and FUS1-lacZ reporter gene assays. The growth arrest assay measures the ability of the receptor to promote α-factor-induced cell division arrest. It is a rigorous test of receptor function that can be used to compare the relative sensitivity to α-factor. This assay also monitors the ability of the cell to maintain the pheromone response for a relatively long time period (18-24 hours). On the other hand, the pheromone-induced FUS1-lacZ reporter gene assay is a short term measurement of signaling activity of the receptor during the early (1 to 2 hours) response of the yeast cells to pheromone. The strains used in this study were engineered to contain a reporter gene construct consisting of a fusion between FUS1 promoter and the lacZ gene encoding the enzyme β-galactosidase [65]. The FUS1-lacZ assay allows for fast, sensitive detection of mating pathway activation by assessing the induction of β-galactosidase activity in response to mating pheromone. The growth arrest and FUS1-lacZ activity of the mutants ranged from ∼30-90% of the ICL1-Xa2 control. Although all mutants were active for signaling in both assays, some mutants exhibited dissimilar activities in the two assays (Table 1). For example, compared to the ICL1-Xa2 control, the S108C mutant exhibited a 35% reduction in growth arrest and 70% reduction in FUS1-lacZ activity. In contrast, the Y111C mutant exhibited only a 10% reduction in growth arrest and a 70% reduction in FUS1-lacZ activity. The double mutant Y26C/V109C exhibited 66% reduction in growth arrest activity, but only a 28% reduction in FUS1-lacZ activity. The differences in functional readouts between the FUS1-lacZ activity and growth arrest activity of various Ste2p mutants have been observed in many studies and has been explained on the basis of the assay used to measure signaling [66-70]. These differences in signaling are similar to those observed in functional selectivity induced by different ligands binding to the same receptor, a phenomenon termed signal bias [71, 72].
Table 1. Biological activities of Cys mutants.
| Receptor | Growth arrest activities (%)1 | Fus1-LacZ Induction (%)2 |
|---|---|---|
| ICL1-Xa2 | 100 | 100 |
| Y26C | 60 | 53 |
| N105C | 79 | 43 |
| Y26C/N105C | 40 | 47 |
| S108C | 65 | 30 |
| Y26C/S108C | 59 | 45 |
| V109C | 84 | 89 |
| Y26C/V109C | 34 | 72 |
| Y111C | 90 | 30 |
| Y26C/Y111C | 70 | 64 |
| T114C | 87 | 57 |
| Y26C/T114C | 75 | 39 |
Relative growth arrest activity compared to that of the ICL1-Xa2 receptor at 1.0 g of α-factor applied to a disk (the halo size of the ICL1-Xa2 receptor was 18 mm). The standard deviation (not shown) of the halo activity for each of the receptors was within 10% (three replicates).
Relative maximal FUS1-lacZ activity compared to that of the induced activity of ICL1-Xa2 at 1.0 μM of α-factor. The standard deviation of the FUS1-lacZ activity (not shown) was within ±5% for each of the receptors (three replicates).
Expression of the Ste2p mutants
The total cellular expression level of each single and double-Cys mutant receptor was determined by Western blot analysis. All mutant receptors showed several bands between ∼44 and ∼55 kDa (Fig. 4A). The multiple bands are typical of Ste2p expression and are due to differences in the glycosylation state, which does not influence receptor function [50]. While all of the Cys constructs were expressed as judged by the Western blot, there was a large variability in the amount of receptor expressed, the glycosylation pattern, and the ratio of monomer to dimer among these mutants (Fig. 4A). For example, the expression of the S108C mutant was the lowest and it showed very little dimerization (Fig 4A; fifth Gel from Left). Similar expression levels for this mutant were found previously [53].
Figure 4.
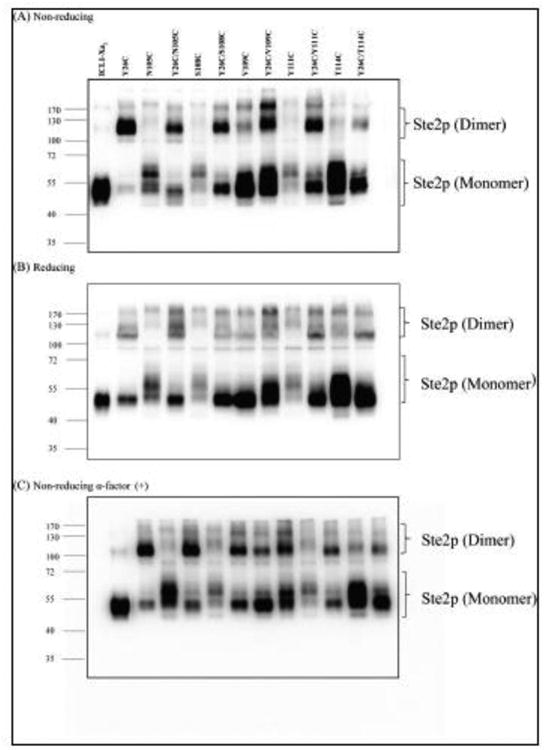
(A) Ste2p expression levels of various Cys mutants. Total membranes from cells expressing various mutants were prepared as described in Materials and Methods. The membrane preparations were fractionated by SDS-PAGE under non-reducing conditions; the gel was blotted and probed using antibody against the C-terminal FLAG epitope tag to detect the presence of Ste2p at either the monomer or dimer positions, (B) Effect of reducing agent (β-mercaptoethanol) on dimerization. (C) Effect of ligand binding on dimerization. Membranes were incubated with α-factor (1μM final concentration) prior to SDS-PAGE gel under non-reducing conditions and immunoblotting. Molecular mass markers (kDa) are indicated on the left-hand side. Blots were overexposed to be able to see the mutants. Blots with shorter exposure time were used for accurate quantification of Ste2p band intensity (Fig. 5).
Although the two intrinsic Cys residues have been substituted with Ser, a weak band at ∼110 kDa, corresponding to a dimerized form of Ste2p, was observed for the ICL1-Xa2 receptor (Lane ICL1-Xa2, Fig. 4A). This band is likely a native, noncovalent dimer which was not disrupted by membrane protein preparation or SDS-PAGE. Such dimers have been observed on SDS-PAGE gels in our lab [51, 59, 60, 73] and those of others working with Ste2p [74-78]. On the other hand, the Y26C mutant exhibited strong dimer formation with only a small amount of monomer detected (Lane Y26C, Fig. 4A). The single Cys mutants N105C, S108C, V109C, Y111C and T114C were present primarily in the monomeric form with just a small amount of dimer. With the exception of V109C, the monomeric banding pattern of the single Cys mutants (N105C, S108C, Y111C and T114C) was different from that of the ICL1-Xa2 control; the main bands were spread over a larger molecular size range in comparison to that of the ICL2-X2a, ranging in size from ∼55 and ∼70 kDa. The diffuse banding pattern of the N105C, S108C, Y111C and T114C mutants was attributed to changes in the glycosylation state of the receptors, as these bands collapsed into a single monomeric band upon treatment with PNGase F [53]. Furthermore, the Ste2p receptor expression levels of the four ECL1 single Cys mutants (N105C, S108C, Y111C and T114C) and the corresponding double Cys mutants (Y26C/N105C, Y26C/S108C, Y26C/Y111C, and Y26C/T114C) were weaker than that of the ICL1-Xa2 (Fig. 4A). Moreover, the banding pattern of the monomeric form of the double Cys mutants was different from that of the ICL1-Xa2 as exhibited by a strong band (at ∼50 kDa) along with a small amount of diffuse bands between ∼50 and ∼70 kDa. The differential banding pattern in the double Cys mutants can be attributed to mutations in ECL1 as observed previously in our lab [53].
The majority of the double Cys mutants exhibited more dimer and less monomer compared to the ICL1-Xa2 under non-reducing condition (Fig. 4A). As stated above, the ICL1-Xa2 and virtually all of the single Cys mutants of Ste2p exhibited no or faint bands at about 110 kDa consistent with a small amount of dimerized Ste2p. Since the dimer was the predominant form for the majority of the double Cys mutants containing Y26C, with weak dimer in the corresponding single Cys mutants, the dimers formed by these double mutants can be attributed to the presence of Y26C. The dimer at ∼110 kDa of all the double Cys mutants decreased in the presence of β-mercaptoethanol indicating the involvement of disulfide bonds in stabilization of the dimer (Compare Figs. 4A & B; See Table 2).
Table 2. Dimer to monomer ratio of Cys mutants in non-reducing and reducing conditions.
| Receptor | Dimer to Monomer Ratio under Non-reducing Conditionsa | Dimer to Monomer Ratio under Reducing Conditionsb |
|---|---|---|
| ICL1-Xa2 | 1.0 | 1.0 |
| Y26C* | 27.9 | 12.3 |
| N105C | 2.2 | 2.3 |
| Y26C/N105C* | 8.0 | 5.9 |
| S108C | 3.1 | 2.3 |
| Y26C/S108C* | 8.3 | 4.4 |
| V109C | 3.7 | 2.2 |
| Y26C/V109C* | 5.7 | 3.4 |
| Y111C | 4.4 | 3.2 |
| Y26C/Y111C* | 8.2 | 4.5 |
| T114C | 1.9 | 1.3 |
| Y26C/T114C* | 4.1 | 2.6 |
Relative dimer to monomer ratio of the Cys mutants as compared to that of the ICL1-Xa2 in the absence of β-ME.
Relative dimer to monomer ratio of the Cys mutants as compared to that of the ICL1-Xa2 in the presence of β-ME.
Significant difference (p<0.05) in dimer to monomer ratio of the receptors in non-reducing and reducing conditions is indicated by “*”.
The dimer to monomer ratio of the mutants of the all the mutants was normalized to that of the ICL1-Xa2. The standard deviation of the relative dimer to monomer ratio for all receptors was within ±0.6.
The dimer to monomer ratio in the gels under non-reducing conditions for Y26C was ∼28 fold greater than that of the ICL1-Xa2 control, whereas that of the double Cys mutants (Y26C/N105C, Y26C/S108C, Y26C/V109C, Y26C/Y111C and Y26C/T114C) ranged from about 4- to 8-fold greater (Table 2). The dimer to monomer ratio of the single Cys mutants ranged from 2-4 fold greater than that of the ICL1-Xa2 control. Under reducing conditions, the dimer to monomer ratio of the Y26C and all the double Cys mutants decreased significantly (p<0.05), whereas the ratio did not change significantly in any of the other single Cys mutants suggesting that the dimerization in the double Cys mutants required Y26C. The dimer to monomer ratios of three double Cys mutants (Y26C/N105C, Y26C/S108C and Y26C/Y111C) under non-reducing conditions ranged from 8.0-8.3, and were not significantly different from each other. On the other hand, under non-reducing conditions the ratios of the two other double Cys mutants, Y26C/V109C and Y26C/T114C, were 5.7 and 4.1, respectively. The differences between the Y26/N105C, Y26C/S108C and Y26C/Y111C double mutants and the Y26C/V109C and Y26C/T114C double mutants is addressed further in the discussion.
Conformational Changes in the N-terminus Upon Ligand Binding
It is generally believed that activation of GPCRs upon ligand binding results in a conformational change involving rearrangement of the various receptor domains [79-82]. Previous studies have also shown that binding of α-factor affected Ste2p dimer formation [52, 60, 78]. Additionally, the N-terminus of Ste2p was reported to be involved in dimerization in several studies [51, 52]. Disulfide crosslinking can also be used to probe ligand-induced conformational change in GPCRs in cysteine-substituted mutant receptors. [78, 83, 84]. We investigated whether incubation with α-factor would influence dimerization of the mutants examined in this study. We observed that dimer formation by the ICL1-Xa2 control and majority of the mutants was not affected by agonist. However, four mutants (Y26C, S108C, Y26C/N105C and Y26C/S108C) exhibited a significant difference in the ratio of dimer to monomer in the absence or presence of agonist (Compare Figs. 4A & C; see Fig. 5 for a graphical representation of the dimer/monomer ratio as determined from densitometry measurements; see Supplemental Fig. S1 and Table S1 for graphical representation of how the information in Fig. 5 was determined). The dimer to monomer ratio of Y26C significantly decreased upon incubation with α-factor. In contrast, the dimer to monomer ratio of S108C, Y26C/N105C and Y26C/S108C increased significantly. These results suggest that α-factor binding induces conformational changes in the N-terminus and ECL1 of Ste2p which alters the availability of the Y26C, S108C, Y26C/N105C and Y26C/S108C residues for cross-linking. These results indicate that the dimer interface formed by the N-terminus of the receptor changes upon receptor activation.
Figure 5.
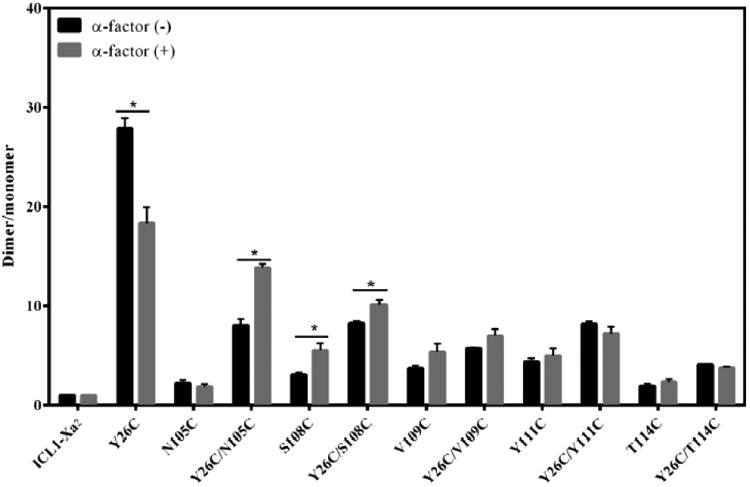
Effect of ligand binding on dimerization. Band intensity of dimer and monomer forms of Ste2p was quantified from Western blots (Figs. 4A & C) using Image Lab (version 4.1). The dimer/monomer ratio of the mutants was normalized to that of the ICL1-Xa2 (Labeled F-H-Xa in figure for Flag-His-Xa2 receptor). Black and grey bars represent the ratio of dimer to monomer in the absence or presence of α-factor, respectively. Statistical significance (p<0.05) in the dimer-monomer ratio is indicated by an asterisk.
Determination of intramolecular interaction between N-terminus and extracellular loop 1
The experiments described above indicated that dimerization mediated by Y26C was hindered by the V109C and T114C mutations in ECL1 suggesting a possible interaction between the N-terminal domain and the first extracellular loop through Y26C and these two positions in Ste2p. To test this idea, we took advantage of the protease (Factor Xa) digestion site engineered into ICL1 of Ste2p. If intramolecular cross-linking between Y26C and V109C or T114C occurred, subsequent Factor Xa digestion would result in a split Ste2p with the N-terminus linked by a disulfide bond to the ECL1 region (Fig. 6A, diagram of expected results). If cross-linking occurred, under non reducing conditions a protein with an apparent molecular weight equivalent to the full-length receptor (∼50 kDa) would be detected by antibody against the C-terminal FLAG epitope tag. Under reducing conditions, the disulfide bond between the two fragments would be reduced and only the 42 kDa fragment would appear on the gel probed with anti-FLAG antibody. If cross-linking did not occur, the receptor would be cut into two fragments, and only a 42 kDa band would be detected on immunoblots under non-reducing or reducing conditions using the C-terminal FLAG antibody (Fig. 6B).
Figure 6.
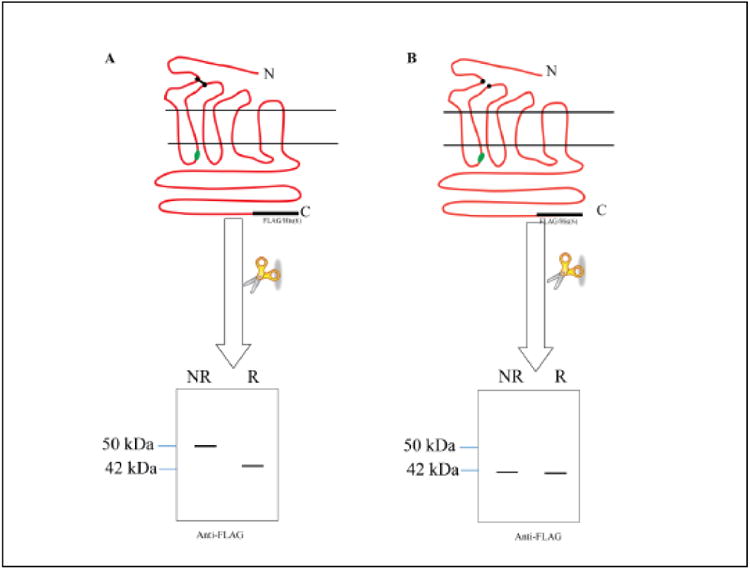
Diagram showing the predicted SDS-PAGE profile of Ste2p with and without disulfide crosslinking between the N-terminus and ECL1 after Factor Xa digestion as detected by immunoblot using antibody against the C-terminal FLAG epitope. Non-reducing and reducing conditions of the sample buffer are indicated by NR and R, respectively. The immunoblot on the left (A) shows the result of crosslinking between ECL1 and the N-terminus. The diagram of immunoblot on the right (B) would result from no N-terminal-ECL1 crosslinking. The N and C-termini of Ste2p are indicated by N and C, respectively. The XX in ICL1 indicates the location of the protease Factor Xa cleavage site. The FLAG and His epitope tags are shown in black.
Digestion of ICL1-Xa2 receptor with Factor Xa led to detection of a ∼42 kDa fragment, although most of the receptor was not cleaved by Factor Xa under these conditions (Fig. 7, see Supplemental Fig. S2 and TableS2 for graphical representation of how comparisons of fragment levels in Fig. 7 was determined.). The total amount of Ste2p detected in both the monomer and dimer (non-disulfide) forms also decreased due to non-specific proteolysis of Ste2p during the overnight incubation with Factor Xa (compare lanes 1 and 2 in Fig. 7A). The monomer bands (∼55 kDa) are due to incomplete Factor Xa digestion. We performed partial digests because a longer incubation led to very extensive degradation of proteins. In contrast, the ∼42 kDa fragment protease digestion fragment was not detected in Y26C/V109C and Y26C/T114C mutants (lanes 4 and 6 in Fig. 7A). Similar results were obtained when the receptors were incubated without (lanes 1-6) or with α-factor (lanes 7-12) before digestion with Factor Xa. In control experiments run under reducing conditions(β-mercaptoethanol) all receptors cut with Factor Xa showed the 42 KDa band (Figure 7B). These results are consistent with the formation of a disulfide bond between Y26C in the extracellular N-terminus and V109C and T114C in ECL1 of Ste2p and demonstrate that these positions are in close proximity in this receptor.
Figure 7.
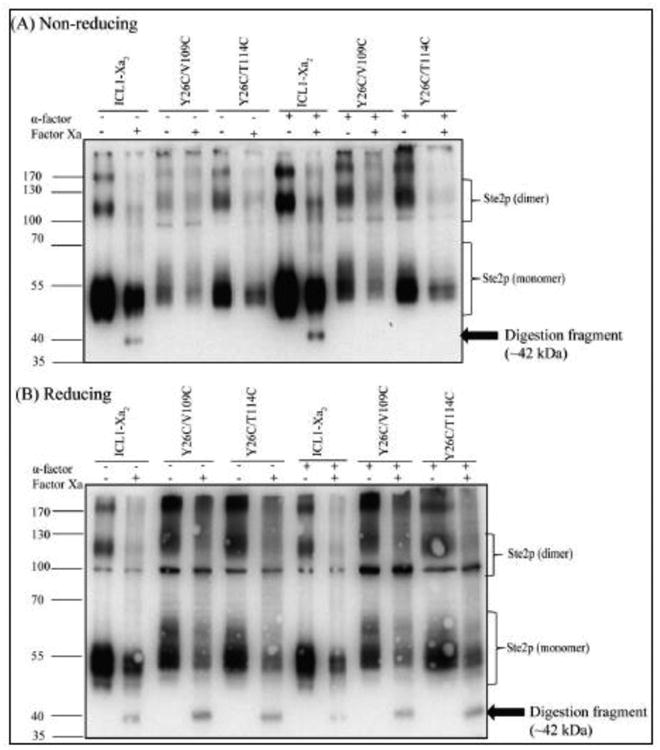
Factor Xa digestion. Membranes prepared from cells expressing the indicated receptors were prepared and digested as described in Materials and Methods. The samples were separated by SDS-PAGE in non-reducing (A) and reducing (B) conditions. The 42 kDa Ste2p fragment detected is marked with an arrow. The molecular markers are shown on the left.
Discussion
Previous studies indicated that the N-terminus of Ste2p is involved in dimerization and contains two glycosylation sites, and that mutation of specific residues in ECL1 affected the glycosylation pattern of the receptor. These results would be consistent with an interaction between the N-terminus and ECL1[51-53]. We now present evidence that a mutation of the highly conserved tyrosine residue to cysteine (Y26C) in the N-terminus will promote receptor dimerization and that Y26 interacts with specific residues (V109 and T114) in the ECL1. We also present data suggesting that the N-terminus-mediated dimer interface of the receptor changes upon receptor activation.
Disulfide cross-linking methodology was used to detect the interaction between two residues of Ste2p in its membrane-bound state. The maximum distance between α-carbons linked by disulfide bonds was shown to be about 7Å [85]. Thus, this method should only identify amino acid side chains that are within this distance. Cysteine residues engineered into GPCRs have been applied to facilitate disulfide bond formation in several GPCRs including Ste2p [60, 73, 78, 81, 86-90].
The mutant receptor Y26C showed significantly increased dimerization (Figs. 4A, 4B, Table 2) over that of the ICL1-Xa2 receptor (Fig. 2.). The finding that Y26C participates in dimer formation is in good agreement with the report that Y26C is solvent-inaccessible to the sulfhydryl reagent MTSEA-Biotin [51] since the Y26C-Y26C interaction likely prevents MTSEA-labeling of this residue. The fact that this residue formed a linkage suggests that the Ste2p-Ste2p interactions involving this region of the N-terminus have significant spatial restrictions, which might make this region relatively rigid. This is consistent with the prediction that this region of the receptor has a β strand [51, 91, 92]. Thus our mutational analysis defines Y26C as a specific residue involved in Ste2p dimerization.
It is interesting that dimerization mediated by Y26C in the N-terminus was hindered by mutations in the ECL1 of the receptor (Table 2). This is consistent with the idea that formation of disulfide bond between Y26C and a residue in the ECL1 decreases the amount of Y26C-Y26C dimer. This results in the reduction of dimer to monomer ratio of the corresponding double Cys mutant receptors. It had been shown previously that mutations in ECL1 affected the glycosylation pattern of the receptor [53], although the glycosylation sites are located in the N-terminus [50]. The mutant receptors Y26C, Y26C/N105C, Y26C/S108C, Y26C/V109C, Y26C/Y111C and Y26C/T114C exhibited markedly increased dimerization over that of the ICL1-Xa2 (Figs. 4A, 4B & Table 2). The dimerization of these mutants was reversed by treatment with β-ME. On the other hand, the single Cys mutants (N105C, S108C, V109C, Y111C and T114C) exhibited weaker dimerization as compared to the double Cys mutants (Figs. 4A, 4B & Table 2) and no significant decrease in dimer to monomer ratio was observed upon treatment with β-ME, indicating that the small amount of dimers formed by these single Cys mutants was due to SDS-resistant association between receptors that is not mediated by disulfide bonds. SDS-resistant dimerization has been observed in previous studies with many proteins including Ste2p [77, 93-101].
Out of the five double Cys mutants tested, two mutants (Y26C/V109C and Y26C/T114C) exhibited a greater decrease in dimerization compared to that of the other three mutants (Y26C/N105C, Y26C/S108C and Y26C/Y111C) indicating that Y26C-mediated dimerization was prevented to a greater extent by cysteine incorporation at positions V109 or T114. These results are consistent with the idea that intra-receptor interactions of Y26C with either V109C or T114C will hinder inter-receptor Y26C-Y26C interactions thus reducing dimerization. The other positions (N105C, S108C, Y111C) appear to interact to a somewhat lesser extent with Y26C and thus the intermolecular Y26C-Y26C interaction is not as greatly affected. These results support the hypothesis that Y26 in the flexible N-terminus interacts with residues of EL1. We note, however, that the flexibility of the N-terminus would change if dimerization occurs and that the competition between intramolecular and intermolecular interactions would influence the flexibility of this region. Since Y26C is located adjacent to a glycosylation site (N25), it is expected that mutations blocking the interaction might influence the glycosylation pattern. Indeed, it was observed previously that mutation in positions T114, N105, S108, and Y111 affect the glycosylation pattern [53].
It is important to note that reduced dimerization of Y26C/V109C and Y26C/T114C might result from non-specific effects of mutation rather than interaction between the N-terminus and ECL1. To ascertain if specific interactions between Y26C and the two residues in ECL1 existed, we used disulfide cross-linking followed by Factor Xa digestion. We found that these two residues (V109C and T114C) in ECL1 indeed cross-link to Y26C (Fig.8). This strategy has been used previously in our lab to determine the involvement of TM regions in dimerization [60]. In a recent report we showed that the N-terminus plays an important role in the regulation of Ste2p signaling [69]. The finding herein begins to reveal at the residue level a possible basis for this regulatory control.
Figure 8.
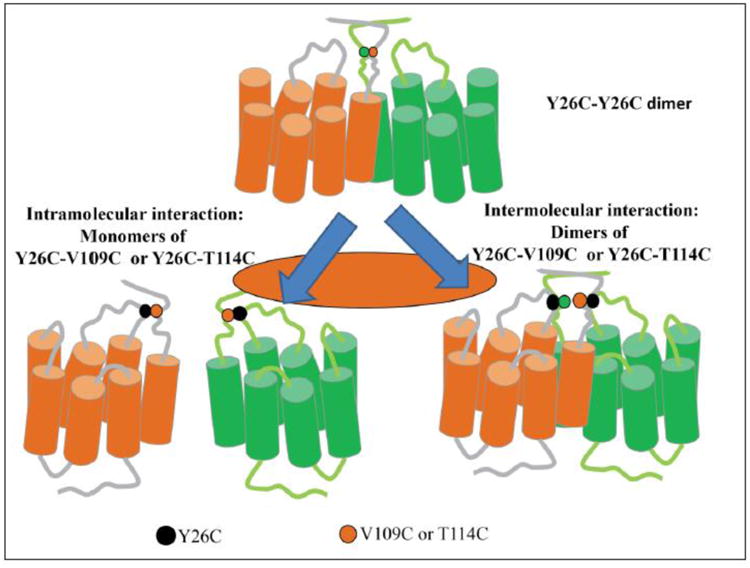
Ste2p dimer mediated by Y26 and its interaction with ECL1. Two Ste2p molecules (orange and green) are shown with the Y26 positions (green and orange dots), V109 (black) and T114 (black) indicated. Intramolecular and intermolecular interactions are shown on the left and right panels, respectively.
We also report here that the N-terminus of Ste2p is involved in a dimer interface that changes upon receptor activation. Specifically, dimerization of Y26C, S108C, Y26C/N105C, and Y26C/S108C was found to change in the presence of α-factor (Fig. 5). The dimerization mediated by Y26C was found to decrease in the presence of α-factor. However, the dimerization mediated by S108C, Y26C/N105C and Y26C/S108C mutants was found to increase. Previous studies in our lab demonstrated that solvent accessibility of several residues (Y101, Y106, and A112) in ECL1 changes upon incubation with α-factor thereby indicating the involvement of this region in receptor activation [53]. A 310 helix was also predicted between residues 106-114 in the ECL1. The results in this study suggest that Y26 interacts with residues that are part of this 310 helix. This is certainly consistent with the periodicity that we observed in the Y26 interactions where mutation at residues 105, 108 and 111 leads to one phenotype and mutation of residue 109 results in a different phenotype. It is possible that in the presence of α-factor, the N-terminus and/or ECL1 undergoes conformational change affecting Y26-mediated dimerization.
A growing number of studies have demonstrated that GPCRs form dimers or higher-ordered oligomers, which have been proposed to be essential for modulation of receptor function [83, 102-108]. In most receptors, the transmembrane domains were reported to be involved in receptor dimerization/oligomerization. However, several studies demonstrated the extracellular N-terminal domain of Ste2p is also associated with dimerization [51, 52]. The residue identified in this study, Y26C, is highly conserved in fungal GPCRs. Conserved residues are often important for structure and function of proteins and conservation is stronger at protein-protein interfaces compared to elsewhere on the protein surface [109-112]. Thus, analysis of sequence conservation in a protein family is a useful strategy to identify key residues that are important for protein function [113-122] as protein-protein interaction sites are subjected to substantial selective pressure throughout the course of evolution [123, 124].
These findings provide valuable information relating to the arrangement of the receptor in which the N-termini of two receptors appear to face each other. In the absence of a crystal structure for Ste2p, the disulfide cross-linking results contributes to understanding structural features of the functional receptor such as inter-amino terminal interactions that may be involved in oligomerization.
Supplementary Material
Highlights.
Dimerization of Ste2p is greatly increased when Y26 in the N-terminus is mutated to Y26C.
The amount of dimer formed by the Y26C mutant is greatly reduced by ligand binding and is consistent with a conformational change in the N-terminus of the receptor upon activation.
Dimerization was decreased by double mutations Y26C/V109C or Y26C/T114C indicating that Y26 is in close proximity to V109 and T1 14 of extracellular loop 1 in native Ste2p.
The results indicate previously unrecognized roles for the N-terminus of Ste2p, and perhaps of GPCRs in general, and pinpoint a specific N-terminus residue involved in GPCR signaling, intramolecular interactions, and receptor dimerization.
Acknowledgments
This work was supported by Grant GM22087 from the National Institute of General Medical Sciences, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 2.O'Hayre M, Vazquez-Prado J, Kufareva I, Stawiski EW, Handel TM, Seshagiri S, Gutkind JS. The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer. Nat Rev Cancer. 2013;13:412–424. doi: 10.1038/nrc3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Periole X. Interplay of G Protein-Coupled Receptors with the Membrane: Insights from Supra-Atomic Coarse Grain Molecular Dynamics Simulations. Chem Rev. 2017;117:156–185. doi: 10.1021/acs.chemrev.6b00344. [DOI] [PubMed] [Google Scholar]
- 4.Granier S, Kobilka B. A new era of GPCR structural and chemical biology. Nature chemical biology. 2012;8:670–673. doi: 10.1038/nchembio.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nature reviews Molecular cell biology. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 6.Thathiah A, De Strooper B. The role of G protein-coupled receptors in the pathology of Alzheimer's disease. Nat Rev Neurosci. 2011;12:73–87. doi: 10.1038/nrn2977. [DOI] [PubMed] [Google Scholar]
- 7.Wang M, Pei L, Fletcher PJ, Kapur S, Seeman P, Liu F. Schizophrenia, amphetamine-induced sensitized state and acute amphetamine exposure all show a common alteration: increased dopamine D2 receptor dimerization. Mol Brain. 2010;3:25. doi: 10.1186/1756-6606-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrieri A, Perez-Nueno VI, Lentini G, Ritchie DW. Recent trends and future prospects in computational GPCR drug discovery: from virtual screening to polypharmacology. Curr Top Med Chem. 2013;13:1069–1097. doi: 10.2174/15680266113139990028. [DOI] [PubMed] [Google Scholar]
- 9.McNeely PM, Naranjo AN, Robinson AS. Structure-function studies with G protein-coupled receptors as a paradigm for improving drug discovery and development of therapeutics. Biotechnology journal. 2012;7:1451–1461. doi: 10.1002/biot.201200076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Latorraca NR, Venkatakrishnan AJ, Dror RO. GPCR Dynamics: Structures in Motion. Chem Rev. 2017;117:139–155. doi: 10.1021/acs.chemrev.6b00177. [DOI] [PubMed] [Google Scholar]
- 11.Heifetz A, James T, Morao I, Bodkin MJ, Biggin PC. Guiding lead optimization with GPCR structure modeling and molecular dynamics. Curr Opin Pharmacol. 2016;30:14–21. doi: 10.1016/j.coph.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Cavasotto CN, Palomba D. Expanding the horizons of G protein-coupled receptor structure-based ligand discovery and optimization using homology models. Chem Commun (Camb) 2015;51:13576–13594. doi: 10.1039/c5cc05050b. [DOI] [PubMed] [Google Scholar]
- 13.Lefkowitz RJ, Shenoy SK. Transduction of receptor signals by beta-arrestins. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- 14.Zhang D, Zhao Q, Wu B. Structural Studies of G Protein-Coupled Receptors. Mol Cells. 2015;38:836–842. doi: 10.14348/molcells.2015.0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobilka BK, Kobilka TS, Daniel K, Regan JW, Caron MG, Lefkowitz RJ. Chimeric alpha 2-,beta 2-adrenergic receptors: delineation of domains involved in effector coupling and ligand binding specificity. Science. 1988;240:1310–1316. doi: 10.1126/science.2836950. [DOI] [PubMed] [Google Scholar]
- 16.Schoneberg T, Liu J, Wess J. Plasma membrane localization and functional rescue of truncated forms of a G protein-coupled receptor. The Journal of biological chemistry. 1995;270:18000–18006. doi: 10.1074/jbc.270.30.18000. [DOI] [PubMed] [Google Scholar]
- 17.Filipek S, Stenkamp RE, Teller DC, Palczewski K. G protein-coupled receptor rhodopsin: a prospectus. Annual review of physiology. 2003;65:851–879. doi: 10.1146/annurev.physiol.65.092101.142611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hargrave PA, Hamm HE, Hofmann KP. Interaction of rhodopsin with the G-protein, transducin. BioEssays : news and reviews in molecular, cellular and developmental biology. 1993;15:43–50. doi: 10.1002/bies.950150107. [DOI] [PubMed] [Google Scholar]
- 19.Khorana HG. Rhodopsin, photoreceptor of the rod cell. An emerging pattern for structure and function. The Journal of biological chemistry. 1992;267:1–4. [PubMed] [Google Scholar]
- 20.Rao VR, Cohen GB, Oprian DD. Rhodopsin mutation G90D and a molecular mechanism for congenital night blindness. Nature. 1994;367:639–642. doi: 10.1038/367639a0. [DOI] [PubMed] [Google Scholar]
- 21.Gether U, Lin S, Ghanouni P, Ballesteros JA, Weinstein H, Kobilka BK. Agonists induce conformational changes in transmembrane domains III and VI of the beta2 adrenoceptor. EMBO J. 1997;16:6737–6747. doi: 10.1093/emboj/16.22.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kristiansen K. Molecular mechanisms of ligand binding, signaling, and regulation within the superfamily of G-protein-coupled receptors: molecular modeling and mutagenesis approaches to receptor structure and function. Pharmacology & therapeutics. 2004;103:21–80. doi: 10.1016/j.pharmthera.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Kobilka BK. G protein coupled receptor structure and activation. Biochimica et biophysica acta. 2007;1768:794–807. doi: 10.1016/j.bbamem.2006.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nature reviews Drug discovery. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 25.Ersoy BA, Pardo L, Zhang S, Thompson DA, Millhauser G, Govaerts C, Vaisse C. Mechanism of N-terminal modulation of activity at the melanocortin-4 receptor GPCR. Nature chemical biology. 2012;8:725–730. doi: 10.1038/nchembio.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coleman JL, Ngo T, Smith NJ. The G protein-coupled receptor N-terminus and receptor signalling: N-tering a new era. Cellular signalling. 2017 doi: 10.1016/j.cellsig.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Pydi SP, Chakraborty R, Bhullar RP, Chelikani P. Role of rhodopsin N-terminus in structure and function of rhodopsin-bitter taste receptor chimeras. Biochem Biophys Res Commun. 2013;430:179–182. doi: 10.1016/j.bbrc.2012.11.029. [DOI] [PubMed] [Google Scholar]
- 28.Beinborn M. Class B GPCRs: a hidden agonist within? Molecular pharmacology. 2006;70:1–4. doi: 10.1124/mol.106.025932. [DOI] [PubMed] [Google Scholar]
- 29.Dong M, Pinon DI, Asmann YW, Miller LJ. Possible endogenous agonist mechanism for the activation of secretin family G protein-coupled receptors. Molecular pharmacology. 2006;70:206–213. doi: 10.1124/mol.105.021840. [DOI] [PubMed] [Google Scholar]
- 30.Kobilka BK, Deupi X. Conformational complexity of G-protein-coupled receptors. Trends in pharmacological sciences. 2007;28:397–406. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 31.Scarborough RM, Naughton MA, Teng W, Hung DT, Rose J, Vu TK, Wheaton VI, Turck CW, Coughlin SR. Tethered ligand agonist peptides. Structural requirements for thrombin receptor activation reveal mechanism of proteolytic unmasking of agonist function. The Journal of biological chemistry. 1992;267:13146–13149. [PubMed] [Google Scholar]
- 32.Vassart G, Pardo L, Costagliola S. A molecular dissection of the glycoprotein hormone receptors. Trends in biochemical sciences. 2004;29:119–126. doi: 10.1016/j.tibs.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 33.Paavola KJ, Stephenson JR, Ritter SL, Alter SP, Hall RA. The N terminus of the adhesion G protein-coupled receptor GPR56 controls receptor signaling activity. The Journal of biological chemistry. 2011;286:28914–28921. doi: 10.1074/jbc.M111.247973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersson H, D'Antona AM, Kendall DA, Von Heijne G, Chin CN. Membrane assembly of the cannabinoid receptor 1: impact of a long N-terminal tail. Molecular pharmacology. 2003;64:570–577. doi: 10.1124/mol.64.3.570. [DOI] [PubMed] [Google Scholar]
- 35.Hague C, Chen Z, Pupo AS, Schulte NA, Toews ML, Minneman KP. The N terminus of the human alpha1D-adrenergic receptor prevents cell surface expression. The Journal of pharmacology and experimental therapeutics. 2004;309:388–397. doi: 10.1124/jpet.103.060509. [DOI] [PubMed] [Google Scholar]
- 36.Dunham JH, Meyer RC, Garcia EL, Hall RA. GPR37 surface expression enhancement via N-terminal truncation or protein-protein interactions. Biochemistry. 2009;48:10286–10297. doi: 10.1021/bi9013775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fay JF, Farrens DL. The membrane proximal region of the cannabinoid receptor CB1 N-terminus can allosterically modulate ligand affinity. Biochemistry. 2013;52:8286–8294. doi: 10.1021/bi400842k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith NJ, Milligan G. Allostery at G protein-coupled receptor homo- and heteromers: uncharted pharmacological landscapes. Pharmacological reviews. 2010;62:701–725. doi: 10.1124/pr.110.002667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chun L, Zhang WH, Liu JF. Structure and ligand recognition of class C GPCRs. Acta pharmacologica Sinica. 2012;33:312–323. doi: 10.1038/aps.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dupre DJ, Hammad MM, Holland P, Wertman J. Role of chaperones in G protein coupled receptor signaling complex assembly. Subcell Biochem. 2012;63:23–42. doi: 10.1007/978-94-007-4765-4_2. [DOI] [PubMed] [Google Scholar]
- 41.Prezeau L, Rives ML, Comps-Agrar L, Maurel D, Kniazeff J, Pin JP. Functional crosstalk between GPCRs: with or without oligomerization. Curr Opin Pharmacol. 2010;10:6–13. doi: 10.1016/j.coph.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 42.Scarselli M, Novi F, Schallmach E, Lin R, Baragli A, Colzi A, Griffon N, Corsini GU, Sokoloff P, Levenson R, Vogel Z, Maggio R. D2/D3 dopamine receptor heterodimers exhibit unique functional properties. The Journal of biological chemistry. 2001;276:30308–30314. doi: 10.1074/jbc.M102297200. [DOI] [PubMed] [Google Scholar]
- 43.Muller TD, Muller A, Yi CX, Habegger KM, Meyer CW, Gaylinn BD, Finan B, Heppner K, Trivedi C, Bielohuby M, Abplanalp W, Meyer F, Piechowski CL, Pratzka J, Stemmer K, Holland J, Hembree J, Bhardwaj N, Raver C, Ottaway N, Krishna R, Sah R, Sallee FR, Woods SC, Perez-Tilve D, Bidlingmaier M, Thorner MO, Krude H, Smiley D, DiMarchi R, Hofmann S, Pfluger PT, Kleinau G, Biebermann H, Tschop MH. The orphan receptor Gpr83 regulates systemic energy metabolism via ghrelin-dependent and ghrelin-independent mechanisms. Nat Commun. 2013;4:1968. doi: 10.1038/ncomms2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stephens B, Handel TM. Chemokine receptor oligomerization and allostery. Prog Mol Biol Transl Sci. 2013;115:375–420. doi: 10.1016/B978-0-12-394587-7.00009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bardwell L. A walk-through of the yeast mating pheromone response pathway. Peptides. 2005;26:339–350. doi: 10.1016/j.peptides.2004.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alvaro CG, Thorner J. Heterotrimeric G Protein-coupled Receptor Signaling in Yeast Mating Pheromone Response. The Journal of biological chemistry. 2016;291:7788–7795. doi: 10.1074/jbc.R116.714980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dowell SJ, Brown AJ. Yeast assays for G protein-coupled receptors. Methods Mol Biol. 2009;552:213–229. doi: 10.1007/978-1-60327-317-6_15. [DOI] [PubMed] [Google Scholar]
- 48.King K, Dohlman HG, Thorner J, Caron MG, Lefkowitz RJ. Control of yeast mating signal transduction by a mammalian beta 2-adrenergic receptor and Gs alpha subunit. Science. 1990;250:121–123. doi: 10.1126/science.2171146. [DOI] [PubMed] [Google Scholar]
- 49.Yin D, Gavi S, Shumay E, Duell K, Konopka JB, Malbon CC, Wang HY. Successful expression of a functional yeast G-protein-coupled receptor (Ste2) in mammalian cells. Biochem Biophys Res Commun. 2005;329:281–287. doi: 10.1016/j.bbrc.2005.01.130. [DOI] [PubMed] [Google Scholar]
- 50.Mentesana PE, Konopka JB. Mutational analysis of the role of N-glycosylation in alpha-factor receptor function. Biochemistry. 2001;40:9685–9694. doi: 10.1021/bi0108507. [DOI] [PubMed] [Google Scholar]
- 51.Uddin MS, Kim H, Deyo A, Naider F, Becker JM. Identification of residues involved in homodimer formation located within a beta-strand region of the N-terminus of a Yeast G protein-coupled receptor. Journal of receptor and signal transduction research. 2012;32:65–75. doi: 10.3109/10799893.2011.647352. [DOI] [PubMed] [Google Scholar]
- 52.Overton MC, Blumer KJ. The extracellular N-terminal domain and transmembrane domains 1 and 2 mediate oligomerization of a yeast G protein-coupled receptor. The Journal of biological chemistry. 2002;277:41463–41472. doi: 10.1074/jbc.M205368200. [DOI] [PubMed] [Google Scholar]
- 53.Hauser M, Kauffman S, Lee BK, Naider F, Becker JM. The first extracellular loop of the Saccharomyces cerevisiae G protein-coupled receptor Ste2p undergoes a conformational change upon ligand binding. The Journal of biological chemistry. 2007;282:10387–10397. doi: 10.1074/jbc.M608903200. [DOI] [PubMed] [Google Scholar]
- 54.Marsh L. Substitutions in the hydrophobic core of the alpha-factor receptor of Saccharomyces cerevisiae permit response to Saccharomyces kluyveri alpha-factor and to antagonist. Molecular and cellular biology. 1992;12:3959–3966. doi: 10.1128/mcb.12.9.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Son CD, Sargsyan H, Naider F, Becker JM. Identification of ligand binding regions of the Saccharomyces cerevisiae alpha-factor pheromone receptor by photoaffinity cross-linking. Biochemistry. 2004;43:13193–13203. doi: 10.1021/bi0496889. [DOI] [PubMed] [Google Scholar]
- 56.Turcatti G, Nemeth K, Edgerton MD, Meseth U, Talabot F, Peitsch M, Knowles J, Vogel H, Chollet A. Probing the structure and function of the tachykinin neurokinin-2 receptor through biosynthetic incorporation of fluorescent amino acids at specific sites. The Journal of biological chemistry. 1996;271:19991–19998. doi: 10.1074/jbc.271.33.19991. [DOI] [PubMed] [Google Scholar]
- 57.Sherman F. Getting started with yeast. Methods in enzymology. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 58.Huang LY, Umanah G, Hauser M, Son C, Arshava B, Naider F, Becker JM. Unnatural amino acid replacement in a yeast G protein-coupled receptor in its native environment. Biochemistry. 2008;47:5638–5648. doi: 10.1021/bi701866e. [DOI] [PubMed] [Google Scholar]
- 59.Umanah GK, Huang L, Ding FX, Arshava B, Farley AR, Link AJ, Naider F, Becker JM. Identification of residue-to-residue contact between a peptide ligand and its G protein-coupled receptor using periodate-mediated dihydroxyphenylalanine cross-linking and mass spectrometry. The Journal of biological chemistry. 2010;285:39425–39436. doi: 10.1074/jbc.M110.149500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim H, Lee BK, Naider F, Becker JM. Identification of specific transmembrane residues and ligand-induced interface changes involved in homo-dimer formation of a yeast G protein-coupled receptor. Biochemistry. 2009;48:10976–10987. doi: 10.1021/bi901291c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.David NE, Gee M, Andersen B, Naider F, Thorner J, Stevens RC. Expression and purification of the Saccharomyces cerevisiae alpha-factor receptor (Ste2p), a 7-transmembrane-segment G protein-coupled receptor. The Journal of biological chemistry. 1997;272:15553–15561. doi: 10.1074/jbc.272.24.15553. [DOI] [PubMed] [Google Scholar]
- 62.Pinson B, Chevallier J, Urban-Grimal D. Only one of the charged amino acids located in membrane-spanning regions is important for the function of the Saccharomyces cerevisiae uracil permease. The Biochemical journal. 1999;339(Pt 1):37–42. [PMC free article] [PubMed] [Google Scholar]
- 63.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 64.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular systems biology. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trueheart J, Boeke JD, Fink GR. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Molecular and cellular biology. 1987;7:2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McCaffrey G, Clay FJ, Kelsay K, Sprague GF., Jr Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. Molecular and cellular biology. 1987;7:2680–2690. doi: 10.1128/mcb.7.8.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dube P, DeCostanzo A, Konopka JB. Interaction between transmembrane domains five and six of the alpha -factor receptor. The Journal of biological chemistry. 2000;275:26492–26499. doi: 10.1074/jbc.M002767200. [DOI] [PubMed] [Google Scholar]
- 68.Parrish W, Eilers M, Ying W, Konopka JB. The cytoplasmic end of transmembrane domain 3 regulates the activity of the Saccharomyces cerevisiae G-protein-coupled alpha-factor receptor. Genetics. 2002;160:429–443. doi: 10.1093/genetics/160.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Uddin MS, Hauser M, Naider F, Becker JM. The N-terminus of the yeast G protein-coupled receptor Ste2p plays critical roles in surface expression, signaling, and negative regulation. Biochimica et biophysica acta. 2016;1858:715–724. doi: 10.1016/j.bbamem.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marsh L, Neiman AM, Herskowitz I. ignal transduction during pheromone response in yeast. Annu Rev Cell Biol. 1991;7:699–728. doi: 10.1146/annurev.cb.07.110191.003411. [DOI] [PubMed] [Google Scholar]
- 71.Kenakin T, Christopoulos A. Signalling bias in new drug discovery: detection, quantification and therapeutic impact. Nature reviews Drug discovery. 2013;12:205–216. doi: 10.1038/nrd3954. [DOI] [PubMed] [Google Scholar]
- 72.Luttrell LM. GPCR Signaling Rides a Wave of Conformational Changes. Cell. 2016;167:602–603. doi: 10.1016/j.cell.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 73.Umanah GK, Huang LY, Maccarone JM, Naider F, Becker JM. Changes in conformation at the cytoplasmic ends of the fifth and sixth transmembrane helices of a yeast G protein-coupled receptor in response to ligand binding. Biochemistry. 2011;50:6841–6854. doi: 10.1021/bi200254h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gehret AU, Bajaj A, Naider F, Dumont ME. Oligomerization of the yeast alpha-factor receptor: implications for dominant negative effects of mutant receptors. The Journal of biological chemistry. 2006;281:20698–20714. doi: 10.1074/jbc.M513642200. [DOI] [PubMed] [Google Scholar]
- 75.Blumer KJ, Reneke JE, Thorner J. The STE2 gene product is the ligand-binding component of the alpha-factor receptor of Saccharomyces cerevisiae. The Journal of biological chemistry. 1988;263:10836–10842. [PubMed] [Google Scholar]
- 76.Shi C, Paige MF, Maley J, Loewen MC. In vitro characterization of ligand-induced oligomerization of the S. cerevisiae G-protein coupled receptor, Ste2p. Biochimica et biophysica acta. 2009;1790:1–7. doi: 10.1016/j.bbagen.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 77.Yesilaltay A, Jenness DD. Homo-oligomeric complexes of the yeast alpha-factor pheromone receptor are functional units of endocytosis. Mol Biol Cell. 2000;11:2873–2884. doi: 10.1091/mbc.11.9.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang HX, Konopka JB. Identification of amino acids at two dimer interface regions of the alpha-factor receptor (Ste2) Biochemistry. 2009;48:7132–7139. doi: 10.1021/bi900424h. [DOI] [PubMed] [Google Scholar]
- 79.Bukusoglu G, Jenness DD. Agonist-specific conformational changes in the yeast alpha-factor pheromone receptor. Molecular and cellular biology. 1996;16:4818–4823. doi: 10.1128/mcb.16.9.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 81.Zeng FY, Hopp A, Soldner A, Wess J. Use of a disulfide cross-linking strategy to study muscarinic receptor structure and mechanisms of activation. The Journal of biological chemistry. 1999;274:16629–16640. doi: 10.1074/jbc.274.23.16629. [DOI] [PubMed] [Google Scholar]
- 82.Luo X, Zhang D, Weinstein H. Ligand-induced domain motion in the activation mechanism of a G-protein-coupled receptor. Protein engineering. 1994;7:1441–1448. doi: 10.1093/protein/7.12.1441. [DOI] [PubMed] [Google Scholar]
- 83.Bouvier M. Oligomerization of G-protein-coupled transmitter receptors. Nat Rev Neurosci. 2001;2:274–286. doi: 10.1038/35067575. [DOI] [PubMed] [Google Scholar]
- 84.Park PS, Filipek S, Wells JW, Palczewski K. Oligomerization of G protein-coupled receptors: past, present, and future. Biochemistry. 2004;43:15643–15656. doi: 10.1021/bi047907k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Srinivasan N, Sowdhamini R, Ramakrishnan C, Balaram P. Conformations of disulfide bridges in proteins. International journal of peptide and protein research. 1990;36:147–155. doi: 10.1111/j.1399-3011.1990.tb00958.x. [DOI] [PubMed] [Google Scholar]
- 86.Li JH, Han SJ, Hamdan FF, Kim SK, Jacobson KA, Bloodworth LM, Zhang X, Wess J. Distinct structural changes in a G protein-coupled receptor caused by different classes of agonist ligands. The Journal of biological chemistry. 2007;282:26284–26293. doi: 10.1074/jbc.M704875200. [DOI] [PubMed] [Google Scholar]
- 87.Yu H, Kono M, McKee TD, Oprian DD. A general method for mapping tertiary contacts between amino acid residues in membrane-embedded proteins. Biochemistry. 1995;34:14963–14969. doi: 10.1021/bi00046a002. [DOI] [PubMed] [Google Scholar]
- 88.Li JH, Hamdan FF, Kim SK, Jacobson KA, Zhang X, Han SJ, Wess J. Ligand-specific changes in M3 muscarinic acetylcholine receptor structure detected by a disulfide scanning strategy. Biochemistry. 2008;47:2776–2788. doi: 10.1021/bi7019113. [DOI] [PubMed] [Google Scholar]
- 89.Ward SD, Hamdan FF, Bloodworth LM, Siddiqui NA, Li JH, Wess J. Use of an in situ disulfide cross-linking strategy to study the dynamic properties of the cytoplasmic end of transmembrane domain VI of the M3 muscarinic acetylcholine receptor. Biochemistry. 2006;45:676–685. doi: 10.1021/bi051503q. [DOI] [PubMed] [Google Scholar]
- 90.Ward SD, Hamdan FF, Bloodworth LM, Wess J. Conformational changes that occur during M3 muscarinic acetylcholine receptor activation probed by the use of an in situ disulfide cross-linking strategy. The Journal of biological chemistry. 2002;277:2247–2257. doi: 10.1074/jbc.M107647200. [DOI] [PubMed] [Google Scholar]
- 91.Shi C, Kaminskyj S, Caldwell S, Loewen MC. A role for a complex between activated G protein-coupled receptors in yeast cellular mating. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:5395–5400. doi: 10.1073/pnas.0608219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shi C, Kendall SC, Grote E, Kaminskyj S, Loewen MC. N-terminal residues of the yeast pheromone receptor, Ste2p, mediate mating events independently of G1-arrest signaling. Journal of cellular biochemistry. 2009;107:630–638. doi: 10.1002/jcb.22129. [DOI] [PubMed] [Google Scholar]
- 93.Bender AT, Nakatsuka M, Osawa Y. Heme insertion, assembly, and activation of apo-neuronal nitric-oxide synthase in vitro. The Journal of biological chemistry. 2000;275:26018–26023. doi: 10.1074/jbc.275.34.26018. [DOI] [PubMed] [Google Scholar]
- 94.Devost D, Zingg HH. Identification of dimeric and oligomeric complexes of the human oxytocin receptor by co-immunoprecipitation and bioluminescence resonance energy transfer. J Mol Endocrinol. 2003;31:461–471. doi: 10.1677/jme.0.0310461. [DOI] [PubMed] [Google Scholar]
- 95.Gentile F, Amodeo P, Febbraio F, Picaro F, Motta A, Formisano S, Nucci R. SDS-resistant active and thermostable dimers are obtained from the dissociation of homotetrameric beta-glycosidase from hyperthermophilic Sulfolobus solfataricus in SDS. Stabilizing role of the A-C intermonomeric interface. The Journal of biological chemistry. 2002;277:44050–44060. doi: 10.1074/jbc.M206761200. [DOI] [PubMed] [Google Scholar]
- 96.Habisch HJ, Gorren AC, Liang H, Venema RC, Parkinson JF, Schmidt K, Mayer B. Pharmacological interference with dimerization of human neuronal nitric-oxide synthase expressed in adenovirus-infected DLD-1 cells. Molecular pharmacology. 2003;63:682–689. doi: 10.1124/mol.63.3.682. [DOI] [PubMed] [Google Scholar]
- 97.Hebert TE, Moffett S, Morello JP, Loisel TP, Bichet DG, Barret C, Bouvier M. A peptide derived from a beta2-adrenergic receptor transmembrane domain inhibits both receptor dimerization and activation. The Journal of biological chemistry. 1996;271:16384–16392. doi: 10.1074/jbc.271.27.16384. [DOI] [PubMed] [Google Scholar]
- 98.Klatt P, Schmidt K, Lehner D, Glatter O, Bachinger HP, Mayer B. Structural analysis of porcine brain nitric oxide synthase reveals a role for tetrahydrobiopterin and L-arginine in the formation of an SDS-resistant dimer. EMBO J. 1995;14:3687–3695. doi: 10.1002/j.1460-2075.1995.tb00038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kolodziejski PJ, Rashid MB, Eissa NT. Intracellular formation of “undisruptable” dimers of inducible nitric oxide synthase, Proceedings of the National Academy of Sciences of the United States of America. 2003;100:14263–14268. doi: 10.1073/pnas.2435290100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Luders J, Pyrowolakis G, Jentsch S. The ubiquitin-like protein HUB1 forms SDS-resistant complexes with cellular proteins in the absence of ATP. EMBO Rep. 2003;4:1169–1174. doi: 10.1038/sj.embor.7400025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Overton MC, Blumer KJ. G-protein-coupled receptors function as oligomers in vivo. Curr Biol. 2000;10:341–344. doi: 10.1016/s0960-9822(00)00386-9. [DOI] [PubMed] [Google Scholar]
- 102.Milligan G. G protein-coupled receptor dimerisation: molecular basis and relevance to function. Biochimica et biophysica acta. 2007;1768:825–835. doi: 10.1016/j.bbamem.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 103.Devi LA. Heterodimerization of G-protein-coupled receptors: pharmacology, signaling and trafficking. Trends in pharmacological sciences. 2001;22:532–537. doi: 10.1016/s0165-6147(00)01799-5. [DOI] [PubMed] [Google Scholar]
- 104.George SR, O'Dowd BF, Lee SP. G-protein-coupled receptor oligomerization and its potential for drug discovery. Nature reviews Drug discovery. 2002;1:808–820. doi: 10.1038/nrd913. [DOI] [PubMed] [Google Scholar]
- 105.Milligan G. G protein-coupled receptor dimerization: function and ligand pharmacology. Molecular pharmacology. 2004;66:1–7. doi: 10.1124/mol.104.000497.. [DOI] [PubMed] [Google Scholar]
- 106.Milligan G. A day in the life of a G protein-coupled receptor: the contribution to function of G protein-coupled receptor dimerization. British journal of pharmacology. 2008;153(1):S216–229. doi: 10.1038/sj.bjp.0707490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Maurice P, Kamal M, Jockers R. Asymmetry of GPCR oligomers supports their functional relevance. Trends in pharmacological sciences. 2011;32:514–520. doi: 10.1016/j.tips.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 108.Ferre S, Ciruela F, Woods AS, Lluis C, Franco R. Functional relevance of neurotransmitter receptor heteromers in the central nervous system. Trends in neurosciences. 2007;30:440–446. doi: 10.1016/j.tins.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 109.Bordner AJ, Abagyan R. Statistical analysis and prediction of protein-protein interfaces. Proteins. 2005;60:353–366. doi: 10.1002/prot.20433. [DOI] [PubMed] [Google Scholar]
- 110.Caffrey DR, Somaroo S, Hughes JD, Mintseris J, Huang ES. Are protein-protein interfaces more conserved in sequence than the rest of the protein surface? Protein Sci. 2004;13:190–202. doi: 10.1110/ps.03323604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Elcock AH, McCammon JA. Identification of protein oligomerization states by analysis of interface conservation. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:2990–2994. doi: 10.1073/pnas.061411798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Valdar WS, Thornton JM. Conservation helps to identify biologically relevant crystal contacts. J Mol Biol. 2001;313:399–416. doi: 10.1006/jmbi.2001.5034. [DOI] [PubMed] [Google Scholar]
- 113.Manning JR, Jefferson ER, Barton GJ. The contrasting properties of conservation and correlated phylogeny in protein functional residue prediction. BMC bioinformatics. 2008;9:51. doi: 10.1186/1471-2105-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Capra JA, Singh M. Predicting functionally important residues from sequence conservation. Bioinformatics. 2007;23:1875–1882. doi: 10.1093/bioinformatics/btm270. [DOI] [PubMed] [Google Scholar]
- 115.Panchenko AR, Kondrashov F, Bryant S. Prediction of functional sites by analysis of sequence and structure conservation. Protein Sci. 2004;13:884–892. doi: 10.1110/ps.03465504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Berezin C, Glaser F, Rosenberg J, Paz I, Pupko T, Fariselli P, Casadio R, Ben-Tal N. ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics. 2004;20:1322–1324. doi: 10.1093/bioinformatics/bth070. [DOI] [PubMed] [Google Scholar]
- 117.del Sol A, Pazos F, Valencia A. Automatic methods for predicting functionally important residues. J Mol Biol. 2003;326:1289–1302. doi: 10.1016/s0022-2836(02)01451-1. [DOI] [PubMed] [Google Scholar]
- 118.Pupko T, Bell RE, Mayrose I, Glaser F, Ben-Tal N. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics. 2002;18(1):S71–77. doi: 10.1093/bioinformatics/18.suppl_1.s71. [DOI] [PubMed] [Google Scholar]
- 119.Landgraf R, Xenarios I, Eisenberg D. Three-dimensional cluster analysis identifies interfaces and functional residue clusters in proteins. J Mol Biol. 2001;307:1487–1502. doi: 10.1006/jmbi.2001.4540. [DOI] [PubMed] [Google Scholar]
- 120.Armon A, Graur D, Ben-Tal N. ConSurf: an algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J Mol Biol. 2001;307:447–463. doi: 10.1006/jmbi.2000.4474. [DOI] [PubMed] [Google Scholar]
- 121.Mirny LA, Shakhnovich EI. Universally conserved positions in protein folds: reading evolutionary signals about stability, folding kinetics and function. J Mol Biol. 1999;291:177–196. doi: 10.1006/jmbi.1999.2911. [DOI] [PubMed] [Google Scholar]
- 122.Casari G, Sander C, Valencia A. A method to predict functional residues in proteins. Nat Struct Biol. 1995;2:171–178. doi: 10.1038/nsb0295-171. [DOI] [PubMed] [Google Scholar]
- 123.Guharoy M, Chakrabarti P. Conservation and relative importance of residues across protein-protein interfaces. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15447–15452. doi: 10.1073/pnas.0505425102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Biswas S, Guharoy M, Chakrabarti P. Dissection, residue conservation, and structural classification of protein-DNA interfaces. Proteins. 2009;74:643–654. doi: 10.1002/prot.22180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


